Summary
The interplay between a protease and its substrates is controlled at many different levels, including coexpression, colocalization, binding driven by ancillary contacts, and the presence of natural inhibitors. Here we focus on the most basic parameter that guides substrate recognition by a protease, the recognition specificity at the catalytic cleft. An understanding of this substrate specificity can be used to predict the putative substrates of a protease, to design protease activated imaging agents, and to initiate the design of active site inhibitors. Our group has characterized protease specificities of several matrix metalloproteinases using substrate phage display. Recently, we have adapted this method to a semiautomated platform that includes several high-throughput steps. The semiautomated platform allows one to obtain an order of magnitude more data, thus permitting precise comparisons among related proteases to define their functional distinctions.
Keywords: Substrate phage display, Substrate, Protease, Specificity, Proteolysis, Filamentous phage, M13 coat protein, 3 gene protein
1. Introduction
Sequence comparisons from over 100 genomes shows that peptidases comprise about 2% of all gene products (1), and 50 years of research show that proteolysis plays an important role in most biological processes. Yet, we still have little knowledge on which protein substrates are cleaved by a particular protease, and we lack the technology for localizing and quantifying proteolytic events in cells and whole animals. One strategy for addressing these issues is to define the fine substrate recognition profile of individual proteases and then use this information to predict physiologic substrates and to design imaging agents and other probes of that particular protease.
Defining the substrate specificity of a protease involves assignment of amino acid preferences for each of the subsites within the catalytic cleft. These sites are normally referred to as primed (on the right side of the scissile bond) and unprimed (on the left side of the scissile bond) according to the definitions of Berger and Schechter (2). Different approaches have been developed for acquiring this information (for review see ref. 3). These include the use of peptide substrate libraries, as well as biological display using bacteriophage and E. coli . Filamentous phage display (4) was first used for protease specificity profiling by Matthews and Wells in 1993 (5) . Since then, substrate phage display has been used to profile a large number of individual proteases (6–11).
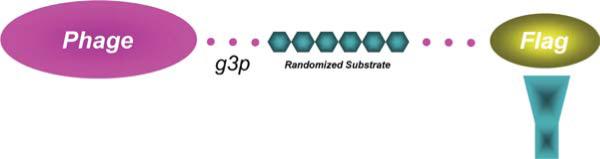
Substrate phage display is typically carried out by displaying a randomized peptide substrate as a fusion protein with the gene 3 protein (g3p) of filamentous M13 bacteriophage. Since g3p is expressed in 3–5 copies per phage, there is a polyvalent display of the substrate peptide. This peptide is flanked on its C-terminal side by g3p and a “spacer” designed to keep the randomized sequence in a disordered conformation. An affinity tag is normally appended to the N-terminal side of the peptide, and this is used to separate cleaved from uncleaved phages during the selection process (Fig. 1). Our group uses a library composed of randomized hexapeptides displayed on g3p and flanked by a FLAG epitope engineered at the NH 2 terminus of the g3p (12). The substrate phage library was generated using a modified version of the fUSE5 phagemid.
Fig. 1.
The affinity tagged randomized peptide substrate fusion protein with the gene 3 protein (g3p) of filamentous M13 bacteriophage bound to anti-FLAG antibody . (See Color Plates)
One of the major limitations of substrate phage display is the length of time required to obtain data that yield a comprehensive substrate recognition profile for a given protease. Recently we have incorporated several automated steps into the substrate phage method that allow substantially higher throughput, and that yield an order of magnitude more data. With this platform in place we believe it possible to define the substrate recognition specificity of every endopeptidase in the human genome within 5–10 years. Here we describe the detailed method linked to the semiautomated platform.
2. Materials
2.1. Phage Propagation and Purification
NZY Broth: 5 g NaCl, 2 g MgSO 4 ·7H2 O, 5 g Bacto-yeast extract, NZ amine (casein hydrolysate), ddH2 O to 1 L. Adjust pH to 7.5 with NaOH and autoclave.
100 mg/mL Kanamycin Sulfate in water, store at –20°C.
20 mg/mL Tetracycline in 50% glycerol store at –20°C.
PEG/NaCl: 100 g PEG 8000 (Sigma catalog # P4463, 116.9 g NaCl, 475 mL water; stir until solutes dissolve (may be necessary to heat to 65° briefly to dissolve the last crystals of PEG). Sterile filter through a 0.22 μm membrane. Store at 4°C (total volume 600 mL).
K91Kan E. coli cells.
Glycerol stock solution 2×: 25 mM Tris, pH 8.0, 0.1 M MgSO4 , 65% Glycerol.
2.2. Phage Substrate Selection
Substrate phage library based on the fUSE5 phagemid with a FLAG epitope engineered at the NH 2 terminus of the gene III protein.
Stock solution of the protease of interest preferably at high concentration.
High affinity inhibitor for active site titration and protease inhibition.
Monoclonal anti-FLAG antibodies M2 (Sigma catalog # F3165).
Monoclonal anti-M13 antibodies (GE Healthcare catalog # 27-9421-01).
Dynabeads M-450 epoxy (Invitorgen catalog # 14011).
Dynal MPC-S magnetic separator (Invitrogen catalog # 120-20D).
Monoclonal anti-M13 antibodies HRP conjugated (GE Healthcare catalog # 27-9420-01).
OPD HRP substrate (Sigma catalog # P3804-100TAB).
HRP substrate buffer: 2.43 mL of 0.1 M Citric Acid, 2.57 mL of 0.2 M Na2 HPO4 , 5 mL of DI water.
30% H2 O2.
4 M H2 SO4.
3 M (NH4)2SO4.
PBS: 50 mM sodium phosphate, pH 7.4, 150 mM NaCl.
100 mM sodium phosphate, pH 7.0.
TBS: 50 mM Tris, pH 7.2, 150 mM NaCl.
TBS-T: TBS, 0.1% Tween 20.
Polystyrene 96-well microplates (Costar catalog # 9018).
50 mL centrifuge tubes (Corning catalog # 430828 or similar).
100 mg/mL BSA (bovine serum albumin, Sigma catalog # A7906) in TBS.
Microplate reader.
Multichannel micropipettor.
PC with Graph Pad Prism or other data fitting software.
2.3. Substrate Identification
K-91Kan E. coli glycerol stock.
