Abstract
Amyloid β peptide (Aβ) plaque in the brain is the primary (post mortem) diagnostic criterion of Alzheimer’s disease (AD). Any physiological role of Aβ constituent is poorly understood. We have previously determined an Aβ interacting domain (AβID) in the promoters of AD–associated genes (Maloney and Lahiri, 2011). This AβID interacts in a DNA–sequence specific manner with Aβ. We now demonstrate novel Aβ activity as a possible transcription factor. Herein, we demonstrate Aβ–chromatin interaction in cell culture by ChIP assay. We observed that human neuroblastoma (SK–N–SH) cells treated with FITC conjugated Aβ1–40 localized Aβ to the nucleus in the presence of H2O2–mediated oxidative stress. Furthermore, primary rat fetal cerebrocortical cultures were transfected with APP and BACE1 promoter–luciferase fusions, and rat PC12 cultures were transfected with polymorphic APP promoter–CAT fusion clones. Transfected cells were treated with different Aβ peptides and/or H2O2. Aβ treatment of cell cultures produced a DNA sequence–specific response in cells transfected with polymorphic APP clones. Our results suggest the Aβ peptide may regulate its own production through feedback on its precursor protein and BACE1, leading to amyloidogenesis in AD.
Keywords: Alzheimer’s disease, amyloid beta, DNA–protein interaction, gene regulation, transcription factor
1. Introduction
Alzheimer’s disease (AD), the most common form of dementia in the elderly (Hebert et al., 2003), is associated with multiple risks that include genetic, epigenetic, dietary, and lifestyle factors (Lahiri and Maloney, 2010b). Neuronal loss, intraneuronal tangles of hyperphosphorylated microtubule–associated τ protein (MAPT), and extracellular deposition of β–amyloid peptide plaque are characteristic of the disorder. Amyloid plaque consists primarily of the 39–42 amino acid amyloid β peptide (Aβ) (Lahiri et al., 2003; De Strooper, 2010), which is cleaved from a larger Aβ precursor protein (APP) by sequenttial actions of the β– and γ–secretase enzymes (Lahiri et al., 2003). Aβ also accumulates within neurons in both human AD cases and transgenic AD models (Gouras et al., 2000; Shie et al., 2003). Non–pathological functions of Aβ include ion channel formation (Jang et al., 2010), kinase activation (Bogoyevitch et al., 2004; Tabaton et al., 2010), cholesterol transport regulation (Yao and Papadopoulos, 2002; Igbavboa et al., 2009), and protection against metal–induced oxidative damage (Zou et al., 2002; Baruch-Suchodolsky and Fischer, 2009). We now report that Aβ may act as a transcription factor with regulatory region(s) of AD–associated genes.
Extracellular Aβ enters the cell under oxidative and heat stress (Ohyagi et al., 2005; Ohyagi and Tabira, 2006; Ohyagi et al., 2007) and is transported to the nucleus by action of the Aβ–related death–inducing protein (AβDIP) (Ohyagi, 2008). Intracellular Aβ induces an increase in levels of the apoptosis–associated tumor protein 53 (p53, gene name TP53) (Ohyagi et al., 2005), the transcription factor achaete–scute complex homolog 1 (ASCL1) (Uchida et al., 2007), and transcription of the β–amyloid cleaving enzyme 1/β–secretase BACE1 gene (Giliberto et al., 2009; Tabaton et al., 2010), while reducing levels of oligodendrocyte lineage transcription factor 2 (OLIG2) (Uchida et al., 2007) and system A glutamine transporter 1 (SAT1/SLC38A1) (Buntup et al., 2008). In the case of p53, this induction has been shown to be from direct action of Aβ upon the TP53 promoter. In that particular case, Aβ activity hinged on binding a known heat shock element (HSE), “GGATTGGGGT” (Ohyagi et al., 2005).
We have recently identified a more general Aβ–interacting domain (AβID) in DNA sequences (Maloney and Lahiri, 2011). The AβID is a decamer with a consensus sequence “KGGRKTGGGG”. Substitution of G→A in the seventh nucleotide of the decamer eliminated DNA–peptide interaction. This substitution corresponds to a single–nucleotide polymorphism (SNP) associated with increased AD risk (Lahiri et al., 2005b).
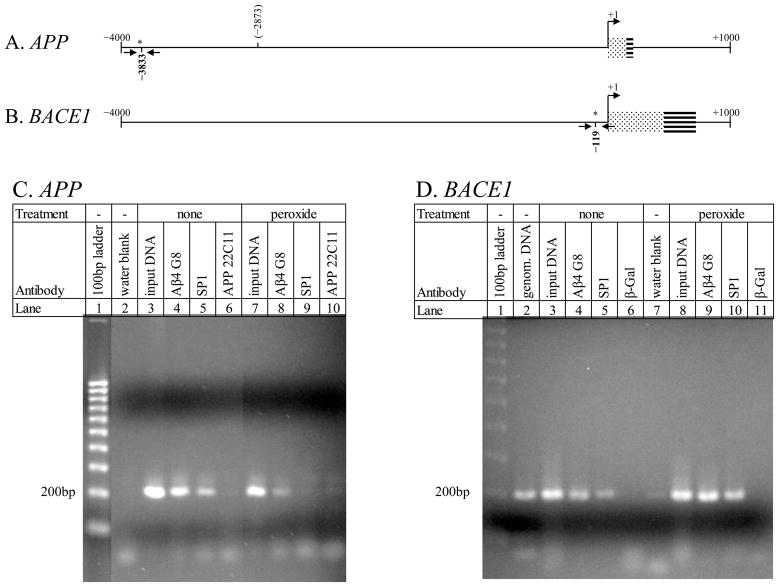
We now turn our attention to investigating functional activity related to Aβ–AβID interaction. We first determined that Aβ associated with DNA in situ in an AβID–related fashion. We did this through chromatin immune precipitation (ChIP) assay. Our ChIP for two chromatin regions that each contained an AβID on the APP gene (APP) and BACE1 promoter sequences (Maloney and Lahiri, 2011) demonstrated in situ Aβ binding to both of the 200 bp regions. PCR signal did not occur when chromatin was precipitated with antibodies against the N–terminal region of APP or against β–galactosidase (β–gal).
To determine induction of Aβ into cell nuclei under oxidative stress, cell cultures were treated with fluorescein isothiocyanate conjugated Aβ1–40 (FITC–Aβ1–40), added to cell cultures in the presence or absence of a oxidative stress induced by H2O2. To determine functional activity of Aβ on 5′–flanking regions of the APP and BACE1 genes, we generated clones containing i) a confirmed AβID within a 3.3 kb BACE1 fragment and ii) lacking any predicted AβID within a 1.2 kb APP fragment. These fragments were each fused to the firefly luciferase coding sequence, and the fusion clones were transfected into primary rat fetal cerebrocortical neuronal (PRCN) cultures. Transfected cultures were treated with Aβ1–42, 1–40, and 1–28 peptides. We found that oxidative stress induces Aβ localization to the nucleus, and that Aβ increased activity in the BACE1 promoter.
In addition, we tested the response to Aβ of two clones previously constructed in our laboratory. Specifically, these two clones correspond to an APP promoter SNP linked to AD risk at −3829 from the +1 TSS (Lahiri et al., 2005b). This polymorphism is within an AβID. We show that oxidative stress and Aβ treatment significantly increase transcriptional activity of the AD–associated SNP. Taken together, these results suggest that the Aβ peptide may possibly function as a transcription factor or co–factor. In addition to other function(s), the Aβ peptide directs normal apoptosis as well as regulating its own production through feedback on its precursor, APP, and the β–secretase enzyme. This would have pathological consequences relevant to AD. We suggest that under a cytotoxic model, normal cytoprotective activity of Aβ, such as protection against metal–induced oxidative stress, would result in increasing Aβ levels crossing a pathogenic threshold, pushing production of APP and BACE1 to pathological levels, initiating a positive feedback loop. Higher levels of BACE1 protein would favor greater production of Aβ, and higher levels of APP protein would provide more substrate. The combination would result in greater Aβ production, which would stimulate the production of proapoptotic proteins. Thus, Aβ’s proposed activity as either TF or cofactor would lead to accumulation of excess Aβ as toxic extracellular amyloid plaque, the hallmark of AD.
2. Materials and Methods
2.1 Chemicals/Reagents
Chemicals and reagents were purchased from Sigma–Aldrich (St. Louis, MO) and were of “molecular biology” or “analytic” quality unless stated otherwise. DNA modifying and restriction enzymes were purchased from Roche (Indianapolis, IN). Different media and sera and other tissue culture reagents were procured from Invitrogen (Carlsbad, CA).
