Abstract
AdpA is a global transcriptional activator triggering morphological differentiation and secondary metabolism in Streptomyces griseus. AdpA influences the expression of >1000 genes; however, the overall picture of the AdpA regulon remains obscure. Here, we took snapshots of the distribution of AdpA across the chromosome in living S. griseus cells using chromatin immunoprecipitation/chromatin affinity precipitation-seq analysis. In both liquid and solid cultures, AdpA bound to >1200 similar sites, which were located on not only in putative regulatory regions (65%), but also in regions (35%) that appeared not to affect transcription. Transcriptome analysis indicated that ∼40% of the AdpA-binding sites in putative regulatory regions were involved in gene regulation. AdpA was indicated to act as a transcriptional repressor as well as an activator. Expression profiles of AdpA-target genes were very different between liquid and solid cultures, despite their similar AdpA-binding profiles. We concluded that AdpA directly controls >500 genes in cooperation with other regulatory proteins. A comprehensive competitive gel mobility shift assay of AdpA with 304 selected AdpA-binding sites revealed several unique characteristics of the DNA-binding property of AdpA. This study provides the first experimental insight into the extent of the AdpA regulon, indicating that many genes are under the direct control of AdpA.
Keywords: ChIP-seq, regulatory cascade, Streptomyces, transcription factor
1. Introduction
The Gram-positive, soil-inhabiting, filamentous bacterial genus Streptomyces is characterized by its ability to produce a wide variety of secondary metabolites. Another characteristic feature of the genus is its complex multicellular development. Spores germinate to form a branched, multinucleoid substrate mycelium, which then produces an aerial mycelium. After septa have been formed at regular intervals along the aerial hyphae, long chains of uninucleoid spores are formed. Generally, secondary metabolites begin to be produced after the onset of morphological differentiation. Many genes required for these two events are coordinately regulated. However, the regulatory networks of such genes are largely unknown.
In the streptomycin producer Streptomyces griseus, a microbial hormone, A-factor (2-isocapryloyl-3-R-hydroxymethyl-γ-butyrolactone), triggers both secondary metabolism and morphological differentiation. We have long studied the A-factor regulatory cascade. A-factor switches on the transcription of adpA by binding to ArpA, the A-factor receptor protein that binds to the promoter of adpA, and dissociating DNA-bound ArpA from the DNA.1 AdpA, an AraC/XylS family transcriptional regulator, activates a number of genes required for morphological differentiation and secondary metabolite formation.2 Thus, an adpA-deleted (ΔadpA) mutant cannot exhibit morphological development or production of many (but not all) secondary metabolites. Our transcriptome analysis showed that 639 and 373 genes were transcriptionally downregulated and upregulated, respectively, in the ΔadpA mutant, when compared with the wild-type strain.3
Up until 2006, we had demonstrated that AdpA directly activates 14 transcripts (17 genes), which include several genes encoding key regulators of secondary metabolism and morphological differentiation, extracellular proteases and an extracellular protease inhibitor protein, and proteins of unknown function.2,4 The 14 AdpA-target promoters are termed ‘category-I’ promoters hereafter. Twenty AdpA-binding sites were identified in the upstream (or just downstream, in the case of adsA) region of the promoters; these are termed ‘category-I’ AdpA-binding sites hereafter. Recent DNA microarray analysis, in which the effect of exogenously supplemented A-factor on the transcriptome in an A-factor-deficient mutant was determined, and comprehensive electrophoretic mobility shift assays (EMSAs) suggested 34 candidates for novel AdpA-target promoters.5 Furthermore, our proteome analysis of extracellular proteins resulted in the identification of 11 novel candidates for AdpA-target promoters, 4 of which were included in the 34 candidates.6 These results indicate that AdpA directly activates a considerable number of genes. We have already determined a consensus AdpA-binding sequence, 5′-TGGCSNGWWY-3′ (S: G or C, W: A or T, Y: C or T, and N: A, C, G, or T),7 which is not strict compared with the consensus binding sequences of well-known bacterial global transcriptional regulators, such as cAMP-receptor protein (CRP),8 LexA,9 and Spo0A.10 Accordingly, a bioinformatics search predicted more than 40 000 AdpA-binding sites in the genome of S. griseus. It is rather unlikely that AdpA binds to all the predicted sites; thus, actual AdpA-binding sites cannot be found by in silico analysis alone.
In the present study, we performed chromatin affinity precipitation (ChAP),11 as well as chromatin immunoprecipitation (ChIP), in combination with high-throughput sequencing technology, to determine the distribution of AdpA across the entire S. griseus chromosome. DNA fragments obtained by ChIP or ChAP were identified with a high-throughput sequencer, which promised higher spatial resolution and sensitivity than microarray-based methods (e.g. ChIP-chip analysis).12 By the ChIP/ChAP-seq analysis, we were able to take snapshots of the distribution of AdpA across the chromosome in living S. griseus cells, not only in liquid culture, but also in solid culture. This information, in combination with transcriptional profiling using DNA microarray, comprehensive EMSA, and in silico prediction of AdpA-binding sites, revealed the extent and complexity of the AdpA regulatory network.
2. Materials and methods
2.1. Bacterial strains and growth conditions
The wild-type strain S. griseus IFO13350 and the adpA mutant were described previously.1 Strains were cultured as described previously.13
2.2. In vivo cross-linking of AdpA and DNA
For in vivo cross-linking of AdpA and DNA in cells of liquid culture, the ΔadpA strain expressing Nhis-adpA was grown in 100 ml YMPD for 18 h at 30°C and treated with formaldehyde (1% final concentration) for 15 min at room temperature. For in vivo cross-linking of AdpA and DNA in cells of solid culture, the same strain was grown on an YMPD-cellophane plate for 1, 2, and 3 days at 28°C. The mycelia were collected with a spatula, resuspended with phosphate-buffered saline (PBS), and treated with formaldehyde (1% final concentration) for 20 min at room temperature. In both cases, formaldehyde was quenched by adding glycine to a final concentration of 250 mM. Finally, the cells were washed with tris-buffered saline buffer (pH 7.5) more than twice and stored at −80°C until use.
2.3. Chromatin immunoprecipitation
Cross-linked cells in 1-ml ChIP buffer (50 mM HEPES–KOH, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.1% sodium deoxycolate, pH 7.5) were sonicated on ice to give an average DNA fragment size of 300–600 bp. After centrifugation at 12 000g for 10 min, the supernatant was collected and diluted with ChIP buffer (a final protein concentration of 3–5 mg/ml). At the same time, 50 μl of anti-mouse IgG dynabeads (Invitrogen), which had been washed twice with PBS containing 5 mg/ml bovine serum albumin (BSA), was mixed with anti-poly-histidine antibody (Qiagen) at 4°C for 4 h, washed twice with 1 ml of PBS–BSA, and resuspended with PBS–BSA. The anti-poly-histidine antibody-conjugated beads were added to the 1 ml of the diluted supernatant, followed by incubation for 6 h at 4°C with gentle rotation. The beads were then washed once with ChIP buffer, once with ChIP high-salt buffer (ChIP buffer containing 300 mM NaCl), twice with ChIP wash buffer (100 mM Tris–HCl, 250 mM LiCl, 1 mM EDTA, 0.5% Nonidet P-40, and 0.5% sodium deoxycolate, pH 8.0), and twice with TE (10 mM Tris and 1 mM EDTA, pH 8.0). Proteins bound to the beads were eluted with 100 μl of elution buffer [50 mM Tris–HCl, 10 mM EDTA, and 1% sodium dodecyl sulphate (SDS), pH 8.0]. After the cross-linking between AdpA and the DNA was destroyed by heating at 65°C overnight, 300 μl of TE was added to the reverse-cross-linked sample. The sample was then treated with RNase A (a final concentration of 0.4 mg/ml) for 2 h at 37°C and with proteinase K (a final concentration of 0.2 mg/ml) for 2 h at 55°C. Finally, DNA fragments were purified by Qiaquick PCR purification kit (Qiagen).