Terrific Broth: 12 g bacto-tryptone, 24 g yeast extract, 4 mL (5.04) g glycerol, DI water 900 mL. Autoclave 90 mL portions in 125 mL polypropylene bottles. When cooled, to each bottle add 10 mL of separately autoclaved potassium phosphate buffer (0.17 M KH2PO4 , 0.72 M K2 HPO4).
LB Agar, 100 μg/mL Kanamycin: 10 g Tryptone, 5 g yeast extract, 5 g NaCl, 200 μL NaOH, 15 g Bacto Agar DI water to 1 L. Autoclave, cool to 50°C, add 1 mL 100 mg/mL Kanamycin – pour 100 mm Petri dishes at 20 mL/dish. Store at 4°C after agar solidifies.
NZY Agar, 100 μg/mL Kanamycin, 40 μg/mL Tetracycline: 5 g NaCl, 2 g MgSO4 ·7H2 O, 5 g Bacto-yeast extract, NZ amine (casein hydrolysate), 15 g Bacto Agar, DI H2 O to 1 L. Adjust pH to 7.5 with NaOH. Autoclave, cool to 50°C, add 1 mL 100 mg/mL Kanamycin – pour into Genetix QTray vented (Genetix catalog # 6023) at 200 mL/tray for automated colony picking using Genetix QPix colony picker or at 20 mL per 100 mm Petri dish. Store at 4°C after agar solidifies.
Multichannel pipettor.
Hamilton LabStar liquid handling robot for HTS phage diplay.
96 deep well plate (Simport Bioblock deep well plate, catalog # T110-10) for HTS phage display or 15 mL sterile disposable round bottom culture tubes for low throughput phage display.
Genetix 96-well flat bottom1/2 height plate (Genetix catalog # X6011) for HTS phage display or sterile 1.5 mL Eppendorf centrifuge tubes for low throughput phage dispay.
Genetix BreatheSeal Film (catalog # E1005).
Aluminum sealing tape, (ISC BioExpress catalog # T-2420-2).
Glycerol stock solution.
Monoclonal anti-M13 antibodies.
Monoclonal anti-FLAG M2 antibodies peroxidase conjugated (Sigma catalog # A8592).
TBS-T: TBS, 0.1% Tween 20.
Polystyrene 96-well microplates.
100 mg/mL BSA in TBS.
HRP Substrate Buffer.
OPD HRP substrate.
30% H2 O2.
Microplate reader.
ATR Multitron Incubator shaker.
2.4. Substrate Sequencing
FUSE5 forward primer: 5′ –TAA TAC GAC TCA CTA TAG GGC AAG CTG ATA AAC CGA TAC AAT T–3′ , 100 μM stock.
FUSE5 Super Reverse primer: 5′ –CCG TAA CAC TGA GTT TCG TC–3 ′ , 100 μM stock.
Platinum PCR Super Mix (Invitrogen catalog # 11306- 016).
96-well PCR plate (Abgene catalog # AB1000).
PCR thermocycler.
- 50× TAE Buffer.
- 242 g Tris base
- 57.1 mL acetic acid
- 100 mL 0.5 M EDTA
- Add ddH2O to 1 L and adjust pH to 8.5
10 mg/mL ethidium bromide (Sigma catalog # E1510).
1% agarose in TAE buffer, 0.5 μg/mL ethidium bromide.
2.5. Identification of Scissile Bonds
NZY Broth.
96 deep well plate (Simport Bioblock deep well plate, catalog # T110-10) for HTS phage display or 15 mL sterile disposable round bottom culture tubes for low throughput phage display.
Costar Assay Block 1 mL plate (catalog # 3958).
Costar Storage Mat III (catalog # 3080).
Monoclonal anti-FLAG antibodies M2.
Monoclonal anti-M13 antibodies.
Dynabeads M-450 epoxy.
Dynabeads M-280 Tosylactivated (Invitrogen catalog # 14204).
2,5-Dihydroxybenzoic acid, 99% (Aldrich catalog # 14,935-7).
Trifluoroacetic acid (Sigma Aldrich catalog # T6508).
MALDI AnchorChip target (Bruker Daltonics catalog # 209515) or other MALDI target.
3 M (NH4)2SO4.
100 mM sodium phosphate, pH 7.0.
TBS-T: TBS, 0.1% Tween 20.
V-bottom polypropylene plates (Costar catalog # 3357).
3. Methods
Prior to delving into procedures for substrate phage display, it is important to mention a few factors and pitfalls that have a significant impact on the success of the procedure. A major factor in the success of substrate phage display is the availability of an ample supply of the test protease, and a reliable method of quantifying its activity. Each time a protease is used for substrate phage its activity should be titrated with an active site inhibitor (13) whenever possible. When the test proteases are expressed and purified in our laboratories, more than 70% of the purified protein is active. With this level of activity, 500 μg to 1 mg of total protease are usually sufficient to complete the phage procedure. We are reluctant to move forward with phage selections for any protease in which less than 30–40% of the total protein has activity. With this quantitative information in hand, we have the basis for quantitative analysis and comparison of any of the data obtained in the course of the experiment.
Another factor that has a major impact on success is the quality of the K91Kan E. coli used for phage infection and propagation. These bacteria must be fresh; generated and used within 24 h. This is a very important requirement and should be followed strictly to obtain consistent results.
An important pitfall in this procedure, as it is with any phage display project, is the possibility of phage contamination. There are really two different manifestations of this problem. The first is the simple contamination of laboratory tools and instruments with a single phage, which then begins to dominate all successive phage amplifications and overrides any selection process. This can be prevented by highly stringent sterile techniques, the use of pipette tips containing filters, and frequent washing of instrumentation. All buffers and solutions should be sterilized either by autoclaving or 0.2 μm filtration.
In another manifestation this problem arises as the propagation of a phage lacking the affinity tag, which then appears as “cleaved” even when it is not (8). This is not a problem for the first round of selection, but can be an issue after the second round, when some phages have adapted to the selection pressure by eliminating the tag from their sequence. To overcome this issue, we include an affinity isolation of the tag-bearing phage prior to the next round of selection.
A protease phage display project involves four basic steps: (1) substrate selection, (2) substrate identification, (3) substrate sequencing, and (4) scissile bond assignment. Steps 2–4 have been automated by our group. The procedure for each step is described in detail later.
3.1. Phage Substrate Selection
These instructions assume that a FLAG-tagged phage library is available for substrate selection.