2.2 Cell culture for transfection and Aβ treatment
Human SK–N–SH neuroblastoma (NB) and rat pheochromocytoma (PC12) cell cultures were acquired from ATCC and cultured in our laboratory as previously described (Ghosh et al., 2000). Briefly, NB cells were seeded into 10 cm culture plates (for ChIP) or 24–well culture plates (for live cell imaging) and maintained in DMEM medium supplemented with 10% fetal bovine serum. PC12 cells were seeded into a collagenated 96–well plate for transfections at 30,000 cells per well and allowed to attach overnight in standard growth medium: RPMI supplemented with 10% horse serum (Invitrogen), 5% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA), and an antibiotic cocktail (Invitrogen).
For primary neuronal cultures, timed–pregnancy Sprague Dawley rats were purchased from Harlan Laboratories (Indianapolis). Animals were treated in accordance to the Indiana University School of Medicine Institutional Animal Use Committee guidelines. PRCN cultures were prepared from E16 rat fetal cerebrocortical tissue using slight modifications from previously published procedures (Brewer et al., 1993; Bailey and Lahiri, 2006). The dam was sacrificed in a CO2 chamber and decapitated. The uterus was removed through an incision in the abdomen and the pups transferred to ice–cold Hanks Buffered Salt Solution (Invitrogen) supplemented with 1 mM sodium pyruvate (Sigma–Aldrich) and antibiotic cocktail (HBSS; Invitrogen). After rinsing, heads were moved to fresh HBSS for dissection. Under a dissecting microscope, overlying tissues and vasculature were removed from the brain surface with fine forceps and whole cortices were separated from subcortical tissue and collected in a separate tube of HBSS on ice. After all cortices were collected, the overlying HBSS was removed and the tissue was homogenized by trituration in 2–3 ml Neurobasal medium supplemented with B27 serum replacement and 5 ng/ml human recombinant basic fibroblast growth factor (Invitrogen). Cells were counted and seeded into poly–D–lysine (Sigma–Aldrich) coated 6– or 12– well plates (Corning, Lowell, MA) at 2,200 cells/mm2 (2×106 cells per well in 6 well plates).
2.3 Aβ peptides and their fragments
Different forward and reverse peptides, Aβ1–42, 1–40, 1–28, 25–35, 29–40, 31–35, 42–1, 40–1, 35–25, FITC–Aβ1–40 were purchased as trifluoroacetic acid salts from Bachem (Torrance, CA) and resuspended at stock concentrations of 1 mg/ml in different solvents per manufacturer’s recommendations. Consultation with the technical staff of the manufacturer indicated that peptides dissolved under these conditions would be dimers or larger aggregates.
2.4 Chromatin immune precipitation (ChIP) of Aβ on specific sequences of the APP and BACE1 promoters
Human NB cells were grown to 70%–80% confluency. One set of cultures was treated with 100 μM H2O2, and all cultures were fixed with 37% formaldehyde, as specified in the ChIP–IT Express kit protocol (ActiveMotif). Chromatin was sheared enzymatically with the Enzymatic ChIP–IT Express kit. Sheared chromatin was then immunoprecipitated overnight with each of the following antibodies: mAb 4G8 (anti–Aβ), mAb 22C11 (anti N–terminus APP), anti–specificity protein 1 (SP1) transcription factor (Santa Cruz Biotechnology, Santa Cruz, CA), or anti–β galactosidase. After immunoprecipitation, PCR was conducted with oligomers flanking the APP −3833 (5′–AGG CCA AAG ATG AGT GAC AGG A–3′, 5′–GGG TTA GGA TCC ATG CTA ATG ACC–3′) or BACE1 −119 (5′–AAG AAA GAC TGA CAG ACG GGA GGT–3′, 5′–GGC GGC TGT CAA AGC CAA A–3) AβIDs. For the APP PCR, the 22C11–precipitated material was used as negative control. For the BACE1 PCR, the anti–β gal precipitated material was used as negative control.
2.5 Aβ treatment of SK–N–SH cells in presence or absence of H2O2
SK–N–SH cells were neuronally differentiated with 10 μM all–trans retinoic acid for 7 days prior to subsequent treatments. Cels were then treated with 1 μM FITC–Aβ1–40 (Bachem) in the presence or absence of 50 μM H2O2 in low serum containing medium (MEM with 1% fetal bovine serum and antibiotics) for 48 hours. Cells were washed in Dulbecco’s PBS and labeled with 10 μM Hoechst 33342 (Sigma) and 1 μM ethidium homodimer (EtHD; Molecular Probes) for 15 minutes at 37°C. Images were taken using a Leica DM IL microscope with an RT–SE SPOT camera, using SPOT Advanced software (Diagnostic Instruments). Experiments were performed in triplicate. Nuclei of live cells, nuclei of dead cells, and nuclei of live cells with positive FITC–Aβ signal were counted, and results analyzed via Fisher’s exact test.
2.6 Construction of promoter–enhancer chloramphenicol transferase (CAT) expression clones
Two clones were previously generated to measure expression effects of a SNP in the APP promoter. This polymorphism is associated with some cases of familial AD. Briefly, 24–mers flanking the −3833G and −3833A sequences of the APP promoter were inserted into the pCAT3P promoter vector (Promega, Madison, WI), which contains the truncated SV40 promoter, to generate two APP polymorphic CAT expression vectors (Lahiri et al., 2005b).
2.6 Construction of luciferase–fusion promoter expression clones
Two clones were generated for evaluation of potential Aβ activity on APP and BACE1 5′–flanking regions. The human BACE1 5′–flanking region clone pBACEP2 (Ge et al., 2004b) was cut with HindIII and XhoI and a 3.3 kb fragment of the BACE1 5′–flanking sequence was gel–purified. This was cloned into the HindIII/XhoI sites of the pGL3–Basic vector (Promega). Additionally, the APP 5′–flanking region clone pAmyl1.2 (Lahiri and Robakis, 1991) was digested with PstI and BamHI. This was blunted with T4 polymerase and cloned into the SmaI site of pGL3–Basic. Clones were verified by appropriate restriction enzyme digest. Transfections of CAT–based constructs were performed using the Lipofectamine with Plus Reagent per manufacturer’s recommended protocol (Invitrogen).
2.8 Aβ exposure of cells transfected with BACE1 or APP promoter constructs
PRCN cells were seeded onto poly–D–lysine coated white–walled 96–well plates (Corning) at 75,000 cells/well in Neurobasal medium supplemented with B27, antibiotic cocktail (Invitrogen) and Normocin (InVivoGen, San Diego, CA). Cells were allowed to differentiate for 14 days prior to transfection, with media changes every fourth day. Cells were transfected with 275 ng/well of the pGL3–Basic plasmid (Promega) containing either the BACE1 3.3 kb or APP 1.2 kb promoter fragment, each fused to the firefly luciferase reporter gene, using 0.75 μl/well of the Transfectin transfection reagent (BioRad, Hercules, CA). Cells were simultaneously transfected with 25 ng/well of the pRL–SV40 Renilla luciferase plasmid (Promega) as an internal control. Twenty–four hours after transfection, 1 μM Aβ1–28, Aβ1–40, or Aβ1–42 was added to the medium. After an additional 24–hour incubation following the addition of Aβ, both firefly and Renilla luciferase activities were measured with the Dual–Luciferase kit and Glowmax luminometer (Promega). Firefly luciferase signal was normalized to Renilla luciferase signal to account for variations in transfection efficiency.
2.9 Aβ exposure of cells transfected with polymorphic APP promoter constructs in presence and absence of H2O2
PC12 cells were seeded and grown as described herein. Transfections were done with 3 μg total DNA, using the Lipofectamine transfection reagent. The pCAT3P empty vector and clones −3833G–CAT, and −3833A–CAT (Lahiri et al., 2005b), were used. At the end of the recovery period, cells were treated with either vehicle, 1 μM Aβ25–35, 50 μM H2O2, or both Aβ and H2O2 at 1 μM and 50 μM, respectively, in 0.5 ml RPMI with 1% horse serum and 0.25% fetal bovine serum (Table 1) and incubated 24 hours. Cells were harvested and lysed. Total lysate protein was measured by Bradford assay (BioRad). Reporter CAT protein levels were measured by ELISA (Roche). CAT signal was adjusted by protein concentration of the cell lysates. Results were analyzed by generalized linear mixed model (Dean and Nielsen, 2007).
Table 1.
Treatment levels for analysis of Aβ and peroxide (H2O2) effects on two different polymorphic APP promoter–CAT reporter fusion clones in neuronal (PC12) cells
| Clone | pCAT3P (Vector alone) | − 3833A | − 3833G | |||
|---|---|---|---|---|---|---|
| Treatment | Aβ | H2O2 | Aβ | H2O2 | Aβ | H2O2 |
| vehicle | 0 | 0 | 0 | 0 | 0 | 0 |
| Aβ | 1 μM | 0 | 1 μM | 0 | 1 μM | 0 |
| peroxide | 0 | 50 μM | 0 | 50 μM | 0 | 50 μM |
| Aβ + perox | 1 μM | 50 μM | 1 μM | 50 μM | 1 μM | 50 μM |
2.10 Data analysis
Transfection assays were performed at N = 4 or N = 5 per treatment. All statistical analysis was carried out using SAS 9.1.3 (SAS Institute, Cary, NC). Expression data was tested for distribution and specific general linear mixed model analysis applied accordingly.