2.4. Chromatin affinity precipitation
Cross-linked cells in 1-ml ChAP buffer [25 mM HEPES–KOH, 10 mM imidazole, 6 M guanidine-HCl, 0.5 M NaCl, 0.5% Triton X-100, and 10% (w/v) glycerol, pH 7.5] were sonicated on ice to give an average DNA fragment size of 300–600 bp. After centrifugation at 12 000g for 10 min, the supernatant was collected and diluted with ChAP buffer (a final protein concentration of 3–5 mg/ml). Fifty microlitres of dynabeads TALON (Invitrogen), which had been washed twice with ChAP buffer, were then added to the 1 ml of the diluted supernatant, followed by incubation for 2 h at room temperature with gentle rotation. The beads were washed four times with ChAP buffer and twice with TE. DNAs bound to the beads were eluted with 100 μl of elution buffer (50 mM Tris–HCl, 0.5 M imidazole, and 0.5% SDS, pH 7.5). After cross-linking between AdpA and DNA was destroyed by heating at 65°C overnight, reverse cross-linked DNA was purified by phenol–chloroform treatment and ethanol precipitation. The DNA pellet was resuspended in 100-μl TE containing 25 μg/ml RNase A, followed by incubation for 2 h at 37°C. DNA fragments were finally purified by Qiaquick PCR purification kit (Qiagen).
2.5. Sequencing of the DNA fragments enriched by ChIP or ChAP
DNA samples were submitted to Takara Bio for sequencing using an Illumina Genome Analyzer II. Libraries were prepared according to the manufacture's instructions. Sequence reads were mapped to the genome of S. griseus using Bowtie.14 Parameters were set to allow mapping of reads to the genome with up to two mismatches. Reads mapped to multiple sites, except for those to terminal inverted repeats and rRNA gene clusters, were excluded from the analysis. Reads were clustered and analysed by CisGenome.15
2.6. Other materials and methods
Other Materials and methods are described in Supplementary Data.
2.7. Accession number
All microarray and ChIP-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE33994.
3. Results
3.1. Expression of polyHis-tagged AdpA in S. griseus
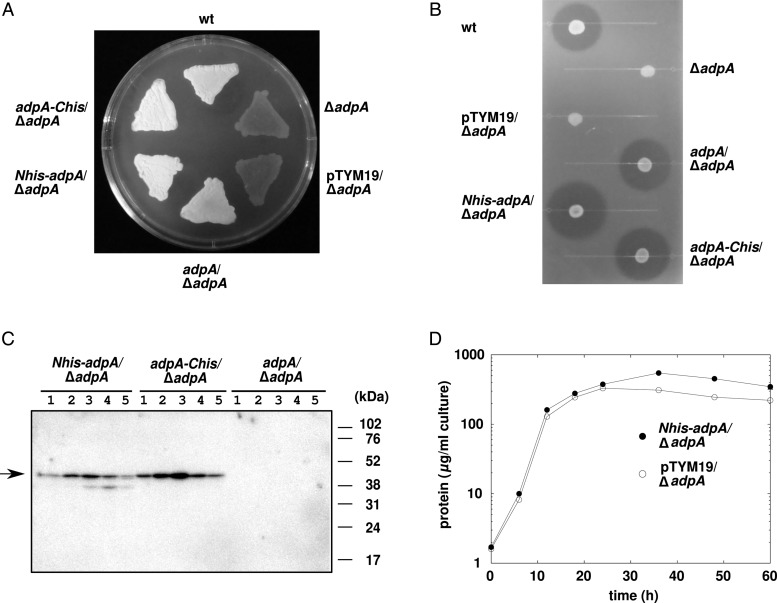
For ChIP and ChAP analyses, ΔadpA strains producing AdpA with a polyHis-tag at either its N- or C-terminus (N-His-AdpA and AdpA-C-His, respectively) were generated. The defects in morphological differentiation (Fig. 1A) and streptomycin production (Fig. 1B) of the ΔadpA strain were completely complemented by the production of N-His-AdpA and AdpA-C-His proteins, as well as by the native AdpA protein, demonstrating that the attached polyHis-tag did not interfere the in vivo function of AdpA. Western blotting analysis with an anti-polyHis-tag antibody showed that the amount of AdpA increased gradually during the exponential growth phase and then decreased gradually during the stationary phase in the mycelium cultured in the nutrient-rich YMPD liquid medium (Fig. 1C). Maximal accumulation of AdpA was observed at 18 h, when the growth became slower than that in the exponential growth phase (Fig. 1D). Western blotting analysis with N-His-AdpA-expressing strain showed that the C-terminal portion of AdpA was partially degraded on solid culture (Supplementary Fig. S1). Degradation of the polyHis-containing C-terminal portion of AdpA-C-His should be also possible; therefore, we used the ΔadpA strain producing N-His-AdpA, not AdpA-C-His, for further analyses.
Figure 1.
The polyHis-tag attached does not interfere with the in vivo function of AdpA. (A) Restoration of morphological differentiation in the ΔadpA mutant by the production of polyHis-tagged AdpA. Strains were grown on YMPD agar for 3 days at 28°C. (B) Restoration of streptomycin production in the ΔadpA mutant by the production of polyHis-tagged AdpA. After the indicated strains were grown on YMP (YMPD without glucose) agar for 4 days at 28°C, an indicator strain was overlaid and incubated overnight at 28°C. (C) Western blotting analysis of polyHis-tagged AdpA. Total protein was extracted at 6 (Lane 1), 12 (Lane 2), 18 (Lane 3), 24 (Lane 4), and 36 (Lane 5) h of liquid culture and 6 μg of each was subjected to electrophoresis. Arrow indicates polyHis-tagged AdpA. Note that western blotting analysis gave stronger signals to purified AdpA-C-His than to purified N-His-AdpA when equal amounts of them were subjected to electrophoresis (data not shown). Therefore, intracellular levels of N-His-AdpA and AdpA-C-His seem to be similar, although AdpA-C-His gave stronger signals than N-His-AdpA. (D) Growth curve in YMPD liquid culture, as determined by the amount of intracellular protein. Closed circles represent the ΔadpA strain producing N-His-AdpA and open circles represent the control ΔadpA strain.
3.2. Genome-wide distribution of AdpA across the chromosome of the mycelium in liquid culture
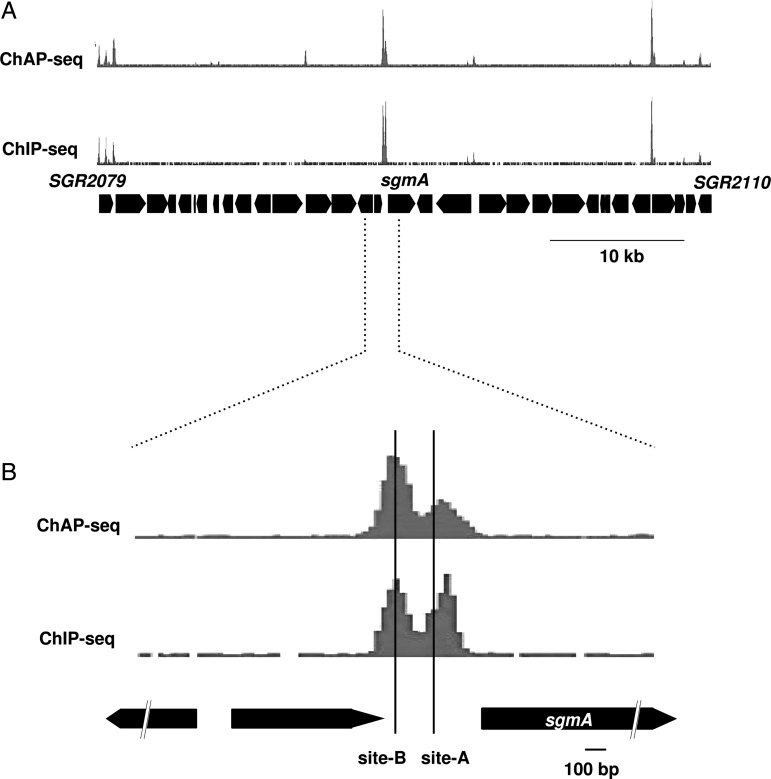
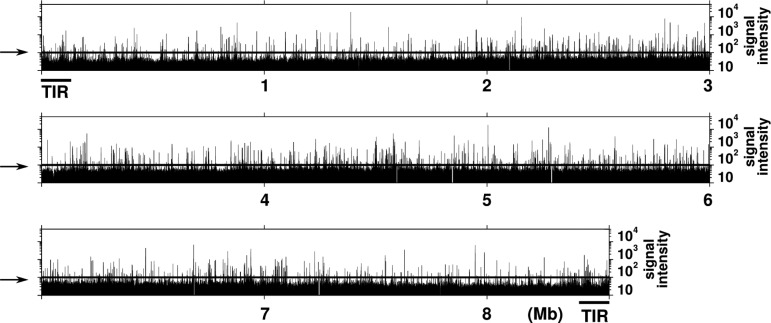
We performed ChAP-seq and ChIP-seq analyses using the N-His-AdpA-producing ΔadpA strain cultured in the YMPD liquid medium for 18 h. Although these two analyses are based on different principles to enrich the AdpA-bound DNA fragments, very similar results were obtained; the correlation coefficient of signal intensity of peaks obtained from ChIP-seq and ChAP-seq was 0.84 (Supplementary Fig. S2). As an example, Fig. 2A shows a small fraction of the result, in which the binding of AdpA on a 50-kb region containing sgmA is depicted. Peaks obtained from both analyses were in good agreement with the known AdpA-binding sites (Sites A and B) for sgmA.7 Regarding Site A, peaks from ChIP-seq and ChAP-seq analyses are shifted from the actual binding site by only 36 and 11 bp, respectively. On the other hand, a peak is located right on Site B (Fig. 2B) in both analyses. Furthermore, we confirmed that a peak was invariably detected within 50 bp from all ‘category-I’ AdpA-binding sites, except the AdpA-binding site of sgiA (AdBS-sgiA), which was not detected (see below). Thus, we successfully obtained a high-resolution map of the in vivo AdpA-binding regions. Figure 3 shows the entire AdpA-binding chromosome map obtained by ChAP; genome-wide distribution of AdpA across the chromosome is obvious.