Before starting substrate selection, prepare M-450 M2 beads. Wash 5 × 10 8 M-450 epoxy magnetic beads three times by 1 mL 100 mM sodium phosphate buffer, pH 7.0 by magnetic separation. Resuspend the beads in 500 μL 0.5 mg/mL M2 antibody and add 250 μL 3 M (NH4)2SO4. Incubate with end-over-end rotation or vortexing overnight at room temperature, making sure the beads are in suspension. In the morning collect the supernatant and measure light absorbance at 280 nm by spectrophotometry. Determine the efficiency of coupling by dividing by the absorbance of the starting antibody solution adjusted by dilution factor of 1.5. Usually 75–90% of the antibody is coupled under these conditions. Resuspend the beads in 1 mL of 1 mg/mL BSA in PBS and incubate with end-over-end rotation or vortexing for 3 h at room temperature. Store the beads at 4°C.
The day of substrate selection, perform active site titration of the protease or specific activity determination by some other method to be able to reproduce the conditions of the experiment.
For proteases requiring extreme pH for optimal activity, determine neutralization conditions.
Determine the concentration of the phage particles in the stock solution of the phage library. Dilute the phage in TBS to a concentration approximately 1012 particles/mL. Scan the diluted sample from 240 to 320 nm in a UV spectrophotometer. There should be a broad peak from 260 to 280 nm with a slight maximum at 269 nm. Measure A269 and A320 and calculate the net A269 by subtracting A320 from A269. Calculate the virion concentration in particles per mL based on the following formula: phage particles/mL = net A269 × 6 × 1016 / N, where N = the number of nucleotides/phage genome. The genome size of a fUSE5 phage = 9,206 bases.
Add 1.2 × 10 12 phage to the final volume of 1.2 mL in protease reaction buffer. Take out two 0.4 mL aliquots for negative control and protease treatment. To the first 0.4 mL aliquot add the protease in the protease buffer, to the final concentration of 0.2 μM of active enzyme. To the second and third aliquots add the same volume of the protease buffer. Incubate the mixtures for 2 h at the optimal temperature for the protease. Upon completion of the incubation add protease inhibitor to all the samples and adjust the pH to 7.2–7.4. Bring the volumes up to 1 mL with TBST.
During the incubation wash 0.8 mL worth of M-450 M2 bead suspension in 1 mL TBST three times using magnetic separator. After the second wash, split the magnetic bead suspension in four 0.25 mL aliquots and remove the supernatant after magnetic separation from two of them. Resuspend the beads with protease treated and 1 aliquot of control phage in one tube each and incubate for 1 h at ambient temperature with rotation end-over-end. The second control aliquot of phage will be used as the before-depletion control. Do not use vortexing to keep the beads in suspension when doing phage separations. After the incubation is over, remove the supernatant from the remaining two tubes of M-450 M2 beads by magnetic separation. Collect the unbound phage from the control and the protease treated phage by magnetic separation and mix with the fresh M-450 M2 beads from the remaining tubes. Incubate at ambient temperature with end-over-end rotation for 3 h. By the end of incubation, collect the supernatants by magnetic separation and determine the efficiency of depletion the following day by ELISA. Store the phage samples at –70°C.
Coat a 96-well microtiter plate with 100 μL/well of 5 μg/mL anti-M13 antibody in TBS at 4°C overnight.
The same night streak an LB agar 100 μg/mL Kanamycin Petri dish with K91Kan E. coli cells and incubate at 37°C overnight. Remove the dish from the incubator in the morning of the following day and place at 4°C until use.
In the morning of the next day block the plate with 120 μL/ well of 100 mg/mL BSA in TBS for 1 h at 37°C.
Prepare seven dilutions of phage with known concentration (standard) from 1011 phage/mL with a step of 1:2 by combining 0.35 mL of each dilution with 0.35 mL TBST. Dilute the before-depletion control, the protease treated and the control M2 depleted phage 1:20 by adding 35 μL of the solution to 665 μL TBST. Serially dilute the phage three times by combining 0.35 mL with 0.35 mL TBST. Add 100 μL/well from each dilution in triplicate. Incubate for 1 h at 37°C. When performing ELISA for subsequent rounds of selection, it is important to use phage from the starting stock used for this round of selection as standard, to obtain consistent results.
After incubation, wash the plate with 120 μL/well TBST three times. Add 100 μL/well anti-M13 monoclonal antibody – HRP conjugate diluted in TBST as recommended by the manufacturer. Incubate for 1 h at 37°C.
Upon completion of the incubation prepare the HRP substrate solution in the HRP substrate buffer. Wash the plate three times with 120 μL/well TBST. Add 4 μL/10 mL of 30% H2O2 to the substrate solution and apply at 100 μL/well to the plate. Incubate with shaking until color develops and add 50 μL 4 M H2SO4 to stop the reaction. Read the plate in a microtiter plate reader at 490 nm.
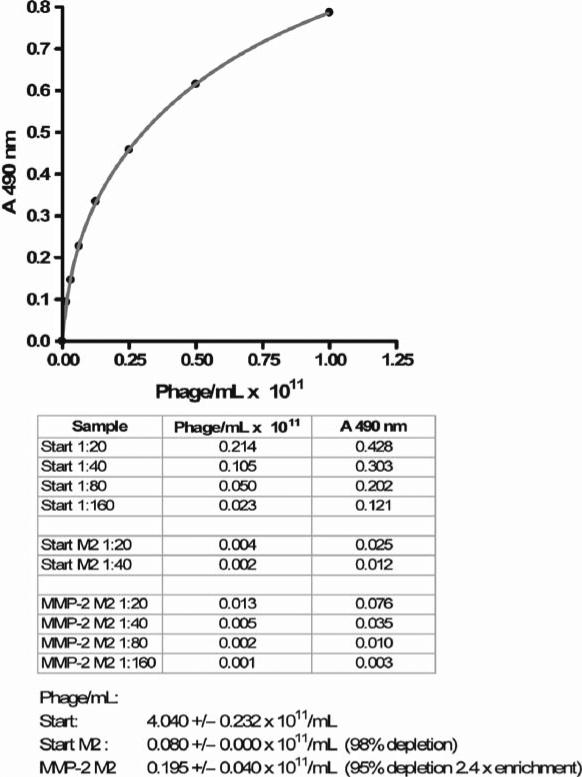
Plot the A490 vs. phage concentration for the standard curve. Fit the data points with the two site binding equation: A490=Bmax1 C /(Kd1 +C)+Bmax2 C /(Kd2+C), where Bmax1 an Bmax2 are the maximum binding per site, and Kd1 and Kd2 are respective dissociation constants, and C is phage concen tration. Apparently, the anti-M13 antibody recognizes two different classes of binding sites on the M13 coat protein expressed by the phage. Determine the phage concentration for the samples from the standard curve. Typically 98–99% of phages are depleted in the untreated sample as observed for the first round of selection, while the difference in depletion between the protease treated and untreated phage represents the enrichment for protease substrates (Fig. 2).