3. Results
3.1 ChIP assay demonstrates Aβ binding to specific sequences within the APP and BACE1 gene promoters on cell culture chromatin
ChIP samples from human NB cells were generated in duplicate, and PCR was carried out. The APP region −3899/−3708 (Fig. 1A), overlapping the AβID at −3833, and BACE1 region −205/−27 (Fig. 1B), overlapping the AβID at −119, were selected for analysis. The APP region (Fig. 1C) had in situ interaction with Aβ. In addition, the AβID at −3833 overlaps a predicted SP1 binding site. Precipitation with anti–SP1 indicated the likelihood that this site actively binds SP1 in human NB cells. Notably, precipitation with 22C11 revealed no signal by PCR. 22C11 was used as a negative control antibody, since it binds the opposite end of the APP protein from the Aβ peptide. Lack of PCR signal from the 22C11 precipitation is strongly suggestive of specificity for the 4G8 anti–Aβ and anti–SP1 antibodies. Peroxide treatment of the cells had reduced but did not eliminate SP1–APP interaction without altering Aβ–APP interaction. For the BACE1 segment, DNA–protein interaction occurred with both Aβ and SP1, as indicated by ChIP (Fig. 1D), with no signal generated by a negative anti–β–galactosidase control precipitation. Aβ and SP1 binding activity was observed both in the presence and absence of H2O2 treatment. The ChIP assay was repeated on separate occasions with different bathes of cells. Results were similar to representative data shown here.
Fig. 1. Chromatin Immunoprecipitation (ChIP) assay of Aβ in NB cell nuclei.
ChIP and PCR were carried out as described in the text. A. ChIP for Aβ binding a 200 bp region containing the predicted APP −3833 AβID. B. ChIP for Aβ binding a 200 bp region containing the BACE1 −119 AβID. C. Precipitated, cross–linked chromatin was probed for presence of Aβ peptide (lanes 4, 8), SP1 (lanes 5, 9), and the N–terminus of the APP protein (lane 6) within the indicated region of the APP promoter. D. Precipitated, cross–linked chromatin was probed for presence of Aβ peptide (lanes 4, 9), SP1 (lanes 5, 10), and β–galactosidase (lanes 6, 11) within the indicated region of the BACE1 promoter. Treatment conditions in the cells and other appropriate controls in the assay were as indicated in the figure.
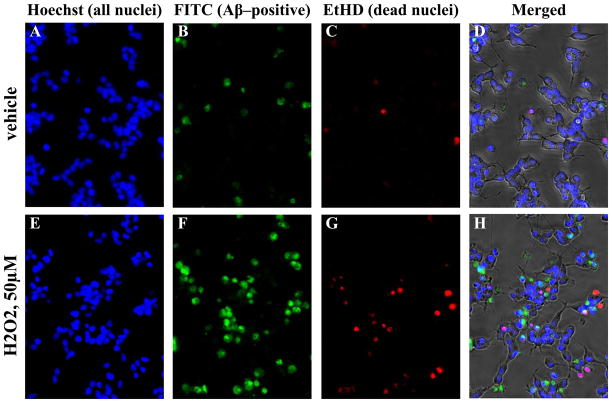
3.2 Treatment of human neuroblastoma cell cultures with H2O2 promotes uptake of Aβ into cell nuclei
SK–N–SH cell cultures were treated under low serum conditions (1% FBS) with vehicle + 1 μM FITC–Aβ1–40 or 50 μM H2O2 + 1 μM FITC–Aβ1–40. After 48 hours, cells were incubated with Hoechst 33342 dye, washed, and incubated with EtHD. Some quenching of FITC fluorescence by EtHD was observed, however brightness adjustment of individual channels confirmed that in H2O2 treated cells, nearly all EtHD positive nuclei observed were also positive for FITC–Aβ. In the vehicle treatment, very few FITC–Aβ1–40 positive cells were observed as well as very few EtHD positive dead cells. Most nuclei in both treatment groups were labeled blue by the Hoechst 33342 dye alone (Fig. 2A, E), indicating that these cells remained intact during this treatment. Nuclei labeled red by EtHD were dead cells with permeabilized membranes (Fig. 2C, G). Nuclei were counted according to EtHD, FITC, and Hoechst 33342 signal presence. Counts were analyzed by Fisher’s exact test. The number of dead nuclei was slightly but significantly higher (p < 10−5, odds ratio 0.576) with H2O2 treatment (Table 2). FITC–Aβ labeling among live cells was significantly more common (p < 10−15, odds ratio 3.118) in H2O2 treated cells (Fig. 2B, F; Table 2) than in vehicle–treated cells. The odds ratio would correspond to a “moderate” effect size if converted to the scale of Cohen’s d (Ferguson, 1966; Rosenthal, 1994; Hopkins, 2000).
Fig. 2. Treatment of SK–N–SH cells with Aβ in the presence or absence of H2O2.
Human SK–N–SH neuroblastoma cells were treated with FITC–Aβ1–40 in presence or absence of H2O2 as described in the text. Cells were incubated with these treatments for 48 hours and visualized with Hoechst 33342 dye, and EtHD, corresponding to all cell nuclei and dead nuclei, respectively. Cells were fluorescently imaged at appropriate wavelengths for FITC, Hoechst treatment, and EtHD treatment. A and E. with all nuclei labeled with Hoechst 33342. B and F. FITC–Aβ1–40 labeled nuclei. C and G. Dead cells labeled with EtHD. D and G. Composite of all three fluorescent signals overlaid on a phase contrast image of the cells.
Table 2.
Treatment of human neuroblastoma (SK–N–SH) cultures with peroxide (H2O2) + Aβ vs. treatment with vehicle +Aβ
| All Nucleia | All Live Nucleib | Live, with FITC signalc | |
|---|---|---|---|
| Vehicle | 1873 | 1736 (92.7%)d | 239 (13.8%)e |
| H2O2f | 1727 | 1519 (88.0%) | 505 (32.2%) |
Counts of nuclei stained by Hoechst 33342 dye.
Counts of nuclei stained by Hoechst 33342 dye minus counts of nuclei stained by Hoechst and EtHD. Fisher’s exact test p < 10−5, odds ratio 0.576 (“small” effect size).
Counts of nuclei not stained by EtHD but positive for Hoechst 33342 and FITC fluorescence. Fisher’s exact test p < 10−15, odds ratio 3.118 (“moderate” effect size).
Percent is live nuclei vs. all nuclei.
Percent is live with FITC vs. live without FITC.
50 μM H2O2.
Signals from all three fluorescent labels are shown overlaid with phase contrast images of the same cells (Fig. 2D, H). Dead nuclei were almost invariably FITC–positive. Given that our criterion for identifying dead cells is membrane permeability, it is not clear in this system whether nuclear localization of FITC–Aβ to the nucleus is part of the apoptotic cascade leading to cell death, or if it leaked into the cell after death. Thus, EtHD positive dead nuclei were excluded from the analysis of FITC–Aβ uptake. We attempted to test the specificity of the Aβ uptake observed in these cells by adding a 5–fold excess of unlabeled Aβ1–40, however this produced increases in cell death as well as apparent aggregation and precipitation, resulting in labeling of non–nuclear structures (data not shown). This phenomenon is the subject of continued investigation. Under the conditions used here, H2O2 mediated oxidative stress more than doubled the number of nuclei that incorporated detectable levels of FITC–Aβ1–40.
