Abstract
Technological advances in both hardware and software have made possible the realization of sophisticated biological imaging experiments using the optical microscope. As a result, modern microscopy experiments are capable of producing complex image data sets. For a given data analysis task, the images in a set are arranged, based on the requirements of the task, by attributes such as the time and focus levels at which they were acquired. Importantly, different tasks performed over the course of an analysis are often facilitated by the use of different arrangements of the images. We present a software framework which supports the use of different logical image arrangements to analyze a physical set of images. Called the Microscopy Image Analysis Tool (MIATool), this framework realizes the logical arrangements using arrays of pointers to the images, thereby removing the need to replicate and manipulate the actual images in their storage medium. In order that they may be tailored to the specific requirements of disparate analysis tasks, these logical arrangements may differ in size and dimensionality, with no restrictions placed on the number of dimensions and the meaning of each dimension. MIATool additionally supports processing flexibility, extensible image processing capabilities, and data storage management.
Index Terms: Microscopy, multi-dimensional data, image analysis, image viewer, software framework
I. Introduction
The optical microscope has been an invaluable tool for the study of biological events at the cellular, the subcellular, and more recently, the single molecule level (e.g., [1], [2], [3]). With advances in both hardware and software technology, the microscopist today is well-equipped to design and operate sophisticated microscopy image acquisition systems. As microscopy imaging experiments have become more creative, however, so have the resulting image data grown in complexity. Therefore, to obtain the desired information from the acquired images in an efficient manner, software is needed that facilitates the analysis of complex image data sets. When designing such a software application, the nature of the image data and of its analysis requirements warrants consideration.
A. Software Design Considerations for Image Data Analysis
Different image arrangements for different analysis tasks
Supported by image acquisition software and hardware components such as optical filters, focusing devices, and image detectors, modern microscopy experiments are capable of producing complex and multi-dimensional image data sets. Throughout the course of a fluorescence microscopy (e.g., [4], [5]) experiment, for example, images of different colors (i.e., different wavelengths) can be captured at different times by one or more cameras at different focus levels. Depending on the nature of an analysis task that needs to be performed, the images are arranged along an appropriate number of dimensions by color, focal position, acquisition time stamp, and/or any other experimental or analytical parameters. Importantly, in an analysis that comprises different types of tasks, different arrangements of the images may be used to facilitate the execution of the component tasks. As necessitated by the specifics of the tasks, these arrangements may differ in the number of dimensions as well as the meaning of the dimensions.
In the most general case, an arrangement is N-dimensional (where N is any positive integer) and the meaning of each dimension is designated as required by an analysis task. For example, a simple linear (i.e., one-dimensional) arrangement with the images sorted in no particular order may be sufficient for a visual inspection of the general image quality. However, a two-dimensional (2D) arrangement with the same images sorted by time in one dimension and color in the other may be more suitable for the purpose of generating overlays of the different colors. In addition, an arrangement is in general not limited to a reordering of the entire set of acquired images, but may comprise only some of the images and/or contain repeated images, also as necessitated by the particular task at hand. For example, to generate the overlay of two large time lapse series acquired simultaneously, but at different rates by two cameras, one might choose to work with only a small time segment of interest, and to repeat within that segment images from the slower camera to temporally align them with the images from the faster camera.
We note that the integer N does not include the x and y dimensions that are intrinsic to an image. An N-dimensional arrangement of images is therefore equivalent to what would commonly be referred to as an (N + 2)-dimensional image data set.
Large numbers of images
Besides the multitude of ways in which it may be arranged, a given data set often consists of a large number of images. Using fast frame rate cameras, for example, microscopists can acquire many images in a relatively short period of time, ending up with thousands or even tens of thousands of images by the end of an experiment. Given the limited hardware resources of a conventional personal computer, data of this size poses a challenge in terms of both storage on the hard disk and processing in random access memory (RAM).
Heterogeneity of images and flexible processing
In general, a microscopy experiment can produce images of different sizes. For example, when using multiple cameras with different specifications, one may be constrained to images of similar, but nevertheless different sizes. In addition, the use of image acquisition software programs from different camera manufacturers may result in a data set composed of images in different file formats. It would therefore be useful for a software application to support the display and processing of a heterogeneous data set comprising differently-sized images of potentially different file formats. The idea of heterogeneity is also important in terms of the processing of the image data. Given an arrangement of images, one should be able to process individual or subsets of the images differently, but at the same time also have the ability to operate on all images uniformly. For example, due to photobleaching of the imaged fluorophore, one might find it necessary to apply different pixel intensity adjustment settings to images in different segments of a time lapse series. On the other hand, to confine an analysis to a region of interest, one might need to crop all the images in the exact same way.
Provision for adding new analysis capabilities
An important point to take into account in the software design is the wide variety of image analysis requirements in microscopy that range from simple tasks such as the cropping of an image to more sophisticated processing such as image deconvolution. Not only is it impractical to support all the existing image processing algorithms for all types of analyses, it is also important to note that analysis requirements are constantly evolving and that customized or new algorithms are always needed. Therefore, it is essential that a general microscopy data analysis application provide some means for the incorporation of new capabilities.
Organized storage of images and associated information
A last point to consider is that, in addition to the images, there are other types of important information that an analysis software application should maintain, potentially on a per-image basis. These include the processing settings (e.g., intensity adjustment settings, crop settings, etc.) which have been applied to the images and which can be stored to provide a history of the processing, the metadata (e.g., acquisition time stamp, imaged fluorophore, etc.) which are essential for certain types of analyses, and analytical results (e.g., computed background intensity, number of identified objects of interest, etc.) which need to be kept. In light of the various types of information that need to be saved and associated with the images, a software application should support a storage management mechanism that helps with the organization of the images and any related experimental or analytical information, both on a temporary basis in RAM and on a permanent basis on the hard disk.
B. Current Software Solutions
Software has been and continues to be developed by various parties to support the analysis of microscopy image data. The Open Microscopy Environment [6], for example, takes an informatics approach to the analysis and storage of microscopy data. This environment defines an extensible data model for the management of not only the images themselves, but also the metadata and the analytical results that are associated with the images. In [7], a data model and architecture are introduced in the context of leveraging grid technologies for the knowledge-based processing of large image data sets. Another example is the popular Java-based application ImageJ [8] which offers an abundance of image analysis capabilities that range from standard functionalities such as intensity adjustment to advanced features such as object tracking. For more information on some of the currently available software packages, see [9].
The typical microscopy image analysis software package today, however, assumes the use of either a single or a few image arrangements throughout the course of an analysis. Moreover, an image data set is limited to a certain number of dimensions, commonly set to five (i.e., limited to a three-dimensional (3D) arrangement), and these dimensions are fixed to represent an image’s x and y dimensions, focal position along the microscope’s optical (z-)axis, color, and acquisition time. Therefore, an analysis consisting of disparate tasks that require arrangements of higher numbers of dimensions with arbitrary representations is in general not readily supported. Some applications also require the loading of an entire set of images into RAM for viewing and processing, and therefore have difficulty supporting the analysis of large numbers of images. The typical software application today also does not readily provide for the arbitrary reordering, repetition, and subset construction of the images in a set. In addition, the images comprising a set are commonly required to have the same size and/or file format, and the storage of processing settings, metadata, and analytical results is not generally supported by all software packages. In general, the typical software solution today addresses some, but not all of the aspects of image analysis discussed in Section I-A.
C. The Microscopy Image Analysis Tool
In this paper, we discuss the design of a microscopy image analysis software framework which takes into account all the points raised in Section I-A regarding the analysis of a complex image data set. This framework is motivated by the recognition that the different tasks involved in the analysis of a data set are potentially facilitated by different arrangements of its images. Importantly, it is based on the central idea that these arrangements can be achieved as different logical views of the same physical images which may reside either in RAM or on the hard disk. In this way, a clear distinction is drawn between the logical data sets (i.e., arrangements) which are used for analysis, and the actual images in RAM or on disk which are looked upon only as physical repositories of the data, and which remain unchanged throughout the course of an analysis. In addition to its underlying support for multiple and arbitrary logical arrangements of images, this framework is a generic one where no restrictions are placed on the dimensionality of an arrangement. Furthermore, the meaning of each dimension of an arrangement can be designated arbitrarily as necessitated by the analysis task at hand.
We note that the framework we introduce here is not meant to supersede other software solutions such as those described in Section I-B. Rather, it is an approach to microscopy image analysis which we find effective in dealing with the challenges posed by a complex data set. In fact, given the wide-ranging problem domains, objectives, and approaches that characterize the different software solutions, it is entirely possible that our framework may be used in conjunction with the other solutions to make certain types of analyses more efficient.
Called the Microscopy Image Analysis Tool, or MIATool for short, our software framework realizes different logical arrangements of the images in a given physical data set via multi-dimensional arrays of pointers to the images. We note that the term “pointer” as used here is distinct from the pointer data type found in programming languages such as C and C++. Rather, it is used in the sense of a general, language-independent data type that stores the address of an image that can reside in RAM or on a hard disk. For example, if in RAM, the value stored could be a memory address. If on disk, the value stored could be a file path.
The use of image pointers provides at least three advantages. First, it eliminates the need to physically replicate the images in RAM or on the hard disk in order to create different arrangements, and therefore helps to save a significant amount of memory and disk space. Also, any reordering, repetition, and subset construction of the images needed to arrive at an arrangement can all be easily achieved by moving, replicating, and selectively creating or removing pointers. Second, since pointers are typically much smaller in size than the images they reference, a pointer array occupies considerably less RAM than an array of actual images. Therefore, pointer arrays allow MIATool to store and process large logical data sets in RAM. Third, since pointers can refer to images of different sizes and file formats, a pointer array can naturally support the analysis of a heterogeneous data set.
To accommodate the analysis of the images specified by an array of pointers, the MIATool framework employs arrays of corresponding size and dimension to manage the various processing settings, metadata, and analytical results that are associated with the pointer-referenced images. The main idea behind these corresponding arrays is that they allow each pointer in a pointer array to be associated with its own processing specification, metadata, and analytical results. Consequently, they provide the flexibility to process each referenced image differently, while at the same time support the uniform processing of some or all of the referenced images through the specification of the same processing settings for appropriate subsets of pointers in a pointer array. Additionally, an advantage offered by corresponding arrays of processing settings is that they can be saved in place of the actual images that result from the processing, which are typically much larger in size than the settings. When this option is used, MIATool is able to make further savings in the usage of the limited hard disk space and at the same time preserve a record of the processing.
Since visual feedback plays an important role in many types of image processing, the MIATool framework specifies an image viewer which supports the visualization of the images referenced by an N-dimensional image pointer array. In addition, it specifies processing tools which allow the manipulation, via a graphical user interface, of the processing settings contained in arrays that correspond to the image pointer array that is currently displayed in a viewer. By interacting with a viewer to provide immediate visual feedback whenever processing settings are changed, these tools make possible the interactive, on-the-fly processing of the pointer-referenced images. While the advantage of immediate visual feedback renders these tools most suitable for types of processing that require relatively little time to complete (e.g., intensity adjustment, cropping, etc.), such tools can in principle be created for all kinds of image processing. Importantly, by defining standard ways of interaction between the viewer, the processing tools, and the corresponding arrays of processing settings, the MIATool framework facilitates the addition of new processing capabilities to its existing repertoire.