Start an overnight culture of K91Kan E. coli by inoculating 3 mL of LB 100 μg/mL Kanamycin with a single colony of a K91Kan cells streaked the night before. Incubate at 37°C overnight with rotation at 250 rpm.
Take 100 μL of the overnight culture and add to 9.9 mL Terrific Broth in a sterile 50 mL centrifuge tube. Incubate at 37°C with shaking at 250 rpm for 2 h. After 2 h start monitoring A600 of the culture by taking out 0.1 mL and adding to 0.9 mL of DI water and measuring A600. Multiply the obtained number by 10 to get the A600 for the culture.
When A600 = 2 AU (1010 cells/mL), slow down the shaking to 50 rpm for 5 min. Add an appropriate amount of the bacterial culture and the correspondingly appropriate volume of Terrific Broth so that after the addition of the substrate enriched phage, both the E. coli and the phage will be at 2 × 10 9 particles/mL. Shake the culture at 100 rpm at 37°C for 15 min. Transfer the infected cells into 10× volume of NZY broth containing 0.22 μg/mL tetracycline. Shake the culture for 35 min at 250 rpm, 37°C.
Add 0.001× volume of 20 mg/mL Tetracycline to the culture and incubate overnight at 250 rpm 37°C.
Determine the number of infected cells. Add 7 μL of culture to 63 μL LB medium. Perform serial dilutions three times by adding 20 μL of bacteria from the previous dilution to 180 μL LB. Plate 100 μL of each dilution onto 100 μg/ mL Kanamycin, 40 μg/mL Tetracycline NZY agar 10 cm Petri dish. Incubate overnight at 37°C. In the morning of the following day, count the colonies on each dish. Multiply by the appropriate dilution factor and then by 10 to get the number of infected cells per mL. Knowing the total number of the infected cells is useful in estimating how many individual phage clones have been obtained during infection.
The following morning centrifuge the culture in a sterile centrifuge tube or bottle at 2,400 × g for 10 min at 4°C. Without disturbing the cell pellet, transfer the phage-containing super-natant into a clean sterile centrifuge container and centrifuge again at 6,200 × g for 10 min at 4°C. Carefully pour the supernatant into a clean sterile container with a screw cap.
Add 0.15× volume of PEG/NaCl solution to the phage, close the container and mix thoroughly by inverting the container. Incubate on ice for at least 4 h or overnight at 4°C. The medium should turn cloudy.
Pellet the phage at 6,200 × g for 40 min at 4°C. Carefully discard the supernatant without disturbing the pellet. Remove the residual supernatant by briefly centrifuging the pellets and carefully aspirating the rest of the liquid.
Add 5 mL of sterile TBS and shake the pellets at 150 rpm at 37°C for approximately 30 min to resuspend the pellet.
Centrifuge the phage solution at 10,100–22,700 × g for 10 min at 4°C. Transfer the supernatant into a fresh sterile centrifuge container.
Add 0.15× volume of PEG/NaCl solution to the container and mix thoroughly by inversion. Allow the phage to precipitate for 1 h on ice. A heavy precipitate should appear.
Centrifuge the phage at 10,100 × g for 40 min at 4°C. Remove the supernatant.
Add 2 mL of sterile TBS to the pellet and allow the pellet to soften for 1 h. Vortex the pellet again to bring the phage into solution
Clear the supernatant by centrifuging at 10,100–22,700 × g for 10 min at room temperature or 4°C.
Collect the supernatant. Determine the phage concentration as described in step 4 above. Store the phage short term at 4°C, long term at –70°C. The first round of selection is complete.
Repeat the above steps for the second round. If more than two rounds of selection are required, as can be the case if poor enrichment for substrate phage is observed after the second round, we recommend enriching the phage from the second round for high FLAG expressers. The reason for this is that after each amplification, there is a loss of tag expressing phage and it can be quite substantial after the second round. We routinely enrich phage for FLAG expression before proceeding to substrate identification.
Take 200 μL M2 M450 beads (10 8 ) and wash three times with TBST by magnetic separation. Add 2.5 × 10 12 phage to the beads and bring the volume up to 1 mL with TBST. Incubate overnight with end-over-end rotation.
The following day, make 10 mL of 0.005% Brij 35 solution in DI water and sterilize it by filtration through a 0.2 μ m filter.
Prepare 0.05% TFA solution in 0.005% Brij 35 solution.
Wash the beads three times with 1 mL TBST by magnetic separation after discarding the supernatant followed by two-time wash with 0.005% Brij solution. Remove any traces of liquid from the beads and add 100 μL of 0.05% TFA to the beads. Shake for 10 min at room temperature making sure the beads stay in suspension. Collect the supernatant by magnetic separation and mix with 100 μL TBS. Determine the phage concentration as described in step 4 above. Multiply the obtained number by the total volume to get the total number of phage. Typically we get around 5 × 1011 phage after enrichment.
Fig. 2.
A typical standard curve for an M13 phage ELISA and the results of the first round of selection of MMP-2 substrates.
3.2. Verification of Substrate Phage
After the final round of selection is complete, one needs to verify that individual phage clones are bearing substrates prior to nucleotide sequencing. This is done by substrate phage ELISA of the media from bacterial cultures grown from single colonies of K91Kan cells bearing a single phage clone. To ensure that we select phage clones with adequate FLAG epitope expression, we use a positive control phage with high FLAG expression level (same or higher than the starting library) and select only those substrate phage clones, whose FLAG expression level is above 0.5 of that of the control. This is done to minimize the error of measurement of the degree of hydrolysis of substrate phage, which is determined by comparing the amount of FLAG with and without exposure to protease. In addition, the level of expression of the FLAG epitope reflects the number of copies of the substrate peptide per phage, thus allowing us to select high expressers for MALDI TOF analysis used for identification of the scissile bond in the substrate peptide.