Figure 2.
ChIP/ChAP-seq analysis of AdpA. The results of ChAP-seq (upper) and ChIP-seq (lower) are shown. (A) A 50-kb region containing sgmA. (B) A 2-kb region containing sgmA. Two AdpA-binding sites (Sites A and B) determined by in vitro experiments7 are indicated.
Figure 3.
Genome-wide distribution of AdpA. The result of ChAP-seq analysis is shown. Horizontal lines indicated by arrows represent the threshold for AdpA binding. The chromosome of S. griseus has terminal inverted repeats (TIR), which are identical 13920-bp sequences, at both ends. AdpA binding cannot be distinguished between the right and left TIR sequences in ChIP/ChAP-seq analysis. Therefore, the intensity of signals detected on TIR is equally divided between right and left TIR sequences.
In both analyses, the signal intensity of the AdpA-binding site for adsA (AdBS-adsA) was lowest among all of the ‘category-I’ AdpA-binding sites, except AdBS-sgiA. Therefore, we used this value as a threshold to exclude background noise in each analysis; peaks with signal intensities equal to or more than that of AdBS-adsA were considered as candidates for AdpA-binding sites (Fig. 3). Finally, 1355 sites, which were detected in both analyses, were regarded as the in vivo AdpA-binding sites.
We classified these sites into three groups by their location in relation to the surrounding genes. First, ‘putative regulatory region’: from −600 to +100 of protein-coding genes (taking a putative translational start site as +1) and −300 to +50 for RNA genes (taking a transcriptional start point of RNA gene as +1). Secondly, ‘coding region’: from +101 to the 3′-terminus (excluding AdpA-binding sites that can be considered as ‘putative regulatory region’ for downstream genes). The last group contained the remainder, which was classified as ‘intergenic region’ (Table 1). This classification suggested that 69% of the AdpA-binding sites were located in ‘putative regulatory regions’ and 31% of them were on regions that appeared not to affect transcription (i.e. ‘coding’ and ‘intergenic’ regions).
Table 1.
The number of AdpA-binding sites in liquid and solid cultures
| Liquid | Solid (days) |
Overlappedd (days) |
|||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | ||
| ‘Regulatory region’ | |||||||
| Proteina | 892 | 772 | 732 | 768 | 706 | 662 | 662 |
| tRNAb | 23 | 11 | 5 | 5 | 11 | 5 | 5 |
| rRNAc | 7 | 7 | 7 | 7 | 7 | 7 | 7 |
| sRNA | 7 | 4 | 3 | 2 | 4 | 3 | 2 |
| Total | 929 | 794 | 747 | 782 | 728 | 677 | 676 |
| ‘Coding region’ | |||||||
| Proteina | 393 | 415 | 425 | 479 | 341 | 334 | 338 |
| rRNAc | 7 | 7 | 7 | 7 | 7 | 7 | 7 |
| Total | 400 | 422 | 432 | 486 | 348 | 341 | 345 |
| ‘Intergenic region’ | |||||||
| Total | 26 | 23 | 23 | 24 | 17 | 17 | 17 |
| Total | 1355 | 1239 | 1202 | 1292 | 1093 | 1035 | 1038 |
aAdpA-binding sites in the TIR region are counted only once.
bStreptomyces griseus has 66 tRNA genes, including ‘duplicated’ gene sets. Duplicated genes cannot be distinguished from each other in ChIP/ChAP analysis; therefore, they were excluded from the analysis. Only 26 transcriptional units containing 27 tRNA genes can be discriminated from others; AdpA was shown to bind to the upstream regions of 23 transcriptional units containing 24 tRNA genes.
cAdpA-binding sites in the common region of six rRNA gene clusters are counted only once.
dThe number of AdpA-binding sites detected in both liquid and solid cultures.
3.3. Relationship between AdpA binding and transcriptional regulation
We examined whether AdpA binding to each ‘putative regulatory region’ could affect transcription of the target gene. For protein-coding genes, gene expression profiles were compared by DNA microarray analysis between the N-His-AdpA-producing ΔadpA strain and the ΔadpA strain containing the empty integration vector pTYM19 on the chromosome. The transcriptome analysis showed that 736 and 457 genes were expressed at higher and lower levels, respectively, in the N-His-AdpA-producing strain when compared with the control ΔadpA strain (fold change ≥2 and ≤0.5, respectively, and P-value <0.05). We obtained expression data of 662 genes from among the 796 genes that have one or more in vivo AdpA-binding sites in their ‘putative regulatory region’. Thus, we could evaluate the apparent significance of 743 of the 892 in vivo AdpA-binding sites in ‘putative regulatory region’; 214 sites (for 175 transcriptional units) and 89 sites (for 81 transcriptional units) were apparently involved in transcriptional activation and repression, respectively. We also examined apparent significance of several AdpA-binding sites in the ‘putative regulatory region’ for tRNA genes and rRNA genes (Supplementary Fig. S3). Expression of only bldA (trn42) was higher in the N-His-AdpA-producing strain when compared with the control ΔadpA strain. We have confirmed that transcription of bldA is activated by AdpA.13
Altogether, we evaluated the apparent significance of 773 sites of the 922 in vivo AdpA-binding sites in ‘putative regulatory regions’. Of 773 sites, 215 sites (28%, for 176 transcriptional units) and 89 sites (12%, for 81 transcriptional units) were possibly involved in transcriptional activation and repression, respectively. In this study, we termed these 304 sites as ‘apparently relevant’ sites. Although we demonstrated that AdpA represses transcription of its own gene,16 it was unexpected that AdpA appears to repress more than 80 other transcriptional units (see Discussion). The in vivo binding of AdpA to the remaining 469 ‘putative regulatory region’ sites seemed not to affect gene expression, at least under the growth conditions examined. We termed these 469 sites as ‘apparently irrelevant’ sites.