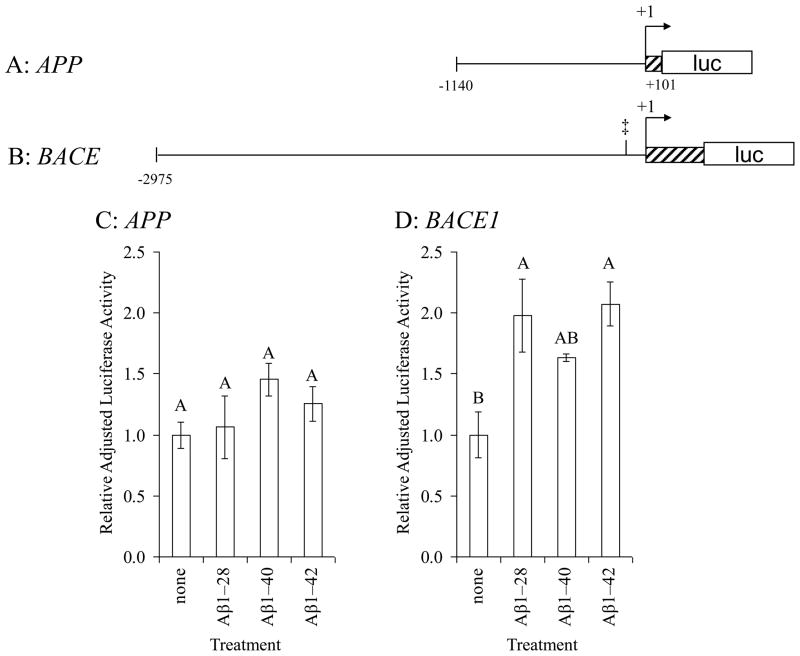
3.3 Aβ treatment altered transient expression of reporter fused to BACE1 5′–flanking region that contains an AβID
PRCN cell cultures were transiently transfected with a 1.2 kb fragment of the APP promoter fused to firefly luciferase (Fig. 3A) and treated with 1 μM Aβ1–28, Aβ1–40, and Aβ1–42. Firefly luciferase activity was measured and adjusted to levels of co–transfected internal control plasmid expressing Renilla luciferase. PRCN cells transfected with the proximal APP promoter fragment showed little response to Aβ treatment (Fig. 3C). No AβIDs were predicted within the APP 1.2 kb fragment. In cells transfected with the BACE1–CAT reporter clone (Fig. 3B), treatment with all three Aβ peptides corresponded to higher levels of adjusted luciferase activity (Fig. 3D). Interaction between Aβ and the BACE1 AβID site at −119 has been confirmed by electrophoretic mobility shift assay (EMSA) (Maloney and Lahiri, 2011).
Fig. 3. Effects of Aβ treatment on APP and BACE1 5′–flanking sequence activity.
A. A 1.2 kb fragment of the APP 5′–flanking region (Song and Lahiri, 1998) was cloned into the pGL3 luciferase expression vector as described in the text. The APP fragment contains a putative site for Aβ binding. B. A 3.3 kb fragment of the human BACE1 5′–flanking region (Ge et al., 2004b) was cloned into the pGL3 luciferase expression vector as described in the text. The BACE1 fragment contains a confirmed site for Aβ binding, determined herein. Primary rat cerebrocortical neuronal (PRCN) cultures were transiently transfected with a Renilla luciferase control vector and C. APP/pGL3 firefly luciferase fusion clone or D. BACE1/pGL3 luciferase fusion clone. Transfected cells were left untreated or treated with Aβ1–28, 1–40, or 1–42, all 1 μM. Cells were harvested and activities of firefly and Renilla luciferases were measured. Firefly luciferase activity within each treatment was normalized to corresponding Renilla luciferase activity. All within each cell type was then normalized to the average firefly/Renilla ratio for untreated APP/luciferase fusion activity. Letters above data bars indicate general linear mixed model multiple range categories. Samples sharing a letter do not significantly differ at p < 0.05. Thus, no differences in APP promoter activity were found between any treatment group, while there were significant increases in BACE1 promoter activity in the Aβ1–28 and Aβ1–42 treatment groups relative to untreated cells and Aβ1–40 was similar to vehicle.
3.4 Gene regulatory activity of Aβ on the APP promoter is altered by a single–nucleotide polymorphism
PC12 cell cultures were transfected with different polymorphic promoter/enhancer clones that contained the AβID at APP −3833, either the “G” or “A” variant, inserted into the pCAT3P vector or with unmodified pCAT3P vector. Transfected cultures were treated with either vehicle, H2O2, Aβ25–35, or both treatments, as described herein. Cells were harvested and protein was extracted. CAT protein levels were measured by ELISA and adjusted for total protein concentration in each respective extract. Adjusted data were analyzed by the SAS GLIMMIX procedure.
Type III tests of fixed effects for the data (Table 3) indicated that each of the three main effects: Clone (pCAT3P, 3833A, 3833G), Aβ treatment (−/+), and H2O2 treatment (−/+); exerted significant individual effects (p < 0.05) on adjusted CAT levels. In addition, a significant interaction was found between H2O2 and Aβ treatment and among all three treatments taken together. Effect sizes were estimated by appropriate generalized ω2 ( ) or partial for a three fixed–effects model, for each individual effect, and for effects taken two at a time (Olejnik and Algina, 2003). Effect sizes were categorized presuming sufficient analogy between η2 and (Cohen, 1988; Rosenthal, 1994; Hopkins, 2000). This may have actually underestimated the effect size, since ω2 is a more conservative estimator than η2 (Warner, 2008).
Table 3.
Type III analysis of treatment by H2O2 and Aβ on polymorphic APP promoter–CAT reporter fusion clones in PC12 cells
| Effect | Num DFa | Den DFb | F Value | Pr > F | Effect Size | ||
|---|---|---|---|---|---|---|---|
| Clone | 2 | 43 | 149.70 | <0.001 | 0.874 | extremely large | |
| Aβ | 1 | 43 | 5.26 | 0.027 | 0.090 | moderate | |
| H2O2 | 1 | 43 | 23.93 | <0.001 | 0.348 | large | |
| Aβ × H2O2 | 1 | 43 | 14.04 | <0.001 | 0.233 | moderate | |
| Clone × Aβ | 2 | 43 | 3.25 | 0.048 | 0.095 | moderate | |
| Clone × H2O2 | 2 | 43 | 1.71 | 0.193 | 0.032 | small | |
| Clone × Aβ × H2O2 | 2 | 43 | 7.13 | 0.002 | 0.222 | moderate |
Numerator degrees of freedom
Denominator degrees of freedom
Effect size category is based on Cohen’s categories for effect sizes, as refined by Hopkins, presuming strong analogy between and η2.
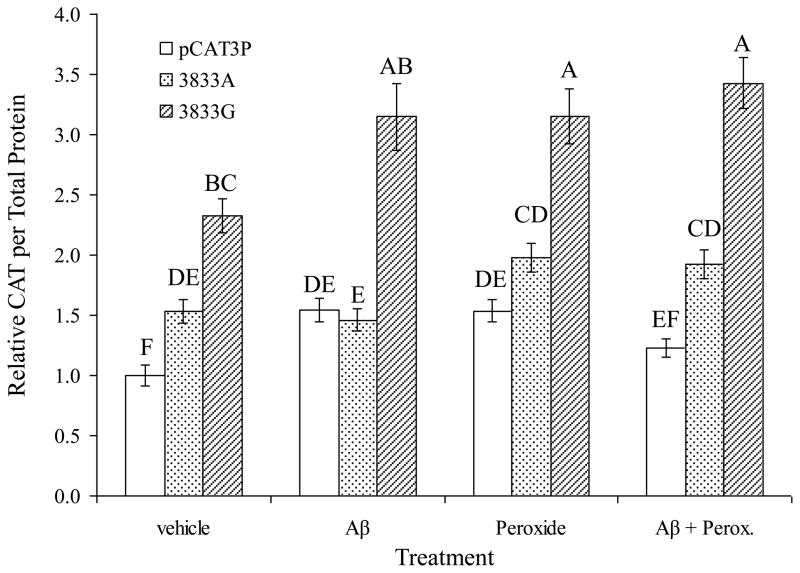
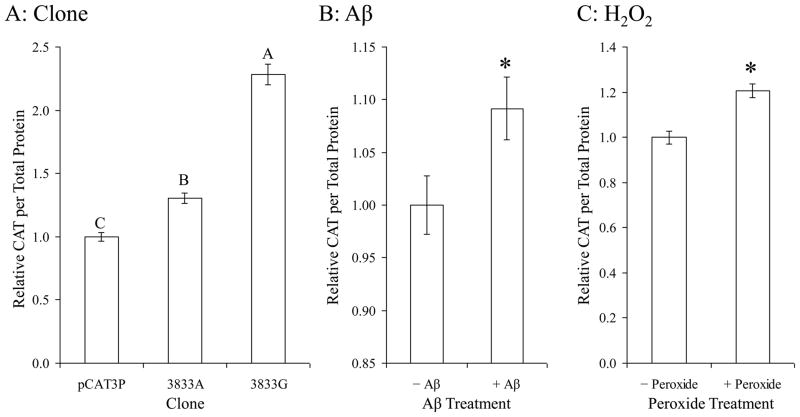
Treatments altered expression in all three clones, including pCAT3P in a complex manner (Fig. 4). However, deeper analysis of results showed differential effects by clone and each treatment. Specifically, when analyzed by individual effects, instead of as a whole (Fig. 5). Each insert resulted in higher levels of CAT reporter protein when compared to unmodified pCAT3P (Fig. 5A). The clone identified herein as −3833G–CAT produced greater levels of CAT protein as measured by ELISA than did −3833A–CAT. This agrees with our previous findings regarding transfection of these clones without other treatment (Lahiri et al., 2005b). Treatment with Aβ25–35 (Fig. 5B) or peroxide (Fig. 5C) also resulted in significant single–effect differences.
Fig. 4. Differential response of a single–nucleotide polymorphism of the APP promoter to Aβ treatment, all three effects combined.