Perform enrichment of the phage from the last round of selection for FLAG epitope expression as described in Subheading 3.1, steps 30–33.
Prepare NZY agar QPix trays or 10 cm Petri dishes by pouring 200 or 20 mL of NZY agar with 100 μg/mL Kanamycin, 40 μg/mL Tetracycline per tray/dish for HTS or LTS versions of the procedure respectively. Store the NZY agar trays/dishes at 4°C in a plastic bag.
Start an overnight 3 mL culture in LB 100 μg/mL Kanamycin from a freshly streaked colony. Incubate overnight at 37°C, shaking at 250 rpm.
Next morning add 100 μL of the overnight culture to 10 mL of Terrific Broth. Incubate at 37°C, shaking at 250 rpm.
After 2 h of incubation start monitoring A600 of 1:10 diluted culture in DI water until it reaches 0.2 AU. Slow the shaking down to 50 rpm for 5 min.
Place the Qtrays or Petri Dishes in a 37°C incubator.
Add 1 mL of the K91Kan culture (10 10 cells/mL at A600 = 2.0) to 9 mL Terrific Broth and supplement with 10 9 FLAG enriched phage from the last round of selection. Continue incubation at slow shaking for 15 min.
Prepare 10 mL NZY broth with 0.22 μg/mL tetracycline from 20 mg/mL stock. Add 1 mL of infected culture to 10 mL of NZY broth 0.22 μg/mL Tetracycline. Shake 35 min at 250 rpm, 37°C.
Add 10 μL of 20 mg/mL stock of Tetracycline to the culture.
Dilute the culture 50-fold in NZY Broth and plate 0.75 mL per QPix tray or 75 μL per 10 cm Petri dishes. Place the trays in a 37°C incubator with 70% humidity. The Petri dishes can go in a 37°C incubator without humidification.
The next morning pick colonies using the QPix robot in the case of QPis trays or manually from the Petri dishes into 96-well deep-well 2 mL plates filled with 1 mL per well of NZY broth supplemented with 20 μg/mL Tetracycline. After picking, put the BreatheSeal film on the plates and incubate in the ATR Multitron incubator shaker at 750 rpm, 37°C, 70% humidity. For a small scale project, a regular shaking incubator can be used at 250 rpm, 37°C.
The same day, coat 96 well ELISA plates with 100 μL/well of 5 μg/mL anti-M13 antibody in PBS at 4°C overnight.
In the morning, block the plates with 120 μL of 100 mg/mL BSA in PBS for 1 h at 37°C.
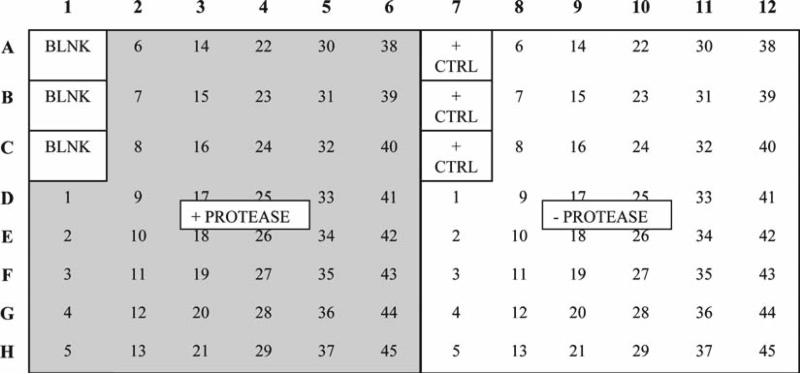
Remove the BSA and add 100 μL/well of the bacterial cultures from the deep well plates as shown in Fig. 3: To the BLNK wells add 100 μL NZY broth. To the + CTRL wells add 100 μL of 1012 phage/mL of the positive control clone in NZY broth.
Incubate the plates for 1 h at 37°C.
Wash the plates with 120 μL/well TBST four times and remove the residual buffer from the plate by gently tapping the plates face down against a paper towel.
Add 100 μL of (50 nM active enzyme) protease solution per well to the left half of the plate as shown in Fig. 3. To the right half add 100 μL/well of the protease buffer. Incubate for 2 h under the optimal conditions for the protease.
Wash the plates four times with 120 μL/well TBST and add 100 μL/well of HRP conjugated M2 antibody appropriately diluted in TBST. Incubate for 1 h at 37°C. Dissolve 1 tablet of OPD per 10 mL of HRP substrate buffer right before the end of the incubation period.
Wash the plates four times with 120 μL/well TBST, remove the residual buffer by gently tapping the plates face down against a paper towel. Add 4 μL of 30% H2 O2 per 10 mL of OPD solution, mix thoroughly and dispense 100 μL/well. Shake the plates gently and observe color development. Usually the plates can be read 5 min after addition of the substrate, but one should exercise proper judgment to stop the reaction at an appropriate time, so as not to over or under develop the reaction. A target A490 value of 1 is optimal. Add 50 μL/well of 4 M H2SO4 to stop the reaction. Read the plates at 490 nm in a microtiter plate reader.
Analyze the data by first eliminating from consideration any clones with A490 less than 0.5 of that of the positive control. For the remaining clones calculate the degree of hydrolysis by dividing the A490 of the + protease well by that of the – protease well and subtracting the result from 1. Disregard any clones with the degree of hydrolysis of less than 0.2. The substrate identification step is complete.
Prepare glycerol stocks of the substrate phage by mixing 100 μL of culture with 100 μL of glycerol stock solution in a Genetix 96-well flat bottom1/2 height plate. Store the glycerol stocks at –70°C. In some cases there is a pH requirement for proteolytic activity that is incompatible with phage capture by anti-M13 antibodies. In these cases, we perform protease treatment of PEG precipitated phage in the pro-tease buffer with a pH necessary for optimal protease activity. After that we adjust the pH to neutral and perform the ELISA.
Prepare glycerol stocks of the substrate phage by mixing 100 μL of culture with 100 μL of glycerol stock solution in a Genetix 96-well flat bottom1/2 height plate. Seal the plates with aluminum sealing tape. Store the glycerol stocks at –70°C.