To help keep track of the multiple image pointer arrays and their corresponding arrays of settings and information that may be used over the course of analyzing a physical image data set, a storage management mechanism is provided by the MIATool framework. Capable of managing storage both in RAM and on the hard disk, this storage manager uses a hierarchical structure to associate a physical image data set with the arrays employed for its analysis, and to maintain the relationships among the various arrays. Furthermore, it serves as the standard channel through which the various arrays are stored and retrieved.
The remainder of this paper is organized as follows. In Section II, we give a description of the general architecture of the MIATool software framework. In Section III, we illustrate the use of different image pointer arrays to perform different analysis tasks on a given physical image data set. This is done via examples of some commonly encountered problems in microscopy image analysis. We follow in Section IV with a discussion on how MIATool uses arrays corresponding to an image pointer array to provide flexibility in the processing of the images it references and to maintain the metadata and analytical results that are associated with those images. In Section V, we present the MIATool image viewer and processing tools which together provide a visual, user-interactive means for working with an image pointer array and its corresponding arrays. In Section VI, we describe how MIATool manages the storage of a physical image data set and the various types of arrays that are used for its analysis. Lastly, we conclude our presentation in Section VII.
II. Architecture
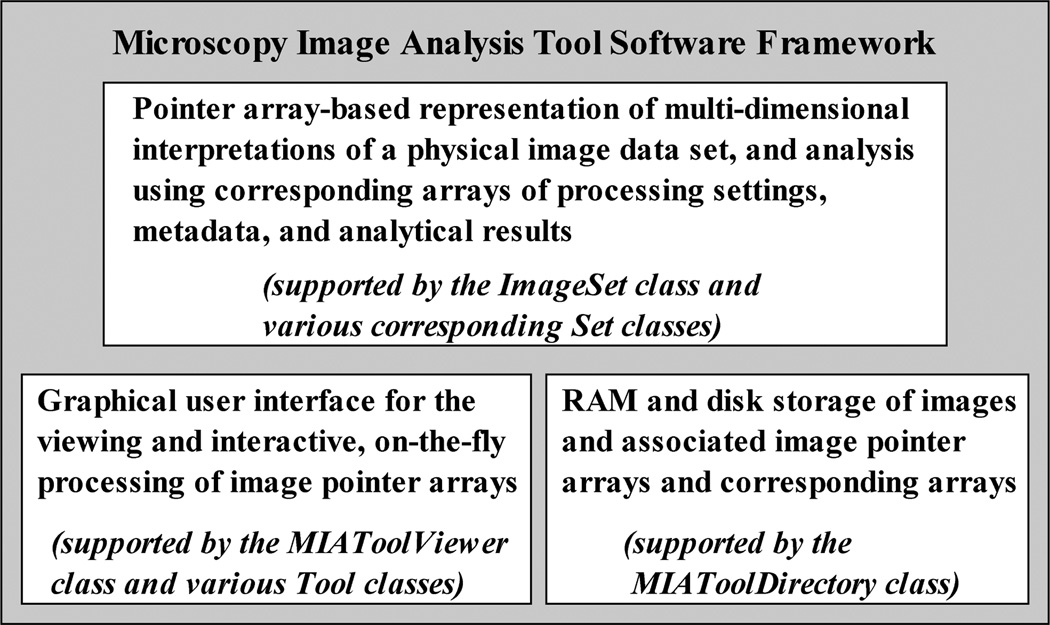
The MIATool software framework comprises three principal components, as shown in Fig. 1. The first component consists of the logical N-dimensional pointer array interpretation of a physical image data set (Section III), along with corresponding N-dimensional arrays of processing settings, metadata, and analytical results that are used to support the analysis of the pointer-referenced images (Section IV). These corresponding arrays are contained in modules which are capable of carrying out the actual processing of the images. The second component includes a viewer and processing tools (Section V) which together provide a graphical user interface for the viewing of the images referenced by an N-dimensional image pointer array, and the interactive, on-the-fly processing of those images by way of modifying the processing settings contained in corresponding arrays. The third component is a storage manager (Section VI) which associates in RAM or on the hard disk a physical image data set with its potentially many pointer and corresponding arrays.
Fig. 1.
The three-component MIATool software framework. The first component provides the underlying representation and analysis of logical image data sets using multi-dimensional image pointer arrays and corresponding arrays of processing settings, metadata, and analytical results. The second component provides a graphical user interface for the viewing and the interactive, on-the-fly processing of the pointer arrays. The third component provides the RAM and disk storage management that associates a physical image data set with the pointer arrays and the corresponding arrays that are used for its analysis.
The underlying pointer array-based representation and processing of a logical data set, the graphical user interface for visual, interactive data analysis, and the image and information storage manager therefore constitute the three main components of the software framework. Provided that standard protocols of interaction are adhered to, this architecture is amenable to the independent development of the components. Based on this design, a prototype software application, MIATool V1.1 [10], has been implemented using the technical programming language of MATLAB (The MathWorks, Inc., Natick, MA) and its image processing toolbox. A preliminary introduction to the prototype implementation MIATool V1.1 was published in the conference paper [11]. In contrast, the current paper focuses on the underlying implementation-independent software framework, including the reference architecture that provides a blueprint for the implementation of the design considerations delineated in Section I-A.
In the sections that follow, we make use of diagrams created using the standard Unified Modeling Language (UML) (e.g., [12]) notation system to help illustrate the design of, and the interaction between, the three main components of the MIATool framework. These diagrams were created using the software package StarUML 5.0 (http://staruml.sourceforge.net).
III. Data analysis using logical arrangements of images
A complex image data set generated by a microscopy experiment is often subjected to multiple types of processing tasks throughout the course of an analysis. As identified in Section I-A to be an important consideration in the design of a data analysis software, these different tasks are often facilitated by different arrangements of the images that may differ in size and dimensionality. To address this important aspect of data analysis, the MIATool software framework supports the realization of different arrangements of a set of images as logical data sets in the form of arrays of pointers to the images. Since the images physically reside either in RAM or on disk, a pointer is simply a memory address or a file path which uniquely identifies an image in a data set.
By creating, manipulating, and storing arrays of image pointers, MIATool avoids having to create or modify physical arrangements of the images which would require the replication or shuffling of the images in RAM or on the hard disk. As a pointer is typically much smaller in size than the image it references, this allows MIATool to make efficient use of the limited amount of RAM and disk space. As a result, this also enables MIATool to accommodate the analysis and storage of large data sets and hence address another important design criterion.
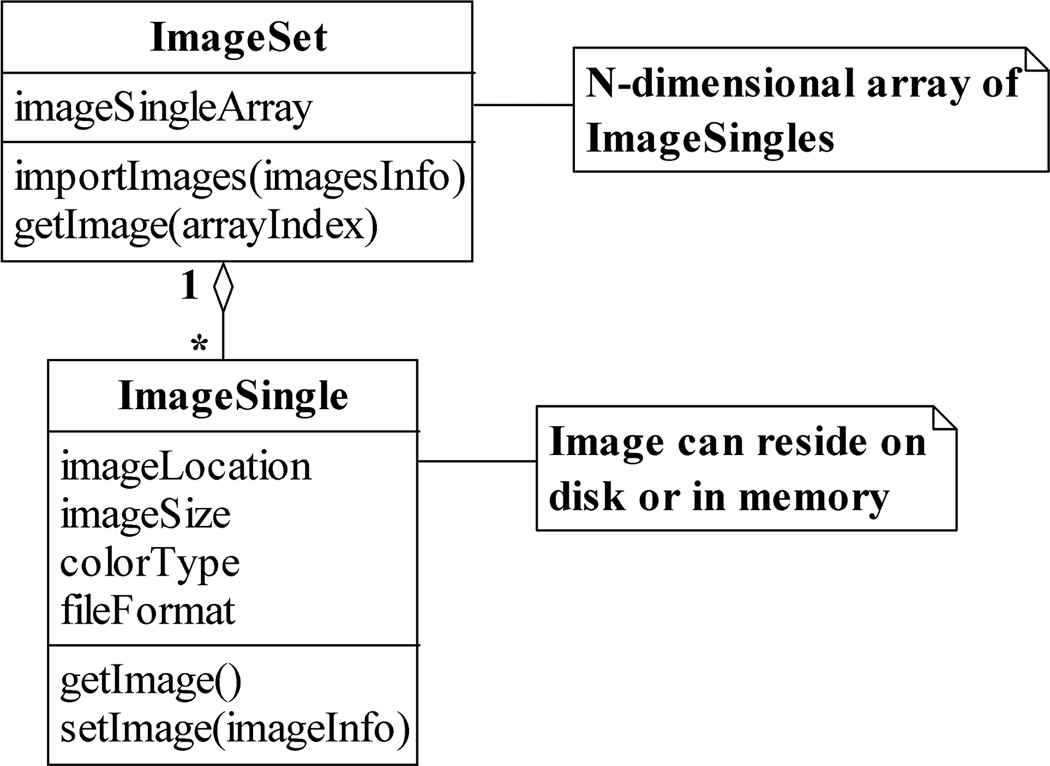
As shown in the UML class diagram of Fig. 2, the concept of a logical image data set is embodied in an ImageSet. The ImageSet class contains in general an N-dimensional image pointer array, and supports operations such as the creation of the array (importImages) and the retrieval of an actual image via a pointer (getImage). Each image pointer in the array takes the form of an ImageSingle, a class which stores not only the physical location (e.g., the file path) of an image, but also information such as the image’s size, color type, and file format. In addition, the ImageSingle class provides operations for creating a pointer to an image (setImage) and for retrieving an image via a pointer (getImage).
Fig. 2.
The ImageSet and ImageSingle classes. An ImageSet contains an N-dimensional array of ImageSingles, where each ImageSingle is a pointer which contains, for example, the location on the hard disk at which an image resides. An ImageSingle also stores information such as the size and color type of the image it references. Both the ImageSet and the ImageSingle support operations for creating pointers to images and for retrieving images via pointers.
The storage of image attributes in an ImageSingle is important in that it allows the ImageSingles belonging to an ImageSet to reference images of, for example, different sizes, color types, and file formats. This directly enables MIATool to support the analysis of heterogeneous image data sets, and thereby address the data heterogeneity design criterion. In particular, the support for heterogeneous image data allows MIATool to deal with data sets of mixed image file formats which can easily arise, for example, from collaborative projects involving different imaging modalities contributed by potentially different research groups (see, e.g., [13], [14]). Note that in order to realize support for such data sets, an implementation of the getImage operation of the ImageSingle class should use the value of the file format attribute to determine the appropriate image reading routine to invoke to retrieve the referenced image from disk.