Polymorphic promoter/enhancer CAT expression clones and pCAT3P were transfected into PC12 cells and the transfected cells were treated with H2O2 and Aβ, singly or in combination, as described in the main text. Reporter CAT ELISA signal was normalized to total cell protein. Results are presented as proportional to untreated pCAT3P vector transfected cells. Letters above data bars indicate general linear mixed model multiple range categories. Statistical significance is indicated by letters above the data bars, with samples sharing a letter do not significantly differ at p < 0.05, and conversely, samples not sharing a letter are significantly different at p < 0.05.
Fig. 5. Differential response of a single–nucleotide polymorphism of the APP promoter to Aβ treatment taken as single effects.
Polymorphic promoter/enhancer CAT expression clones and pCAT3P vector plasmid were transfected into PC12 cells and the transfected cells were treated with H2O2 and Aβ, singly or in combination, as described in the main text. Reporter CAT ELISA signal was normalized to total cell protein. A. Both of the clones tested (A and G) had significantly higher activity than the vector, and the G allele was significantly more active than the A allele. B and C. Both Aβ and H2O2 treatment significantly increased activity of this site, independent of the polymorphism. Results are presented as proportional to untreated pCAT3P vector transfected cells. Letters above data bars indicate generalized linear mixed model multiple range categories. Samples sharing a letter do not significantly differ at p < 0.05. For Aβ treatment and H2O2, “*” indicates significant difference at p < 0.05.
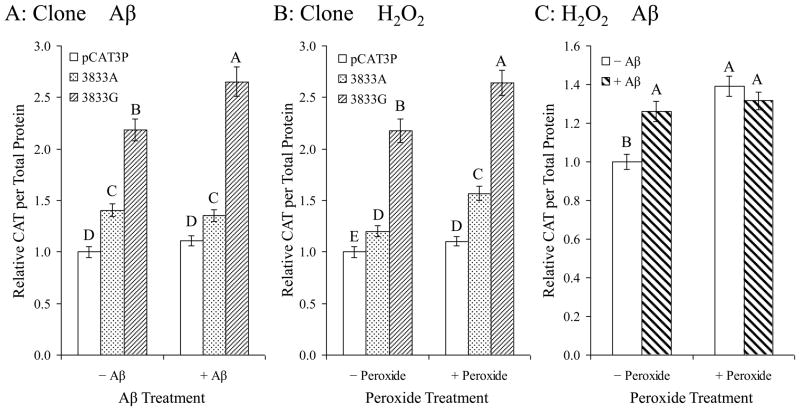
When considered in interactions of two effects (Fig. 6), the most interesting combination was clone × Aβ. Specifically, while Aβ treatment (holding effects of H2O2 treatment constant) significantly increased CAT protein levels for −3833G–CAT transfected cells, no similar response occurred in −3833A–CAT or pCAT3P transfected cells (Fig. 6A). When peroxide treatment vs. clone was examined, H2O2 treatment resulted in increased CAT protein levels regardless of specific promoter fragment insert (Fig. 6B). This is a notable contrast to the different effects of Aβ on each clone. DNA sequence specificity for Aβ treatment response was observed, while H2O2 treatment produced a nonspecific response that was not altered by changes in the Aβ–binding target sequence. Finally, interaction between Aβ and H2O2 treatments (Fig. 6C) had some significance (p = 0.048), possibly explained by effect of H2O2 in facilitating uptake of external Aβ into the cell.
Fig. 6. Differential response of a single–nucleotide polymorphism of the APP promoter to Aβ treatment analyzed as pairs of effects.
Polymorphic promoter/enhancer CAT expression clones and pCAT3P were transfected into PC12 cells and the transfected cells were treated with H2O2 and Aβ, singly or in combination, as described in the main text. CAT ELISA signal was normalized to total cell protein. Results were then normalized to adjusted signal for untreated pCAT3P transfected cells and expressed as marginal means estimated by holding a single effect (clone, Aβ treatment, or H2O2 treatment) constant. A. Aβ treatment increased transcriptional activity of the G but not the A clone. Both clones were significantly more active than the pCAT3P vector. B. H2O2 treatment increased the activity of both clones, indicating that this effect is not sequence–specific. C. Aβ treatment in the absence of H2O2 increased promoter activity, and H2O2 increased promoter activity significantly both in the presence or absence of Aβ. Letters above data bars indicate generalized linear mixed model multiple range categories. Samples sharing a letter do not significantly differ at p < 0.05. Although the −H2O2/−Aβ treatment had a significant difference from combinations that were treated with H2O2,, Aβ, or H2O2 + Aβ, when examining Aβ × H2O2, the interaction was not significant overall by Type III test.
4. Discussion
In addition to forming extracellular plaque in AD, Aβ is neurotoxic in monomers and oligomers (Walsh et al., 2002; Morgan et al., 2004; Ono et al., 2009). Aβ does play a multifaceted physiological role, including kinase induction (Bogoyevitch et al., 2004), reducing metal–induced oxidative damage (Baruch-Suchodolsky and Fischer, 2009), regulating cholesterol transport (Igbavboa et al., 2009), and altering glutamatergic transmission in the basal forebrain (Chin et al., 2007).
While Aβ is normally a secreted peptide, oxidative stress can induce intracellular localization of the peptide (Ohyagi et al., 2005; Ohyagi and Tabira, 2006). Structural studies have indicated that Aβ in its non–pathogenic α conformation, may have a structure common to certain transcription factors and Aβ1–42 binds DNA in vitro (Durell et al., 1994; Barrantes et al., 2007). Extending such findings, Ohyagi et al demonstrated Aβ peptide effects on TP53 apoptosis–associated protein gene expression (Ohyagi et al., 2005). Aβ enhanced p53 levels by direct interaction with a proximal 5′–region of the TP53 promoter/5′–UTR sequence. Transcription factors ASCL1 and OLIG2 are also regulated in cell culture by Aβ (Uchida et al., 2007). Native BACE1 gene transcription is upregulated in cell culture by addition of Aβ1–42 (Tamagno et al., 2009). Such activity has been demonstrated specifically with the Aβ25–35 peptide for the SLC38A1 gene (Buntup et al., 2008). Another cleavage product of the APP protein, APP protein intracellular domain fragment (AICD), forms part of the AICD/KAT5/APBB1 “transcription factory” complex (ATF) (Konietzko et al., 2010). ATF can regulate APP gene transcription (von Rotz et al., 2004). However, Aβ stimulation of BACE1 RNA transcription occurred independently of addition of AICD (Giliberto et al., 2009), suggesting that APP protein processing may generate multiple transcriptionally active peptides. Indeed, Aβ was recently shown to stimulate its own secretion (Marsden et al., 2011), and this study suggested a mechanism involving direct interaction with the APP and BACE1 regulatory regions.
Regulation of APP protein expression occurs at several levels. In addition to regulation through SP1 sites and GC box binding proteins in the proximal promoter region (La Fauci et al., 1989; Pollwein et al., 1992; Dosunmu et al., 2009), and through other proximal (Ge et al., 2004a) and distal promoter elements (Lahiri et al., 1999), APP mRNA translation is regulated at both the 5′–UTR (Ge et al., 2004b; Lahiri et al., 2005a) and 3′–UTR (Long and Lahiri, 2010). In addition, promoter–like activity has been found within the CDS regions of the APP gene (Vostrov et al., 2010). Our work suggests an additional mechanism of APP regulation that involves polymorphism–specific feedback between Aβ levels and transcriptional activity of its precursor gene.
We have recently demonstrated the existence of a generalized AβID that has a consensus of “KGGRKTGGGG”, primarily through electrophoretic mobility shift assays (Maloney and Lahiri, 2011). However, binding of Aβ to DNA in vitro would be moot if the peptide did not exist intracellularly and interact with chromatin. Intracellular Aβ has been previously shown to exist (Ohyagi et al., 2005; Ohyagi and Tabira, 2006). We confirmed such presence further when comparing cells treated with FITC–Aβ1–40 + H2O2 to cells treated with FITC–Aβ alone. FITC signal was observed in a minority of nuclei in either case, but it was significantly more common in H2O2 treated cells. We also observed that dead cells treated with H2O2 were almost invariably labeled with FITC–Aβ1–40. This suggests that Aβ internalization could be involved in the process of oxidative stress mediated cell death. Previous observations of TP53 activation by intracellular Aβ (Ohyagi et al., 2005) and the antioxidant role of p53 (Sablina et al., 2005) are consistent with a cell death pathway involving Aβ and the p53 pathway.