Centrifuge the remaining overnight cultures at 4,000 × g for 10 min at 4°C. Collect the supernatants into fresh deep 96-well plates. Add 0.15× volume of PEG/NaCl solution, cover the plate with a storage mat, mix by inversion end-over-end about 100 times, spin briefly at 300 × g and put at 4°C overnight. Centrifuge at 4,600 × g for 90 min at 4°C. Carefully discard the supernatant and remove residual liquid by gently tapping the plates face down against a paper towel.
Add 120 μL/well of the protease buffer and shake the plates at 750 rpm in a ATR Multitron shaker incubator or similar for 30 min.
Transfer 50 μL of the phage solution to 50 μL of protease buffer per well on the right side of a 96-well uncoated ELISA plate (Fig. 3). Transfer another 50 μL of the phage solution to 50 μL of 100 nM protease in protease buffer in the counterpart wells on the left side of the plate. Incubate at appropriate temperature for 2 h.
Neutralize the samples by addition of an appropriate amount of neutralization solution (determined earlier). Transfer the samples to an anti-M13 coated BSA blocked plate. Incubate for 1 h at 37°C.
Proceed as described in steps 17–19 above.
Fig. 3.
Template for substrate phage ELISA. Bacterial cultures are applied to anti-M13 antibody coated 96-well plate at 100 μL/well. Each culture is applied to the plate twice: on the left half of the plate and symmetrically on the right half. The phage captured on the left half of the plate is treated with protease ( shaded area ), while their counterparts on the right are not. The degree of hydrolysis is determined by subtracting the ratios of A490 between the protease treated and nontreated samples from 1.
3.3. Sequencing Phage Substrates
Once the protease substrate phage clones have been identified, the nucleotide sequence of the random insert is determined. This is done by PCR amplification of the region of the g3p bearing the randomized insert followed by sequencing of the PCR amplicons. The PCR procedure is performed in-house in a 96-well format and the sequencing of the resulting amplicons is outsourced.
Thaw the glycerol stocks of the substrate phage at room temperature. Cherry pick the substrate clones identified by substrate phage ELISA and inoculate 1,250 μL of LB 20 μg/mL Tetracycline per well of 2 mL 96-deep well plates, using QPix colony picking robot for an HTS phage display or a sterile Pasteur loop for an LTS one. Seal the plates with BreatheSeal film. Grow the inoculates overnight at 750 rpm, 37°C, 70% humidity in an ATR Multitron shaker incubator. For a small scale project, a regular shaking incubator can be used at 250 rpm, 37°C.
- The following morning prepare the PCR master mix:
- 2 μL/sample 5 μM FUSE5 forward primer
- 2 μL/sample 5 μ M FUSE5 Super Reverse primer
- 45 μL Platinum PCR Super Mix
- Dispense 49 μL of the master mix per well of a 96-well PCR plate. Add 2 μL per well of the overnight culture. Run the PCR as follows:
- 72°C 10 min
- 94°C 3 min
- 34 × (94°C 50 s, 50°C 1 min, 72°C 1 min)
- 72°C 6 min
- Hold at 4°C.
To determine the quality of the PCR, run 10 PCR products per plate at 10 μL/lane on 1% agarose gel.
Freeze the PCR products and send out for sequencing.
3.4. Identification of Scissile Bonds
Once the sequences of phage substrates are known from nucleotide sequencing, the position of the scissile bond within the substrate peptide is determined directly from purified phage using MALDI TOF MS. We take advantage of the fact that the phage express sufficient levels of g3p, so that the peptide hydrolyzed from the phage by protease is present in adequate quantity to analyze by MS. Once the mass of the cleaved portion of the peptide substrate is known, this information can be combined with sequence information to pinpoint the position of the scissile bond.
Prepare the necessary amount of M-450 anti-M13 beads at 10 8 beads per sample by following the protocol described in Subheading 3.1 . Use anti-M13 instead of M2 antibodies.
Prepare M2 coupled M-280 beads. Wash 1.29 × 10 9 beads three times with 1 mL of 100 mM sodium phosphate pH 7.0 by magnetic separation. Resuspend the beads in 1 mL of 250 μg/mL M2 antibody in 100 mM sodium phosphate, pH 7.0.
Incubate with end-over-end rotation overnight at 37°C.
In the morning of the following day measure the A280 of the supernatant after magnetic separation. Determine the amount of antibody coupled. Typically 80–90% of antibody gets coupled to M-280 beads.
Remove the supernatant by magnetic separation and resuspend the beads in 1 mL of sterile 1 mg/mL BSA in TBS. Incubate with end-over-end rotation for 4 h at room temperature. Store at 4°C.
Thaw the glycerol stocks of substrate phage and inoculate 1.2 mL cultures of substrate phage in 2 mL 96-deep well plates and grow overnight as described in Subheading 3.3 of step 1.
The following morning wash the M-450 anti-M13 beads three times with TBST. Resuspend in the original volume of TBST and dispense into an Assay Block 1 mL 96-well plate at 200 μL/well.
Spin down the cultures at 4,000 × g, 4°C for 10 min.
Remove TBST from the M-450 anti-M13 beads by magnetic separation and resuspend the beads in 1 mL of culture super-natant. Seal the plates with the storage mats and incubate with rotation end-over-end for 3 h at room temperature.
Wash the beads with 1 mL of the protease incubation buffer three times by magnetic separation and resuspend in 100 μL/well of 50–200 nM active protease. Incubate with vortex shaking overnight at the optimal temperature.
- Alternatively, one can use PEG precipitation for buffer exchange, especially if the protease buffer has acidic or alkaline pH incompatible with the antibody capture of the phage. This approach provides good results as well, although there will be residual PEG left over. One has to test if PEG has an effect on the protease activity before using this method.
- Add 0.15× volume of PEG/NaCl solution, cover the plate with a storage mat, mix by inversion end-over-end about 100 times and incubate at 4°C overnight. Centrifuge at 4,600 × g for 90 min at 4°C. Carefully discard the supernatant and remove residual liquid by gently tapping the plates face down against a paper towel.
- Add 100 μL/well of the protease solution and shake the plates at 750 rpm in a ATR Multitron shaker incubator or similar overnight.
- The next morning, add the appropriate amount of neutralization buffer.