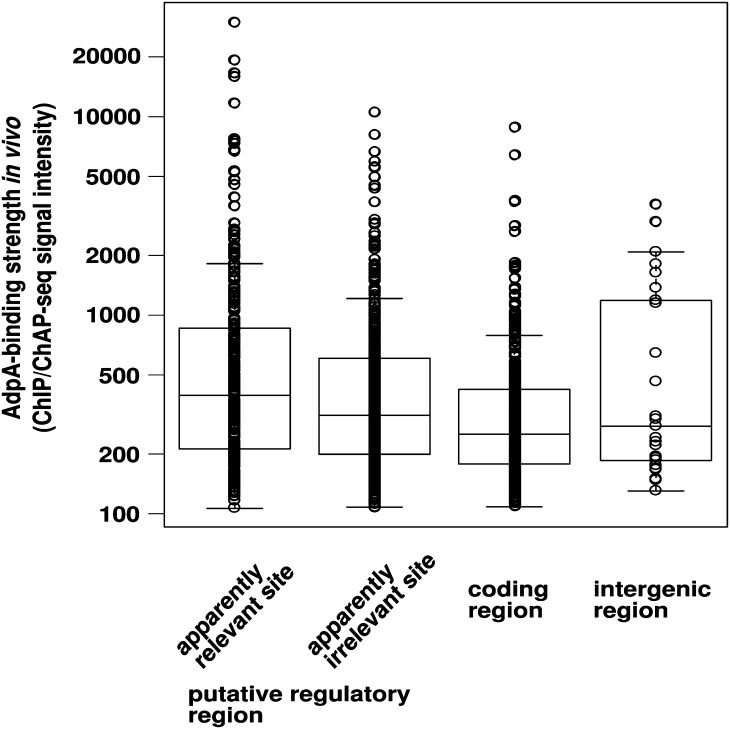
We then compared the distribution of ChIP/ChAP-seq signal intensities (average values), which reflects the strength of AdpA–DNA interaction in vivo, between apparently relevant and irrelevant sites (Fig. 4). No significant difference was observed between them. Furthermore, there was no significant difference in distribution of ChIP/ChAP-seq signal intensities among the three positional classifications of AdpA-binding sites (‘putative regulatory’, ‘coding’, and ‘intergenic’ regions) (Fig. 4). These results indicated that in vivo AdpA-binding strength is not related to the functional significance of particular AdpA-binding sites.
Figure 4.
In vivo AdpA-binding strength is not related to the functional significance of particular AdpA-binding sites. One-dimensional scatter plots of AdpA-binding strength in vivo are shown for four respective classifications of AdpA-binding sites. The means of peak signals derived from ChIP-seq and ChAP-seq are plotted. Box plots are also shown. In the box plot, the bottom and top of the box are the 25th and 75th percentile (the lower and upper quartiles), respectively, and the band near the middle of the box is the 50th percentile (the median). The ends of the whiskers represent the lowest datum still within 1.5 IQR of the lower quartile, and the highest datum still within 1.5 IQR of the upper quartile (IQR, inter-quartile range, is equal to the difference between the upper and lower quartiles).
3.4. Confirmation of in vitro AdpAbinding to the in vivo AdpA-binding sites detected by ChIP/ChAP-seq
We examined whether the AdpA-C-His protein purified from Escherichia coli binds to the in vivo AdpA-binding sites by comprehensive competitive EMSA. We tested all 304 ‘apparently relevant’ AdpA-binding sites. ChIP/ChAP-seq signal peaks were detected within 50 bp from all of the ‘category-I’ AdpA-binding sites, except AdBS-sgiA; therefore, 130–200 bp DNA fragments containing a centrally positioned predicted AdpA-binding site (ChIP/ChAP-seq peak) were amplified by PCR. These DNA fragments were used as the competitors in EMSA using AdpA-C-His. In the competitive EMSA, we used two DIG-labelled probes with different AdpA-binding affinities: the high-affinity sgmA probe and the low-affinity sgiA probe.5 We detected 247 DNA fragments that at the least competed with the sgiA probe; however, 57 DNA fragments did not. AdpA frequently shows relatively low DNA-binding strength to short DNA fragments6 and true AdpA-binding sites may be separated from the ChIP/ChAP signal peaks by more than 50 bp in some cases; we increased the lengths of these 57 AdpA-unbound DNA fragments to 200–250 bp and re-examined their AdpA binding by competitive EMSA. As a result, we detected 37 DNA fragments that at least competed with the sgiA probe, but the remaining 20 DNA fragments did not. We identified at least one sequence that is similar to the AdpA-binding consensus sequence on each of the 284 DNA fragments to which AdpA bound in vitro. We regarded the consensus-like sequences as probable AdpA-binding sequences. When more than two possible AdpA-binding sequences existed in a DNA fragment, we selected the sequence closest to the ChIP/ChAP-seq signal peak in this study. In summary, of 304 ‘apparently relevant’ in vivo AdpA-binding sites, 284 sites (93%) were confirmed to be bound by AdpA in vitro. The remaining 20 AdpA-binding sites may not have resulted from false-positive signals, because the peaks were detected similarly by both ChIP-seq and ChAP-seq analyses, and because there was an AdpA-binding consensus-like sequence around each peak. It is possible that AdpA binds to these sites cooperatively with some other DNA-binding protein in vivo (see Discussion).
3.5. Comparison of the strength of the AdpA–DNA interaction between in vivo and in vitro
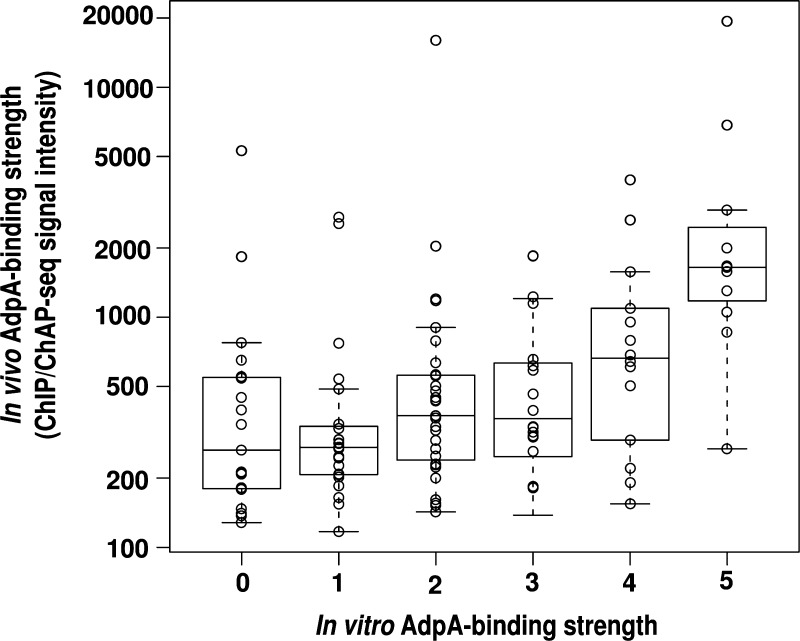
We next determined whether there was a correlation in the strength of the AdpA–DNA interaction between in vivo and in vitro for the 304 ‘apparently relevant’ AdpA-binding sites. To roughly evaluate in vitro AdpA-binding strength, we rated the binding strength of AdpA to each DNA fragment in six ranks (rank ‘0’ to ‘5’) as described previously.5 Briefly, the strength of the AdpA–DNA interaction in vitro was assessed by competitive EMSA using two DIG-labelled probes with different AdpA-binding affinities. In vitro AdpA-binding strength is often underestimated when an AdpA-binding sequence is located at the end of the DNA fragment.6 Therefore, we selected 115 AdpA-binding sequences, which were located within 30 bp from the centre of each DNA fragment (130–200 bp), and compared in vitro AdpA-binding strength to in vivo AdpA-binding strength deduced from ChIP/ChAP-seq signal intensities (average values) (Fig. 5). Contrary to our expectation, there was very poor correlation of AdpA-binding strength between in vitro and in vivo conditions (correlation coefficient was 0.43). However, the sequences that were strongly bound by AdpA in vitro (those classified into ranks ‘5’ and ‘4’) generally showed relatively high in vivo AdpA-binding strength.
Figure 5.
Comparison of the strength of the AdpA–DNA interaction in vivo and in vitro. AdpA binding to 115 regions are examined (see text). One-dimensional scatter plots of in vivo AdpA-binding strength are shown for six respective ranks of in vitro AdpA-binding strength. Box plots (see the legend for Fig. 4) are also shown.