We note that in our MATLAB implementation of the ImageSingle class in MIATool V1.1, the address of an image on disk is a file path that is straightforwardly stored as a string field. However, since MATLAB does not support a native pointer data type such as that in C or C++, the address of an image in RAM is stored in two basic ways. One, the image can reside in a field of the ImageSingle itself, in which case its address is simply given by the field. Two, the image can reside in the “UserData” property of a MATLAB figure, in which case its address is given by the handle of the figure. Note that these details are implementation-specific choices which we made in the development of MIATool V1.1. Other methods are certainly possible and can be used in a different implementation of the framework.
Depending on the specific requirements of an analysis, an image pointer array can be of any dimensionality and can contain pointers arranged to represent an arbitrary reordering of the physical set of images. A pointer array can also contain repeated pointers to the same image, and at the same time consist of pointers that refer to just a subset of the images in the complete data set. In what follows, we illustrate, using concrete examples of problems frequently faced in microscopy image analysis, the use of different pointer arrays (and hence different ImageSets) to analyze a physical image data set. These examples represent a small sample of the broad range of pointer arrangements that may be employed to carry out different analysis tasks on a data set. Importantly, though a particular example might in and of itself be a relatively simple image processing task, together they demonstrate the high level of data analysis customization that MIATool is designed to support. (For more data analysis examples using pointer arrangements, see [11].)
Let us consider a fluorescence microscopy (e.g., [4], [5]) live cell imaging experiment in which a cell is labeled with two differently-colored fluorescent dyes. The green dye is attached to the protein of interest, and the red dye to endosomes with which vesicles containing the protein of interest interact. The objective is to track the trajectories of the fast-moving vesicles and observe their associations with the relatively stationary endosomes. Accordingly, two simultaneously running cameras are used to image the same focal plane within the cell, but each captures the fluorescence of a different dye, and writes sequentially numbered images in the order of acquisition to its own designated directory on the hard disk. In general, the two cameras may not be synchronized in time, and hence the total number of images acquired by each camera may be different at the end of the experiment. Furthermore, the images captured by the two cameras can potentially be of different sizes and saved in different file formats.
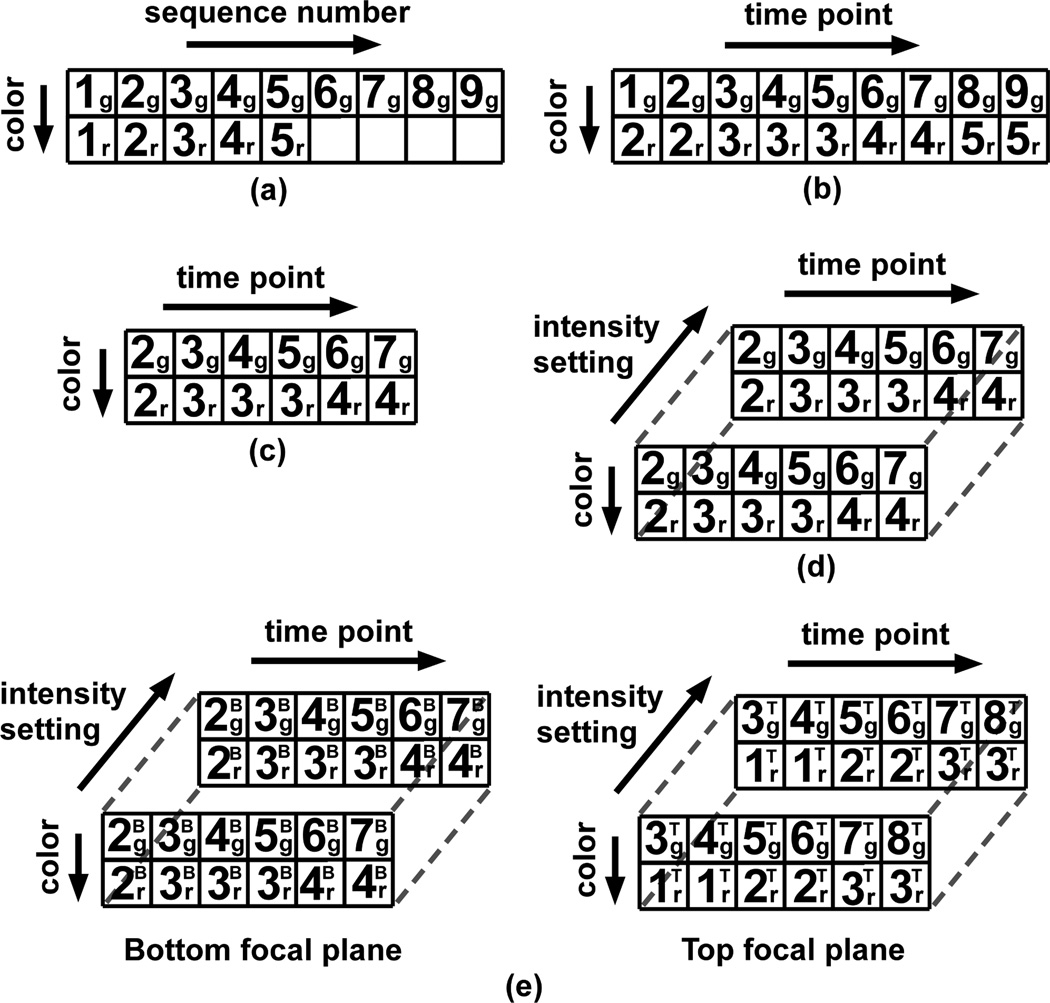
To visually assess the quality of the acquired images, we may first want to step through and view the image files contained in the two camera output directories. For this purpose, we can construct an ImageSet containing a 2D array of image pointers that mirrors the physical arrangement of the images on disk (Fig. 3(a)). Specifically, this array has two rows of potentially different lengths, each containing pointers to images in a different directory (or equivalently, from a different camera, of a different color) that are ordered by the sequence numbers of the images they reference. The MIATool viewer (Section V) can then be used to traverse this 2D pointer array and display the referenced images of possibly different sizes and file formats. Conferred by pointers that can reference images with different attributes, this ability of the MIATool framework to handle heterogeneous data sets is an important advantage over software designs that restrict data sets to consist of images of uniform size and file format.
Fig. 3.
(a)–(d) Logical arrangements (image pointer arrays) of different sizes and dimensionalities for performing different analysis tasks on the same set of physical images. An image pointer is represented by a number followed by a subscript. The number refers to the sequence number of the physical image that is referenced by the pointer, and the subscript “g” or “r” refers, respectively, to the green or red color of that image. These arrangements are suitable for (a) the simple visual inspection of, (b) the temporally-synchronized overlay of, (c) the logical extraction of an event of interest from, and (d) the differential processing (e.g., intensity adjustment) of “duplicates” of, the physical images. (e) A 4D arrangement which references images acquired from two different focal planes. The superscripts “B” and “T” associated with the pointers refer to the “bottom” and “top” focal planes, respectively.
After the initial verification of image quality, we may want to overlay the images from the two cameras in order to visualize the interactions between the green vesicles and the red endosomes. To ensure that these interactions are properly observed, overlays need to be performed with pairs of images that were captured at approximately the same time. Assuming that the two cameras were not temporally synchronized, we then need to form pairs of images based on their acquisition time stamps. This can be done using another ImageSet which contains a pointer array that is similar to the one used for the initial visualization, but whose columns represent time points rather than sequence numbers (Fig. 3(b)). Compared to the array of Fig. 3(a), the pointers in one row of the array of Fig. 3(b) might be shifted with respect to the pointers in the other row as per the time stamp information. In addition, if images of the relatively immobile endosomes were acquired at a slower rate, pointers to these images may be repeated to synchronize them against the pointers to the faster acquired images of the vesicles. Given this time-synchronized pointer array, overlaid images can be generated by moving through the columns and processing the images one pair at a time. It is important to note that instead of creating it from scratch, the array of Fig. 3(b) can be derived by shifting and replicating the pointers in the array of Fig. 3(a). In general, it is often the case that the creation of a new ImageSet can be made more efficient by deriving it from an existing ImageSet.
Now suppose that thousands of images were taken by each camera, and that from the viewing of the overlaid images, we identify a small sequence of a few hundred time points which contain the trajectory of a representative green vesicle. To logically isolate this particular sequence, we can create a third ImageSet which contains a 2D pointer array that references only the images confined to this time frame (Fig. 3(c)). As an example of how one pointer array can sometimes be easily derived from another, the creation of this new 2D array simply requires that we keep a contiguous portion of the array of Fig. 3(b) and discard the rest. The significantly smaller pointer array of Fig. 3(c) allows us to focus our analysis on just a particular segment of the image data. A collection of such arrays can be generated to “mark” all the events of interest within a large physical data set.
Let us now assume that the red dye used to label the endosomes also attaches to other cellular organelles, but at much lower quantities. Due to the significant difference in the amount of labeling, an appropriate intensity setting for viewing the strongly labeled endosomes will not allow the weakly labeled organelles to be seen clearly. Therefore, a different intensity setting is needed in order to observe any potential interactions between the green vesicles and the weakly labeled organelles. A method that supports the simultaneous existence of two intensity adjustment settings per image, and yet requires only a single pointer array, is to stack two copies of a given 2D array to form a 3D array. This 3D pointer array will therefore contain two pointers to each image along its third dimension, and each of the two pointers can be associated with a different intensity setting (Section IV). Fig. 3(d) shows an example of such a 3D array of duplicate pointers that has been derived by stacking two copies of the array of Fig. 3(c). In order to work with it, a fourth ImageSet would be created. As other arrangements of possibly different dimensionalities can be made of the same set of duplicate pointers, an advantage of this particular 3D arrangement is that, for any given image, one can go from one intensity setting to the other by simply toggling the value of the third dimension.
To illustrate the concept of logical data sets, we have thus far demonstrated MIATool’s use of different image pointer arrays to perform different analysis tasks on a relatively simple physical image data set. To show how easily a change to the experiment can make the resulting data more complex, let us assume that two additional cameras were used to capture the same green and red fluorescence, but from a focal plane within the cell that is located higher along the microscope’s z-axis. Due to the additional view of the cell that is provided by the images from this “top” plane, this multifocal plane imaging setup [15] allows us to detect the fast-moving green vesicles of interest at locations along the z-axis that we would otherwise not be able to detect by imaging at only the “bottom” focal plane. Therefore, by using four cameras to simultaneously capture images from two distinct focal planes, we can better visualize the trajectories of the green vesicles in three dimensions.
Since MIATool places no limits on the number and meaning of the dimensions of an image pointer array, the same analysis tasks can be performed on the more complex data set using the same type of logical arrangements as before, but with an additional dimension to distinguish images from the “top” and “bottom” focal planes. At the end of the same sequence of tasks, we would obtain a four-dimensional (4D) pointer array as depicted in Fig. 3(e). Note that in Fig. 3(e), the two 3D arrays corresponding to the “top” and “bottom” focal planes are intended to represent a 4D array where the fourth dimension allows us to toggle between images of the same color and from the same time point, but from different focal planes and with potentially different intensity settings.