Wash an appropriate amount of M2 M-280 magnetic beads three times in TBST by magnetic separation. Resuspend in the original volume of TBST and dispense into an Assay Block 1 mL 96-well plate at 200 μL/well. Remove the TBST after magnetic separation. Resuspend the beads in the protease treated phage and incubate with vortex mixing for 3 h. Wash three times with 100 μL/well of 5 mM Tris, 0.1 mg/mL BSA by magnetic separation. Wash three times with 100 μL/well of 5 mM Tris by magnetic separation. Remove the residual liquid.
Elute the bound peptides by addition of 10 μL/well of 0.1% TFA and vortex mixing for 5 min at room temperature.
Prepare the matrix solution by mixing 10 mg DHB with 0.333 mL acetonitrile and 0.667 mL 0.1% TFA. Add 5 μL of the matrix solution to a v-bottom polypropylene plate.
Remove 5 μL of the eluted peptides after magnetic separation and mix with the matrix solution in the v-bottom polypropylene plate.
Place 1 μL of the peptide/matrix solution per spot of an AnchorChip 400 target. Let dry at room temperature.
Determine the masses of the released peptides by MALDI TOF.
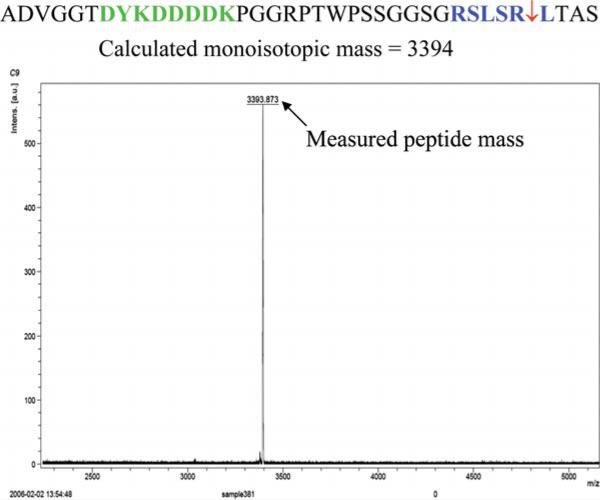
Determine the scissile bond position by matching the mass of the released peptide to the amino acid sequence obtained by translation of the DNA sequencing data of the PCR products (Fig. 4).
Fig. 4.
Mapping of the position of the scissile bond by MALDI TOF MS. The amino acid sequence was determined by DNA sequencing of a PCR amplicons from an MMP-2 substrate phage infected bacterial culture followed by translation of the DNA sequence. The mass of the peptide released from 10 12 MMP-2 substrate phage by MMP-2 was determined by MALDI TOF MS performed using Bruker Daltonics UltraFlex II mass spectrometer. (See Color Plates)
3.5. Data Analysis
The simplest and at the same time the most informative way of representing positional preferences of a given amino acid in a binding subsite is to use graphical sequence logos. This method was developed by Scheider and Stephens (14) and was implemented as a Web service, called WebLogo, by Crooks et al. (15). It is available at: http://weblogo.berkeley.edu/. It displays the nucleic acid bases or amino acids patterns as a result of multiple sequence alignment. The patterns consist of stacks of characters representing the sequence. Each stack represents a single position in the sequence. The characters that occur most commonly are located at the top of the stack. The height of each character is proportional to its frequency at that position. The overall height of the stack reflects the sequence conservation at that position. Instead of using frequencies as a way of scaling the height of the entire stack, that height can be adjusted according to the information content (measured in bits) of the sequence at that position.
According to Schneider and Stephens (14), the information content is calculated in the following way. First, the sequence conservation at a specific position in the alignment: Rseq is calculated as the difference between the maximum possible entropy and the entropy of the observed symbol distribution:
where pn is the observed frequency of symbol n at a specific position, N is a number of distinct symbols used in a sequence type, which is 4 for nucleic acids and 20 for proteins., yielding the maximum value of entropy (log2 N) equal to 2 or 4.32, for nucleic acids and proteins, respectively. The sum of sequence conservation (Rseq) at each position of the logo measures the information content of the logo. This is true under the assumption that there is no inter-positional correlation and the background distribution of symbols is uniform.
In our calculation for determining patterns in the set of substrates for matrix metalloproteinases studied in the phage display experiments, we use the frequency-based instead of information-based logos. To prepare input for the WebLogo program, e.g., the set of aligned sequences, we use an approach that is based on Position Weight Matrix (PWM). A PWM assumes independence between positions in the pattern and allows one to take into account the background distribution. An element in a PWM is calculated as mi,j = log(pi,j/bi), where pi,j is the probability of observing symbol (amino acid) i at position j of the motif, and bi is the probability of observing the symbol i in a background model. The value of particular element of the PWM is the log-odds of a given symbol being found in the current pattern vs. being found in the background distribution. From the PWM matrix one can rederive the set of sequences that correspond to appropriate log-odds values, which in turn can be used as input to WebLogo program.
