Abstract
The evolutionarily conserved homeotic (Hox) genes are organized in clusters and expressed collinearly to specify body patterning during embryonic development. Chromatin reorganization and decompaction are intimately connected with Hox gene activation. Linker histone H1 plays a key role in facilitating folding of higher order chromatin structure. Previous studies have shown that deletion of three somatic H1 subtypes together leads to embryonic lethality and that H1c/H1d/H1e triple knockout (TKO) embryonic stem cells (ESCs) display bulk chromatin decompaction. To investigate the potential role of H1 and higher order chromatin folding in the regulation of Hox gene expression, we systematically analyzed the expression of all 39 Hox genes in triple H1 null mouse embryos and ESCs by quantitative RT-PCR. Surprisingly, we find that H1 depletion causes significant reduction in the expression of a broad range of Hox genes in embryos and ESCs. To examine if any of the three H1 subtypes (H1c, H1d and H1e) is responsible for decreased expression of Hox gene in triple-H1 null ESCs, we derived and characterized H1c−/−, H1d−/−, and H1e−/− single-H1 null ESCs. We show that deletion of individual H1 subtypes results in down-regulation of specific Hox genes in ESCs. Finally we demonstrate that, in triple-H1- and single-H1- null ESCs, the levels of H3K4 trimethylation (H3K4me3) and H3K27 trimethylation (H3K27me3) were affected at specific Hox genes with decreased expression. Our data demonstrate that marked reduction in total H1 levels causes significant reduction in both expression and the level of active histone mark H3K4me3 at many Hox genes and that individual H1 subtypes may also contribute to the regulation of specific Hox gene expression. We suggest possible mechanisms for such an unexpected role of histone H1 in Hox gene regulation.
Introduction
The Hox genes, encoding a family of evolutionarily conserved transcription factors that contain a DNA binding homeodomain, play fundamental roles in specifying anterior-posterior body patterning during development and are critical for cell fate determination [1]–[3]. The expression levels of Hox genes are tightly controlled throughout embryonic development, and aberrant expression and mutation of Hox genes can lead to body malformations and multiple types of malignancies [4], [5].
Hox genes are organized into genomic clusters and their physical order within the cluster corresponds to their expression order along the anterior-posterior axis. In mammals, there are 39 Hox genes arranged in four genomic clusters of thirteen paralog groups (A-D) [6], which are thought to derive from tandem duplication of ancestral genes [7], [8]. Progressive transition of histone modifications and local chromatin decondensation have been found to associate with sequential expression of Hoxb and Hoxd loci during embryonic development and/or stem cell differentiation [9]–[13]. Hox gene clusters are spatially compartmentalized and the transition in their 3D structure corresponds with the changes of H3K4me3 and H3K27me3 [14]. The temporal collinearity of the order of Hox gene activation along their physical sequence at genomic loci [15], stepwise transition of chromatin status and spatial configuration [9], [14], and the necessity of the cluster organization for full repression of the entire cluster suggest an important role of chromatin structure in regulation of Hox genes [9]-[13]. However, it remains to be determined whether the change of chromatin structure is a contributing factor or a consequence of Hox gene activation.
Linker histone H1 is the major chromatin structural protein involved in folding of chromatin into high order structure. H1 binds to the nucleosome and the linker DNA between nucleosomes to promote compaction of nucleosome arrays [16], [17]. Multiple H1 subtypes exist in mammals, providing additional levels of modulation on chromatin structure and function. Among the 11 mammalian H1 subtypes identified, 5 somatic H1 subtypes (H1a-e) are present in abundance in all dividing and non-dividing cells, whereas the replacement H1 (H10) and the 4 germ cell specific H1s are expressed in differentiating cells and germ cells, respectively [18]. Depletion of three somatic H1 subtypes (H1c, H1d, and H1e) together results in embryonic lethality at midgestation, demonstrating the necessity of H1 for mammalian development [19]. We have previously shown that H1c, H1d, and H1e triple knockout (H1 TKO) embryos and embryonic stem cells (ESCs) have marked reduction of total H1 levels and that H1 TKO ESCs display changes in bulk chromatin, including chromatin decondensation, a decreased nucleosome repeat length, as well as reduced levels of histone modifications H3K27me3 and H4K12Ac [19], [20]. Thus H1 TKO embryos and ESCs offer a unique opportunity to examine how the changes in chromatin structure influence Hox gene expression.