In practice, we have used the same types of logical arrangements to analyze complex image data of a similar nature. In [16] and [17], for example, 3D trajectories of vesicles and single molecules inside live cells were determined from images acquired with multifocal plane imaging setups. In those experiments, multiple cameras were operated at different speeds to capture images of different colors from up to four distinct focal planes.
To close this section, we provide some concrete numbers regarding the size of the image data set on which our examples have been based. These numbers are representative of the actual image data we analyzed in [16] and [17] using our prototype implementation of MIATool, and will illustrate the significant advantage gained with the use of image pointer arrays in terms of RAM usage.
Suppose that 6000 16-bit grayscale images, each of 420×400 pixels, are acquired by the camera which captures the fluorescence of the green dye from the “bottom” focal plane. This amounts to approximately 328 kilobytes (KB) per image, and approximately 1.88 gigabytes (GB) for all 6000 images. At the same time, suppose 4000 16-bit grayscale images, each of 420×420 pixels, are acquired by the camera that captures the fluorescence of the red dye from the “bottom” focal plane. This equates to approximately 345 KB per image, and approximately 1.31 GB for all 4000 images. The total size for all 10000 images from the “bottom” focal plane is then approximately 3.19 GB. Now suppose that the same amounts of data are acquired by the two cameras that image the “top” focal plane. The size of the entire set of 20000 images then becomes approximately 6.38 GB.
Given a conventional personal computer with 1, 2, or even 4 GB of RAM, software applications that require the loading of all 6.38 GB of images into RAM would either not be able to handle this data set, or at a minimum cause a substantial slowdown of the application itself and the rest of the computer system. Using the pointer array representation of MIATool, however, an ImageSet containing a pointer array that references the 20000 images would require a significantly smaller amount of RAM. In MIATool V1.1, for example, an ImageSet containing a 3D array of pointers to the 20000 images would only take up approximately 145 megabytes (MB) of RAM. This 3D array is constructed by stacking two copies of the type of 2D pointer array depicted in Fig. 3(a) along a third dimension. One copy contains pointers that reference the 10000 images from the “bottom” focal plane, and the other contains pointers that reference the 10000 images from the “top” focal plane.
We note that the significant savings in RAM provided by an ImageSet will translate to similar savings in disk space. Therefore, instead of physically replicating images to form various arrangements such as those illustrated in Fig. 3, the saving of ImageSets containing pointer arrangements could save a nontrivial amount of disk space. Also, it is important to point out that, regardless of how large or small the data set, a software application that requires all images to be of the same size would not readily support the analysis of the type of heterogeneous data described.
IV. Corresponding arrangements of processing settings, metadata, and analytical results
In Section III, we illustrated MIATool’s use of different image pointer arrays to facilitate the different processing tasks that comprise a data analysis. Here, we describe the means by which the framework supports the execution of a given task. In general, a task may require that each of the images referenced by its pointer array be processed differently. Moreover, the processing of the referenced images may potentially be based on metadata that is specific to each image, and may produce analytical results that need to be maintained on a per-image basis. Specified as a crucial design consideration in Section I-A, MIATool supports this processing flexibility by making use of arrays of processing settings, metadata, and analytical results which are constructed in parallel to an image pointer array. In what follows in this section, we give our main focus to arrays of processing settings, but end with a discussion on arrays of metadata and analytical results which can conceptually be seen as simple special cases of arrays of processing settings.
Given an N-dimensional image pointer array, one should on the one hand be able to process each of the referenced images differently, and on the other hand have the option to process all referenced images in a uniform way. In order to accommodate both extremes and all permutations in between, MIATool uses arrays corresponding in size and dimension to a pointer array to support the flexible processing of its referenced images. More specifically, an image referenced by a pointer belonging to element (x1, x2,…, xN) of a pointer array is processed according to the settings that are stored in element (x1, x2,…, xN) of a corresponding array. With such a parallel design, custom processing is possible on a per-image basis, while at the same time uniform processing for all or subsets of the images can be achieved by simply specifying identical settings in the appropriate elements of a corresponding array.
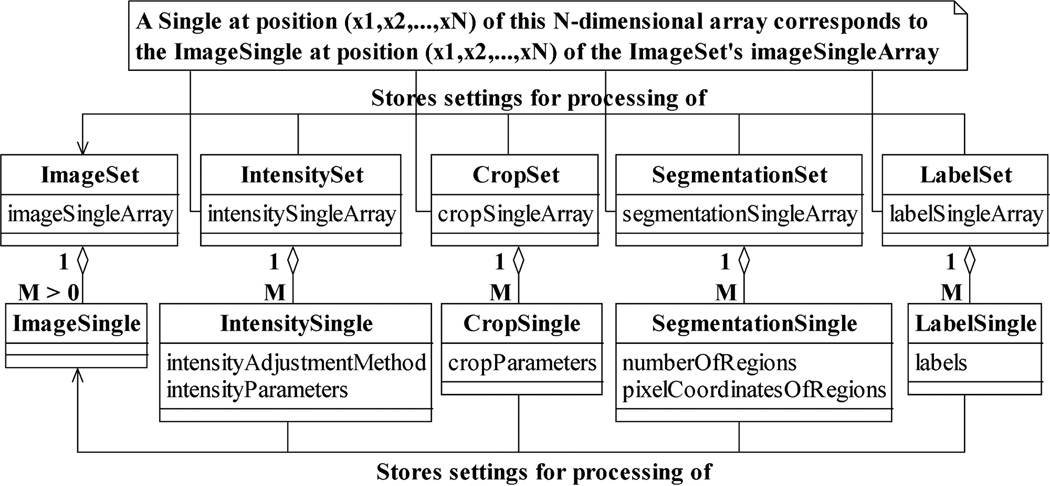
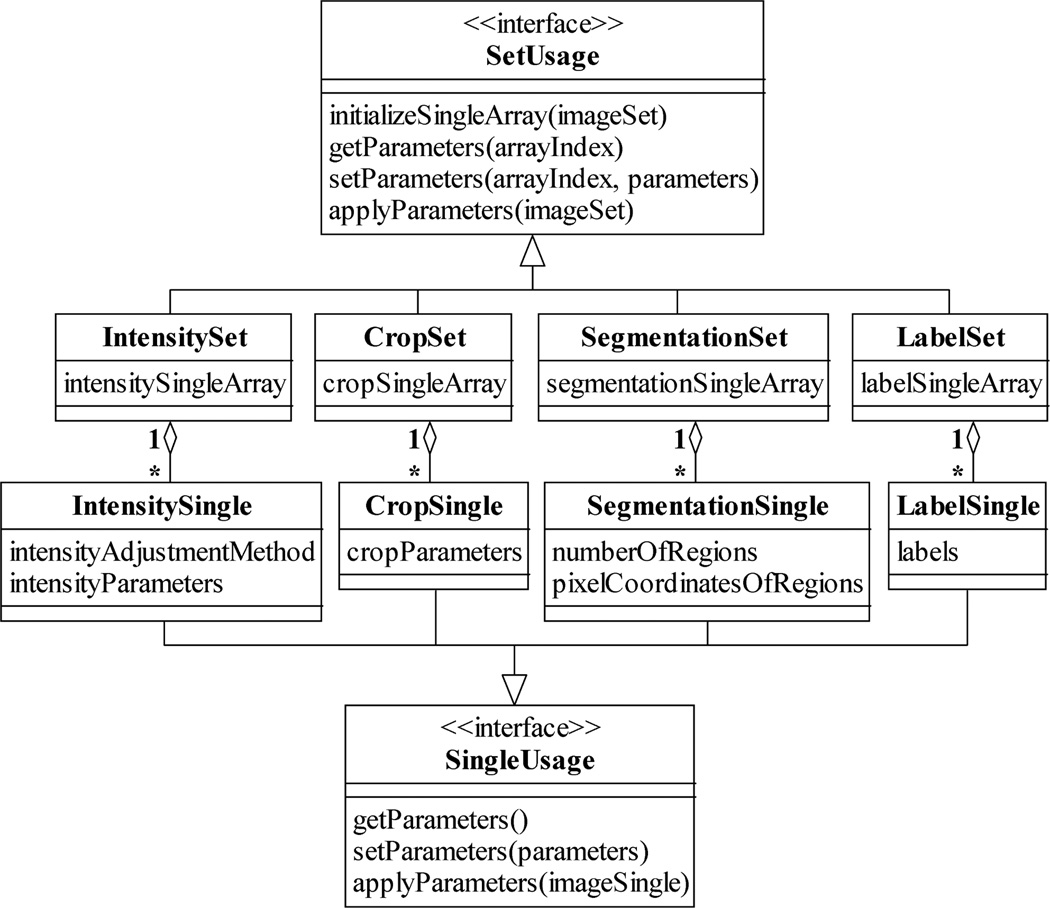
Different corresponding arrays of processing settings are used for different processing tasks. Just as an image pointer array is stored in and managed by an ImageSet, these corresponding arrays are kept in and handled by various Set classes, as illustrated in Fig. 4. Arrays that store intensity adjustment and crop settings, for example, reside in the classes IntensitySet and CropSet. Moreover, analogous to image pointers being instances of the ImageSingle class, each element of a corresponding array is an instance of a Single class that contains the processing settings for a specific image. As shown in Fig. 4, an IntensitySingle, for instance, keeps information such as the intensity adjustment method to use and the values of the associated adjustment parameters. Similarly, a CropSingle stores parameters whose values describe the region of the image to retain while the rest is trimmed.
Fig. 4.
Examples of Set and Single classes for the processing of ImageSets and ImageSingles. Analogous to the ImageSet-ImageSingle relationship, a Set contains an N-dimensional array of its associated type of Singles. A Single at a particular position in the N-dimensional array stores the settings for the processing of the ImageSingle located at the same position in the array contained in a corresponding ImageSet. An IntensitySingle and a CropSingle contain, respectively, the settings for the intensity adjustment and cropping of the image referenced by a corresponding ImageSingle. Similarly, a SegmentationSingle and a LabelSingle store, respectively, settings for the partitioning and labeling of the referenced image.
Importantly, Fig. 5 shows that all Set classes support a common repertoire of operations specified by the interface SetUsage. In addition to operations for the storing and retrieving of processing settings to and from the elements of the array that is managed by a Set, this interface requires a Set class to implement two important operations. Given an image pointer array by way of an ImageSet, the initializeSingleArray operation creates an array of processing settings that corresponds in size and dimension to that pointer array, and hence initializes the Set for working with the supplied ImageSet. The applyParameters operation accepts the same ImageSet as input, and processes each of its referenced images according to the settings stored in the Set. The applyParameters operation shows that Sets do not only store the processing settings, but also carry out the actual tasks of intensity adjustment, cropping, segmentation, labeling, etc. In an analogous manner, all Single classes implement the interface SingleUsage, which specifies analogous operations at the level of a single image.