Next, we calculated the ‘match score’ of these 115 AdpA-binding sequences to the consensus AdpA-binding sequence by Mast17 and examined a relationship between the match score (which represented the degree of similarity to the AdpA-binding consensus sequence) and AdpA-binding strength. There was almost no correlation between the match score and in vivo AdpA-binding strength (correlation coefficient was 0.23) (Supplementary Fig. S4A). However, the sequences that were strongly bound by AdpA in vivo tended to show relatively high match scores. There was also a very poor correlation between the match score and in vitro AdpA-binding strength (correlation coefficient was 0.36), but the sequences that were strongly bound by AdpA in vitro (especially those classified into ranks ‘5’ and ‘4’) showed relatively high match scores (Supplementary Fig. S4B). Note that AdpA binds DNA in two different ways. Type I binding occurs when both subunits of the AdpA dimer bind each of the two divergent AdpA-binding sequences and type II, where only one subunit binds a single AdpA-binding sequence to anchor the AdpA dimer. However, we did not consider any possible type-I AdpA binding in the analyses described above; only single AdpA-binding consensus-like sequences found in each AdpA-binding region were used for the analyses (see Discussion). Among the 115 AdpA-binding sequences, there were six kinds of duplicate sequences and four kinds of triplicate sequences. For example, the AdpA-binding sequences 5′-TGGCCGAAAA-3′ and 5′-TGTCCGAAAA-3′ were found in two and three different AdpA-binding sites, respectively. Interestingly, in 7 of the 10 cases, in vitro AdpA-binding strength significantly differed between two (or three) DNA fragments containing an identical AdpA-binding sequence (Supplementary Table S2). This result indicated that not only the 10 nucleotides of the AdpA-binding consensus sequence, but also the neighbouring nucleotide sequences affect the AdpA-binding strength in vitro (see Discussion).2,7
3.6. In vivo AdpA-binding profiles on solid culture
Our previous DNA microarray analysis showed that S. griseus had significantly different gene expression profiles between liquid and solid cultures (unpublished data). There had also been a suggestion that there should be a large difference in the genes regulated by AdpA between liquid and solid cultures (unpublished data). Therefore, we were interested in the in vivo difference in the AdpA-binding profiles between liquid and solid cultures.
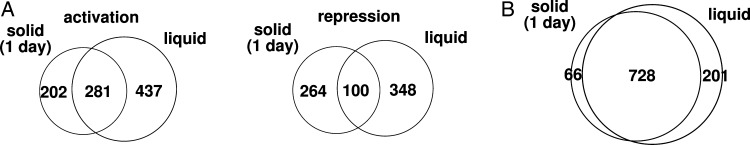
First, we compared the transcriptomes using a DNA microarray between the N-His-AdpA-producing ΔadpA strain and the control ΔadpA strain containing the empty vector pTYM19. RNA was extracted from both strains grown on YMPD agar at 28°C for 1–3 days. In this culture, the N-His-AdpA-producing ΔadpA strain grew as substrate mycelium at day 1, as substrate mycelium plus aerial mycelium at day 2, and as substrate mycelium plus aerial mycelium with spores at day 3. The control ΔadpA strain could not make aerial mycelium and spores throughout culture. At day 1, 531 and 457 genes were expressed at higher and lower levels, respectively, in the N-His-AdpA-producing strain when compared with the control ΔadpA strain (fold change ≥2 and ≤0.5, respectively, and P-value <0.05). This indicated that 531 and 457 genes were activated and repressed, respectively, by AdpA (directly or indirectly) at day 1. Similarly, the numbers of apparently AdpA-activated and repressed genes were 509 and 394 (at day 2), and 611 and 606 (at day 3), respectively. The mycelium at day 1 on solid culture should roughly correspond to the mycelium of 18-h liquid culture, in that both mycelia were in the late exponential growth phase; therefore, we compared AdpA-regulated genes between the 18-h liquid culture and 1-day solid culture. When genes whose transcriptional data were statistically available in both the transcriptome analyses were compared, only 281 genes were apparently activated by AdpA in both liquid and solid cultures, while 437 and 202 genes were activated specifically in liquid and solid cultures, respectively (Fig. 6A). On the other hand, only 100 genes were apparently repressed by AdpA in both liquid and solid cultures, while 348 and 264 genes were specifically repressed in liquid and solid cultures, respectively (Fig. 6A). Thus, we confirmed that there was indeed a large difference in the genes regulated by AdpA between liquid and solid cultures.
Figure 6.
Venn diagrams showing different expressions of AdpA-regulated genes (A) and similar AdpA-binding profiles (B) between 18-h liquid and 1-day solid cultures. (A) Numbers of genes activated (left) and repressed (right) by AdpA directly or indirectly. (B) Numbers of AdpA-binding sites in ‘putative regulatory regions’.
The AdpA-binding profiles at days 1, 2, and 3 were then examined by ChAP-seq. Similar to the ChIP/ChAP-seq analysis in liquid culture, the signal intensity of AdBS-adsA was used as a threshold to exclude background noise. Contrary to our expectation, the AdpA-binding profiles did not change significantly among 18-h liquid culture and 1-, 2-, and 3-day solid cultures (Table 1). When in vivo AdpA-binding sites on ‘putative regulatory regions’ were compared between 18-h liquid culture and 1-day solid culture, 728 sites were shared, and 201 and 66 sites were specifically detected in the liquid and solid cultures, respectively (Fig. 6B). Interestingly, many of these specifically detected sites have relatively low ChIP-seq and/or ChAP-seq signal intensities. Transcriptome analysis indicated that only 3 of the 201 sites specifically detected in the liquid culture were apparently involved in gene regulation (Supplementary Fig. S5). In other words, only three liquid culture-specific AdpA-binding sites were ‘apparently relevant’. Similarly, only 3 of the 66 solid culture-specific sites were ‘apparently relevant’ (Supplementary Fig. S5). Biological significance of the liquid or solid culture-specific regulation of these genes by AdpA is unknown. However, from these results, we concluded that the large difference in the genes regulated by AdpA between liquid and solid cultures resulted from regulatory elements other than AdpA binding; AdpA-binding profiles did not change significantly between cultures.
3.7. Extraction of putative AdpA-target genes
ChAP-seq and transcriptome analyses in solid culture suggested 150 new ‘apparently relevant’ AdpA-binding sites in addition to the 304 sites described above. In total, of 1001 in vivo AdpA-binding sites on ‘putative regulatory region’ of protein-coding genes in liquid and/or solid cultures, we identified 262 sites (26%, for 217 transcriptional units, 342 genes) and 172 sites (17%, for 156 transcriptional units, 224 genes) that were apparently involved in transcriptional activation and repression, respectively, in either liquid or solid culture. In addition, 20 AdpA-binding sites that apparently had contradictory effects on gene regulation among 18-h liquid culture and 1-, 2-, and 3-day solid cultures were detected (e.g. SGR4671 was apparently activated in 18-h liquid culture by AdpA and repressed in 2- and 3-day solid culture). These sites were excluded from the following lists (Tables 2 and 3; Supplementary Tables S3 and S4). We then catalogued candidates for direct target genes of AdpA. AdpA presumably activates the promoters of 88 transcriptional units containing 145 genes in both liquid and solid cultures, which include 7 ‘category-I’ AdpA-target promoters (adsA, amfR, strR, sprB, sgmA, AdBS2-orf1, and AdBS4-orf1) and 20 probable AdpA-target promoters suggested by Hara et al.5 and/or Akanuma et al.6 (Table 2; Supplementary Table S3). In accordance with the proposed function of AdpA, these AdpA-target genes included many genes probably involved in secondary metabolism and morphological differentiation. On the other hand, AdpA presumably represses 40 transcriptional units containing 54 genes in both liquid and solid cultures (Table 3). We also listed 54 AdpA-target transcriptional units (79 genes) that are activated specifically on solid culture (Supplementary Table S4). They included four ‘category-I’ AdpA-target genes (ssgA, sprU, sprT, and AdBS3-orfA) and four known candidates for AdpA-target transcriptional units5,6; however, many of them (56%) were categorized as function-unknown genes. Note that we estimated transcriptional units from transcriptome data, gene synteny, and functional relationship with its neighbouring genes.
Table 2.