Investigation of chromatin–protein interaction within live cells via ChIP suggested that the Aβ peptide interacts with the chromatin of SK–N–SH human neuroblastoma cells, which strongly supports the intra-nuclear localization of Aβ1–40 suggested by the live-cell imaging data. More specifically, it may bind within two 200 bp regions of chromatin corresponding to −3899/−3708 in the APP gene promoter and to −205/−27 in the BACE1 gene promoter. Both of these regions include Aβ binding oligomer sites, at 3833 in the APP promoter at and at −119 in the BACE1 promoter. We also determined that Aβ binding signal within the APP promoter is unlikely to result from binding of by full–length APP to DNA, as immunoprecipitation with the N–terminal binding negative control antibody 22C11 did not produce a PCR signal. To date, no positive ChIP result has been reported using the 22C11 antibody. The APP and BACE1 promoter regions found to contain AβIDs that have in situ activity in human neuroblastoma cells are also produced a signal consistent with SP1 binding, which suggests the possibility that Aβ and SP1 may interact. The “G” vs. “A” SNP status at −3829 (within the AβID at −3833/−3824) in the APP promoter of SK–N–SH cells is currently unknown. However, in situ transcription factor interaction with a potential target site is more complex than is indicated by the “naked DNA” model of EMSA. We, therefore, consider our ChIP data to be strongly supportive rather than conclusive.
To demonstrate activity associated with Aβ binding to selected promoter sequences, we constructed luciferase fusion clones with a 3.3 kb fragment of the BACE1 5′–flanking region and a 1.2 kb fragment of the APP 5′–flanking region. The BACE1 sequence contained a single confirmed AβID while the APP sequence contained no predicted AβIDs. PRCN cultures were transiently transfected with each of these luciferase fusion reporter clones, and the transfected cells were treated with Aβ peptides 1–28, 1–40, and 1–42. In our system, all three peptides had some apparent effect on BACE1 promoter activity, as measured by reporter activity levels. This is in contrast to Ohyagi et al, who determined that Aβ1–42 altered expression levels of the TP53 promoter but Aβ1–40 did not (Ohyagi et al., 2005). In addition, our Aβ1–28 did not show DNA binding capacity as measured by EMSA (Maloney and Lahiri, 2011) but was associated with changes in reporter gene activity levels. The apparent effect on promoter activity without direct DNA binding suggests that, in addition to direct Aβ–DNA interaction, Aβ may regulate BACE1 promoter activity through other mechanisms.
We have noted that a potential Aβ binding motif starting at −3833 in the APP promoter had Aβ binding activity, and this activity was greatly reduced by a naturally occurring G→A SNP. Response to Aβ treatment differed between polymorphic insert variants, suggesting specific differences in Aβ binding activity at the polymorphic site. Both variants and the pCAT3P vector responded to H2O2 stimulation, suggesting that response to Aβ treatment is specific to Aβ–DNA interaction at the polymorphic site and not a general oxidative stress reaction from Aβ activity as an oxidizing species. We, therefore, suggest that this particular site in the APP promoter may be an active site for Aβ activity as a transcription factor in vivo and that differences in Aβ interaction at the site may correspond to differential risk of AD depending on the SNP. The peptide used for this experiment was Aβ25–35, which induces transcription of the BACE1 gene promoter (Tabaton et al., 2010) and reduces transcription of the SLC38A1 gene (Buntup et al., 2008). Our own work did not show that this peptide had different binding affinity based on the polymorphism in EMSA (Maloney and Lahiri, 2011), but our assay used a great excess of Aβ25–35, which could have overcome an incomplete reduction in DNA–protein affinity. Alternately, segments of the Aβ peptide outside the 25–35 range could also contribute to sequence specificity such that part of the peptide that might interfere with binding to the −3833A version of the SNP is excluded from this truncated peptide.
The −3833G variant is associated with reduced AD risk in a test population (Lahiri et al., 2005b). Our in vitro and cell culture work, herein, has indicated that this allele confers both greater affinity for and activity of full–length 1–40 and 1–42 Aβ peptide than with −3833A. While it is tempting to associate increased transcription of the APP gene with greater risk of AD, APP is neuroprotective when processed through the α–secretase cleavage pathway (Fahrenholz and Postina, 2006; Postina, 2008). Since the end products of APP metabolism can be either neurotrophic or neurotoxic, these data alone do not fully explain the protective effect of the −3829G SNP but rather add to our expanding knowledge of the varied function of Aβ. Potential pathological effects of Aβ activity may, therefore, include stimulation of BACE1 overexpression and stimulation of other apoptotic pathways, rather than through upregulation of APP gene expression.
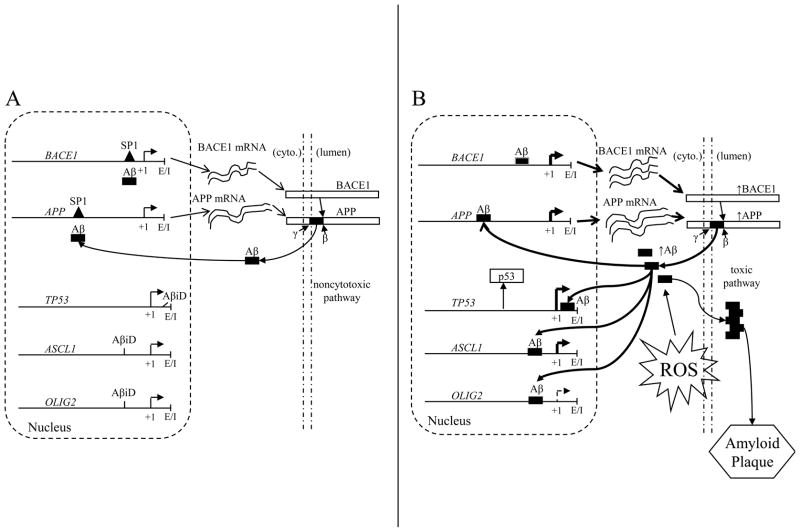
The many functions of the Aβ peptide may include acting as a transcription factor that directs apoptosis and regulates its own production through feedback on expression of its precursor protein and the β–secretase enzyme (Fig. 7A). Aβ production may be stimulated as a neuroprotective mechanism under cytotoxic conditions (Fig. 7B), until a pathogenic threshold is crossed, and in turn increasing transcription of APP and BACE1 genes to pathological levels, further stimulating its own production in a positive feedback loop. Additionally, increased Aβ would result in a “cytotoxic pair response” involving increased ASCL1 and decreased OLIG2 gene products. The combination would result in greater Aβ production, stimulating the production of apoptotic proteins such as p53
Fig. 7. Models of Aβ feedback and activity as a transcription factor.
A. Non–cytotoxic pathway. Aβ levels are insufficient to significantly activate apoptotic genes such as TP53 and the “cell death pair response” of upregulated ASCL1 and downregulated OLIG2 through their AβID sequences. These levels maintain APP and BACE1 at non–pathogenic levels. B. Cytotoxic pathway. Aβ levels and resulting APP and BACE1 production are stimulated by conditions such as metal–induced oxidation (ROS), which ultimately results in increased production of Aβ peptides. The levels of Aβ are sufficient to significantly increase levels of p53 and induce the “cell death pair response” of upregulated ASCL1 and downregulated OLIG2. Additional Aβ is produced beyond what is required for physiological function, and the excess Aβ becomes deposited as amyloid plaque.
Current treatments for AD concentrate upon remediation of cholinergic loss or other specific receptor–based treatments (Lahiri et al., 2003). However, even if restricting AD etiology to effects of Aβ, evidence exists to suggest a wide variety of pathways that are worth exploration (Lahiri and Maloney, 2010a). Furthermore, other pathways, such as those that lead to loss of synaptic markers, may not be directly caused by Aβ and could be targeted through separate mechanisms (Bailey and Lahiri, 2010). Several promising compounds with varied modes of action have recently failed in clinical trials, including phenserine, metrifonate, tarenflurbil, dimebon, and most recently, semagacestat, highlighting the difficulties in slecting effective targets for potential AD treatments (Sambamurti et al., 2011). These failures highlight both the difficulty in translating a compound from pre–clinical conceptualization to clinical use, and the potential need to re–examine the methodologies applied in these clinical trials (Becker and Greig, 2010a; Becker and Greig, 2010b).
Nevertheless, given our data and the work of others cited herein, we propose that Aβ’s very broad range of potential functions could be extended to include activity as a DNA transcription factor, modifying expression of AD–related genes, such as APP and BACE1 and glutamatergic pathway genes, such as SLC38A1. We have confirmed that oxidative stress can induce uptake of extracellular Aβ peptide into neuronal cells. Finally, we have determined that the Aβ peptide alters promoter–reporter clone activity in a DNA sequence–specific manner. Our own work has concentrated on gene sequences intimately tied with pathogenesis of AD. However, preliminary examination of sequences downloaded from the Eukaryotic Promoter Database (Schmid et al., 2006) produced multiple instances in other gene promoters of high–quality matches for the AβID weight matrix that we developed (Maloney and Lahiri, 2011). Determination of functional or structural gene families in which the AβID may be over– or underrepresented will serve to further elucidate the overall role of Aβ as a transcription factor both within a healthy organism and potentially point to pathogenic activity of Aβ in associated disorders.
Acknowledgments
This work was supported by the Alzheimer’s Association and National Institutes of Health AG18379 and AG18884.
