Abstract
The covalent marking of proteins by methyl group addition to arginine residues can promote their recognition by binding partners or can modulate their biological activity. A small family of gene products that catalyze such methylation reactions in eukaryotes (PRMTs) work in conjunction with a changing cast of associated subunits to recognize distinct cellular substrates. These reactions display many of the attributes of reversible covalent modifications such as protein phosphorylation or protein lysine methylation; however, it is unclear to what extent protein arginine demethylation occurs. Physiological roles for protein arginine methylation have been established in signal transduction, mRNA splicing, transcriptional control, DNA repair, and protein translocation.
Chemistry and Enzymology of Protein Arginine Methylation and Demethylation
Biology relies upon the enlarged repertoire of interactions that occur when proteins are posttranslationally modified. In recent years, it has become clear that methyl groups stand beside phosphate groups as major controlling elements in protein function. A wide variety of methylation (and in some cases demethylation) reactions occur at the side chains of a number of amino acid residues and at protein N- and C-termini. These modifications generate distinct sets of chemical interactions that play roles in a multitude of regulatory pathways (Clarke and Tamanoi, 2006).
The modification of arginine side chain guanidino groups is probably the most quantitatively extensive protein methylation reaction in mammalian cells (Gary and Clarke, 1998). The number of distinct modified proteins is also large (Pahlich et al., 2006). Arginine is unique among amino acids as its guanidino group contains five potential hydrogen-bond donors that are positioned for favorable interactions with biological hydrogen-bond acceptors. In protein-DNA complexes, arginine residues are the most frequent hydrogen bond donors to backbone phosphate groups and to thymine, adenine, and guanine bases (Luscombe et al., 2001). Specific networks of hydrogen bonds can form with arginine residues and adjacent phosphate groups in RNA loops (Calnan et al., 1991), and the arginine-aspartate two H-bond interaction is especially stable in proteins (Mitchell et al., 1992). Each addition of a methyl group to an arginine residue not only changes its shape, but also removes a potential hydrogen bond donor. Such chemistry could promote the preferential inhibition by methylation of some, but not all, binding partners. For example, arginine methylation of the Sam68 proline-rich motifs can inhibit its binding to SH3, but not WW domains (Bedford et al., 2000). Methylation of arginine residues might also increase their affinity to aromatic rings in cation-pi interactions (Hughes and Waters, 2006). Such interactions are seen in the aromatic cage of the SMN tudor domain that likely interacts with the methylated tail of the SmD splicing factor (Sprangers et al., 2003). Thus, modification of arginine residues in proteins can readily modulate their binding interactions and thus can regulate their physiological functions.
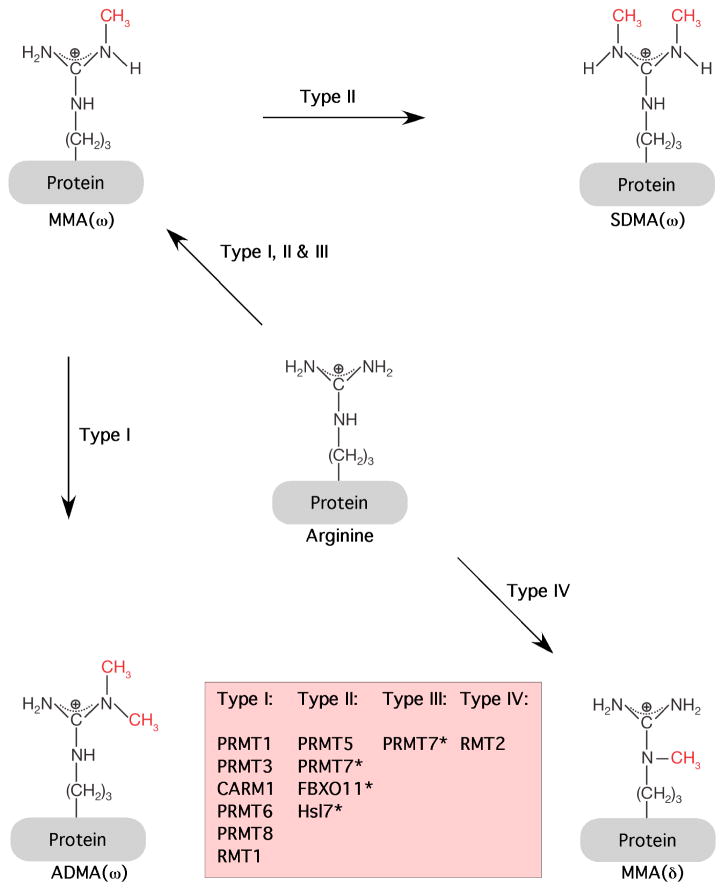
Three distinct types of methylated arginine residues occur in mammalian cells. The most prevalent is omega-NG,NG-dimethylarginine (Paik and Kim, 1980). Here, two methyl groups are placed on one of the terminal nitrogen atoms of the guanidino group; this derivative is commonly referred to as asymmetric dimethylarginine (ADMA) (Figure 1). Two other derivatives occur at levels of about 20% to 50% that of ADMA. These include the symmetric dimethylated derivative, where one methyl group is placed on each of the terminal guanidino nitrogens (omega-NG,N′G-dimethylarginine; SDMA) and the monomethylated derivative with a single methyl group on the terminal nitrogen atom (omega-NG-monomethylarginine; MMA). These three derivatives are present on a multitude of distinct protein species in the cytoplasm, nucleus, and organelles of mammalian cells (Bedford and Richard, 2005). Methylated arginine residues in proteins are often flanked by one or more glycine residues (Gary and Clarke, 1998), but there are many exceptions to this general rule.
Figure 1. Types of methylation on arginine residues.
Types I, II and III PRMTs generate monomethylarginine (MMAω) on one of the terminal (ω) guanidino nitrogen atoms. These two nitrogen atoms are equivalent. The subsequent generation of asymmetric dimethylarginine (ADMA) is catalyzed by type I enzymes, and the production of symmetric dimethylarginine (SDMA) is catalyzed by type II enzymes. Type III PRMTs only monomethylate. Type IV enzyme activity that catalyzes the monomethylation of the internal guanidino nitrogen atom has only been described in yeast. The type of methylation reactions catalyzed by PRMT7, FBXO11, and Hsl7 are still being established, and are thus marked with an asterisk (see text).
The formation of MMA, ADMA, and SDMA in mammalian cells is performed by a sequence-related family of catalytic subunits of protein arginine methyltransferases termed PRMTs (Figure 2). The exact number of genes encoding these catalytic subunits is under current investigation; six genes are known to encode enzymes with well-characterized activities (PRMT1, 3, 4 (CARM1), 5, 6, and 8), and another three genes encode sequence-related proteins with possible or probable methyltransferase activities (PRMT2, 7, 9 (4q31)) (Bedford, 2007). Each PRMT species harbors the characteristic motifs of seven-beta strand methyltransferases (Katz et al., 2003), as well as additional “double E” and “THW” sequence motifs particular to the PRMT subfamily (Cheng et al., 2005). It has also been proposed that the FBXO11 and FBXO10 proteins, which do not harbor these signature motifs, represent a second family of protein arginine methyltransferases; however these activities require validation (Cook et al., 2006; Krause et al., 2007). The gene products with well-characterized activities, generally purified as fusion proteins, catalyze MMA formation; PRMT1, 3, 4 (CARM1), 6 and 8 additionally catalyze ADMA formation, whereas PRMT5 additionally catalyzes SDMA formation. Enzymes that form ADMA are designated “type I”; those that form SDMA are designated “type II” (Gary and Clarke, 1998). To date, no enzyme has been found that forms both ADMA and SDMA derivatives.
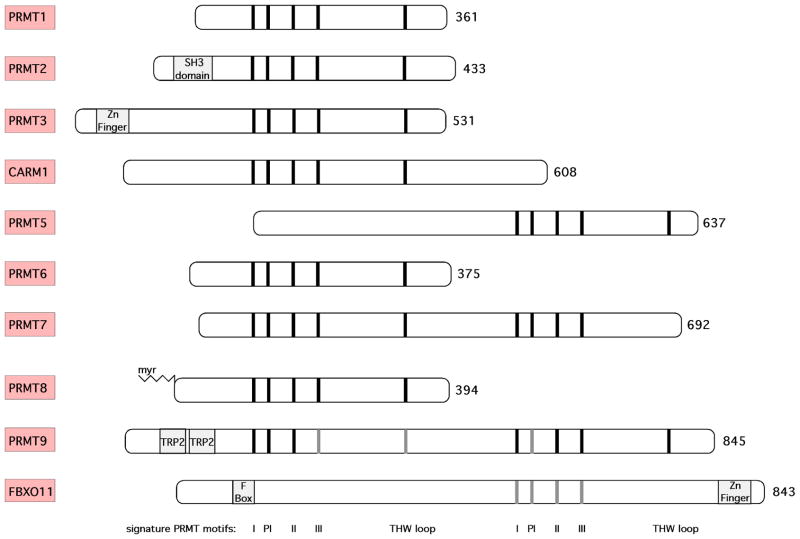
Figure 2. The protein arginine methyltransferase family.
The mammalian PRMT family currently contains nine highly related members; all harbor signature motifs I, post-I, II, and III, and the conserved THW loop (in black bars). FBXO11 might have arginine methyltransferase activity, but is not structurally related to the other PRMTs and required “forced” alignments to identify putative signature motifs (in gray bars). Protein domains that might assist in substrate recognition are marked in gray boxes. The accession numbers for the PRMTs are as follows: hPRMT1 (AAF62893), hPRMT2 (AAH00727), hPRMT3 (AAC39837), hCARM1 (NP_954592), hPRMT5 (AAF04502), hPRMT6 (Q96LA8), hPRMT7 (NP_061896), mPRMT8 (DAA01382), hPRMT9 (AAH64403) and mFBXO11 (AAI28480).