List of genes that are probably activated directly by AdpA in both liquid and solid cultures (selecteda)
| Transcriptional unit | ID | Description | Memob | Identified previouslyc |
|---|---|---|---|---|
| SGR745 | SGR745 | Putative M23-family secreted peptidase | p | |
| SGR919–SGR916 | SGR919 | Hypothetical protein | 2 | |
| SGR918 | Putative subtilisin-like serine protease | p | ||
| SGR917 | Hypothetical protein | |||
| SGR916 | Hypothetical protein | |||
| SGR1063–SGR1059 | SGR1063 | rarA | m, s | |
| SGR1062 | rarB | m, s | ||
| SGR1061 | rarC | m, s | ||
| SGR1060 | rarD | m, s | ||
| SGR1059 | rarE | m, s | ||
| SGR2079 | SGR2079 | terpene cyclase (gcoA) | s | 2 |
| SGR2095 | SGR2095 | sgmA | p | 1 |
| SGR2393 | SGR2393 | amfR | m | 1 |
| SGR2446–SGR2447 | SGR2446 | Tyrosinase co-factor protein (melC1–2) | s | 2, 3 |
| SGR2447 | Tyrosinase (melC2–2) | s | ||
| SGR3307–SGR3306 | SGR3307 | Anti-sigma factor antagonist (bldG) | m | |
| SGR3306 | Putative anti-sigma factor | |||
| SGR3340 | SGR3340 | Putative WhiB-family transcriptional regulator (wblA) | m, s | 2 |
| SGR3902 | SGR3902 | Sporulation associated protein ORF1590 | m | 2 |
| SGR4151 | SGR4151 | adsA | m | 1 |
| SGR4809 | SGR4809 | Putative lantibiotic modifying enzyme | s | |
| SGR5762 | SGR5762 | sprB | p | 1 |
| SGR5914 | SGR5914 | strU | s | 2 |
| SGR5922–SGR5923 | SGR5922 | stsB | s | |
| SGR5923 | stsA | s | ||
| SGR5931–SGR5932 | SGR5931 | strR | s | 1, 2 |
| SGR5932 | aphD | s | ||
| SGR6071 | SGR6071 | Putative LAL-subfamily transcriptional regulator (AdBS4-orf1) | s | 1 |
| trn42 | trn42 | bldA | m, s |
aAn extract of genes involved in morphological differentiation, secondary metabolism, and extracellular protease. See Supplementary Table S3 for full list. bm, involved in morphological differentiation; s, involved in secondary metabolism; p, extracellular protease. c1, ‘category-I’ target; 2, suggested by Hara et al.5; 3, suggested by Akanuma et al.6
Table 3.
List of genes that are probably repressed directly by AdpA in both liquid and solid cultures
| Transcriptional unit | ID | Description | Memo |
|---|---|---|---|
| SGR291 | SGR291 | Hyaluronidase | |
| SGR955 | SGR955 | Putative M23-family secreted peptidase | |
| SGR966–SGR960 | SGR966 | Putative squalene/phytoene synthase (hopE) | |
| SGR965 | Putative squalene/phytoene synthase (hopD) | ||
| SGR964 | Putative squalene/phytoene dehydrogenase (hopC) | ||
| SGR963 | Putative polyprenyl diphosphate synthase (hopB) | ||
| SGR962 | Putative squalene-hopene cyclase (hopA) | ||
| SGR961 | Conserved hypothetical protein | ||
| SGR960 | Conserved hypothetical protein | ||
| SGR1311–SGR1309 | SGR1311 | Conserved hypothetical protein | |
| SGR1310 | Conserved hypothetical protein | ||
| SGR1309 | Putative uricase | ||
| SGR1869 | SGR1869 | Hypothetical protein | |
| SGR1903 | SGR1903 | Putative acylphosphatase | pm |
| SGR2045–SGR2046 | SGR2045 | Putative glycine cleavage system protein T | pm |
| SGR2046 | Putative glycine cleavage system protein H | pm | |
| SGR2195 | SGR2195 | Putative Ku70/Ku80 protein | |
| SGR2702 | SGR2702 | Putative tryptophanyl-tRNA synthetase | pm |
| SGR2910 | SGR2910 | Putative methylated-DNA–protein-cysteine S-methyltransferase | |
| SGR3022 | SGR3022 | Putative TetR-family transcriptional regulator | |
| SGR3072 | SGR3072 | Putative iron dependent regulatory protein | |
| SGR3524–SGR3525 | SGR3524 | Hypothetical protein | |
| SGR3525 | Conserved hypothetical protein | ||
| SGR3620 | SGR3620 | Conserved hypothetical protein | |
| SGR3646–SGR3645 | SGR3646 | Putative cytochrome D ubiquinol oxidase subunit I | pm |
| SGR3645 | Putative cytochrome D ubiquinol oxidase subunit II | pm | |
| SGR3675 | SGR3675 | Conserved hypothetical protein | |
| SGR3782 | SGR3782 | Hypothetical protein | |
| SGR3897 | SGR3897 | Putative M23-family secreted peptidase | |
| SGR3928–SGR3929 | SGR3928 | Putative Pit accessory protein | pm |
| SGR3929 | Putative low-affinity inorganic phosphate transporter (pitH) | pm | |
| SGR4276 | SGR4276 | RNA polymerase principal sigma factor (hrdD) | |
| SGR4380 | SGR4380 | Putative membrane protein | |
| SGR4383 | SGR4383 | Putative ribose-phosphate pyrophosphokinase | pm |
| SGR4455 | SGR4455 | Hypothetical protein | |
| SGR4456 | SGR4456 | Putative RpiR-family transcriptional regulator | |
| SGR4482 | SGR4482 | Putative acyl-CoA dehydrogenase | pm |
| SGR4489 | SGR4489 | Putative acyl-CoA hydrolase | pm |
| SGR4623 | SGR4623 | Putative ATP-dependent Clp protease adaptor protein ClpS | |
| SGR4652 | SGR4652 | Putative secreted protein | |
| SGR4803 | SGR4803 | Hypothetical protein | |
| SGR4919 | SGR4919 | Conserved hypothetical protein | |
| SGR4930 | SGR4930 | Putative nucleoside diphosphate kinase | pm |
| SGR4995 | SGR4995 | Putative 2-nitropropane dioxygenase | |
| SGR5104 | SGR5104 | Conserved hypothetical protein | |
| SGR5469 | SGR5469 | Putative indol-3-glycerol phosphate synthase | pm |
| SGR5674 | SGR5674 | Putative molybdenum cofactor biosynthesis protein A | |
| SGR5721 | SGR5721 | Putative CTP synthetase | pm |
| SGR6046 | SGR6046 | Pyrimidine operon regulatory protein | pm |
| SGR6069–SGR6070 | SGR6069 | Putative hydrolase | |
| SGR6070 | Putative amino oxidase | ||
| SGR6240 | SGR6240 | Conserved hypothetical protein | |
| SGR6353–SGR6354 | SGR6353 | Hypothetical protein | |
| SGR6354 | Hypothetical protein |
pm, presumably involved in primary metabolism.
Of the 14 ‘category-I’ AdpA-target transcriptional units, two genes (sprA and sprD) were not detected as genes activated by AdpA in the present DNA microarray analysis, probably because of technical limitations of the analysis. However, in vivo AdpA binding to their ‘category-I’ AdpA-binding sites was detected. In contrast, the DNA microarray analysis showed that the ‘category-I’ AdpA-target gene sgiA4 was activated by AdpA, but AdpA binding to its AdpA-binding site in vivo was not detected by ChIP/ChAP-seq analysis for an unknown reason. It should be noted that the sgiA locus has been suggested to have some high-order DNA structure. The putative high-order DNA structure of the sgiA locus may hamper the efficiency of ChIP/ChAP with N-His-AdpA. In summary, of the 14 ‘category-I’ AdpA-target transcriptional units, only 3 units were dropped from the candidate lists. Therefore, a majority of AdpA-target genes were listed in this study.
4. Discussion
In this study, ChIP/ChAP-seq technology was used to take snapshots of distribution of AdpA across the chromosome in living S. griseus cells. In combination with transcriptome analysis, the results indicated that AdpA directly controls >500 genes; AdpA regulon seems to be more extensive than previously suggested. Genes that are likely to be under the direct control of AdpA were also extensively catalogued (Tables 2 and 3; Supplementary Tables S3 and S4), which should contribute to future studies of the regulation of secondary metabolism and morphological differentiation in S. griseus. In fact, bldA was identified as an AdpA-target gene by this study, revealing a unique positive feedback loop between two global regulators, AdpA and BldA, in S. griseus.13 In addition to the identification of AdpA regulon, certain unique characteristics of AdpA regulation were revealed, as discussed separately below.
4.1. Numbers of in vivo AdpA-binding sites and transcriptional units belonging to the AdpA regulon
Although the AdpA-binding consensus sequence (5′-TGGCSNGWWY-3′) has four alternative nucleotides and one free nucleotide, the ‘category-I’ AdpA-binding sites usually contain a few inconsistent nucleotides within the consensus sequence. Thus, the DNA-binding specificity of AdpA is much lower than that of other well-known regulators in bacteria. ChIP/ChAP analysis of 18-h liquid culture and ChAP analysis of 1-, 2-, and 3-day solid cultures indicated ∼1500 AdpA-binding sites. Approximately 35% of them were located on regions that appeared not to affect transcription (i.e. ‘coding’ and ‘intergenic’ regions). Furthermore, transcriptome analysis indicated that ∼60% of the AdpA-binding sites in ‘putative regulatory regions’ should not affect transcription. Taken together, ∼75% of the in vivo AdpA-binding sites seemed not to be involved in gene regulation, and only ∼25% of them were ‘apparently relevant’. We found 262 and 172 AdpA-binding sites that are apparently involved in transcriptional activation (217 transcriptional units, 342 genes) and repression (156 transcriptional units, 224 genes), respectively, in either liquid or solid culture. To confirm the AdpA dependency of these transcriptional units, the effect of mutation of AdpA-binding sites on transcription of each of the putative target genes should be examined. We previously revealed that the bldK operon is indirectly activated by AdpA, in spite of AdpA binding to the upstream region of bldK.18 Therefore, it is possible that some of the putative AdpA-dependent transcriptional units described above are actually regulated indirectly by AdpA (see the legend for Supplementary Table S3). It is also possible that many of them are directly regulated by AdpA, like the ‘category-I’ AdpA-target genes.