Abbreviations: The abbreviations used are
- AD
Alzheimer’s disease
- AICD
APP protein intracellular domain fragment
- APP
Aβ–precursor protein
- APP
Aβ–precursor protein gene
- ASCL1
achaete–scute complex homolog 1 gene
- ATF
AICD/KAT5/APBB1 transcription factory complex
- Aβ
amyloid beta–peptide
- BACE1
β–amyloid cleaving enzyme 1/β–secretase gene
- EMSA
electrophoretic mobility shift assay
- FITC
fluorescein isothiocyanate
- HSE
heat shock element
- OLIG2
oligodendrocyte lineage transcription factor 2 gene
- p53
tumor protein 53
- PC12
rat pheochromocytoma cells
- PRCN
primary rat cerebrocortical neuronal (culture)
- SNP
single–nucleotide polymorphism
- SLC38A1
solute carrier family 38 member 1 gene
- SP1
specificity protein 1
- TP53
tumor protein 53 gene
Footnotes
Portions of this work have been presented as part of the proceedings of the 21st American Peptide Symposium (Lahiri et al., 2009).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey JA, Lahiri DK. Neuronal differentiation is accompanied by increased levels of SNAP-25 protein in fetal rat primary cortical neurons: implications in neuronal plasticity and Alzheimer’s disease. Ann N Y Acad Sci. 2006;1086:54–65. doi: 10.1196/annals.1377.001. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Lahiri DK. A novel effect of rivastigmine on pre-synaptic proteins and neuronal viability in a neurodegeneration model of fetal rat primary cortical cultures and its implication in Alzheimer’s disease. J Neurochem. 2010;112:843–53. doi: 10.1111/j.1471-4159.2009.06490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrantes A, Rejas MT, Benitez MJ, Jimenez JS. Interaction between Alzheimer’s Abeta1–42 peptide and DNA detected by surface plasmon resonance. J Alzheimers Dis. 2007;12:345–55. doi: 10.3233/jad-2007-12408. [DOI] [PubMed] [Google Scholar]
- Baruch-Suchodolsky R, Fischer B. Abeta40, either soluble or aggregated, is a remarkably potent antioxidant in cell-free oxidative systems. Biochemistry. 2009;48:4354–70. doi: 10.1021/bi802361k. [DOI] [PubMed] [Google Scholar]
- Becker RE, Greig NH. Lost in translation: neuropsychiatric drug development. Sci Transl Med. 2010a;2:61rv6. doi: 10.1126/scitranslmed.3000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker RE, Greig NH. Why so few drugs for Alzheimer’s disease? Are methods failing drugs? Curr Alzheimer Res. 2010b;7:642–51. doi: 10.2174/156720510793499075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoyevitch MA, Boehm I, Oakley A, Ketterman AJ, Barr RK. Targeting the JNK MAPK cascade for inhibition: basic science and therapeutic potential. Biochim Biophys Acta. 2004;1697:89–101. doi: 10.1016/j.bbapap.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 1993;35:567–76. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- Buntup D, Skare O, Solbu TT, Chaudhry FA, Storm-Mathisen J, Thangnipon W. Beta-amyloid 25–35 peptide reduces the expression of glutamine transporter SAT1 in cultured cortical neurons. Neurochemical Research. 2008;33:248–56. doi: 10.1007/s11064-007-9527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JH, Ma L, MacTavish D, Jhamandas JH. Amyloid beta protein modulates glutamate-mediated neurotransmission in the rat basal forebrain: involvement of presynaptic neuronal nicotinic acetylcholine and metabotropic glutamate receptors. J Neurosci. 2007;27:9262–9269. doi: 10.1523/JNEUROSCI.1843-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2. Erlbaum; Hillsdale, NJ: 1988. [Google Scholar]
- De Strooper B. Proteases and proteolysis in Alzheimer disease: a multifactorial view on the disease process. Physiol Rev. 2010;90:465–94. doi: 10.1152/physrev.00023.2009. [DOI] [PubMed] [Google Scholar]
- Dean CB, Nielsen JD. Generalized linear mixed models: a review and some extensions. Lifetime Data Anal. 2007;13:497–512. doi: 10.1007/s10985-007-9065-x. [DOI] [PubMed] [Google Scholar]
- Dosunmu R, Wu J, Adwan L, Maloney B, Basha MR, McPherson CA, Harry GJ, Rice DC, Zawia NH, Lahiri DK. Lifespan profiles of Alzheimer’s disease-associated genes and products in monkeys and mice. J Alzheimers Dis. 2009;18:211–30. doi: 10.3233/JAD-2009-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durell SR, Guy HR, Arispe N, Rojas E, Pollard HB. Theoretical models of the ion channel structure of amyloid beta-protein. Biophys J. 1994;67:2137–2145. doi: 10.1016/S0006-3495(94)80717-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenholz F, Postina R. Alpha-secretase activation--an approach to Alzheimer’s disease therapy. Neurodegener Dis. 2006;3:255–61. doi: 10.1159/000095264. [DOI] [PubMed] [Google Scholar]
- Ferguson GA. Statistical Analysis in Psychology & Education. McGraw-Hill; New York: 1966. [Google Scholar]
- Ge YW, Ghosh M, Song W, Maloney B, Lahiri D. Mechanism of promoter activity of the beta-amyloid precursor protein gene in different cell types. Identification of a specific 30 bp fragment in the proximal promoter region. J Neurochem. 2004a;90:1432–44. doi: 10.1111/j.1471-4159.2004.02608.x. [DOI] [PubMed] [Google Scholar]
- Ge YW, Maloney B, Sambamurti K, Lahiri DK. Functional characterization of the 5′ flanking region of the BACE gene: identification of a 91 bp fragment involved in basal level of BACE promoter expression. FASEB J. 2004b;18:1037–1039. doi: 10.1096/fj.03-1379fje. [DOI] [PubMed] [Google Scholar]
- Ghosh C, Song W, Lahiri DK. Efficient DNA transfection in neuronal and astrocytic cell lines. Mol Biol Rep. 2000;27:113–121. doi: 10.1023/a:1007173906990. [DOI] [PubMed] [Google Scholar]
- Giliberto L, Borghi R, Piccini A, Mangerini R, Sorbi S, Cirmena G, Garuti A, Ghetti B, Tagliavini F, Mughal MR, Mattson MP, Zhu X, Wang X, Guglielmotto M, Tamagno E, Tabaton M. Mutant presenilin 1 increases the expression and activity of BACE1. J Biol Chem. 2009;284:9027–38. doi: 10.1074/jbc.M805685200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouras GK, Tsai J, Naslund J, Vincent B, Edgar M, Checler F, Greenfield JP, Haroutunian V, Buxbaum JD, Xu H, Greengard P, Relkin NR. Intraneuronal Abeta42 accumulation in human brain. Am J Pathol. 2000;156:15–20. doi: 10.1016/s0002-9440(10)64700-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert LE, Scherr PA, Bienias JL, Bennett DA, Evans DA. Alzheimer disease in the US population: prevalence estimates using the 2000 census. Arch Neurol. 2003;60:1119–22. doi: 10.1001/archneur.60.8.1119. [DOI] [PubMed] [Google Scholar]
- Hopkins WG. A new view of statistics. Internet Society for Sport Science. 2000 http://www.sportsci.org/resource/stats/
- Igbavboa U, Sun GY, Weisman GA, He Y, Wood WG. Amyloid beta-protein stimulates trafficking of cholesterol and caveolin-1 from the plasma membrane to the Golgi complex in mouse primary astrocytes. Neuroscience. 2009;162:328–38. doi: 10.1016/j.neuroscience.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Arce FT, Ramachandran S, Capone R, Azimova R, Kagan BL, Nussinov R, Lal R. Truncated beta-amyloid peptide channels provide an alternative mechanism for Alzheimer’s Disease and Down syndrome. Proc Natl Acad Sci USA. 2010;107:6538–43. doi: 10.1073/pnas.0914251107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konietzko U, Goodger ZV, Meyer M, Kohli BM, Bosset J, Lahiri DK, Nitsch RM. Co-localization of the amyloid precursor protein and Notch intracellular domains in nuclear transcription factories. Neurobiol Aging. 2010;31:58–73. doi: 10.1016/j.neurobiolaging.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Fauci G, Lahiri DK, Salton SR, Robakis NK. Characterization of the 5′-end region and the first two exons of the beta-protein precursor gene. Biochem Biophys Res Commun. 1989;159:297–304. doi: 10.1016/0006-291x(89)92437-6. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Farlow MR, Sambamurti K, Greig NH, Giacobini E, Schneider LS. A critical analysis of new molecular targets and strategies for drug developments in Alzheimer’s disease. Curr Drug Targets. 2003;4:97–112. doi: 10.2174/1389450033346957. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Ge YW, Maloney B. Characterization of the APP proximal promoter and 5′-untranslated regions: identification of cell-type specific domains and implications in APP gene expression and Alzheimer’s disease. FASEB J. 2005a;19:653–655. doi: 10.1096/fj.04-2900fje. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Maloney B. Beyond the signaling effect role of amyloid-beta(42) on the processing of APP, and its clinical implications. Exp Neurol. 2010a;225:51–54. doi: 10.1016/j.expneurol.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri DK, Maloney B. The “LEARn” (Latent Early-life Associated Regulation) model integrates environmental risk factors and the developmental basis of Alzheimer’s disease, and proposes remedial steps. Exp Gerontol. 2010b;45:291–6. doi: 10.1016/j.exger.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri DK, Maloney B, Bailey J, Ge Y-W. Role of alzheimer’s amyloid-beta peptide as a putative transcription factor. In: Lebl M, editor. Peptides: Breaking Away, Proceedings of the Twenty-First American Peptide Symposium; Bloomington, Indiana, USA. 2009. pp. 185–186. [Google Scholar]