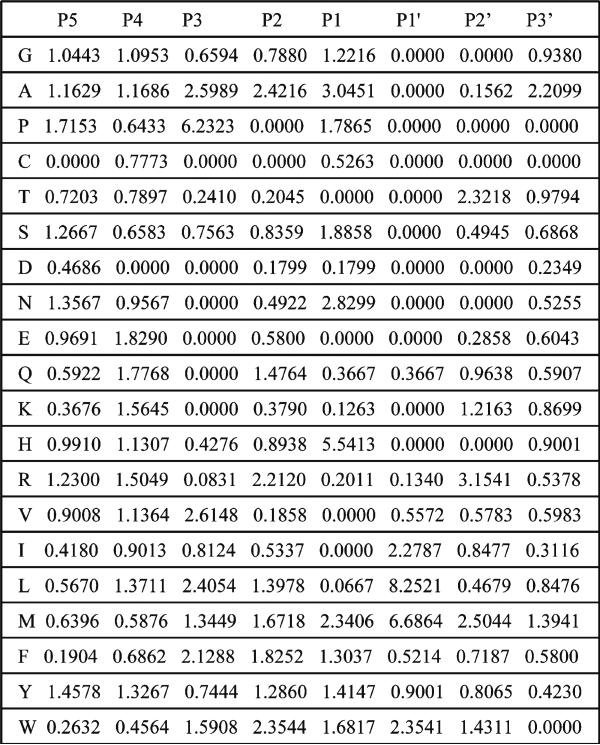
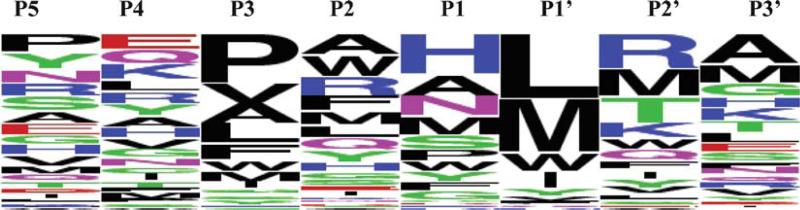
The specific procedure of deriving PWM for substrates of metalloproteinases is as follows. First, we aligned the sequences of all substrates for a given enzyme along the hydrolyzed peptide bond. Each sequence of the substrate in the phage display experiment consists of six amino acids variable regions and the N-terminal and C-terminal constant tags. Since a hydrolysis site can occur at any position in the six amino acids variable region, we included some portion of the constant tags in the sequence alignment. All aligned sequences have been uniformly trimmed to the length of eight amino acids; only those amino acids that span P5-P3 positions have been left for further consideration. In the next step, we derived the position weight matrices (PWMs), specific for each metalloproteinase, using the frequencies of each individual amino acid at each position in the alignment. The PWMs have been normalized by distribution of amino acids in the background sequences. In deriving distribution of the background sequences we included also short, three and two amino acid long fragments of the constant tags at the N- and C-terminals, respectively. In this way, we took into account biasing of the background and substrate distributions by constant fragments of the sequences. The background sequences are chosen randomly and are predicted to be hydrolyzed with less than 5% probability. In the last step, each position of the PWM is renormalized to the values in the range between 0 and 1 over all amino acids. Then each value for a given amino acid at a given position is multiplied by 100. This is the final number each amino acid would appear in the set of sequences, which are used as input for the WebLogo program. An example of the PWM for the 234 substrates of MMP-16 metalloproteinase and the corresponding sequence logo are shown in Figs. 5 and 6.
Fig. 5.
Position weight matrix of 234 substrates of MMP-16 .
Fig. 6.
WebLogo representation of the position weight matrix depicted in Fig. 5. (See Color Plates)
3.6. Concluding Remarks
Protease substrate phage display is a powerful tool for delineation of protease substrate specificity at the level of amino acid sequence. Our group adapted this approach for high throughput data acquisition and analysis, thereby making it capable of studying the evolution of substrate specificity in closely related families of proteases. These studies may shed light on the biological processes affected by different members of the family as a result of differential substrate specificities. As a bonus, one gets information on unique and selective peptide substrates that can be used for such applications as activity-based probes, selective inhibitors, and so on. We feel that the ambitious goal of characterizing substrate specificities of all human proteases is now well within reach. This work was supported by Grant Number RR020843 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH) and its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
References
- 1.Barrett AJ, Rawlings Neil D., Woessner J. Fred. Handbook of Proteolytic Enzymes. Elsevier Academic Press; San Diego: 2004. [Google Scholar]
- 2.Berger A, Schechter I. Mapping the active site of papain with the aid of peptide substrates and inhibitors. Philos Trans R Soc Lond B Biol Sci. 1970;257(813):249–64. doi: 10.1098/rstb.1970.0024. [DOI] [PubMed] [Google Scholar]
- 3.Diamond SL. Methods for mapping protease specificity. Curr Opin Chem Biol. 2007;11(1):46–51. doi: 10.1016/j.cbpa.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 4.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228(4705):1315–7. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 5.Matthews DJ, Wells JA. Substrate phage: selection of protease substrates by monovalent phage display. Science. 1993;260(5111):1113–7. doi: 10.1126/science.8493554. [DOI] [PubMed] [Google Scholar]
- 6.Ohkubo S, Miyadera K, Sugimoto Y, Matsuo K, Wierzba K, Yamada Y. Identification of substrate sequences for membrane type-1 matrix metalloproteinase using bacteriophage peptide display library. Biochem Biophys Res Commun. 1999;266(2):308–13. doi: 10.1006/bbrc.1999.1816. [DOI] [PubMed] [Google Scholar]
- 7.Cloutier SM, Chagas JR, Mach JP, Gygi CM, Leisinger HJ, Deperthes D. Substrate specificity of human kallikrein 2 (hK2) as determined by phage display technology. Eur J Biochem. 2002;269(11):2747–54. doi: 10.1046/j.1432-1033.2002.02960.x. [DOI] [PubMed] [Google Scholar]
- 8.Smith MM, Shi L, Navre M. Rapid identification of highly active and selective substrates for stromelysin and matrilysin using bacteriophage peptide display libraries. J Biol Chem. 1995;270(12):6440–9. doi: 10.1074/jbc.270.12.6440. [DOI] [PubMed] [Google Scholar]
- 9.Hills R, Mazzarella R, Fok K, et al. Identification of an ADAMTS-4 cleavage motif using phage display leads to the development of fluorogenic peptide substrates and reveals matrilin-3 as a novel substrate. J Biol Chem. 2007;282(15):11101–9. doi: 10.1074/jbc.M611588200. [DOI] [PubMed] [Google Scholar]
- 10.Pan W, Arnone M, Kendall M, et al. Identification of peptide substrates for human MMP-11 (stromelysin-3) using phage display. J Biol Chem. 2003;278(30):27820–7. doi: 10.1074/jbc.M304436200. [DOI] [PubMed] [Google Scholar]
- 11.Deng SJ, Bickett DM, Mitchell JL, et al. Substrate specificity of human collagenase 3 assessed using a phage-displayed peptide library. J Biol Chem. 2000;275(40):31422–7. doi: 10.1074/jbc.M004538200. [DOI] [PubMed] [Google Scholar]
- 12.Kridel SJ, Chen E, Kotra LP, Howard EW, Mobashery S, Smith JW. Substrate hydrolysis by matrix metalloproteinase-9. J Biol Chem. 2001;276(23):20572–8. doi: 10.1074/jbc.M100900200. [DOI] [PubMed] [Google Scholar]
- 13.Bieth JG. Theoretical and practical aspects of proteinase inhibition kinetics. Methods Enzymol. 1995;248:59–84. doi: 10.1016/0076-6879(95)48007-2. [DOI] [PubMed] [Google Scholar]
- 14.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18(20):6097–100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]