In the present study, we firstly analyzed the expression changes of all Hox genes in H1 TKO embryos and ESCs, and found reduced expression of a distinct set of Hox genes in embryos and ESCs, respectively. Furthermore, by characterizing H1c−/−; H1d−/−; and H1e−/− single-H1 null ESCs established in this study, we showed that individual H1 subtypes regulate specific Hox genes in ESCs. Finally we demonstrated that the levels of H3K4me3 were significantly diminished at the affected Hox genes in H1 TKO- and single-H1 KO- ESCs, whereas H3K27me3 occupancy was modestly increased at specific Hox genes. These results suggest that the marked reduction of H1 levels and decondensation of bulk chromatin cause repression of many Hox genes in embryos and ESCs, which may be in part mediated through individual H1 subtypes as well as changes in H3K4me3 and H3K27me3.
Results
Loss of H1c, H1d and H1e Leads to Decreased Expression of Hox Genes in Embryos and Embryonic Stem Cells
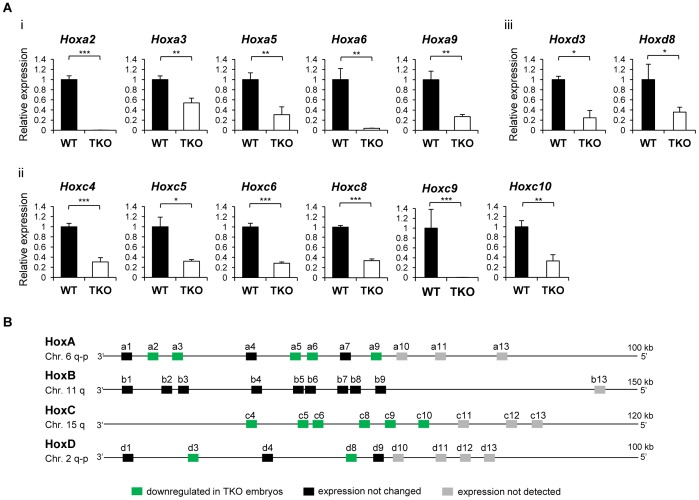
To gain a comprehensive view of the effects histone H1 depletion and changes in bulk chromatin on the regulation of Hox gene clusters, we designed a full set of quantitative reverse-transcription PCR assays (qRT-PCR) to measure the expression levels of all 39 murine Hox genes across the 4 Hox gene clusters in H1 TKO embryos. H1c/H1d/H1e triple heterozygotes were intercrossed to obtain H1 TKO and wild-type (WT) littermate embryos. Most of the H1 TKO embryos display growth retardation and various defects at E9.5 [19]. To minimize the secondary effects caused by broad defects of H1 TKO embryos, we chose to analyze Hox gene expression at E8.5 when H1 TKO embryos with comparable size to WT embryos can be recovered. We selected two littermate pairs of WT and H1 TKO embryos at E8.5, and examined the expression patterns of all 39 Hox genes using the highly sensitive qRT-PCR assays. As expected, most Hox genes were expressed in E8.5 embryos, except the most posterior genes within each cluster (Figure 1). However, surprisingly, many Hox genes were expressed at reduced levels in H1 TKO embryos, including Hoxa2, Hoxa3, Hoxa5, Hoxa6, Hoxa9, Hoxc4, Hoxc5, Hoxc6, Hoxc8, Hoxc9, Hoxc10, Hoxd3, and Hoxd8 (Figure 1). This effect is especially prominent in Hoxa and Hoxc clusters, in which nearly all of the expressed genes were reduced 3-fold or more (Figure 1A). Interestingly, we did not find increased expression among any of the Hox genes (Figure 1B), and none of the Hoxb genes were affected in H1 TKO embryos in comparison with WT embryos.
Figure 1. Reduction of Hox gene expression in H1 TKO embryos.
(A) Relative expression of Hox genes with altered mRNA levels in H1 TKO embryos compared with WT. Down-regulated Hox genes are located in HoxA (i), HoxC (ii), and HoxD (iii) clusters. Expression levels of Hox genes were analyzed by qRT-PCR and normalized over GAPDH and represented as a fold change between H1 TKO and WT embryos at E8.5. *: P<0.05, **: P<0.01, ***: P<0.001. Error bars: S.D. (B) The schematic representation of Hox gene clusters with expression patterns in H1 TKO embryos compared with WT.