A clear understanding of how PRMT catalytic polypeptides can be assembled into active complexes with other protein subunits in cells is needed. Although PRMT1, 3, 4(CARM1), 5, 6, and 8 are active methyltransferases in the absence of other polypeptide species in vitro, they might also bind additional partners for their functions in vivo. It now appears that interactions between PRMTs and their binding partners (and presumed regulatory subunits) can be transient or permanent (Table 1). It is possible that the species with poorly defined activities require the presence of other subunits for catalytic activity.
TABLE 1.
Interacting Protein Partners of Mammalian PRMTs
| PRMT species | Interacting Species |
|---|---|
| PRMT1 | poly(ADP-ribose) polymerase and NF-kappaB |
| Sam68 and the MLL complex | |
| Btg1 and Tis2/Btg2 | |
| intracytoplasmic domain of the type I interferon receptor | |
| hCAF1 | |
| PRMT2 | RB |
| PRMT8 (via SH3 domain) | |
| PRMT3 | DAL-1/4.1B |
| Zn-finger domain – binds 40S rpS2 | |
| CARM1/PRMT4 | p160 familly coactivators |
| SWI/SNF complex of transcription | |
| NUMAC complex | |
| PRMT5 | * nucleolin |
| hIws1 transcription elongation factor | |
| Histone-binding protein COPR5 | |
| * LIM protein AJUBA | |
| SWI-SNF | |
| methylosome complex with pICln and MEP50 | |
| Blimp1 | |
| DAL-1/4.1B tumor suppressor | |
| RNA polymerase II FCP phosphatase | |
| * SPT4/SPT5 transcription elongation factor | |
| PRMT7 | |
| PRMT7 | CTCFL testis-specific factor |
| PRMT5 | |
| PRMT8 | Ewing Sarcoma protein |
| PRMT1 | |
| SH3 domains of PRMT2, Fyn, Plcγ & P85 |
PRMT5-interacting species that may be compromised by the use of FLAG-tagged fusion proteins (Nishioka and Reinberg, 2003).
An important question under current investigation is whether protein arginine demethylation reactions occur to reverse the effects of the modifications. Initial studies indicated that methyl groups were stable on arginine residues. The apparent absence of protein arginine demethylases suggested that the only way to reverse the effects of the modification would be to degrade the protein to its component amino acids and then make a new unmodified version by protein synthesis. However, two types of enzymes that can remove methyl groups from arginine residues in proteins were recently identified. MMA residues in proteins can be deiminated to citrulline residues by the PAD4 peptidylarginine deiminase (Thompson and Fast, 2006). It is unknown whether the citrulline residue might be converted to an arginine residue to complete the demethylation process; however it has been suggested that this enzyme is unlikely to play a physiological demethylation role. Indeed, peptides containing MMA are more slowly deiminated than those containing arginine residues and those containing ADMA are not deiminated at all; thus methylation might in fact inhibit a reaction that normally converts arginine residues to citrulline residues (Raijmakers et al., 2007). Further work will be needed to assess the role of these enzymes in demethylation. Additionally, recent work has suggested that a second type of enzyme, the Jumonji domain-containing proteins, which was originally identified as a family of lysine demethylases, can also demethylate arginine residues. Indeed, JMJD6 can directly regenerate arginine residues from methylated histone species (Chang et al., 2007). It will be important to determine if there are additional enzymes that catalyze similar reactions, and to assess the biological significance of each of these demethylation pathways in regulating protein arginine methylation.
The Mammalian PRMTs
In the section below, we describe the conserved family of mammalian PRMTs 1-9 as well as a second family of putative enzymes related to the F-box only proteins (Figure 2). There does not appear to be major redundancy between these enzymes; mouse knockouts display generally clear and dramatic phenotypes (Table 2). The diversity of these enzymes is enhanced by alternative splicing reactions that lead to amino acid sequence variants (Goulet et al., 2007).
TABLE 2.
Phenotypes of mice lacking protein arginine methyltransferases
| Gene | Phenotype(s) |
|---|---|
| Prmt1 | Embryonic lethal, but ES cells can survive |
| Trapping mutant generates a hypomorphic allele | |
| Prmt2 | Mice are viable |
| Mouse embryo fibroblasts have increased activity of NF-kappaB and are more resistant to apoptosis than wild type cells | |
| Prmt3 | Mice are viable, but mutant embryos are slightly smaller |
| Trapping mutant generates a hypomorphic allele | |
| Carm1/Prmt4 | Newborns small and die shortly after birth |
| T cell development blocked in thymus | |
| Lipid metabolism altered in embryos |
The major type I enzyme, PRMT1 – broad specificity with multiple interacting partners
PRMT1 was the first mammalian protein arginine methyltransferase identified as a single gene product and was initially characterized as a GST-fusion protein expressed in bacterial cells (Lin et al., 1996). Previous purifications of these enzymes from mammalian tissues was complicated by their low abundance and the presence of a multitude of polypeptides in the final preparations, making it difficult to discern which were contaminants and which were truly associated with the enzymatic activity (Gary and Clarke, 1998). PRMT1 is responsible for the bulk (about 85%) of total protein arginine methylation activity in cultured RAT1 fibroblast cells as well as in mouse liver (Tang et al., 2000).
PRMT1 has a very wide substrate specificity, with a preference for methylating arginine residues that are flanked by one or more glycine residues (Gary and Clarke, 1998; Lee and Bedford, 2002). Structural studies have identified three different peptide-binding channels, suggesting that different PRMT1 substrates might approach the active site from different angles (Zhang and Cheng, 2003). This view is supported by surface-scanning mutational analysis that identified certain mutations which selectively block the binding of distinct substrates (Lee et al., 2007). Recent kinetic studies demonstrated that distal substrate residues influence arginine methylation, and the available evidence suggests a partially processive mechanism (Osborne et al., 2007). The three-dimensional structure of PRMT1 suggests that it is active as a homodimer (Zhang and Cheng, 2003). However, PRMT1 appears to be associated with a number of proteins in the cell (Table 1). Gel filtration analyses of the enzyme in mammalian cell extracts suggest sizes ranging from 275 kDa to 450 kDa; by contrast the PRMT1 dimer is expected to be 80 kDa (Gary and Clarke, 1998; Hung et al., 2007). This variation could represent the range of different oligomers that are present in the cell.
Two minor type I enzymes with more defined cellular localization – PRMT 3 and PRMT8
PRMT3 is located exclusively in the cytosol and has a zinc-finger domain that appears to anchor it to its substrates (Frankel and Clarke, 2000). A major methyl-accepting substrate in mammalian cells is the S2 protein of the small ribosomal subunit; much of PRMT3 is in fact associated with ribosomes via an interaction between its zinc-finger domain and the S2 protein (Swiercz et al., 2007).
PRMT8 is an unusual catalytic subunit for two reasons. Although its amino acid sequence is closely related to PRMT1, it has a relatively narrow tissue distribution, being limited mainly to brain (Lee et al., 2005a). It is also the only PRMT known to be membrane associated; PRMT8 is attached to the plasma membrane via N-terminal myristoylation (Lee et al., 2005a). The in vitro activity of the full-length recombinant enzyme is low; however, removal of the N-terminal domain by truncation or proteolysis results in a large activation (Sayegh et al., 2007). The N-terminal region contains two proline-rich sequences that can bind a number of SH3 domains, including that of PRMT2. It is possible that binding of proteins to this N-terminal domain can result in physiological PRMT8 activation and/or changes in its cellular location. Recently, a number of PRMT8-binding proteins were identified in cultured cells of probable neuronal origin (Pahlich et al., 2008).
Type I enzymes with restricted substrate specificities – CARM1/PRMT4 and PRMT6
CARM1/PRMT4 can be distinguished from PRMT1 because it catalyzes the methylation of only a few distinct substrates (Lee and Bedford, 2002). It binds the steroid receptor coactivators (SRC1-3) and has clear transcriptional coactivator activity itself, thus its name – the coactivator associated arginine methyltransferase 1 (CARM1) (Chen et al., 1999). As it was the fourth arginine methyltransferase described, it is also referred to as PRMT4. CARM1 loss is not compatible with life (Yadav et al., 2003). In cells, CARM1 forms a complex with ATP-remodeling (SWI/SNF) factors (Xu et al., 2004). CARM1 recruitment to transcriptional promoters feeds into the “histone code”, resulting in elevated levels of H3R17 (histone H3 Arg-17) and H3R26 methylation, which are associated with transcriptional activation. It also methylates other transcriptional coactivators and a subset of splicing factors (Cheng et al., 2007; Feng et al., 2006). The CARM1 crystal structure was solved by two groups, providing insight into its mechanism of action (Troffer-Charlier et al., 2007; Yue et al., 2007). A comparison of the apo and holo states of CARM1 reveals SAH-induced conformational changes of the cofactor pocket, which likely generate an access channel for the target arginine (Yue et al., 2007). Structural analysis reveals that the CARM1 N-terminus assumes a PH domain-like fold (Troffer-Charlier et al., 2007).
PRMT6 is a nuclear enzyme characterized by its specificity for distinct methyl-accepting substrates and by its automethylation (Frankel et al., 2002). PRMT6-specific substrates include the nuclear scaffold protein HMGA1a/b (Miranda et al., 2005; Sgarra et al., 2006), DNA polymerase beta (El-Andaloussi et al., 2007), the HIV Tat protein (Boulanger et al., 2005), and histone H3 (Guccione et al., 2007; Hyllus et al., 2007; Iberg et al., 2008). PRMT6 is the major PRMT responsible for histone H3R2 methylation and it has a clearly defined role in antagonizing the MLL-complex dependent methylation of the Lys-4 residue. Kinetic studies have shed light on how the enzyme catalyzes the dimethylation of arginine residues (Lakowski and Frankel, 2008). The use of a peptide substrate with a single arginine residue (WGGYSRGGYGGW) demonstrated that the monomethylated derivative is recognized with higher affinity and methylated more rapidly than the unmodified derivative. These results suggest a distributive, rather than processive mechanism. It remains unknown if this also occurs for protein substrates, or for substrates with multiple methylatable arginine residues.