- Lahiri DK, Nall C, Ge YW. Promoter activity of the beta-amyloid precursor protein gene is negatively modulated by an upstream regulatory element. Brain Res Mol Brain Res. 1999;71:32–41. doi: 10.1016/s0169-328x(99)00150-3. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Robakis NK. Characterization of two blocks of cis–acting regulatory elements modulating the expression of the gene encoding the Alzheimer’s Amyloid Precursor Proteins. In: Iqbal K, editor. Alzheimer’s Disease: Basic Mechanisms, Diagnosis and Therapeutic Strategies. J. Wiley & Sons; New York: 1991. pp. 444–450. [Google Scholar]
- Lahiri DK, Wavrant De-Vrieze F, Ge YW, Maloney B, Hardy J. Characterization of two APP gene promoter polymorphisms that appear to influence risk of late-onset Alzheimer’s disease. Neurobiol Aging. 2005b;26:1329–1341. doi: 10.1016/j.neurobiolaging.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Long JM, Lahiri DK. MicroRNA-101 downregulates Alzheimer’s amyloid-β precursor protein levels in human cell cultures and is differentially expressed. Biochem Biophys Res Commun. 2010 doi: 10.1016/j.bbrc.2010.12.053. IN PRESS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney B, Lahiri DK. The Alzheimer’s amyloid β–peptide (Aβ) binds a specific DNA Aβ–interacting domain (AβID) in the APP, BACE1, and MAPT promoters in a sequence–specific manner: characterizing a new regulatory motif. Gene. 2011 doi: 10.1016/j.gene.2011.06.004. IN PRESS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden IT, Minamide LS, Bamburg JR. Amyloid-beta-Induced Amyloid-beta Secretion: A Possible Feed-Forward Mechanism in Alzheimer’s Disease. J Alzheimers Dis. 2011;24:681–91. doi: 10.3233/JAD-2011-101899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C, Colombres M, Nunez MT, Inestrosa NC. Structure and function of amyloid in Alzheimer’s disease. Prog Neurobiol. 2004;74:323–49. doi: 10.1016/j.pneurobio.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Ohyagi Y. Intracellular amyloid beta-protein as a therapeutic target for treating Alzheimer’s disease. Curr Alzheimer Res. 2008;5:555–61. doi: 10.2174/156720508786898514. [DOI] [PubMed] [Google Scholar]
- Ohyagi Y, Asahara H, Chui DH, Tsuruta Y, Sakae N, Miyoshi K, Yamada T, Kikuchi H, Taniwaki T, Murai H, Ikezoe K, Furuya H, Kawarabayashi T, Shoji M, Checler F, Iwaki T, Makifuchi T, Takeda K, Kira JI, Tabira T. Intracellular Abeta42 activates p53 promoter: a pathway to neurodegeneration in Alzheimer’s disease. FASEB J. 2005;19:255–257. doi: 10.1096/fj.04-2637fje. [DOI] [PubMed] [Google Scholar]
- Ohyagi Y, Tabira T. Intracellular amyloid beta-protein and its associated molecules in the pathogenesis of Alzheimer’s disease. Mini Rev Med Chem. 2006;6:1075–80. doi: 10.2174/138955706778560175. [DOI] [PubMed] [Google Scholar]
- Ohyagi Y, Tsuruta Y, Motomura K, Miyoshi K, Kikuchi H, Iwaki T, Taniwaki T, Kira J. Intraneuronal amyloid beta42 enhanced by heating but counteracted by formic acid. J Neurosci Methods. 2007;159:134–8. doi: 10.1016/j.jneumeth.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Olejnik S, Algina J. Generalized eta and omega squared statistics: measures of effect size for some common research designs. Psychol Methods. 2003;8:434–47. doi: 10.1037/1082-989X.8.4.434. [DOI] [PubMed] [Google Scholar]
- Ono K, Condron MM, Teplow DB. Structure-neurotoxicity relationships of amyloid beta-protein oligomers. Proc Natl Acad Sci U S A. 2009;106:14745–50. doi: 10.1073/pnas.0905127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollwein P, Masters CL, Beyreuther K. The expression of the amyloid precursor protein (APP) is regulated by two GC-elements in the promoter. Nucleic Acids Res. 1992;20:63–68. doi: 10.1093/nar/20.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postina R. A closer look at alpha-secretase. Curr Alzheimer Res. 2008;5:179–86. doi: 10.2174/156720508783954668. [DOI] [PubMed] [Google Scholar]
- Rosenthal R. Parametric measures of effect size. In: Cooper H, Hedges LV, editors. The Handbook of Research Synthesis. Sage; New York: 1994. p. 239. [Google Scholar]
- Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–13. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambamurti K, Greig NH, Utsuki T, Barnwell EL, Sharma E, Mazell C, Bhat NR, Kindy MS, Lahiri DK, Pappolla MA. Targets for AD treatment: conflicting messages from gamma-secretase inhibitors. J Neurochem. 2011;117:359–74. doi: 10.1111/j.1471-4159.2011.07213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid CD, Perier R, Praz V, Bucher P. EPD in its twentieth year: towards complete promoter coverage of selected model organisms. Nucleic Acids Res. 2006;34:D82–5. doi: 10.1093/nar/gkj146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shie FS, LeBoeuf RC, Jin LW. Early intraneuronal Abeta deposition in the hippocampus of APP transgenic mice. Neuroreport. 2003;14:123–9. doi: 10.1097/01.wnr.0000051151.87269.7d. [DOI] [PubMed] [Google Scholar]
- Song W, Lahiri DK. Functional identification of the promoter of the gene encoding the Rhesus monkey beta-amyloid precursor protein. Gene. 1998;217:165–176. doi: 10.1016/s0378-1119(98)00340-0. [DOI] [PubMed] [Google Scholar]
- Tabaton M, Zhu X, Perry G, Smith MA, Giliberto L. Signaling effect of amyloid-beta(42) on the processing of AbetaPP. Exp Neurol. 2010;221:18–25. doi: 10.1016/j.expneurol.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagno E, Guglielmotto M, Giliberto L, Vitali A, Borghi R, Autelli R, Danni O, Tabaton M. JNK and ERK1/2 pathways have a dual opposite effect on the expression of BACE1. Neurobiol Aging. 2009;30:1563–73. doi: 10.1016/j.neurobiolaging.2007.12.015. [DOI] [PubMed] [Google Scholar]
- Uchida Y, Nakano S, Gomi F, Takahashi H. Differential regulation of basic helix-loop-helix factors Mash1 and Olig2 by beta-amyloid accelerates both differentiation and death of cultured neural stem/progenitor cells. J Biol Chem. 2007;282:19700–9. doi: 10.1074/jbc.M703099200. [DOI] [PubMed] [Google Scholar]
- von Rotz RC, Kohli BM, Bosset J, Meier M, Suzuki T, Nitsch RM, Konietzko U. The APP intracellular domain forms nuclear multiprotein complexes and regulates the transcription of its own precursor. J Cell Sci. 2004;117:4435–48. doi: 10.1242/jcs.01323. [DOI] [PubMed] [Google Scholar]
- Vostrov AA, Taheny MJ, Izkhakov N, Quitschke WW. A nuclear factor-binding domain in the 5′-untranslated region of the amyloid precursor protein promoter: Implications for the regulation of gene expression. BMC Res Notes. 2010;3:4. doi: 10.1186/1756-0500-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–9. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- Warner RM. Applied Statistics: From Bivariate through Multivariate Techniques. Sage Publications; Thousand Oaks, CA: 2008. [Google Scholar]
- Yao ZX, Papadopoulos V. Function of beta-amyloid in cholesterol transport: a lead to neurotoxicity. FASEB J. 2002;16:1677–9. doi: 10.1096/fj.02-0285fje. [DOI] [PubMed] [Google Scholar]
- Zou K, Gong JS, Yanagisawa K, Michikawa M. A novel function of monomeric amyloid beta-protein serving as an antioxidant molecule against metal-induced oxidative damage. J Neurosci. 2002;22:4833–41. doi: 10.1523/JNEUROSCI.22-12-04833.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]