Fig. 5.
The SetUsage and SingleUsage interfaces. The SetUsage interface is supported by all Sets and specifies standard Set operations such as the creation of an N-dimensional array of Singles, the assignment and the retrieval of processing settings to and from a particular Single, and the actual processing of the images referenced by a corresponding ImageSet according to the stored processing settings. Analogously, the SingleUsage interface is implemented by all Singles and specifies similar standard operations at the level of a Single.
In general, the output of the applyParameters operation of a Set is a new ImageSet containing an array of pointers to the (e.g., intensity-adjusted, cropped, segmented, or labeled) images generated by its processing. These resulting images can again reside either in RAM or on the hard disk, and if need be, they can be subjected to further processing by the next Set in a sequence of processing operations. In this model of processing, actual images and an ImageSet that refers to them are generated at each intermediate processing step. While this is a good way to proceed in cases where images created at intermediate steps are desired, in other cases it could present problems when there is insufficient RAM or disk space.
An alternative model of processing is therefore to iterate through the initial array of pointers and process one referenced image at a time from beginning to finish. In this way, only a single set of final images is created at the end of the processing sequence. This alternative approach can be readily realized by carrying out the processing at the level of Singles instead of Sets. That is, given an image (i.e., an ImageSingle) from the initial ImageSet, we can process it from beginning to end by invoking in proper order the applyParameters operation on each of its corresponding Singles. The two models of processing need not be mutually exclusive, and depending on the available hardware resources and the intermediate results that are desired, they can be applied as appropriate to different segments of a sequence of processing steps.
In addition to processing flexibility, the use of corresponding arrays provides at least three further advantages. First, the parallel design allows the straightforward propagation of processing settings when one image pointer array is derived from another as illustrated by the sequence of analysis tasks described in Section III. Due to the one-to-one correspondence between a pointer array and its associated arrays of processing settings, the arrays of settings corresponding to an existing pointer array can be manipulated in exactly the same way as the pointer array to arrive at arrays of settings that not only correspond in size and dimension to a derived pointer array, but also retain the same processing settings for the images referenced by the derived pointer array. This carryover of processing settings is useful since in many situations, preexisting settings apply just as well to a derived image pointer array.
Second, by adhering to the paradigm of Sets and their associated Singles, the design criterion of software extensibility is accounted for as new image processing capabilities can be incorporated into MIATool with relative ease in the form of new types of Sets and Singles. As we will see in Section V, this paradigm also forms the basis for the extensibility of the image display and the interactive, on-the-fly processing framework adopted by the MIATool viewer and the various image processing tools. Third, the saving of Sets and Singles provides a practically useful alternative to the saving of the images that result from the processing. A Single that contains processing settings is typically much smaller than the image that results from the processing, and therefore occupies significantly less disk space. This is another way by which MIATool addresses the design consideration of accommodating the analysis of large image data sets. Furthermore, the saved settings readily provide a record of the processing that can be used at a later time to generate the desired images.
Besides processing settings, corresponding arrays can be used for the storage of information such as the metadata and the analytical results that are associated with the images referenced by a pointer array. For example, new types of Sets and Singles can be created to maintain for each referenced image of a pointer array metadata such as its acquisition time stamp, focus level, and color (i.e., wavelength). Analytical results such as the objects of interest identified in a tracking application and their computed attributes (e.g., size, centroid, fluorescence intensity, etc.) can also be stored on a per-image basis using the Set and Single paradigm. Note that since these Sets and Singles are used purely for information maintenance and do not perform any processing on images, the applyParameters operations specified by the SetUsage and SingleUsage interfaces can be trivially defined to either do nothing or to simply return the stored information.
V. Image viewer and processing tools
In Section IV, we discussed MIATool’s use of corresponding arrays (i.e., Sets) of processing settings, metadata, and analytical results for the processing of the images referenced by an array of pointers (i.e., an ImageSet). There, the nature of the processing described assumes that the precise processing specifications are already known, and that therefore no visual feedback or user interactivity is necessary during the execution of an analysis task. For many types of image processing, however, the ability to visualize the images as well as the changes made to the images is desirable if not crucial. A simple intensity adjustment, for example, often requires the user to manually try out and visually assess different adjustment methods and/or intensity settings before deciding on the best choice. In this section, we describe the viewer and tools specified by the MIATool framework to support image visualization and user-interactive, on-the-fly processing with visual feedback.
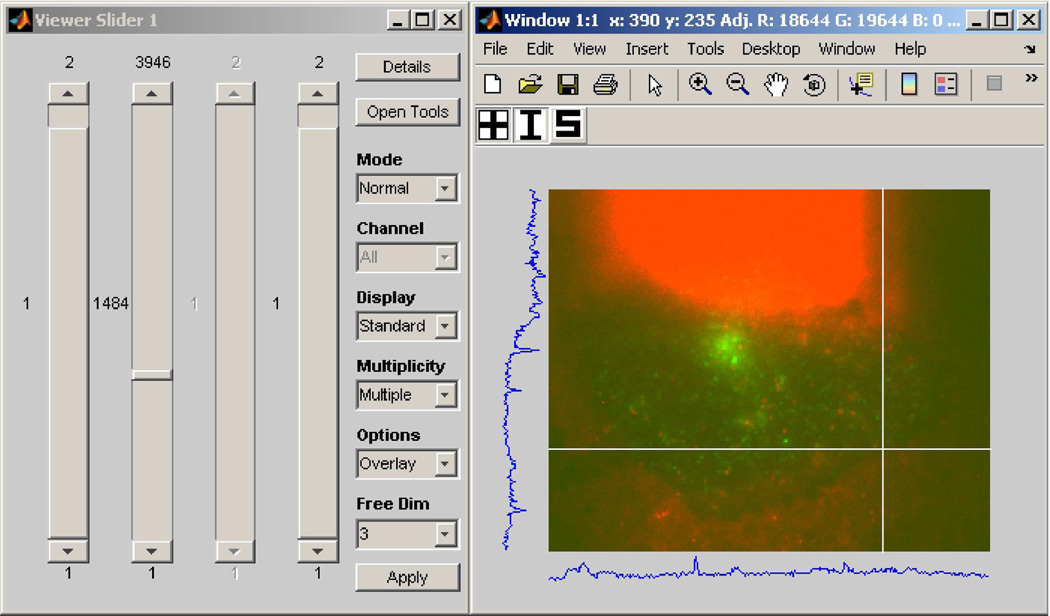
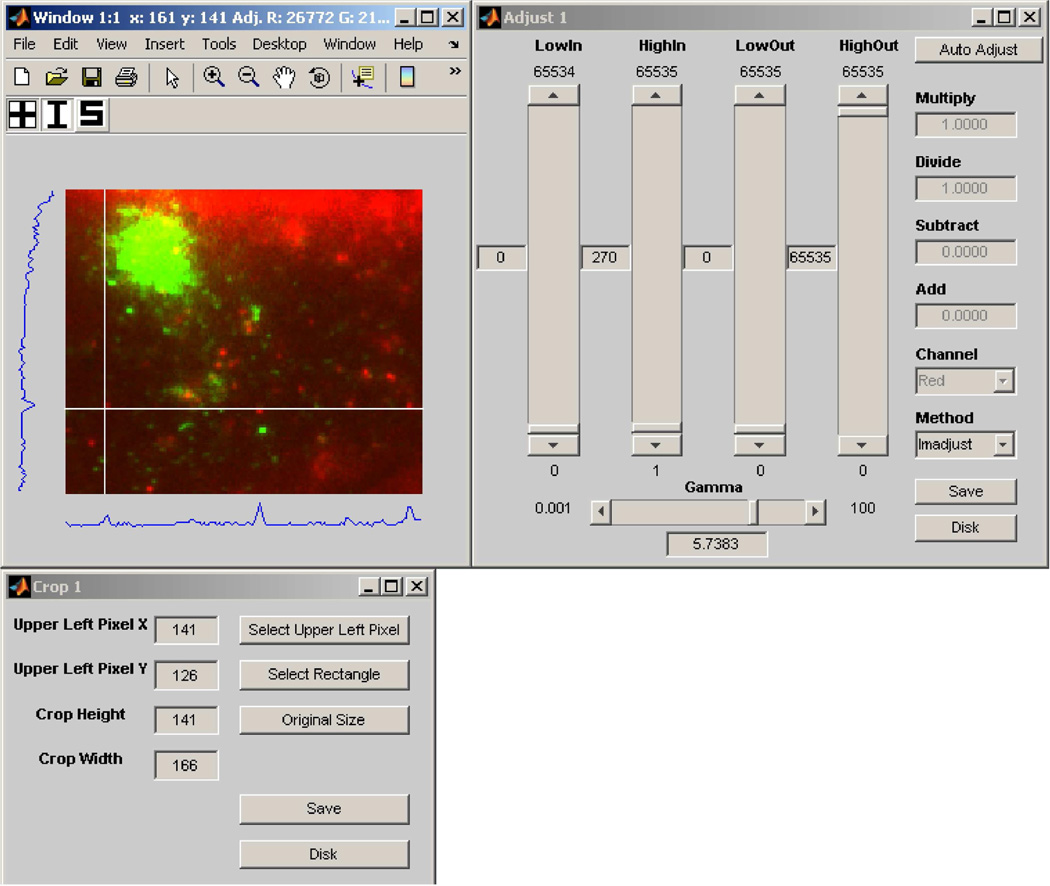
As we alluded to in the discussion of Fig. 3(a), perhaps the most basic of necessities when given an N-dimensional image pointer array is to be able to traverse the array and view the referenced images. To address this requirement, MIATool provides as a basic component of its graphical user interface an image viewer that supports the traversal and display of the images referenced by a pointer array. Since each pointer in an N-dimensional array is uniquely identified by an N-tuple (x1, x2,…, xN), the MIATool viewer allows the selection of a pointer with a set of N controls such as sliders, each specifying the value of a different dimension. The screen capture of Fig. 6, for example, shows an instance of the MIATool V1.1 [10] viewer that has been opened with four sliders for the traversal of a 4D image pointer array.
Fig. 6.
An instance of the MIATool V1.1 viewer that has been opened with four sliders for the traversal of a 4D image pointer array. The array contains replicated pointers that reference a physical data set of 11321 images on the hard disk, acquired using a two-plane multifocal plane microscopy imaging setup. The dimensions of the pointer array are given by 2 (focal planes) × 3946 (time points) × 2 (colors/fluorophores) × 2 (intensity settings). The displayed image is an RGB overlay, formed on the fly, of two grayscale images of a human microvascular endothelial cell acquired at different wavelengths corresponding to QD 655-labeled IgG (red channel) and pHluorin-labeled FcRn (green channel). The two grayscale images were acquired by two cameras that simultaneously imaged the “bottom” focal plane of the two-plane imaging setup.