A well-characterized type II enzyme – PRMT5
PRMT5 appears to be the major type II mammalian enzyme activity. It likely functions with multiple binding partners, but the identification of these partners has been greatly complicated by a technical problem caused by the αFLAG antibody recognizing PRMT5 (Nishioka and Reinberg, 2003). Nevertheless, PRMT5 seems to function in several types of complexes in both the cytoplasm and the nucleus. In the nucleus, binding to COPR5 (cooperator of PRMT5) can result in the preferential methylation of H4R3 over H3R8 (Lacroix et al., 2008). Moreover, COPR5 binding appears to be responsible for PRMT5 transcriptional corepressor activities. Like CARM1, PRMT5 can complex with hSWI/SNF ATP-dependent chromatin remodeling proteins, and in this context it functions as a transcriptional coactivator (Dacwag et al., 2007). It also associates with regulators of transcriptional elongation (Liu et al., 2007). In the cytoplasm, PRMT5 is involved in snRNP biogenesis through its ability to methylate a number of Sm proteins (Neuenkirchen et al., 2008). In these modifications, PRMT5 might function in conjunction with PRMT7 (Gonsalvez et al., 2007; see below).
Additional members of the seven-beta strand PRMT family with no or poorly characterized activities – PRMT2, PRMT7, and PRMT9(4q31)
In comparison to the well described type I and II PRMTs, no activity has been shown for PRMT2 or PRMT9(4q31); the evidence for PRMT7 activity remains controversial. It is possible that these species function only when complexed to additional polypeptide subunits; alternatively, the specific substrates for these enzymes have not been identified.
PRMT2 is of interest because it is a clear coactivator of gene expression much like PRMT1 and CARM1, and this activity appears to rely upon the integrity of the methyltransferase domain. PRMT2 is a coactivator of both the androgen receptor and the estrogen receptor alpha (Meyer et al., 2007). In addition, PRMT2 can promote apoptosis and inhibit NF-kappaB transcription by blocking IkappaB-alpha nuclear export (Ganesh et al., 2006). Interestingly, its SH3 domain binds the PRMT8 N-terminal domain (Sayegh et al., 2007).
PRMT7 has been described as having type III activity (Figure 1) towards arginine-containing peptides (Miranda et al., 2004) or type II activity towards peptide and protein substrates (Lee et al., 2005b). Neither of these activities is particularly robust. Indirect evidence correlates PRMT7 activity with either resistance or sensitivity to DNA damaging agents (Gros et al., 2006; Verbiest et al., 2008), and sensitivity of the kidney to damage caused by certain antibiotics (Zheng et al., 2005). The physical interaction of the CTCFL protein with PRMT7 also suggests a role in imprinting control region (ICR) DNA methylation (Jelinic et al., 2006). Both PRMT5 and PRMT7 are involved in the methylation of the C-terminal tails of Sm proteins, and both are required for normal snRNP biogenesis in human cells (Gonsalvez et al., 2007). This result opens the possibility that a PRMT5/PRMT7 heterodimer could be the active core of the enzyme complex involved in this Sm protein methylation reaction. PRMT7 might also play a role in embryonic stem cell (ESC) pluripotency, as its expression is lost when ESCs are induced to differentiate, much like the pluripotency marker OCT4 (Buhr et al., 2008). More work is needed to establish PRMT7 interacting partners and to characterize its enzymatic activities.
PRMT9(4q31) was identified based on homology with other family members as a product of a gene on human chromosome 4q31 (Lee et al., 2005a). We refer to this protein with the human chromosomal location because the PRMT9 designation has also been used for the product of the FBXO11 gene in humans on chromosome 2p16 (Cook et al., 2006) (see below). PRMT9(4q31) has not been biochemically characterized to date. Interestingly, it contains two tetratricopeptide repeats (TPR), motifs which often mediate protein-protein interactions (Bedford, 2007).
Are F-Box only family members protein arginine methyltransferases?
Two F-box only family members have been suggested to be protein arginine methyltransferases (Cook et al., 2006; Krause et al., 2007). Notably, neither appear to belong to the family of seven-beta strand methyltransferases that includes PRMT1-9 (Katz et al., 2003). The first, FBXO11/PRMT9(2p16) was identified from a “forced” alignment of its amino acid sequence to the PRMT family (Cook et al., 2006). The identification of type II methylation activity (Cook et al., 2006) is questionable as the HeLa-cell expressed FLAG-tagged fusion protein preparation was likely to have been contaminated with PRMT5 (Nishioka and Reinberg, 2003). In an independent study, no activity was reported for the human or worm FBXO11 proteins (Fielenbach et al., 2007). A second F-box only protein FBXO10 has been tentatively identified as PRMT11, but no biochemical characterization has been done (Krause et al., 2007). Further work is needed to conclusively determine whether F-box only proteins can be protein arginine methyltransferases.
Biological Roles of Arginine Methylation
Protein substrates for PRMTs make up an increasingly large fraction of the proteome. As new substrates are characterized, a broadening spectrum of PRMT-regulated cellular processes has been realized. Importantly, PRMT knockout mice provided a key resource for the in vivo validation of many of these in vitro findings.
Transcriptional Coactivators
PRMTs can methylate and regulate transcription factors, other coactivators, and histones. Their ability to methylate histones provides a direct route for input into the epigenetic regulation of gene expression. The arginine methylation of coactivators (e.g. p300, CBP & SRC3) provides an indirect mechanism to impact a gene’s epigenetic state by regulating the competence of these histone acetyltransferases. The first evidence implicating PRMTs in transcriptional regulation came with the identification of CARM1 as a nuclear receptor coactivator (Chen et al., 1999). In transient transfection assays, CARM1 and PRMT1 synergistically enhanced steroid hormone receptor-mediated reporter gene activation (Bedford and Richard, 2005). Strong synergy between these two PRMTs is further supported by siRNA-mediated single and double knockdowns and subsequent transcriptome analysis, which unmasked a group of STAT5 controlled genes whose expression is down-regulated only when both CARM1 and PRMT1 are removed (Kleinschmidt et al., 2008). Although PRMTs were initially identified as estrogen and androgen receptor coactivators, they are now emerging as coregulators of a large number of transcription factors including p53, YY1, NF-κB, PPARγ, RUNX1, E2F1 (Yadav et al., 2008; Zhao et al., 2008). Thus, PRMT1 and CARM1 function as general transcriptional coactivators, much like p300/CBP. Because they coactivate a large number of transcription factors, one would expect there to be a unifying mechanism that would explain this phenomenon – like histone methylation, which in turn recruits methyl-binding effector proteins that facilitate transcription. To date, no such methylarginine binding effectors have yet been identified. However, transcription factor specific mechanisms have been identified. RUNX1, for example, is methylated by PRMT1, and this methylation triggers the dissociation of the transcriptional repressor SIN3A, thereby promoting RUNX1 transcriptional activity (Zhao et al., 2008).
Transcriptional Corepressors
The synergy between PRMT5 and E2F1 in cyclin E1 corepression provided the first demonstration for PRMT repressor activity (Fabbrizio et al., 2002). PRMT5 has since been identified as a general transcriptional repressor that functions with numerous transcription factors and repressor complexes, including BRG1 and hBRM (Pal et al., 2003), Blimp1 (Ancelin et al., 2006), and Snail (Hou et al., 2008). The mechanism by which PRMT5 mediates transcriptional repression remains to be elucidated. However, this enzyme, once recruited to a promoter, symmetrically dimethylates H3R8 and H4R3 (Pal et al., 2004). Importantly, the H4R3 site is a major target for PRMT1 methylation (ADMA) and is generally regarded as a transcriptional activating mark. Thus, both H4R3me2s (repressive; me2s indicates SDMA modification) and H4R3me2a (active; me2a indicates ADMA modification) marks are produced in vivo. No effector has yet been identified that will bind to these isolated histone methyl-marks. However, the RAG2-PHD finger binds H3R2me2sK4me3 slightly better than either H3R2me2aK4me3 or H3K4me3 (Ramon-Maiques et al., 2007), suggesting that arginine methylation sites on the histone tails can be recognized in combination with other marks. The specificity of PRMT5 for H3R8 and H4R3 can be altered by its interaction with COPR5 (Lacroix et al., 2008), and this could perhaps play an important role in determining PRMT5 corepressor status. Indeed, the general assumption that PRMT5 functions as a transcriptional repressor is not cut-and-dry, as there are at least two examples where it functions as a coactivator. First, reduced PRMT5 levels (by RNAi) results in decreased IL-2 expression (Richard et al., 2005), and second, PRMT5 is required for MyoD-induced muscle differentiation, in particular for myogenin transcription (Dacwag et al., 2007).