The reduction of expression of many Hox genes may cause the growth retardation often observed in H1 TKO embryos at E9.5. However, it remained a formal possibility that the decreased expression of Hox genes in H1 TKO embryos was a result of the slight growth retardation presented in the KO embryos, although the H1 TKO embryos used for this analysis were indistinguishable from their WT and heterozygous littermate controls in size and developmental stage. In order to analyze the effects of H1 on a homogeneous cell population, we gauged the effects of H1 depletion on Hox gene expression in H1 TKO ESCs. Hox genes are repressed by polycomb repressive complexes (PRCs) in ESCs [21]–[26]. Loss of components of either PRC1 or PRC2 in ESCs leads to upregulation of Hox genes, presumably due to respective loss of chromatin compaction and H3K27 trimethylase activity [13], [22], [27]. We have shown previously that H1 TKO ESCs have decondensed local chromatin and reduced levels of H3K27m3 in bulk chromatin [20]. We surmise that these changes may lead to elevated levels of expression of specific Hox genes. Examination of previous expression data from microarray assays showed that the microarray used for hybridization only contained 11 Hox genes, most of which were undetectable in ESCs by the array [20].
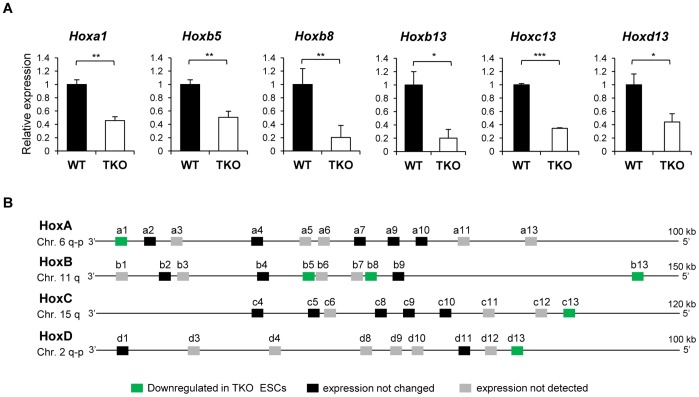
We thus applied the qRT-PCR assays to compare the expression levels of all 39 Hox genes in WT and TKO ESCs. Consistent with the finding that pluripotent ESCs possess a hyperactive transcriptome [28], we detected expression of 21 Hox genes, albeit at low levels, in either or both of WT and H1 TKO ESCs. These genes include Hoxa1, Hoxa2, Hoxa4, Hoxa7, Hoxa9, Hoxa10, Hoxb2, Hoxb4, Hoxb5, Hoxb8, Hoxb9, Hoxb13, Hoxc4, Hoxc5, Hoxc8, Hoxc9, Hoxc10, Hoxc13, Hoxd1, Hoxd11, and Hoxd13 (Figure 2). Unexpectedly, no increased expression in any of the Hox genes was found in H1 TKO ESCs. Instead, the expression levels of 6 Hox genes, Hoxa1, Hoxb5, Hoxb8, Hoxb13, Hoxc13, and Hoxd13, were reduced, with an average of 2–3 fold less in H1 TKO ESCs compared with WT (Figure 2A). Other Hox genes did not show consistent changes in expression by loss of H1c, H1d and H1e in ESCs (Figure 2B).
Figure 2. Decreased expression of Hox genes in H1 TKO ESCs.
(A) Expression analysis of Hox genes in WT and H1 TKO ESCs. Y axis and data normalization are as described in the legend to Figure 1. *: P<0.05, **: P<0.01, ***: P<0.001. Error bars: S.D. (B) Expression patterns of Hox genes in H1 TKO in comparison with WT ESCs.