Recently, den Hengst et al.19 used ChIP-chip technology to identify in vivo binding regions of BldD, which is a global transcriptional repressor for both morphological differentiation and secondary metabolism in Streptomyces coelicolor A3(2). They identified 167 putative BldD-binding sites across the chromosome. Transcriptome analysis with a bldD mutant and its parent strain confirmed that at least 29 of the 167 BldD-binding sites were relevant. Similarly, Molle et al.10 identified 54 transcriptional units (121 genes) that are under the direct control of Spo0A, the master regulator for entry into sporulation in Bacillus subtilis. Our current study revealed that AdpA directly controls many more genes than these global transcriptional regulators for bacterial differentiation. To the best of our knowledge, the AdpA regulon seems to be the largest one in bacteria.
4.2. The strength of the AdpA–DNA interaction is not related to the biological significance of AdpA-binding sites
Approximately 75% of the in vivo AdpA-binding sites apparently have no function in the regulation of gene expression. Similar results have been observed for E. coli RutR,20 Bacillus subtilis AbrB,21 and Streptomyces coelicolor BldD;19 binding of them to most of their in vivo binding sites did not apparently affect gene expression. We speculate that AdpA binding to such regions has no or little biological significance. ChIP-chip analysis by Grainger et al.22 revealed that CRP in E. coli bound to 68 high-affinity sites, including 29 known CRP-dependent promoter regions, and interacted with thousands of weaker sites across the chromosome. They suggested that CRP should be considered as a chromosome-shaping protein contributing to the compaction of the chromosome (because DNA-bound CRP bends its target sharply), in addition to its function as a promoter-specific regulator. We cannot exclude the possibility that AdpA also functions as a chromosome-shaping protein in S. griseus. However, a striking difference between the DNA-binding profiles of CRP and AdpA concerns their DNA-binding strengths. As described above, CRP binds strongly to its target promoter regions and interacts weakly with ‘background’ regions. In contrast, AdpA binds with a similar strength to ‘apparently irrelevant’ regions similarly to ‘apparently relevant’ regions. Thus, the strength of the AdpA–DNA interaction is not related to the functional significance of particular AdpA-binding sites. The broad range of DNA-binding strengths to a variety of genuine target promoter regions, as well as ‘apparently irrelevant’ regions, is one of the distinctive properties of AdpA. We hypothesize that the position of the AdpA-binding site relative to the transcriptional start point, rather than the binding strength, is important for transcriptional regulation by AdpA. In fact, our previous studies have shown that AdpA activates 10 of 14 ‘category-I’ AdpA-target genes by binding to –50 to –70 regions in respect to their transcriptional start points.
4.3. Complexity of the DNA-binding properties of AdpA
Regarding the selected 115 probable AdpA-binding sequences, we showed poor correlation of AdpA-binding strength between in vitro and in vivo conditions, while the sequences that were strongly bound by AdpA in vitro generally showed relatively high AdpA-binding strength in vivo. Furthermore, of the 304 in vivo AdpA-binding regions examined, 20 regions (6.6%) were not bound by AdpA in vitro, although they contain AdpA-binding consensus-like sequences. The ChIP-chip analyses of several bacterial global regulators, such as CtrA in Caulobacter crescentus23 and LexA and FNR in E. coli,24,25 have indicated that these regulators bind dozens of ‘non-canonical’ sites in vivo, which are not similar to their consensus binding sequences. Interestingly, these unconventional targets were not bound by the respective regulators in vitro. Wade et al.24 suggested that binding of LexA to these unconventional targets requires additional factors, such as cooperative interaction with other proteins and/or unusual DNA conformation (e.g. bent or supercoiled DNA). Thus, AdpA may also require similar additional factors to bind some of the AdpA-binding sites detected by ChIP/ChAP-seq. Such additional factors may explain the poor correlation of AdpA-binding strength between in vitro and in vivo conditions. We postulated that AdpA might frequently interact with other DNA-binding proteins to regulate target genes. This assumption is also supported by the observation that the expression profiles of apparently AdpA-dependent genes were very different between liquid and solid cultures, despite the similar AdpA-binding profiles in both cultures. Thus, in vivo, AdpA seems to exert its regulatory functions cooperatively with other proteins in many cases. Detailed analysis of cooperative regulation between AdpA and other regulatory proteins will form a very important part of our future studies.
Importantly, the degree of similarity to the AdpA-binding consensus sequence was not necessarily consistent with the strength of the AdpA–DNA interaction of each AdpA-binding site (both in vivo and in vitro), although the sequences that were strongly bound by AdpA in vitro showed a relatively high similarity to the consensus sequence. This is a striking contrast to the binding mode of LexA to its conventional targets, in which the strength of the LexA–DNA interaction can be well approximated by the degree of similarity to the LexA-binding consensus sequence.24 However, there are analytical limitations that should be taken into consideration, as well as our incomplete understanding of the molecular mechanism of DNA recognition by AdpA. First, in this analysis, type-I AdpA binding, in which both subunits of the AdpA dimer bind each of the two divergent AdpA-binding sequences, was not considered, because we could not conclusively identify type-I-binding sites without DNase I footprinting. When AdpA binds to a type-I site, another AdpA-binding sequence in the vicinity should affect the strength of the AdpA–DNA interaction. Therefore, regarding genuine type-I sites, we could not correctly estimate the relationship between the strength of the AdpA–DNA interaction and the degree of similarity to the consensus sequence. Secondly, it is possible that some DNA fragments used for the competitive EMSA had two (or more) AdpA-binding sites. Our ChIP/ChAP-seq analysis clearly detected two peaks that are separated by 200 bp in the upstream region of sgmA (Fig. 2B). However, we failed to discriminate two AdpA-binding sites13 that are separated by 50 bp in the upstream region of bldA (data not shown). When AdpA binds to two (or more) sites in a DNA fragment, it may lead to overestimation of the binding strength of the identified AdpA-binding sequence. Finally, in some cases, the additional eight nucleotides 3′ to the consensus sequence may contribute to the DNA-binding strength more significantly than expected. AdpA, which belongs to the AraC/XylS family, has two helix-turn-helix DNA-binding motifs. Based on the DNA-binding characteristics of some AraC/XylS family regulators, such as AraC, XylS, and SoxS, we proposed that the helix-turn-helix at the N-terminal side (HTH-1) recognizes the relatively highly conserved TGGCS sequence at the 5′-end of the consensus sequence.2 The other helix-turn-helix at the C-terminal side (HTH-2) recognizes the other nucleotides in the consensus sequence with the next eight nucleotides.2 Although nucleotide recognition by HTH-2 seems to be less important than that by HTH-1, it may significantly affect the binding strength in some cases.
4.4. AdpA as a transcriptional repressor
It was unexpected that AdpA could directly repress more than 100 transcriptional units in ether liquid or solid culture. Our previous study indicated that AdpA represses transcription of its own gene by binding to three sites in the 5′ upstream region of adpA, probably forming a loop structure of the DNA.16 However, ChIP/ChAP-seq analysis detected only one AdpA-binding site in the ‘putative regulatory region’ of most (>90%) of the transcriptional units that were apparently repressed by AdpA. In such cases, AdpA may bind to the −35 and/or −10 sequences to hamper RNA polymerase from binding to them to initiate transcription. Interestingly, several genes that are presumably involved in primary metabolism are included in the list of possible genes directly repressed by AdpA in both liquid and solid cultures (Table 3). To reveal a new aspect of global gene regulation by AdpA, further detailed analyses of the repressor function of AdpA is necessary.