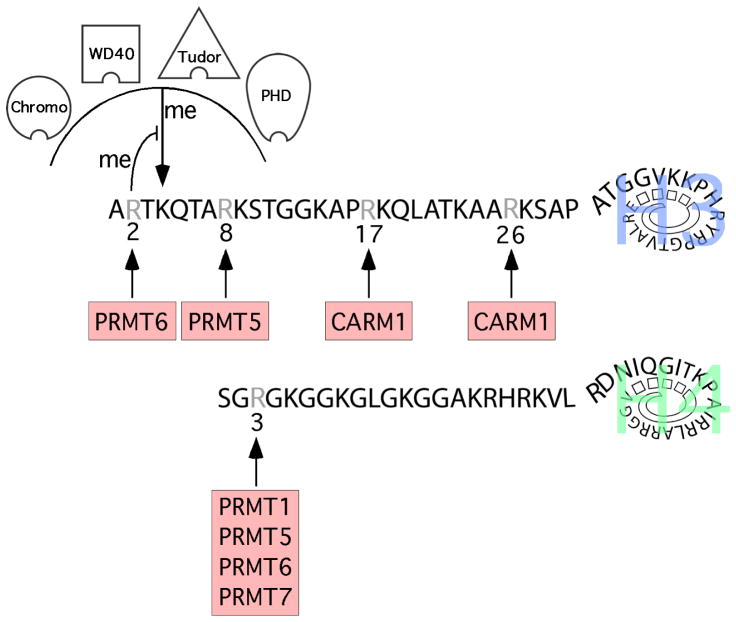
PRMT6 also feeds into the epigenetic code and possesses transcriptional repressor activities. It is the primary enzyme responsible for H3R2 methylation in mammalian cells, (Guccione et al., 2007; Hyllus et al., 2007; Iberg et al., 2008). PRMT6 can strongly methylate H3K4me1 and H3K4me2 peptides, and weakly methylate a H3K4me3 peptide (Iberg et al., 2008). However, prior methylation of the H3R2 site essentially prevents the MLL1 complex from methylating H3K4 (Hyllus et al., 2007). H3K4 methylation is found at the 5′ end of active genes and is responsible for recruiting chromatin-remodeling enzymes to establish and maintain a transcriptionally active state. The effectors that bind H3K4me3 harbor methyl-specific binding domains, including PHD, Chromo and Tudor domains. H3R2 methylation blocks the ability of most of these domains to bind the histone H3 N-terminal tail in in vitro assays (Iberg et al., 2008). In cells, WDR5 chromatin binding is sensitive to PRMT6 expression levels (Guccione et al., 2007; Hyllus et al., 2007; Iberg et al., 2008). Thus, PRMT6 functions as a transcriptional repressor by blocking the recruitment of transcriptional activators to the methylated H3K4 mark (Figure 3). PRMT6 also methylates the N-terminal tails of histones H4 and H2A (their first 5 residues are the same) at R3 (Hyllus et al., 2007); however the role of methylation at these sites is unknown.
Figure 3. Arginine methylation of histone H3 and H4 N-terminal tails.
The sites of PRMT-mediated methylation on the histone tails are noted. H3R2 methylation antagonizes the docking of a number of effector proteins, including those containing chromo, PHD, Tudor, and WD40 domains. H3R2 methylation structurally impedes the binding of H3K4me3 recognition domains and also prevents H3K4 methylation by the MLL1 complex. The first 5 residues of histones H2A and H4 are identical, and H2A is also methylated at the R3 position.
PRMT7, which has been reported to deposit the H4R3me2s mark, is a key player in DNA methylation of the imprint control region (ICR) (Jelinic et al., 2006). CTCFL binds ICR through its zinc finger region. The CTCFL N-terminal end interacts with the PRMT7 C-terminal end, and the CTCFL/PRMT7 complex is proposed to methylate H4R3, thereby facilitating the recruitment of the DNA methyltransferases Dnmt3a and Dnmt3b. This finding provides the first link between genomic imprinting and arginine methylation.
mRNA Splicing
The vast majority of PRMT substrates are associated with RNA (Pahlich et al., 2006). Thus, arginine methylation has been implicated in all aspects of RNA metabolism, including mRNA transcription, splicing, transport, translation, and turnover. Direct evidence for a role of arginine methylation in splicing first came from studies showing that in vitro splicing reactions could be blocked using pan SDMA-specific antibodies (Boisvert et al., 2002). In addition, splicing efficiency is reduced in hypomethylated nuclear extracts. CARM1, which deposits an ADMA mark, has also been clearly identified as a regulator of alternative splicing (Cheng et al., 2007; Ohkura et al., 2005). CARM1 methylates the splicing factors CA150, SAP49, SmB and U1C. Of note, CA150 methylation generates a docking motif for the SMN Tudor domain. The use of splicing reporter assays demonstrates that CARM1 promotes exon skipping in an enzyme-dependent manner.
Nuclear/Cytoplasmic Shuttling
The first identified biological role for arginine methylation involved nucleocytoplasmic shuttling in yeast (Shen et al., 1998). RNA binding proteins (RBPs), like snRNPs and hnRNPs, account for over 60% of the ADMA found in the nucleus (Bedford and Richard, 2005). These RBPs contain GAR motifs and PGM (proline-glycine-methionine) motifs, which are the prime sites of PRMT1 and CARM1 methylation, respectively (Cheng et al., 2007; Najbauer et al., 1993). Importantly, many of these RBPs shuttle between the nucleus and the cytoplasm, and two members (Npl3 and Hrp1) of this protein family are trapped in the nucleus in cells lacking Hmt1/Rmt1, the primary Type 1 PRMT in yeast (Shen et al., 1998). In mammalian cells, the inhibition of methylation of the high molecular weight forms of FGF2 results in a decrease in their nuclear accumulation (Pintucci et al., 1996), and this is also the case for Sam68 (Bedford and Richard, 2005). The Xenopus laevis RNA binding protein CIRP2 is methylated by XlPRMT1, and XlPRMT1 overexpression forces hypermethylated CIRP2 out of the nucleus (or blocks its return), resulting in its cytoplasmic accumulation (Aoki et al., 2002). Finally, although the methylated form of the adenovirus 100K protein is exclusively nuclear, methylation inhibitors prevent 100K nuclear accumulation (Iacovides et al., 2007). Interestingly, some hypomethylated proteins accumulate in the nucleus and others in the cytoplasm. The mechanism(s) by which arginine methylation regulates nucleocytoplasmic shuttling has not been elucidated.
DNA Repair
In higher eukaryotes, DNA double strand breaks (DSBs), induced by ionizing radiation or during replication, are repaired through homologous recombination repair (HRR) or non-homologous end joining (NHEJ). The mammalian Mre11/Rad50/NBS1 (MRN) complex plays a critical role in HRR. Using methyl-specific antibodies, the Richard laboratory isolated methylated protein complexes from HeLa cell extracts and, in so doing, identified the components of the MRN complex (Boisvert et al., 2003). Mre11 is the only protein in this complex that harbors a GAR motif and follow up studies indicated that this region of Mre11 was indeed methylated by PRMT1 in vitro and in vivo (Boisvert et al., 2005a). Further investigation showed that the GAR motif regulates the 3′->5′ exonuclease activity of Mre11 on dsDNA. In cells treated with a global methyltransferase inhibitor followed by the DNA damaging agent etoposide, few γH2AX foci formed and Mre11 was not recruited to DSBs, implying that methylation regulates MRN relocalization to DSBs (Boisvert et al., 2005b). Indeed, the methylated MRN GAR motif, on its own, can colocalize with γH2AX foci (Dery et al., 2008). Like Mre11, 53BP1 is also involved in DSB repair. 53BP1 contains a GAR motif that is methylated by PRMT1 in vivo; however methylation does not increase after DNA damage and a 53BP1 GAR motif mutant showed normal foci formation (Adams et al., 2005; Boisvert et al., 2005c).
DNA polymerase β (Pol-β) is implicated in base excision repair. Pol-β harbors two functional domains – a N-terminal lyase domain and a C-terminal domain that contains the nucleotidyltransferase activity of the polymerase. Recent findings indicate that the Pol-β lyase domain can associate with PRMT6 and PRMT1 (El-Andaloussi et al., 2007; El-Andaloussi et al., 2006). These two PRMTs play distinct, non-redundant, roles in regulating Pol-β function. PRMT6 methylates Pol-β R83 and R152, and augments its polymerase activity by enhancing its DNA binding and processivity (El-Andaloussi et al., 2006). PRMT1 methylates R137, which in turn blocks the Pol-β–PCNA interaction (El-Andaloussi et al., 2007). As yet, there is no evidence that the levels of arginine methylation on histone tails are altered, either in the vicinity of DNA damage or as part of a more global DNA damage response.
Signal transduction
Arginine methylation plays very clear roles in the nucleus, where it impacts transcription and splicing. However, the functions that arginine methylation plays in conveying information from the cell surface, through the cytoplasm and into the nucleus have remained elusive, particularly because it appears to be a stable mark. There are, nevertheless, a number of findings that point towards a role for PRMTs in signal transduction. PRMT8, owing to its myristoylation and plasma membrane association, could impact a signaling pathway near its start (Lee et al., 2005a). In addition, PRMT8 harbors a proline-rich domain that can interact with SH3 domains, suggesting that it could serve as a signaling hub (Sayegh et al., 2007). PRMT1 binds the cytoplasmic region of the type I interferon receptor and has also been implicated in NGF receptor signaling (reviewed in Bedford and Richard, 2005). PRMT activities are also regulated by the T-cell antigen receptor signaling pathway (Blanchet et al., 2005). Recent findings indicate that insulin treatment of cultured myotubes promotes PRMT1 accumulation in the membrane fraction (Iwasaki and Yada, 2007), thus providing the first evidence that arginine methylation might be involved in insulin signaling and glucose metabolism. Recently it was reported that PRMT1 plays a role in nongenomic estrogen signaling (Le Romancer et al., 2008). In this process the estrogen receptor (ER) forms a multiprotein cytoplasm complex that activates a number of downstream signaling molecules, including PKC, ras, Akt and MAPK. Estrogen-ER triggers the rapid, reversible, receptor methylation by PRMT1, which in turn facilitates ER binding to Src, FAK and p85. Thus, PRMT1 recruitment by ER occurs in both the cytoplasm and the nucleus, generating two distinct complexes; a signal transducing component and an epigenetic-modulating component.
The Regulation of Arginine Methylation
Most PRMTs are active as recombinant proteins, suggesting that these enzymes might also be constitutively active in cells unless regulated by post-translational modifications or through interactions with other proteins and protein complexes. In addition, PRMT activity can be restricted by subcellular compartmentalization. Finally, enzymes that counteract PRMT activity can reverse or block substrate methylation. Indeed, all these methods of PRMT regulation have been reported; however their relative importance remains to be established.