Upon the selection of a pointer, the MIATool viewer retrieves on the fly the referenced image from RAM or the hard disk and displays it to a window. The currently displayed image is always overwritten with a newly retrieved image. In this way, the viewing of a large image data set which physically resides on disk is made possible without having to first load all the referenced images into RAM, which can often be a problem when the amount of RAM is very limited. The physical image data set displayed by the viewer of Fig. 6, for example, consists of 11321 16-bit grayscale images, each of 320×390 pixels. The size of each image is therefore approximately 244 KB, and that of the entire data set is approximately 2.63 GB. The loading of all 2.63 GB of data into RAM could already prove difficult with a conventional personal computer. However, as we explain next, the 4D image pointer array (i.e., ImageSet) that is loaded in the viewer of Fig. 6 actually references 31568 images by virtue of replicated pointers to the 11321 physical images on the disk. Whereas this 4D ImageSet as implemented in MIATool V1.1 only takes up approximately 231 MB of RAM, an equivalent 4D arrangement of actual replicated images would require around 7.34 GB of RAM.
The set of 11321 physical images was produced by the type of live cell fluorescence imaging experiment we carried out in [17], where multifocal plane microscopy [15] was used to image and track in three dimensions the itineraries of the neonatal Fc receptor (FcRn) and its ligand immunoglobulin G (IgG) in a human microvascular endothelial cell. Similar to the experiment described in Section III, four cameras were used to simultaneously acquire time sequences of images from two distinct focal planes, and the images from each camera were written to the camera’s own separate directory in the order they were acquired. In the “bottom” focal plane, the first camera captured the fluorescence from pHluorin-labeled FcRn, and the second camera captured the fluorescence from quantum dot (QD) 655-labeled IgG. In the “top” focal plane, the third camera captured the fluorescence from monomeric red fluorescence protein (mRFP)-labeled FcRn, and the fourth camera captured the fluorescence from QD 655-labeled IgG. (For more details concerning the experiment, see [17].)
Since the four cameras acquired images at different rates, the resulting data set of 11321 images consists of four directories of image sequences of different lengths. This 2D physical arrangement of images is therefore sorted by camera and sequence number, and the images acquired by the different cameras are not temporally synchronized with one another. However, to properly visualize the trajectories of, and the interactions between FcRn and IgG, we needed to view overlays of temporally synchronized images from each focal plane with different intensity settings. To this end, we made use of the pointer array manipulations illustrated in Fig. 3 to temporally synchronize the images and introduce the necessary dimensions. The resulting 4D pointer array is the 2 (focal planes) × 3946 (time points) × 2 (colors/fluorophores) × 2 (intensity settings) array loaded in the viewer of Fig. 6, and it is similar to the 4D array depicted in Fig. 3(e).
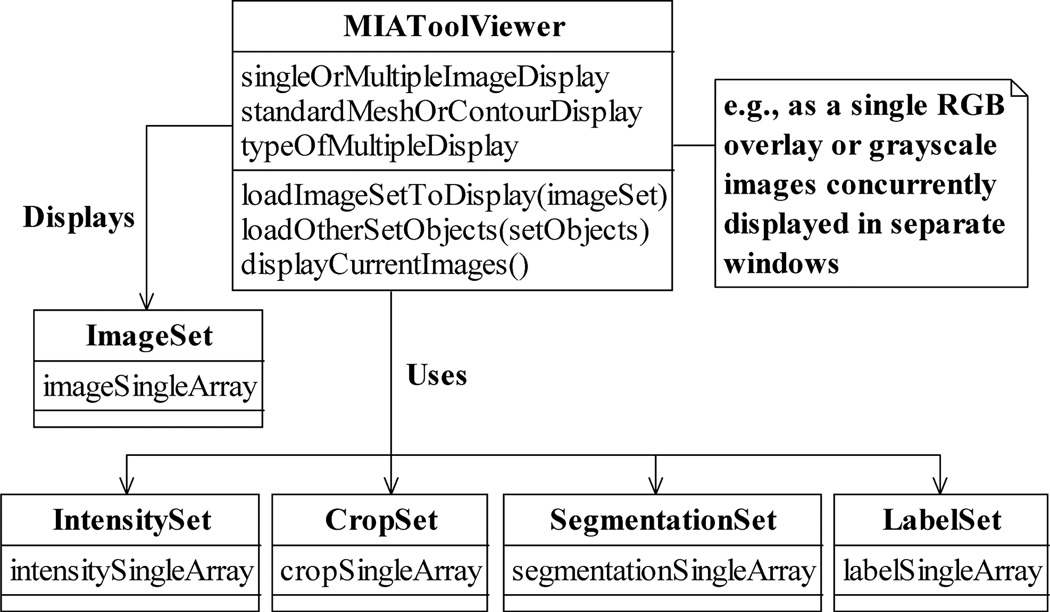
Abstracted by the class MIAToolViewer as shown in Fig. 7, the MIATool viewer supports various display modes that are useful for microscopy image analysis. Besides the standard 2D display, an image can be presented, for example, as a 3D mesh or as one of the color channels in an overlay (as in Fig. 6) with other images. Note that modes such as the overlay display require the selection of multiple images instead of just one, and the mechanism for making such a selection in a viewer is implementation-specific. In MIATool V1.1, for example, viewing of RGB overlays is achieved by scrolling through one dimension of an ImageSet and viewing the images in another dimension in groups of up to three as different color channels of a single RGB image. In Fig. 6, the dimension from which images are taken to form the overlays is indicated by the disabled (i.e., grayed out) slider.
Fig. 7.
The MIAToolViewer class. A MIAToolViewer supports the interactive traversal and viewing of the images referenced by an ImageSet. Optionally, it uses the processing settings contained in the various types of Sets to process (e.g., intensity-adjust, crop, etc.) the images on the fly before displaying them in processed form. A MIAToolViewer supports operations for loading the ImageSet to view and the Sets to use, and for displaying the currently selected images. Different types of displays can be specified. A grayscale image, for example, can be visualized individually as a standard 2D image, a 3D mesh, or a contour plot, or it can be displayed simultaneously with other grayscale images, either in parallel but in its own window, or as a color channel in an overlay.
It is important to point out that the specification of the simultaneous display of multiple images in a MIAToolViewer is a general concept that readily includes the 3D visualization of the images. Just as up to three images can be selected to form an RGB overlay, up to M (where M is an integer greater than 1) images can be selected and displayed as a 3D volume of, for instance, a time lapse sequence or a z-stack. Such visualization options in three dimensions can be useful, if not essential, in the analysis of complex biological structures (see, e.g., [18], [19]).
Importantly, Fig. 7 also illustrates that, given an image pointer array in the form of an ImageSet, the viewer can additionally be supplied with corresponding Sets. By carrying out on-the-fly processing of an image according to its associated settings contained in these Sets, the viewer enables the viewing of processed (e.g., intensity-adjusted, cropped, segmented, etc.) images without requiring that they pre-exist in RAM or on the hard disk. However, in order that the viewer is shielded from the specifics of the various processing tasks, this on-demand processing of images relies on its interaction with the SetUsage and SingleUsage interfaces (Fig. 5). As such, new processing capabilities can be added to the viewer by way of new types of Sets and Singles that support operations which conform to their respective interfaces.
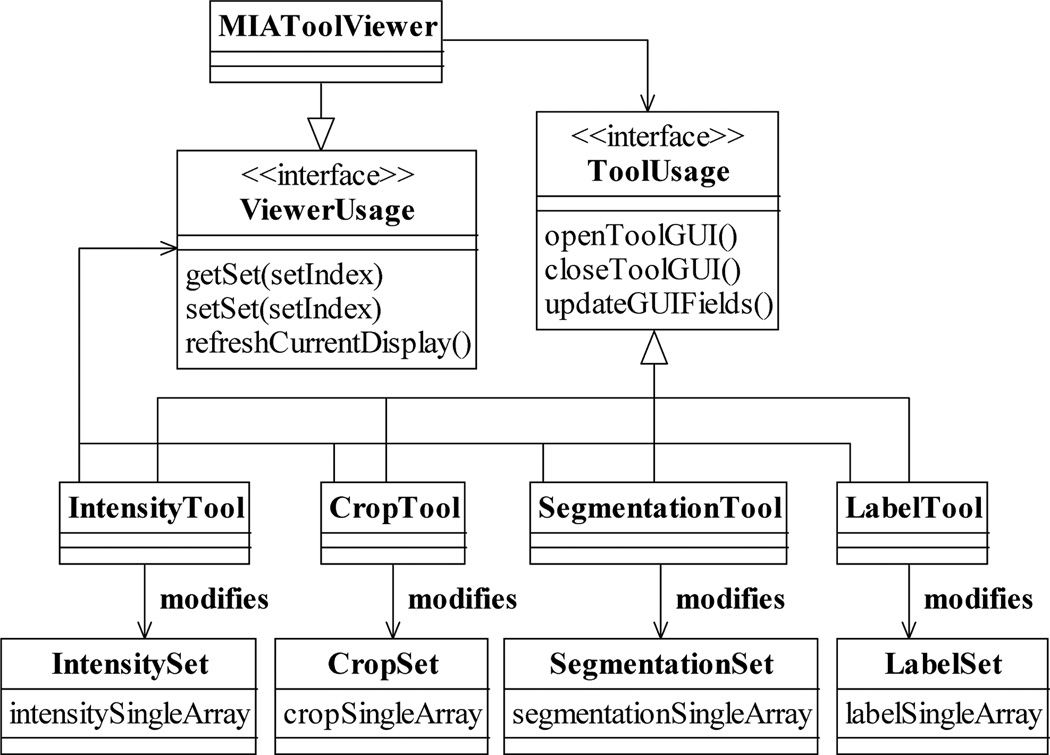
To allow the user to view and to interactively specify and modify processing settings, MIATool provides graphical user interfaces for displaying and manipulating the contents of the various types of Sets. The graphical user interface to each type of Set is managed by a different image processing tool. For example, as shown in Fig. 8, an IntensityTool is responsible for mediating access to an IntensitySet, while a CropTool is the intermediary that facilitates the manipulation of a CropSet. As it is important for the user to receive immediate visual feedback on the effects of the changed settings on the images, an image processing tool is designed to be able to work with the MIATool viewer. Through the interface ViewerUsage (Fig. 8) that is supported by the viewer, a Tool can retrieve a Set from the viewer, return to it a modified version of the Set based on the user input, and “ask” the viewer to re-process and re-display the current image. Any changes due to the modified settings are then reflected immediately in the refreshed display. As an example, Fig. 9 shows the same viewer as in Fig. 6, but displaying an altered version of the same image that has been specified interactively via the intensity adjustment tool and the crop tool shown.
Fig. 8.