4.5. Biological significance of the low DNA-binding specificity of AdpA
Although bacterial genomes are packaged by histone-like proteins into a nucleoid structure, this structure is not analogous to eukaryotic chromatin; bacterial genomes are permissive to transcription factor binding.21,22,24 Wade et al.24 proposed that bacteria must evolve transcription factors with high DNA-binding specificity because of equal accessibility of the genomes. However, this is obviously not applicable to AdpA; the DNA-binding specificity of AdpA is very low, and AdpA can bind to >1500 sites (including many ‘apparently irrelevant’ sites) across the chromosome. AdpA orthologues are highly conserved (more than 90% amino acid similarity) among many Streptomyces species, and their essential roles in morphological differentiation and/or secondary metabolism have been revealed in various Streptomyces strains.13 Surprisingly, the two helix-turn-helix DNA-binding motifs are completely identical among these proteins, indicating that they have the same DNA-binding specificity. Thus, the low DNA-binding specificity of AdpA has been maintained during evolution, suggesting that this atypical characteristic of AdpA as a bacterial transcription factor should be very important for the biology of Streptomyces. There could be some advantages conferred by the low DNA-binding specificity of AdpA for Streptomyces. First, the low DNA-binding specificity of AdpA enables binding to many sites on the chromosome, which facilitates regulation of many genes. For transition from vegetative growth to reproductive (differentiating) growth, including secondary metabolite production, a drastic change of gene expression is undoubtedly necessary. Therefore, the low DNA-binding specificity seems to be advantageous to AdpA as a pivotal regulator for this process. Secondly, the low DNA-binding specificity of AdpA may generate evolutional flexibility of the regulatory network. It is thought that Streptomyces achieves the productivity of a wide variety of secondary metabolites by acquiring foreign biosynthetic enzyme genes through horizontal gene transfer.26 The low DNA-binding specificity of AdpA may increase the occurrence of an AdpA-binding site(s) in the regulatory region of newly acquired genes, which may increase the chance that AdpA regulates them. Thus, the low DNA-binding specificity of AdpA may enable frequent reconstruction of local gene regulation. This may be advantageous to the evolution of Streptomyces.
4.6. The A-factor regulatory system in Streptomyces griseus
Streptomyces griseus seems to have evolved the highly integrated A-factor regulatory cascade, and the function of AdpA as a global regulator for morphological differentiation and secondary metabolism may be much more significant in S. griseus compared with other Streptomyces species. In the A-factor regulatory system, the hormonal signal triggers the production of only one transcriptional regulator, AdpA, which directly regulates the transcription of more than 500 genes. This ‘highly converged’ regulatory system seems to represent an example of sophisticated global gene regulation in bacteria. This system has the advantage of being able to switch the expression profiles of many genes in a relatively short period. We think that this system may reflect a fundamental aspect of differentiation of Streptomyces.
Supplementary Material
Acknowledgements
We thank N. Nakata for technical assistance with the competitive EMSA.
Footnotes
Edited by Naotake Ogasawara
Supplementary Data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by the Targeted Proteins Research Program (TPRP) of the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) (grant number A2); and a Funding Program for Next Generation World-Leading Researchers from the Bureau of Science, Technology, and Innovation Policy, Cabinet Office, Government of Japan (grant number GS006).
References
- 1.Ohnishi Y., Kameyama S., Onaka H., Horinouchi S. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 1999;34:102–11. doi: 10.1046/j.1365-2958.1999.01579.x. [DOI] [PubMed] [Google Scholar]
- 2.Ohnishi Y., Yamazaki H., Kato J.Y., Tomono A., Horinouchi S. AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 2005;69:431–9. doi: 10.1271/bbb.69.431. [DOI] [PubMed] [Google Scholar]
- 3.Ohnishi Y., Ishikawa J., Hara H., et al. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 2008;190:4050–60. doi: 10.1128/JB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirano S., Kato J.Y., Ohnishi Y., Horinouchi S. Control of the Streptomyces subtilisin inhibitor gene by AdpA in the A-factor regulatory cascade in Streptomyces griseus. J. Bacteriol. 2006;188:6207–16. doi: 10.1128/JB.00662-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hara H., Ohnishi Y., Horinouchi S. DNA microarray analysis of global gene regulation by A-factor in Streptomyces griseus. Microbiology. 2009;155:2197–210. doi: 10.1099/mic.0.027862-0. [DOI] [PubMed] [Google Scholar]
- 6.Akanuma G., Hara H., Ohnishi Y., Horinouchi S. Dynamic changes in the extracellular proteome caused by absence of a pleiotropic regulator AdpA in Streptomyces griseus. Mol. Microbiol. 2009;73:898–912. doi: 10.1111/j.1365-2958.2009.06814.x. [DOI] [PubMed] [Google Scholar]
- 7.Yamazaki H., Tomono A., Ohnishi Y., Horinouchi S. DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 2004;53:555–72. doi: 10.1111/j.1365-2958.2004.04153.x. [DOI] [PubMed] [Google Scholar]
- 8.Kolb A., Busby S., Buc H., Garges S., Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 1993;62:749–95. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 9.Walker G.C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol. Rev. 1984;48:60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molle V., Fujita M., Jensen S.T., et al. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 2003;50:1683–701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa S., Ogura Y., Yoshimura M., et al. Distribution of stable DnaA-binding sites on the Bacillus subtilis genome detected using a modified ChIP-chip method. DNA Res. 2007;14:155–68. doi: 10.1093/dnares/dsm017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park P. J. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–80. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higo A., Horinouchi S., Ohnishi Y. Strict regulation of morphological differentiation and secondary metabolism by a positive feedback loop between two global regulators AdpA and BldA in Streptomyces griseus. Mol. Microbiol. 2011;81:1607–22. doi: 10.1111/j.1365-2958.2011.07795.x. [DOI] [PubMed] [Google Scholar]
- 14.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji H., Jiang H., Ma W., Johnson D.S., Myers R.M., Wong W.H. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008;26:1293–300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato J.Y., Ohnishi Y., Horinouchi S. Autorepression of AdpA of the AraC/XylS family, a key transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. J. Mol. Biol. 2005;350:12–26. doi: 10.1016/j.jmb.2005.04.058. [DOI] [PubMed] [Google Scholar]
- 17.Bailey T.L., Gribskov M. Methods and statistics for combining motif match scores. J. Comput. Biol. 1998;5:211–21. doi: 10.1089/cmb.1998.5.211. [DOI] [PubMed] [Google Scholar]
- 18.Akanuma G., Ueki M., Ishizuka M., Ohnishi Y., Horinouchi S. Control of aerial mycelium formation by the BldK oligopeptide ABC transporter in Streptomyces griseus. FEMS Microbiol. Lett. 2011;315:54–62. doi: 10.1111/j.1574-6968.2010.02177.x. [DOI] [PubMed] [Google Scholar]
- 19.den Hengst C.D., Tran N.T., Bibb M.J., Chandra G., Leskiw B.K., Buttner M.J. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 2010;78:361–79. doi: 10.1111/j.1365-2958.2010.07338.x. [DOI] [PubMed] [Google Scholar]
- 20.Shimada T., Ishihama A., Busby S.J., Grainger D.C. The Escherichia coli RutR transcription factor binds at targets within genes as well as intergenic regions. Nucleic Acids Res. 2008;36:3950–5. doi: 10.1093/nar/gkn339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chumsakul O., Takahashi H., Oshima T., et al. Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation. Nucleic Acids Res. 2011;39:414–28. doi: 10.1093/nar/gkq780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grainger D.C., Hurd D., Harrison M., Holdstock J., Busby S.J. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl. Acad. Sci. USA. 2005;102:17693–8. doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laub M.T., Chen S.L., Shapiro L., McAdams H.H. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle , Proc. Natl Acad. Sci. USA. 2002;99:4632–7. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wade J.T., Reppas N.B., Church G.M., Struhl K. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Dev. 2005;19:2619–30. doi: 10.1101/gad.1355605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grainger D.C., Aiba H., Hurd D., Browning D.F., Busby S.J. Transcription factor distribution in Escherichia coli: studies with FNR protein. Nucleic Acids Res. 2007;35:269–78. doi: 10.1093/nar/gkl1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chater K.F., Biró S., Lee K.J., Palmer T., Schrempf H. The complex extracellular biology of Streptomyces. FEMS Microbiol. Rev. 2010;34:171–98. doi: 10.1111/j.1574-6976.2009.00206.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.