First, PRMT-binding proteins can regulate methyltransferase activity. These binding proteins have the capacity to inhibit, activate, or change the substrate specificity of PRMTs. The related proteins, BTG1 and TIS2/BTG2, bind PRMT1 and stimulate its activity towards selected substrates (Lin et al., 1996). hCAF1, a BTG1 binding protein, also regulates PRMT1 activity (Robin-Lespinasse et al., 2007). Binding of the tumor suppressor DAL-1 inhibits PRMT3 enzymatic activity, both in in vitro reactions and in cell lines (Singh et al., 2004). CARM1 is found in the nucleosomal methylation activator complex (NUMAC), a complex of at least 10 proteins (Xu et al., 2004). Within NUMAC, CARM1 acquires the ability to methylate nucleosomal histone H3, whereas recombinant CARM1 preferentially methylates free histone H3. Likewise, when associated with COPR5, PRMT5 changes its balance of activity from H3R8 towards H4R3 (Lacroix et al., 2008). PRMT5 also complexes with the hSWI/SNF chromatin remodelers, BRG and BRM, and this association enhances PRMT5 methyltransferase activity (Pal et al., 2004). CTCFL (a protein that associates with the imprinting control region) binding has also been reported to elevate PRMT7 activity (Jelinic et al., 2006).
Second, PRMT activity will likely be regulated by posttranslational modifications. Some evidence shows that PRMT1, 4, 6 and 8 are automethylated; however the functional consequence of this methylation remains unknown (Sayegh et al., 2007). In addition, CARM1 can be phosphorylated during mitosis by an as yet unidentified kinase (Higashimoto et al., 2007). This phosphorylation event prevents CARM1 homodimerization and decreases its PRMT activity.
Third, adjacent posttranslational modifications can mask arginine methylation motifs. Histone N-terminal tails are rich in various posttranslational modifications and provide a number of examples of this phenomenon. There is an interplay between H3K9 lysine acetylation and PRMT5-mediated H3R8 arginine methylation: H3K9ac blocks H3R8me (Pal et al., 2004). The existence of a H3R8me2a or me2s mark will in-turn block G9a-mediated H3K9 methylation (Rathert et al., 2008). In addition, PRMT6-mediated H3R2 methylation is inhibited, but not totally blocked, by H3K4me3 (Guccione et al., 2007; Iberg et al., 2008). Interestingly, H3K4me1 and H3K4me2 peptides are just as good a substrate for PRMT6 as the unmethylated peptide (Iberg et al., 2008). H3K4 methylation is also blocked by prior H3R2 methylation (Hyllus et al., 2007). It is very likely that similar regulatory scenarios play out on non-histone PRMT substrates, where diverse signaling pathways may be impacted.
Fourth, arginine residues, within the context of a protein, can be converted to citrulline by deimination. The core histones H2A, H3 and H4 comprise a major group of deiminated proteins (Nakashima et al., 2002). Peptidyl arginine deiminases (PADs) catalyze the deimination of arginine, but not MMA or DMA, to citrulline (Raijmakers et al., 2007). Thus, PADs are not demethylases. However, these enzymes might carryout a preemptive strike on key sites of arginine methylation, thereby preventing subsequent methylation. Indeed, PADs can block methylation on an arginine residue by converting it to citrulline (Thompson and Fast, 2006).
Lastly, the first arginine demethylase was recently reported (Chang et al., 2007). This enzyme, JMJD6, a Jumonji domain-containing protein, demethylates H3R2me2 and H4R3me2. Of note, it appears that JMJD6 can demethylate both the symmetric and asymmetric forms of H4R3me2 (Chang et al., 2007). Further work is required in this field, especially the characterization of additional proteins in the jumonji-only family of enzymes, of which JMJD6 is a member. The possibility also exists that other types of arginine demethylases have yet to be discovered.
Methylarginine-regulated protein-protein interactions
Arginine methylation facilitates the interaction of GAR and PGM motifs with Tudor domains. The symmetric dimethylation of SmB by PRMT5 is required for an interaction with the Tudor domains of SMN, SPF30 and TDRD3 (Cote and Richard, 2005). The asymmetric dimethylation of CA150 by CARM1 also provides a docking site for the SMN Tudor domain (Cheng et al., 2007). Thus, motifs harboring either ADMA or SDMA residues bind a subset of Tudor domain-containing proteins. It is likely that a conserved aromatic “cage” in Tudor domains is the methylarginine-binding pocket (Sprangers et al., 2003), as is the case for other methyl-binding domains (Taverna et al., 2007). Tudor domain containing proteins such as JMJD2a, 53BP1 and PHF20 also bind methyllysine motifs (Kim et al., 2006); thus there are two distinct classes of methyl-binding Tudor domains. However, the propensity of Tudor domains to bind either methylarginine or methyllysine motifs cannot be predicted from their primary sequence, and thus their binding selectivity must be determined empirically.
Arginine methylation can also negatively regulate protein-protein interactions. The methylation of arginine residues adjacent to a proline-rich motif can block the binding to SH3, but not WW, domain protein modules (Bedford et al., 2000). A second example of arginine methylation blocking a protein-protein interaction is the CARM1-mediated modification of the p300 GRIP1 binding domain (Lee et al., 2005c). Finally, recent structural studies have revealed that histone H3R2 forms critical interactions with an array of H3 N-terminal tail binding domains (Taverna et al., 2007). PRMT6-mediated H3R2 methylation completely blocks the interaction of WDR5 with histone H3 (Couture et al., 2006). Moreover, this modification reduces the binding affinity of the CHD1 double chromodomains, fourfold relative to H3K4me3 alone (Flanagan et al., 2005). Further work has demonstrated that, as a rule, H3R2 methylation blocks the binding of most H3 N-terminal tail binding domains (chromo, PHD, WD40 & Tudor) (Iberg et al., 2008) (Figure 3). Future studies are needed to determine if there are any protein domains that can preferentially interact with the H3R2me mark.
Arginine Methylation and Disease
PRMTs are ubiquitously expressed (except for PRMT8), they are found in both the nucleus and the cytoplasm and they methylate a host of cellular proteins. Thus, deregulation of these enzymes or their substrates will likely be implicated in the pathogenesis of a number of different diseases. Clearly, PRMTs are not only epigenetic regulators, as free methylarginine regulates nitric oxide synthase, viral proteins are heavily methylated and genetics implicates a prominent “reader” of methylarginine marks with spinal muscular atrophy. In this section, we begin to scratch the surface of what will likely develop into a very active field of research.
Cancer
Initial studies linking the PRMTs to cancer were circumstantial, but we are now gaining a better mechanistic understanding of how deregulated PRMT function might transform cells. Both PRMT1 and CARM1 are general transcriptional coactivators, and as such, their aberrant expression or altered regulation likely affects many transcriptional pathways. PRMT1 mRNA levels have been reported to be higher in a panel of breast cancer cell lines than in normal controls (Goulet et al., 2007). The first concrete evidence that PRMT1 plays a role in oncogenesis came from studies demonstrating that mouse primary haematopoietic cells that are transduced with the MLL-EEN gene fusion product display enhanced self-renewal abilities and can form compact CFU-GEMM-like colonies in vitro (Cheung et al., 2007). The portion of EEN that is fused to MLL harbors a SH3 domain that can bind the proline-rich molecule Sam68. Sam68 is a well-characterized PRMT1 substrate. When either Sam68 or PRMT1 are fused directly to MLL, in place of EEN, these fusions also can activate MLL oncogenic properties, implying that methylated Sam68 is a crucial player here (Cheung et al., 2007). Interestingly, CARM1 does not possess this ability.
CARM1 levels are elevated in castration-resistant prostate cancer (Hong et al., 2004; Majumder et al., 2006), as well as in aggressive breast tumors that also express high-levels of the oncogenic coactivator AIB1 (El Messaoudi et al., 2006). Importantly, CARM1 methylates AIB1, thereby regulating its activity and stability (Feng et al., 2006; Naeem et al., 2007). CARM1 recruitment to ERα regulated promoters relies on the presence of AIB1, and CARM1 is essential for estrogen-induced proliferation of the MCF-7 breast cancer cell line (Frietze et al., 2008).
The normal developmental process of epithelial-mesenchymal transition (EMT) plays an important role in tumor progression. As a consequence of EMT, cancer cells can migrate and invade much more efficiently, and this process likely plays an active role in the metastatic spread of a tumor. A hallmark of EMT is the loss of E-cadherin expression; E-cadherin expression is actively repressed by the transcription factor SNAIL. A recent report demonstrated that PRMT5 interacts with SNAIL, through a bridging molecule (AJUBA) (Hou et al., 2008). Importantly, siRNA-mediated PRMT5 knockdown results in elevated E-cadherin expression. These data indicate that PRMT5 is a critical SNAIL corepressor. In addition, PRMT5 over-expression promotes anchorage-independent cell growth (Pal et al., 2004), supporting the notion that PRMT5 might be an oncoprotein. Consistent with this idea, PRMT5 levels are elevated in gastric cancer (Kim et al., 2005), and in lymphoma and leukemia cells (Pal et al., 2007).
Cardiovascular disease
Nitric oxide (NO), which is synthesized from L-arginine by nitric oxide synthase (NOS), plays multiple roles in the cardiovascular system, the immune system and as a neurotransmitter in the brain. ADMA and MMA, but not SDMA, are endogenously produced amino acids that inhibit NOS (reviewed in Bedford and Richard, 2005). These free methylarginine species are generated by the proteolysis of methylated proteins. Thus, PRMTs are responsible for generating NOS inhibitors in vivo. To maintain a steady pool size, free methylarginines are metabolized by dimethylarginine dimethylaminohydrolase (DDAH) enzymes. An imbalance in this pool, by PRMT or DDAH dysfunction, might increase cardiovascular risk. Recent studies of Ddah1 knockout mice provide genetic support for this hypothesis (Leiper et al., 2007). These knockout mice accumulate ADMA and display reduced NO signaling, which manifests itself in vascular pathophysiology, including elevated blood pressure.
Viral Pathogenesis
A number of different viral proteins have been characterized as PRMT substrates, including the herpes simplex virus 1 nuclear regulatory protein ICP27, the hepatitis C virus protein NS3, the Epstein-Barr virus nuclear antigen 2 (EBNA2), adenovirus E1B-AP5 and L4-100K, and the HIV-1 proteins Rev, Tat and the nucleocapsid protein (reviewed in Bedford and Richard, 2005). Each of these methylated viral proteins contains GAR-motifs that are targeted for methylation by either PRMT1 or PRMT6. Studies using global methyltransferase inhibitors have revealed that methylation contributes to maximal virus infectivity in an HIV-1 infection cell culture model (Willemsen et al., 2006) and is critical for productive adenovirus infection (Iacovides et al., 2007). Thus, the methylation of viral proteins may provide a means by which they can enhance their complexity and expand their repertoire of interacting partners, and in the process possibly disrupt normal cellular functions.