Interaction of the MIATool viewer with various image processing tools via the ViewerUsage and the ToolUsage interfaces. The processing settings contained in a Set can be displayed and modified through the graphical user interface provided by a corresponding Tool. The settings in an IntensitySet, for example, can be manipulated using an IntensityTool. Via the ViewerUsage interface supported by the MIATool viewer, any type of Tool can retrieve its corresponding Set, return a potentially modified version of the Set, and for immediate visual feedback request the viewer to re-display the current image which may have been altered by the changed settings. On the other hand, all types of Tools implement the ToolUsage interface which allows the viewer to, for example, open and close a Tool, and to request a Tool to update its settings display whenever a new current image is selected.
Fig. 9.
An instance of the MIATool V1.1 intensity adjustment tool (upper right panel) and an instance of the crop tool (lower left panel) that have been opened for the display and modification of, respectively, an IntensitySet and a CropSet that are loaded in an instance of the MIATool viewer (upper left panel). The viewer instance is the same as the one in Fig. 6 and is displaying a version of the same image that has been intensity-adjusted and cropped on the fly according to altered intensity and crop settings specified via the two tools. The settings displayed in the tools reflect that of the modified image currently displayed.
Conversely, all image processing tools implement a common interface ToolUsage (Fig. 8) which is used by the MIATool viewer. Relying on the operations specified by this interface, the viewer can, for example, open and close the graphical user interface that is provided by a Tool without knowing the Tool’s implementation details. Through this interface, it can also request the various Tools to display the processing settings that correspond to the currently displayed image (e.g., the settings displayed in the intensity adjustment tool and the crop tool of Fig. 9 reflect that of the displayed image). This capability is essential as the viewer uses it to ensure that the information reported by the tool graphical user interfaces stays updated whenever a new current image is selected by the user. Importantly, by adhering to the ToolUsage and ViewerUsage interfaces, the design criterion of software extensibility specified in Section I-A is also fulfilled for the visual and interactive component of MIATool as viewer-compatible image processing tools can be created with relative ease to support new types of Sets and Singles.
Along with the Sets they modify, the image processing tools we have discussed thus far constitute a simple and extensible means of supporting processing on a per-image basis. While it makes sense to realize many kinds of image processing in this manner, there is a different category of processing that operates on multiple images at a time. Just as 3D visualization displays multiple images together in one form or another, 3D processing such as movie making and particle tracking operates on multiple images at a time. Analogous to how 3D visualization can be realized, an implementation of the MIATool framework can take advantage of the MIATool viewer’s multiple image selection feature to create 3D processing tools. Internally in our laboratory, for example, a movie making tool and other types of 3D processing tools have been implemented which, analogous to the way RGB overlays are displayed in MIATool V1.1, operate on images along a particular dimension of an image pointer array.
VI. Storage Management of Images and Associated Information
In Section III, we gave practical examples of microscopy image analysis which illustrate that different image pointer arrays (ImageSets) may be employed for performing various analysis tasks on images from the same physical data set (Fig. 3). In Section IV, we discussed corresponding arrays of settings of various types (IntensitySets, CropSets, etc.), as well as arrays of metadata and analytical results, which may be associated with a given image pointer array. Consequently, a physical set of images in RAM or on the hard disk can be associated with several ImageSets, each of which can in turn be associated with several corresponding Sets. All things combined, a physical image data set can potentially be associated with many ImageSets and corresponding Sets of different types. Even more Sets could be involved if, for example, multiple Sets of the same type are associated with the same image pointer array. One can imagine, for instance, the use of multiple CropSets to define different regions of interest within the referenced images.
In addition to being potentially large in quantity, the Sets associated with a physical data set are related to one another in different ways. For example, while all ImageSets are “peers” in the sense that they represent independent logical data sets derived from the same physical image data, a given IntensitySet is “attached” to the particular ImageSet with which it is associated.
Due to the potentially large amount of differently-related information that can be associated with a physical image data set, a storage management mechanism that provides organization is needed as pointed out in Section I-A as a software design criterion. That is, not only is it important for this mechanism to group data and information that are related, it is essential that it organizes them in a way that encodes their relationships. To this end, MIATool employs a manager which enforces storage in a hierarchical directory structure to help with the organization of a physical image data set with the potentially many and differently-related ImageSets and corresponding Sets that are used for its analysis. The hierarchical directory structure importantly allows the manager to capture the relationships between the various ImageSets and corresponding Sets, and it can be applied to both storage in RAM and storage on the hard disk.
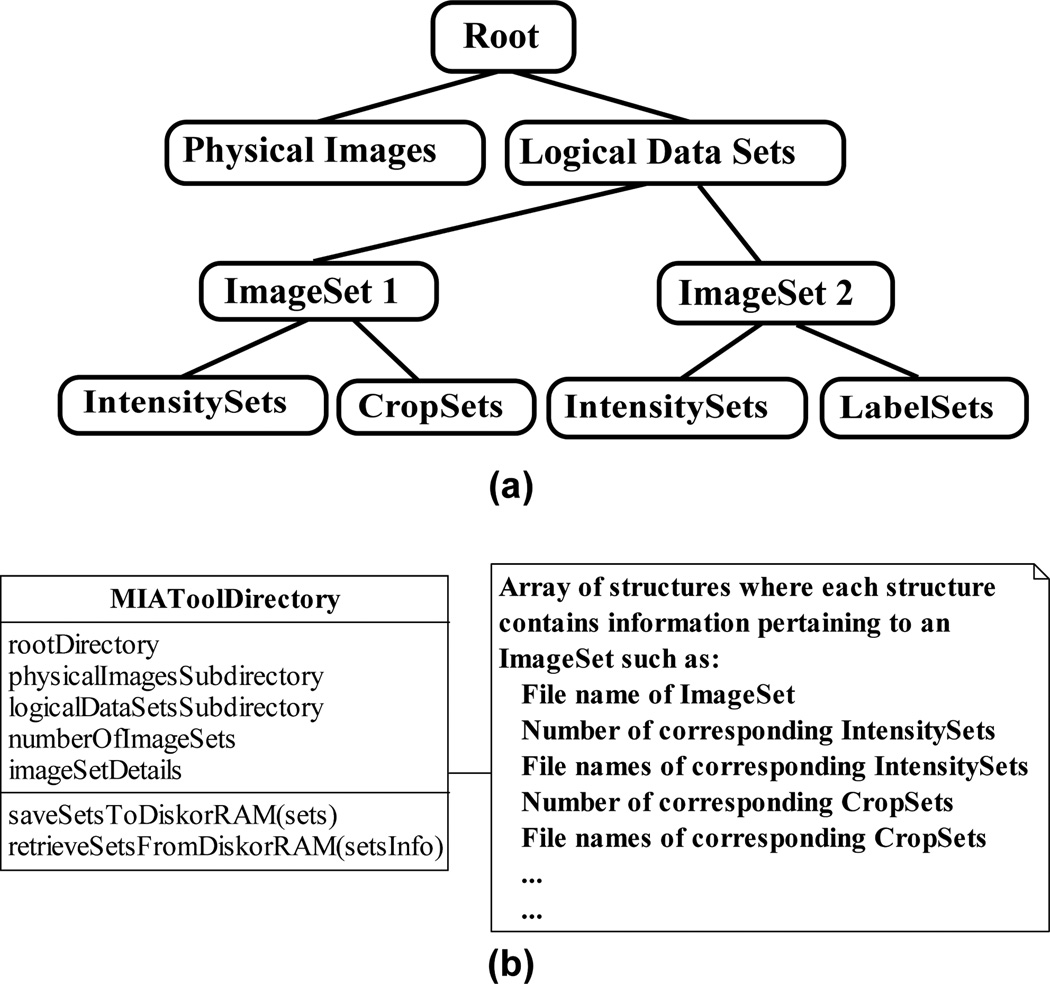
As illustrated in Fig. 10(a), the MIATool storage manager stores all ImageSets and their corresponding Sets in the same directory as the physical image data with which they are associated, but under a subdirectory structure of their own to denote a clear separation between the physical images and the logical data sets used to analyze them. Within this subdirectory, each ImageSet is given its own subdirectory, underneath which all of its corresponding IntensitySets, CropSets, etc., are grouped by type and stored in separate subdirectories of their own. For storage on disk, this hierarchy of directories is literally created on the hard disk. For storage in RAM, however, the implemented directory structure would only be logical in nature.
Fig. 10.
MIATool’s image and Set storage management. (a) Sketch of a representative hierarchical directory structure used by the MIATool storage manager. Underneath a root directory are two subdirectories, one containing the physical set of images and the other the logical data sets used to analyze those images. Within the latter subdirectory, each logical data set (i.e., ImageSet) occupies its own subdirectory. Underneath each ImageSet subdirectory are subdirectories which store the Sets (e.g., IntensitySets, CropSets, etc.) that correspond to that ImageSet. (b) The storage manager abstracted by the MIAToolDirectory class. A MIAToolDirectory manages a directory structure like the one depicted in (a). It keeps track of the location of the root directory and the names of all of its subdirectories, and maintains information such as the number of saved ImageSets, their file names, and similar details pertaining to any saved Sets corresponding to each ImageSet. A MIAToolDirectory also supports operations for the saving and retrieval of the various Sets to and from the directory structure it manages.
The MIATool storage manager is abstracted by the class MIAToolDirectory, diagrammed in Fig. 10(b). An instance of MIAToolDirectory acts as a table of contents for the directory structure it manages. It keeps track of information such as the location of the top level directory and the names of the subdirectories that contain the physical image data and the ImageSets. It also records information such as the number of saved ImageSets and their file names, and maintains details such as the number and file names of corresponding Sets of each type that are associated with each ImageSet. (Note that when the storage is in RAM, details such as directory and file names are still relevant as they provide a means for uniquely identifying each ImageSet and its associated Sets.) Importantly, a MIAToolDirectory provides operations for the saving and the retrieval of ImageSets and the various Sets to and from its managed directory structure. These operations ensure that ImageSets and their associated Sets are saved to and retrieved from the correct locations within the hierarchy of directories, and that the information contained in a MIAToolDirectory is updated properly (e.g., that an appropriate counter is incremented when a new Set is saved).
Though most straightforwardly interpreted and implemented as a directory structure that resides on a single hard drive of a single computer, it is important to note that the hierarchical structure managed by a MIAToolDirectory can be realized as one that spans multiple networked computers running potentially different operating systems. As long as all the images and all the associated ImageSets and their corresponding Sets are uniquely identifiable and retrievable across the network and the various platforms, a MIAToolDirectory can be implemented that manages subdirectories of images, ImageSets, and other Sets that reside on different computers. A MIAToolDirectory implementation that supports such network-spanning, cross-platform image data sets can be particularly useful for large-scale collaborative projects (see, e.g., [13], [14]).
In a collaborative project, it is also important that different users are able to access the same image data set simultaneously, and yet manipulate it differently. This sharing of data can help to avoid the replication of the large amounts of data that are especially typical of large-scale collaborations. To this end, read-only image data can be stored on shared drives across the network, such that multiple users can access the images at the same time, but are not able to overwrite them. Given a shared data set, two general approaches can be used to support the analyses performed by the users. With either approach, each user can create and work with his or her own ImageSets that reference the same shared images. However, the two schemes differ in the way the access to the shared images and the storage of results are realized.