Spinal Muscular Atrophy
Spinal muscular atrophy (SMA) is an autosomal recessive disease resulting from the loss of SMN1 gene function. SMA is among the leading genetic causes of infant death, with a prevalence of ~1 in 6000 live births. SMN acts as a molecular chaperone for arginine-methylated proteins that participate in RNA metabolism. The SMN protein contains a Tudor domain that is a well-characterized methylarginine-dependent binding module. This Tudor domain can bind both ADMA and SDMA motifs, and interacts with a number of PRMT5 and CARM1 substrates (Cheng et al., 2007; Tadesse et al., 2008). Point mutations within the SMN Tudor domain have been identified in SMA patients, establishing a clear link between the methyl-binding requirements of SMN and SMA pathogenesis (Cusco et al., 2004).
Challenges in analyzing PRMTs
It is remarkable how much we have learned about protein arginine methyltransferases from studying the properties of recombinant PRMTs. However, these species, generally expressed as tagged fusion proteins, can have different catalytic and substrate recognition properties than the endogenous protein and can lack the necessary accessory subunits. The fact that PRMTs often form cellular complexes with other polypeptides that can affect their activities (Table 1) also emphasizes the need for caution in interpreting results from studies with recombinant fusion proteins. This situation could be further complicated because some protein-protein interactions might be permanent and some might be transitory. Purifications of PRMTs from cells and tissues should result in the retention of at least the more tightly bound partners (Hung et al., 2007). However, little progress has been made in characterizing what are probably multiple endogenous complexes of each PRMT.
Several approaches can be used to distinguish the MMA, ADMA, and SDMA PRMT products (Pahlich et al., 2006). Chromatographic methods can detect very small amounts (sub-femtomole) of radiolabeled derivatives. The use of high-capacity, high resolution, cation exchange chromatography columns based on sulfonate cross-linked polystyrene resins allows the near-baseline resolution of the three major mammalian arginine derivatives, MMA, ADMA, and SDMA (Branscombe et al., 2001). For additional confirmation, it is possible to do thin-layer chromatography on each of the radiolabelled peak fractions (Branscombe et al., 2001; Lakowski and Frankel, 2008). However, it is more difficult to use thin-layer chromatography as an initial separation step of crude hydrolysates because the resolution is much more limited than in ion-exchange chromatography and the presence of other radiolabeled materials can be mistaken for methylated arginine residues. Recent advances in mass spectrometry instrumentation and techniques have opened a new window in the non-isotopic analysis of enzyme products (Hsieh and Tam, 2006; Pesavento et al., 2008). In many cases, it is now possible to detect monomethyl and dimethylarginine derivatives, and it is possible to distinguish symmetric from asymmetric dimethylation by breakdown products (Brame et al., 2004; Gehrig et al., 2004).
Emerging Themes
A number of areas in the PRMT field will likely receive attention in the coming years. These include the structural determination of the PRMTs in complexes with their interacting partners, substrates, and small molecule inhibitors. Also of interest will be studies that address the integration of signal transduction and arginine methylation pathways. Specifically, how is PRMT activity regulated by posttranslational modifications such as phosphorylation, acetylation, ubiquitination, and trans- or automethylation? We also clearly need to gain a better understanding of the enzyme(s) that demethylate arginine residues. Finally, only three mammalian proteins (SMN, SPF30 and TDRD3) have been demonstrated to bind methylarginine motifs through their Tudor domains. The quest for additional methylarginine-binding modules and proteins will likely be a high priority for a number of research groups over the coming years.
Acknowledgments
We apologize to those researchers whose original work could not be cited due to space constraints. MTB is supported by NIH grant - DK62248. SGC is supported by NIH grant -GM026020. We would like to thank the following people for reading over this review: Stéphane Richard, Laurie Read, Adam Frankel and members of the Clarke and Bedford Laboratories.
References
- Adams MM, Wang B, Xia Z, Morales JC, Lu X, Donehower LA, Bochar DA, Elledge SJ, Carpenter PB. 53BP1 oligomerization is independent of its methylation by PRMT1. Cell Cycle. 2005;4:1854–1861. doi: 10.4161/cc.4.12.2282. [DOI] [PubMed] [Google Scholar]
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- Aoki K, Ishii Y, Matsumoto K, Tsujimoto M. Methylation of Xenopus CIRP2 regulates its arginine- and glycine-rich region-mediated nucleocytoplasmic distribution. Nucleic Acids Res. 2002;30:5182–5192. doi: 10.1093/nar/gkf638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT. Arginine methylation at a glance. J Cell Sci. 2007;120:4243–4246. doi: 10.1242/jcs.019885. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Frankel A, Yaffe MB, Clarke S, Leder P, Richard S. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J Biol Chem. 2000;275:16030–16036. doi: 10.1074/jbc.M909368199. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Blanchet F, Cardona A, Letimier FA, Hershfield MS, Acuto O. CD28 costimulatory signal induces protein arginine methylation in T cells. J Exp Med. 2005;202:371–377. doi: 10.1084/jem.20050176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, Cote J, Boulanger MC, Cleroux P, Bachand F, Autexier C, Richard S. Symmetrical dimethylarginine methylation is required for the localization of SMN in Cajal bodies and pre-mRNA splicing. J Cell Biol. 2002;159:957–969. doi: 10.1083/jcb.200207028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, Cote J, Boulanger MC, Richard S. A Proteomic Analysis of Arginine-methylated Protein Complexes. Mol Cell Proteomics. 2003;2:1319–1330. doi: 10.1074/mcp.M300088-MCP200. [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Dery U, Masson JY, Richard S. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 2005a;19:671–676. doi: 10.1101/gad.1279805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, Hendzel MJ, Masson JY, Richard S. Methylation of MRE11 regulates its nuclear compartmentalization. Cell Cycle. 2005b;4:981–989. doi: 10.4161/cc.4.7.1830. [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Rhie A, Richard S, Doherty AJ. The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle. 2005c;4:1834–1841. doi: 10.4161/cc.4.12.2250. [DOI] [PubMed] [Google Scholar]
- Boulanger MC, Liang C, Russell RS, Lin R, Bedford MT, Wainberg MA, Richard S. Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J Virol. 2005;79:124–131. doi: 10.1128/JVI.79.1.124-131.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brame CJ, Moran MF, McBroom-Cerajewski LD. A mass spectrometry based method for distinguishing between symmetrically and asymmetrically dimethylated arginine residues. Rapid Commun Mass Spectrom. 2004;18:877–881. doi: 10.1002/rcm.1421. [DOI] [PubMed] [Google Scholar]
- Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, Clarke S. Prmt5 (janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem. 2001;276:32971–32976. doi: 10.1074/jbc.M105412200. [DOI] [PubMed] [Google Scholar]
- Buhr N, Carapito C, Schaeffer C, Kieffer E, Van Dorsselaer A, Viville S. Nuclear proteome analysis of undifferentiated mouse embryonic stem and germ cells. Electrophoresis. 2008;29:2381–2390. doi: 10.1002/elps.200700738. [DOI] [PubMed] [Google Scholar]
- Calnan BJ, Tidor B, Biancalana S, Hudson D, Frankel AD. Arginine-mediated RNA recognition: the arginine fork. Science. 1991;252:1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- Cheng D, Cote J, Shaaban S, Bedford MT. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell. 2007;25:71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Cheng X, Collins RE, Zhang X. Structural and sequence motifs of protein (histone) methylation enzymes. Annu Rev Biophys Biomol Struct. 2005;34:267–294. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung N, Chan LC, Thompson A, Cleary ML, So CW. Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol. 2007;9:1208–1215. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- Clarke S, Tamanoi F, editors. The Enzymes. 3. XXIV. Academic Press; 2006. Protein Methyltransferases. [Google Scholar]
- Cook JR, Lee JH, Yang ZH, Krause CD, Herth N, Hoffmann R, Pestka S. FBXO11/PRMT9, a new protein arginine methyltransferase, symmetrically dimethylates arginine residues. Biochem Biophys Res Commun. 2006;342:472–481. doi: 10.1016/j.bbrc.2006.01.167. [DOI] [PubMed] [Google Scholar]
- Cote J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J Biol Chem. 2005;280:28476–28483. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Trievel RC. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat Struct Mol Biol. 2006;13:698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- Cusco I, Barcelo MJ, del Rio E, Baiget M, Tizzano EF. Detection of novel mutations in the SMN Tudor domain in type I SMA patients. Neurology. 2004;63:146–149. doi: 10.1212/01.wnl.0000132634.48815.13. [DOI] [PubMed] [Google Scholar]
- Dacwag CS, Ohkawa Y, Pal S, Sif S, Imbalzano AN. The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol Cell Biol. 2007;27:384–394. doi: 10.1128/MCB.01528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dery U, Coulombe Y, Rodrigue A, Stasiak A, Richard S, Masson JY. A glycine-arginine domain in control of the human MRE11 DNA repair protein. Mol Cell Biol. 2008 doi: 10.1128/MCB.02025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Messaoudi S, Fabbrizio E, Rodriguez C, Chuchana P, Fauquier L, Cheng D, Theillet C, Vandel L, Bedford MT, Sardet C. Coactivator-associated arginine methyltransferase 1 (CARM1) is a positive regulator of the Cyclin E1 gene. Proc Natl Acad Sci U S A. 2006;103:13351–13356. doi: 10.1073/pnas.0605692103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Andaloussi N, Valovka T, Toueille M, Hassa PO, Gehrig P, Covic M, Hubscher U, Hottiger MO. Methylation of DNA polymerase beta by protein arginine methyltransferase 1 regulates its binding to proliferating cell nuclear antigen. FASEB J. 2007;21:26–34. doi: 10.1096/fj.06-6194com. [DOI] [PubMed] [Google Scholar]
- El-Andaloussi N, Valovka T, Toueille M, Steinacher R, Focke F, Gehrig P, Covic M, Hassa PO, Schar P, Hubscher U, Hottiger MO. Arginine methylation regulates DNA polymerase Beta. Mol Cell. 2006;22:51–62. doi: 10.1016/j.molcel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Fabbrizio E, El Messaoudi S, Polanowska J, Paul C, Cook JR, Lee JH, Negre V, Rousset M, Pestka S, Le Cam A, Sardet C. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 2002;3:641–645. doi: 10.1093/embo-reports/kvf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Yi P, Wong J, O’Malley BW. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol. 2006;26:7846–7857. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N, Guardavaccaro D, Neubert K, Chan T, Li D, Feng Q, Hutter H, Pagano M, Antebi A. DRE-1: an evolutionarily conserved F box protein that regulates C. elegans developmental age. Dev Cell. 2007;12:443–455. doi: 10.1016/j.devcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- Frankel A, Clarke S. PRMT3 is a distinct member of the protein arginine N-methyltransferase family. Conferral of substrate specificity by a zinc-finger domain. J Biol Chem. 2000;275:32974–32982. doi: 10.1074/jbc.M006445200. [DOI] [PubMed] [Google Scholar]
- Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S, Bedford MT. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem. 2002;277:3537–3543. doi: 10.1074/jbc.M108786200. [DOI] [PubMed] [Google Scholar]
- Frietze S, Lupien M, Silver PA, Brown M. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 2008;68:301–306. doi: 10.1158/0008-5472.CAN-07-1983. [DOI] [PubMed] [Google Scholar]
- Ganesh L, Yoshimoto T, Moorthy NC, Akahata W, Boehm M, Nabel EG, Nabel GJ. Protein methyltransferase 2 inhibits NF-kappaB function and promotes apoptosis. Mol Cell Biol. 2006;26:3864–3874. doi: 10.1128/MCB.26.10.3864-3874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- Gehrig PM, Hunziker PE, Zahariev S, Pongor S. Fragmentation pathways of N(G)-methylated and unmodified arginine residues in peptides studied by ESI-MS/MS and MALDI-MS. J Am Soc Mass Spectrom. 2004;15:142–149. doi: 10.1016/j.jasms.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Gonsalvez GB, Tian L, Ospina JK, Boisvert FM, Lamond AI, Matera AG. Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J Cell Biol. 2007;178:733–740. doi: 10.1083/jcb.200702147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet I, Gauvin G, Boisvenue S, Cote J. Alternative splicing yields protein arginine methyltransferase 1 isoforms with distinct activity, substrate specificity, and subcellular localization. J Biol Chem. 2007;282:33009–33021. doi: 10.1074/jbc.M704349200. [DOI] [PubMed] [Google Scholar]
- Gros L, Renodon-Corniere A, de Saint Vincent BR, Feder M, Bujnicki JM, Jacquemin-Sablon A. Characterization of prmt7alpha and beta isozymes from Chinese hamster cells sensitive and resistant to topoisomerase II inhibitors. Biochim Biophys Acta. 2006;1760:1646–1656. doi: 10.1016/j.bbagen.2006.08.026. [DOI] [PubMed] [Google Scholar]
- Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- Higashimoto K, Kuhn P, Desai D, Cheng X, Xu W. Phosphorylation-mediated inactivation of coactivator-associated arginine methyltransferase 1. Proc Natl Acad Sci U S A. 2007;104:12318–12323. doi: 10.1073/pnas.0610792104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Kao C, Jeng MH, Eble JN, Koch MO, Gardner TA, Zhang S, Li L, Pan CX, Hu Z, et al. Aberrant expression of CARM1, a transcriptional coactivator of androgen receptor, in the development of prostate carcinoma and androgen-independent status. Cancer. 2004;101:83–89. doi: 10.1002/cncr.20327. [DOI] [PubMed] [Google Scholar]
- Hou Z, Peng H, Ayyanathan K, Yan KP, Langer EM, Longmore GD, Rauscher FJ., 3rd The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol. 2008;28:3198–3207. doi: 10.1128/MCB.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CH, Tam MF. Detection of dimethylarginines in protein hydrolysates by matrix-assisted laser desorption/ionization mass spectrometry. Anal Biochem. 2006;350:151–155. doi: 10.1016/j.ab.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Hughes RM, Waters ML. Arginine methylation in a beta-hairpin peptide: implications for Arg-pi interactions, DeltaCp(o), and the cold denatured state. J Am Chem Soc. 2006;128:12735–12742. doi: 10.1021/ja061656g. [DOI] [PubMed] [Google Scholar]
- Hung CJ, Chen DH, Shen YT, Li YC, Lin YW, Hsieh M, Li C. Characterization of protein arginine methyltransferases in porcine brain. J Biochem Mol Biol. 2007;40:617–624. doi: 10.5483/bmbrep.2007.40.5.617. [DOI] [PubMed] [Google Scholar]
- Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovides DC, O’Shea CC, Oses-Prieto J, Burlingame A, McCormick F. Critical role for arginine methylation in adenovirus-infected cells. J Virol. 2007;81:13209–13217. doi: 10.1128/JVI.01415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iberg AN, Espejo A, Cheng D, Kim D, Michaud-Levesque J, Richard S, Bedford MT. Arginine methylation of the histone h3 tail impedes effector binding. J Biol Chem. 2008;283:3006–3010. doi: 10.1074/jbc.C700192200. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Yada T. Protein arginine methylation regulates insulin signaling in L6 skeletal muscle cells. Biochem Biophys Res Commun. 2007;364:1015–1021. doi: 10.1016/j.bbrc.2007.10.113. [DOI] [PubMed] [Google Scholar]
- Jelinic P, Stehle JC, Shaw P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006;4:e355. doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz JE, Dlakic M, Clarke S. Automated identification of putative methyltransferases from genomic open reading frames. Mol Cell Proteomics. 2003;2:525–540. doi: 10.1074/mcp.M300037-MCP200. [DOI] [PubMed] [Google Scholar]
- Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Sohn HY, Yoon SY, Oh JH, Yang JO, Kim JH, Song KS, Rho SM, Yoo HS, Kim YS, et al. Identification of gastric cancer-related genes using a cDNA microarray containing novel expressed sequence tags expressed in gastric cancer cells. Clin Cancer Res. 2005;11:473–482. [PubMed] [Google Scholar]
- Kleinschmidt MA, Streubel G, Samans B, Krause M, Bauer UM. The protein arginine methyltransferases CARM1 and PRMT1 cooperate in gene regulation. Nucleic Acids Res. 2008;36:3202–3213. doi: 10.1093/nar/gkn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause CD, Yang ZH, Kim YS, Lee JH, Cook JR, Pestka S. Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther. 2007;113:50–87. doi: 10.1016/j.pharmthera.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Lacroix M, Messaoudi SE, Rodier G, Le Cam A, Sardet C, Fabbrizio E. The histone-binding protein COPR5 is required for nuclear functions of the protein arginine methyltransferase PRMT5. EMBO Rep. 2008;9:452–458. doi: 10.1038/embor.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski TM, Frankel A. A kinetic study of human protein arginine N-methyltransferase 6 reveals a distributive mechanism. J Biol Chem. 2008;283:10015–10025. doi: 10.1074/jbc.M710176200. [DOI] [PubMed] [Google Scholar]
- Le Romancer M, Treilleux I, Leconte N, Robin-Lespinasse Y, Sentis S, Bouchekioua-Bouzaghou K, Goddard S, Gobert-Gosse S, Corbo L. Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Mol Cell. 2008;31:212–221. doi: 10.1016/j.molcel.2008.05.025. [DOI] [PubMed] [Google Scholar]
- Lee DY, Ianculescu I, Purcell D, Zhang X, Cheng X, Stallcup MR. Surface-scanning mutational analysis of protein arginine methyltransferase 1: roles of specific amino acids in methyltransferase substrate specificity, oligomerization, and coactivator function. Mol Endocrinol. 2007;21:1381–1393. doi: 10.1210/me.2006-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Bedford MT. PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 2002;3:268–273. doi: 10.1093/embo-reports/kvf052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J Biol Chem. 2005a;280:32890–32896. doi: 10.1074/jbc.M506944200. [DOI] [PubMed] [Google Scholar]
- Lee JH, Cook JR, Yang ZH, Mirochnitchenko O, Gunderson SI, Felix AM, Herth N, Hoffmann R, Pestka S. PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J Biol Chem. 2005b;280:3656–3664. doi: 10.1074/jbc.M405295200. [DOI] [PubMed] [Google Scholar]
- Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci U S A. 2005c;102:3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiper J, Nandi M, Torondel B, Murray-Rust J, Malaki M, O’Hara B, Rossiter S, Anthony S, Madhani M, Selwood D, et al. Disruption of methylarginine metabolism impairs vascular homeostasis. Nat Med. 2007;13:198–203. doi: 10.1038/nm1543. [DOI] [PubMed] [Google Scholar]
- Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhou Z, Chen G, Bao S. A putative transcriptional elongation factor hIws1 is essential for mammalian cell proliferation. Biochem Biophys Res Commun. 2007;353:47–53. doi: 10.1016/j.bbrc.2006.11.133. [DOI] [PubMed] [Google Scholar]
- Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder S, Liu Y, Ford OH, 3rd, Mohler JL, Whang YE. Involvement of arginine methyltransferase CARM1 in androgen receptor function and prostate cancer cell viability. Prostate. 2006;66:1292–1301. doi: 10.1002/pros.20438. [DOI] [PubMed] [Google Scholar]