In the first approach, each user has his or her own storage manager (i.e., MIAToolDirectory) through which he or she accesses the shared images and saves the results of analysis. In this scenario, each user’s own ImageSets and corresponding Sets are saved under his or her own directory structure. This approach avoids the sharing of a storage manager by the users, but lacks a central mechanism that keeps track of the storage locations of the results of all analyses that are associated with the shared image data. (Note that since they are read-only, it is possible to have multiple storage managers that provide potentially concurrent access to the shared images.)
In the second approach, all users go through a shared storage manager to access the shared images and save their results. In this scheme, each user’s ImageSets and corresponding Sets are saved under a single central directory structure. The single storage manager adopted by this approach provides a means to centrally locate the results of all analyses that are performed on the shared image data. However, while the storage manager can allow concurrent access to the read-only images, the saving of the users’ analysis results must be done in a sequential way to ensure that the manager always has accurate knowledge of the contents of the central directory structure.
VII. Conclusion
We have described the MIATool software framework which has been built based on several design considerations pertaining to the analysis of complex image data sets produced by modern optical microscopy experiments. A central design criterion is support for the use of different arrangements of a set of images to facilitate the execution of the different processing tasks that comprise a microscopy data analysis. To this end, MIATool supports data analysis that is based on logical image arrangements in the form of arrays of pointers to the physical images. These image pointer arrays can be of arbitrary size and dimension, thus allowing MIATool to accommodate analysis tasks with disparate requirements.
The use of image pointer arrays also allows MIATool to support the storage and analysis of large image data sets, and thereby address another important software design consideration. Pointer arrays are typically significantly smaller in size than the sets of images they reference. Therefore, by enabling the realization, manipulation, and storage of different image arrangements without the need to replicate and shuffle the actual images in their physical storage medium, pointer arrays make for the space-efficient usage of RAM and the hard disk, and naturally permit MIATool to handle data sets containing large numbers of images. An additional advantage of using a pointer array is that its pointers can refer to images of different sizes and file formats. Consequently, MIATool can easily support sets of different-sized images of possibly different file formats, and hence account for the data heterogeneity design consideration.
To address the design criterion of flexibility in processing, the idea of image pointer arrays is complemented by corresponding arrays of processing settings, metadata, and analytical results which provide the ability to perform differential processing on a per-image basis. Importantly, the construction of these corresponding arrays is described by a simple paradigm that, when adhered to, allows the relatively easy incorporation of new image processing capabilities. A crucial design consideration, the idea of software extensibility also plays an important role in the design of MIATool’s image viewer and processing tools. The viewer supports the visualization of the images referenced by a multi-dimensional image pointer array, while the tools support their interactive, on-the-fly processing via the modification of the processing settings stored in corresponding arrays. By specifying the viewer and the processing tools to interact through well-defined interfaces, the framework allows the straightforward addition of new viewer-compatible processing tools.
Lastly, in accordance with the storage management design criterion, MIATool specifies a storage manager which enforces, either in RAM or on the hard disk, the association of a physical image data set with the pointer and corresponding arrays that are used for its analysis. Importantly, this manager plays an organizing role in using a hierarchical directory structure to maintain the relationships among the various arrays.
The MIATool framework and its current implementation [10] have been developed over the course of several years based on design elements we have found to be essential for working with microscopy image data. In our laboratory, it has been, and continues to be, employed for projects of varying sophistication. Taken together, we find that the various features of MIATool make it a suitable software framework for a research environment where microscopy imaging experiments produce constantly evolving data analysis requirements.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (R01 GM071048, R01 AI050747, R01 AI039167, and R01 GM085575).
Biographies

Jerry Chao received the B.S. and M.S. degrees in computer science from the University of Texas at Dallas, Richardson, in 2000 and 2002, respectively. He is currently working toward the Ph.D. degree in the Department of Electrical Engineering at the same institution.
From 2003 to 2005, he developed software for microscopy image acquisition and analysis at the University of Texas Southwestern Medical Center, Dallas. He is currently a Research Assistant in the Department of Electrical Engineering, University of Texas at Dallas, Richardson. His research interests include image and signal processing for cellular microscopy and the development of software for bioengineering applications.
Mr. Chao is a Student Member of the Biophysical Society.

E. Sally Ward received the Ph.D. degree from the Department of Biochemistry, Cambridge University, Cambridge, U.K., in 1985.
From 1985 to 1987, she was a Research Fellow at Gonville and Caius College while working at the Department of Biochemistry, Cambridge University. From 1988 to 1990, she held the Stanley Elmore Senior Research Fellowship at Sidney Sussex College and carried out research at the MRC Laboratory of Molecular Biology, Cambridge. In 1990, she joined the University of Texas Southwestern Medical Center, Dallas, as an Assistant Professor. Since 2002, she has been a Professor in the Department of Immunology at the same institution, and currently holds the Paul and Betty Meek-FINA Professorship in Molecular Immunology. Her research interests include antibody engineering, molecular mechanisms that lead to autoimmune disease, questions related to the in vivo dynamics of antibodies, and the use of microscopy techniques for the study of antibody trafficking in cells.

Raimund J. Ober (S’87-M’87-SM’95) received the Ph.D. degree in engineering from Cambridge University, Cambridge, U.K., in 1987.
From 1987 to 1990, he was a Research Fellow at Girton College and the Engineering Department, Cambridge University. In 1990, he joined the University of Texas at Dallas, Richardson, where he is currently a Professor with the Department of Electrical Engineering. He is also Adjunct Professor at the University of Texas Southwestern Medical Center, Dallas. He is an Associate Editor of Multidimensional Systems and Signal Processing and Mathematics of Control, Signals, and Systems, and a past Associate Editor of IEEE Transactions on Circuits and Systems and Systems and Control Letters. His research interests include the development of microscopy techniques for cellular investigations, in particular at the single molecule level, the study of cellular trafficking pathways using microscopy investigations, and signal/image processing of bioengineering data.
Contributor Information
Jerry Chao, Department of Electrical Engineering, University of Texas at Dallas, Richardson, TX 75080 USA, and also with the Department of Immunology, University of Texas Southwestern Medical Center, Dallas, TX 75390 USA (jcscy@utdallas.edu)..
E. Sally Ward, Department of Immunology, University of Texas Southwestern Medical Center, Dallas, TX 75390 USA (sally.ward@utsouthwestern.edu)..
Raimund J. Ober, Department of Electrical Engineering, University of Texas at Dallas, Richardson, TX 75080 USA, and also with the Department of Immunology, University of Texas Southwestern Medical Center, Dallas, TX 75390 USA (ober@utdallas.edu)..
References
- 1.Lacey AJ, editor. Light Microscopy in Biology: A Practical Approach. 2nd ed. Oxford, UK: Oxford University Press; 1999. [Google Scholar]
- 2.Inoué S, Spring KR. Video Microscopy: The Fundamentals. 2nd ed. New York: Plenum Press; 1997. [Google Scholar]
- 3.Moerner WE. New directions in single-molecule imaging and analysis. Proc. Natl. Acad. Sci. USA. 2007;vol. 104(no. 31):12 596–12 602. doi: 10.1073/pnas.0610081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herman B. Fluorescence Microscopy. 2nd ed. Oxford, UK: BIOS Scientific Publishers; 1998. [Google Scholar]
- 5.Lichtman JW, Conchello J. Fluorescence microscopy. Nat. Methods. 2005;vol. 2(no. 12):910–919. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]
- 6.Swedlow JR, Goldberg I, Brauner E, Sorger PK. Informatics and quantitative analysis in biological imaging. Science. 2003;vol. 300(no. 5616):100–102. doi: 10.1126/science.1082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmed WM, Lenz D, Liu J, Robinson JP, Ghafoor A. XML-based data model and architecture for a knowledge-based grid-enabled problem-solving environment for high-throughput biological imaging. IEEE Trans. Inf. Technol. Biomed. 2008 Mar.vol. 12(no. 2):226–240. doi: 10.1109/TITB.2007.904153. [DOI] [PubMed] [Google Scholar]
- 8.Rasband WS. ImageJ. [Online]. Available: http://rsb.info.nih.gov/ij.
- 9.Clendenon JL, Byars JM, Hyink DP. Image processing software for 3D light microscopy. Nephron Exp. Nephrol. 2006;vol. 103(no. 2):e50–e54. doi: 10.1159/000090616. [DOI] [PubMed] [Google Scholar]
- 10.MIATool V1.1. [Online]. Available: http://www4.utsouthwestern.edu/wardlab/miatool.
- 11.Chao J, Long P, Ward ES, Ober RJ. Design and application of the Microscopy Image Analysis Tool. Proc. IEEE Engineering in Medicine and Biology Workshop; Dallas, TX. 2007. pp. 94–97. [Google Scholar]
- 12.Schmuller J. Sams Teach Yourself UML in 24 Hours. 3rd ed. Indianapolis, IN: Sams Publishing; 2004. [Google Scholar]
- 13.Lichtman JW, Sanes JR. Ome sweet ome: what can the genome tell us about the connectome? Curr. Opin. Neurobiol. 2008;vol. 18(no. 3):346–353. doi: 10.1016/j.conb.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helmstaedter M, Briggman KL, Denk W. 3D structural imaging of the brain with photons and electrons. Curr. Opin. Neurobiol. 2008;vol. 18(no. 6):633–641. doi: 10.1016/j.conb.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Prabhat P, Ram S, Ward ES, Ober RJ. Simultaneous imaging of different focal planes in fluorescence microscopy for the study of cellular dynamics in three dimensions. IEEE Trans. Nanobiosci. 2004;vol. 3(no. 4):237–242. doi: 10.1109/tnb.2004.837899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prabhat P, Gan Z, Chao J, Ram S, Vaccaro C, Gibbons S, Ober RJ, Ward ES. Elucidation of intracellular recycling pathways leading to exocytosis of the Fc receptor, FcRn, by using multifocal plane microscopy. Proc. Natl. Acad. Sci. USA. 2007;vol. 104(no. 14):5889–5894. doi: 10.1073/pnas.0700337104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ram S, Prabhat P, Chao J, Ward ES, Ober RJ. High accuracy 3D quantum dot tracking with multifocal plane microscopy for the study of fast intracellular dynamics in live cells. Biophys. J. 2008;vol. 95(no. 12):6025–6043. doi: 10.1529/biophysj.108.140392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurrus E, Hardy M, Tasdizen T, Fletcher PT, Koshevoy P, Chien C-B, Denk W, Whitaker R. Axon tracking in serial block-face scanning electron microscopy. Med. Image Anal. 2009;vol. 13(no. 1):180–188. doi: 10.1016/j.media.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, Tapia JC, White OL, Lichtman JW. The interscutularis muscle connectome. PLoS Biol. 2009;vol. 7(no. 2):e1000032. doi: 10.1371/journal.pbio.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]