- Meyer R, Wolf SS, Obendorf M. PRMT2, a member of the protein arginine methyltransferase family, is a coactivator of the androgen receptor. J Steroid Biochem Mol Biol. 2007;107:1–14. doi: 10.1016/j.jsbmb.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Miranda TB, Miranda M, Frankel A, Clarke S. PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J Biol Chem. 2004;279:22902–22907. doi: 10.1074/jbc.M312904200. [DOI] [PubMed] [Google Scholar]
- Miranda TB, Webb KJ, Edberg DD, Reeves R, Clarke S. Protein arginine methyltransferase 6 specifically methylates the nonhistone chromatin protein HMGA1a. Biochem Biophys Res Commun. 2005;336:831–835. doi: 10.1016/j.bbrc.2005.08.179. [DOI] [PubMed] [Google Scholar]
- Mitchell JB, Thornton JM, Singh J, Price SL. Towards an understanding of the arginine-aspartate interaction. J Mol Biol. 1992;226:251–262. doi: 10.1016/0022-2836(92)90137-9. [DOI] [PubMed] [Google Scholar]
- Naeem H, Cheng D, Zhao Q, Underhill C, Tini M, Bedford MT, Torchia J. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol Cell Biol. 2007;27:120–134. doi: 10.1128/MCB.00815-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najbauer J, Johnson BA, Young AL, Aswad DW. Peptides with sequences similar to glycine, arginine-rich motifs in proteins interacting with RNA are efficiently recognized by methyltransferase(s) modifying arginine in numerous proteins. J Biol Chem. 1993;268:10501–10509. [PubMed] [Google Scholar]
- Nakashima K, Hagiwara T, Yamada M. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J Biol Chem. 2002;277:49562–49568. doi: 10.1074/jbc.M208795200. [DOI] [PubMed] [Google Scholar]
- Neuenkirchen N, Chari A, Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 2008;582:1997–2003. doi: 10.1016/j.febslet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Reinberg D. Methods and tips for the purification of human histone methyltransferases. Methods. 2003;31:49–58. doi: 10.1016/s1046-2023(03)00087-2. [DOI] [PubMed] [Google Scholar]
- Ohkura N, Takahashi M, Yaguchi H, Nagamura Y, Tsukada T. Coactivator-associated arginine methyltransferase 1, CARM1, affects pre-mRNA splicing in an isoform-specific manner. J Biol Chem. 2005;280:28927–28935. doi: 10.1074/jbc.M502173200. [DOI] [PubMed] [Google Scholar]
- Osborne TC, Obianyo O, Zhang X, Cheng X, Thompson PR. Protein arginine methyltransferase 1: positively charged residues in substrate peptides distal to the site of methylation are important for substrate binding and catalysis. Biochemistry. 2007;46:13370–13381. doi: 10.1021/bi701558t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahlich S, Zakaryan RP, Gehring H. Protein arginine methylation: Cellular functions and methods of analysis. Biochim Biophys Acta. 2006;1764:1890–1903. doi: 10.1016/j.bbapap.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Pahlich S, Zakaryan RP, Gehring H. Identification of proteins interacting with protein arginine methyltransferase 8: the Ewing sarcoma (EWS) protein binds independent of its methylation state. Proteins. 2008;72:1125–1137. doi: 10.1002/prot.22004. [DOI] [PubMed] [Google Scholar]
- Pal S, Baiocchi RA, Byrd JC, Grever MR, Jacob ST, Sif S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007;26:3558–3569. doi: 10.1038/sj.emboj.7601794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Yun R, Datta A, Lacomis L, Erdjument-Bromage H, Kumar J, Tempst P, Sif S. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol. 2003;23:7475–7487. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento JJ, Bullock CR, LeDuc RD, Mizzen CA, Kelleher NL. Combinatorial modification of human histone H4 quantitated by two-dimensional liquid chromatography coupled with top down mass spectrometry. J Biol Chem. 2008;283:14927–14937. doi: 10.1074/jbc.M709796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintucci G, Quarto N, Rifkin DB. Methylation of high molecular weight fibroblast growth factor-2 determines post-translational increases in molecular weight and affects its intracellular distribution. Mol Biol Cell. 1996;7:1249–1258. doi: 10.1091/mbc.7.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raijmakers R, Zendman AJ, Egberts WV, Vossenaar ER, Raats J, Soede-Huijbregts C, Rutjes FP, van Veelen PA, Drijfhout JW, Pruijn GJ. Methylation of arginine residues interferes with citrullination by peptidylarginine deiminases in vitro. J Mol Biol. 2007;367:1118–1129. doi: 10.1016/j.jmb.2007.01.054. [DOI] [PubMed] [Google Scholar]
- Ramon-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, Yang W. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci U S A. 2007;104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X, Jeltsch A. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard S, Morel M, Cleroux P. Arginine methylation regulates IL-2 gene expression: a role for protein arginine methyltransferase 5 (PRMT5) Biochem J. 2005;388:379–386. doi: 10.1042/BJ20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin-Lespinasse Y, Sentis S, Kolytcheff C, Rostan MC, Corbo L, Le Romancer M. hCAF1, a new regulator of PRMT1-dependent arginine methylation. J Cell Sci. 2007;120:638–647. doi: 10.1242/jcs.03357. [DOI] [PubMed] [Google Scholar]
- Sayegh J, Webb K, Cheng D, Bedford MT, Clarke SG. Regulation of protein arginine methyltransferase 8 (PRMT8) activity by its N-terminal domain. J Biol Chem. 2007;282:36444–36453. doi: 10.1074/jbc.M704650200. [DOI] [PubMed] [Google Scholar]
- Sgarra R, Lee J, Tessari MA, Altamura S, Spolaore B, Giancotti V, Bedford MT, Manfioletti G. The AT-hook of the chromatin architectural transcription factor high mobility group A1a is arginine-methylated by protein arginine methyltransferase 6. J Biol Chem. 2006;281:3764–3772. doi: 10.1074/jbc.M510231200. [DOI] [PubMed] [Google Scholar]
- Shen EC, Henry MF, Weiss VH, Valentini SR, Silver PA, Lee MS. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 1998;12:679–691. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Miranda TB, Jiang W, Frankel A, Roemer ME, Robb VA, Gutmann DH, Herschman HR, Clarke S, Newsham IF. DAL-1/4.1B tumor suppressor interacts with protein arginine N-methyltransferase 3 (PRMT3) and inhibits its ability to methylate substrates in vitro and in vivo. Oncogene. 2004;23:7761–7771. doi: 10.1038/sj.onc.1208057. [DOI] [PubMed] [Google Scholar]
- Sprangers R, Groves MR, Sinning I, Sattler M. High-resolution X-ray and NMR structures of the SMN Tudor domain: conformational variation in the binding site for symmetrically dimethylated arginine residues. J Mol Biol. 2003;327:507–520. doi: 10.1016/s0022-2836(03)00148-7. [DOI] [PubMed] [Google Scholar]
- Swiercz R, Cheng D, Kim D, Bedford MT. Ribosomal protein rpS2 is hypomethylated in PRMT3-deficient mice. J Biol Chem. 2007;282:16917–16923. doi: 10.1074/jbc.M609778200. [DOI] [PubMed] [Google Scholar]
- Tadesse H, Deschenes-Furry J, Boisvenue S, Cote J. KH-type splicing regulatory protein interacts with survival motor neuron protein and is misregulated in spinal muscular atrophy. Hum Mol Genet. 2008;17:506–524. doi: 10.1093/hmg/ddm327. [DOI] [PubMed] [Google Scholar]
- Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, Clarke S, Herschman HR. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J Biol Chem. 2000;275:7723–7730. doi: 10.1074/jbc.275.11.7723. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PR, Fast W. Histone citrullination by protein arginine deiminase: is arginine methylation a green light or a roadblock? ACS Chem Biol. 2006;1:433–441. doi: 10.1021/cb6002306. [DOI] [PubMed] [Google Scholar]
- Troffer-Charlier N, Cura V, Hassenboehler P, Moras D, Cavarelli J. Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains. EMBO J. 2007;26:4391–4401. doi: 10.1038/sj.emboj.7601855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbiest V, Montaudon D, Tautu MT, Moukarzel J, Portail JP, Markovits J, Robert J, Ichas F, Pourquier P. Protein arginine (N)-methyl transferase 7 (PRMT7) as a potential target for the sensitization of tumor cells to camptothecins. FEBS Lett. 2008;582:1483–1489. doi: 10.1016/j.febslet.2008.03.031. [DOI] [PubMed] [Google Scholar]
- Willemsen NM, Hitchen EM, Bodetti TJ, Apolloni A, Warrilow D, Piller SC, Harrich D. Protein methylation is required to maintain optimal HIV-1 infectivity. Retrovirology. 2006;3:92. doi: 10.1186/1742-4690-3-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Cho H, Kadam S, Banayo EM, Anderson S, Yates JR, 3rd, Emerson BM, Evans RM. A methylation-mediator complex in hormone signaling. Genes Dev. 2004;18:144–156. doi: 10.1101/gad.1141704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav N, Cheng D, Richard S, Morel M, Iyer VR, Aldaz CM, Bedford MT. CARM1 promotes adipocyte differentiation by coactivating PPARgamma. EMBO Rep. 2008;9:193–198. doi: 10.1038/sj.embor.7401151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM, Bedford MT. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc Natl Acad Sci U S A. 2003;100:6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue WW, Hassler M, Roe SM, Thompson-Vale V, Pearl LH. Insights into histone code syntax from structural and biochemical studies of CARM1 methyltransferase. EMBO J. 2007;26:4402–4412. doi: 10.1038/sj.emboj.7601856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Cheng X. Structure of the Predominant Protein Arginine Methyltransferase PRMT1 and Analysis of Its Binding to Substrate Peptides. Structure (Camb) 2003;11:509–520. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Jankovic V, Gural A, Huang G, Pardanani A, Menendez S, Zhang J, Dunne R, Xiao A, Erdjument-Bromage H, et al. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008;22:640–653. doi: 10.1101/gad.1632608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Schmidt-Ott KM, Chua S, Foster KA, Frankel RZ, Pavlidis P, Barasch J, D’Agati VD, Gharavi AG. A Mendelian locus on chromosome 16 determines susceptibility to doxorubicin nephropathy in the mouse. Proc Natl Acad Sci U S A. 2005;102:2502–2507. doi: 10.1073/pnas.0409786102. [DOI] [PMC free article] [PubMed] [Google Scholar]