Specific Regulation of Hox Genes in ESCs by Individual H1 Subtypes
To assess the effects of each of the three deleted somatic H1 subtypes in H1 TKO (H1c, H1d and H1e) on Hox gene expression in ESCs, we established ESCs that are null for only one of these three H1 subtypes. H1c−/−; H1d−/−; and H1e−/− mice develop normally and are fertile [29]. Male and female mice homozygous for each single-H1 deletion were bred, H1c−/−; H1d−/−; and H1e−/− blastocysts were harvested from pregnant female mice at 3.5 day post coitum and their respective single-H1 knockout (KO) ESCs were derived from outgrowth of blastocysts. As shown in metaphase chromosome spreads, the single-H1 KO ESCs had normal karyotypes with 40 chromosomes (Figure S1A) and showed colony morphology typical of undifferentiated ESCs when cultured under conditions promoting self-renewal of ESCs (Figure S1B). They expressed high levels of pluripotency factor OCT4, which is absent in differentiated cells, such as mouse embryonic fibroblasts (MEF) (Figure S1Cii). These single-H1 KO ESCs also had comparable growth rate to WT ESCs (data not shown). Upon differentiation, the single-H1 KO ESCs were able to form embryoid bodies (EB) with characteristic cystic structures and differentiated cell morphologies (Figure S1Ci). As expected, these EBs displayed decreased levels of OCT4 (Figure S1Cii), and increased expression of many differentiation markers, such as AFP, Gata4, T (Brachyury), and FLT1, compared with ESCs (Figure S1Ciii). In addition, teratoma formation analysis indicated that the single-H1 KO ESCs formed typical teratomas containing cells differentiated into all three germ layers after injection into immunodeficient mice (data not shown). These data indicate that any one of these three somatic H1 subtypes is dispensable for self-renewal and differentiation of ESCs.
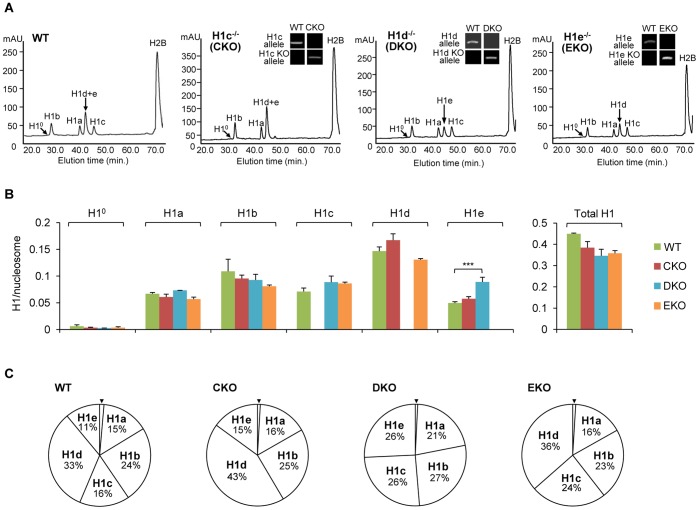
We next analyzed the total H1 levels and composition of H1 subtypes in these single-H1 KO ESCs. HPLC and mass spectrometry analyses of histone extracts from these cells confirmed the lack of the deleted H1 subtype in the respective H1c−/−, H1d−/−, and H1e−/− ESCs (Figure 3A). As described previously and shown here [30], [31], quantification of the peaks of each H1 subtype and H2B allows calculation of the H1 to nucleosome ratio (H1/nuc). Such analysis showed that, except for H1e in H1d-KO ESCs, the absolute levels of the remaining H1 subtypes were largely unchanged in single-H1 null ESCs (Figure 3B), indicating that there was little increase or compensation in the levels of the remaining H1s for the lost H1. As expected, undifferentiated ESCs express negligible amount of H10 (Figure 3A), an H1 subtype enriched in differentiating and non-dividing cells [32], [33]. Although relative proportions of H1 subtypes were altered by single-H1 deletion (Figure 3C), the total H1/nuc ratios of H1c−/−, H1d−/−, and H1e−/− ESCs were comparable with respective values of 0.38, 0.35, and 0.35 (Figure 3B). These ratios were about 25% lower than that of WT ESCs (0.45), but about 50% higher than that of H1 TKO ESCs (0.25) [20]. These single-H1 KO ESCs provide ideal cell resources to ascertain if the effects present in H1 TKO ESCs were caused by any one of the lost H1 subtypes or by the marked reduction in total H1 levels in H1 TKO ESCs.
Figure 3. Generation and reverse-phase HPLC analysis of single-H1 KO ESCs.
(A) RP-HPLC analysis of total histones from WT and the single-H1 KO ESCs. The identity of the histone subtypes is indicated above each peak. mAU, milli-absorbency at 214 nm. Genotype analyses of single-H1 KO ESCs are shown in insets in respective HPLC profiles. (B) The ratios of individual H1 (left) and total H1 (right) to nucleosome for WT and single-H1 KO ESCs. Ratios were determined from the RP-HPLC and mass spectrometry analyses as described in methods. ***: P<0.001 (C) The percentage of each H1 subtype among total H1 histones for WT and single-H1 KO ESCs. % total H1 for H10 (marked with arrowhead) is equal to or less than 1%.
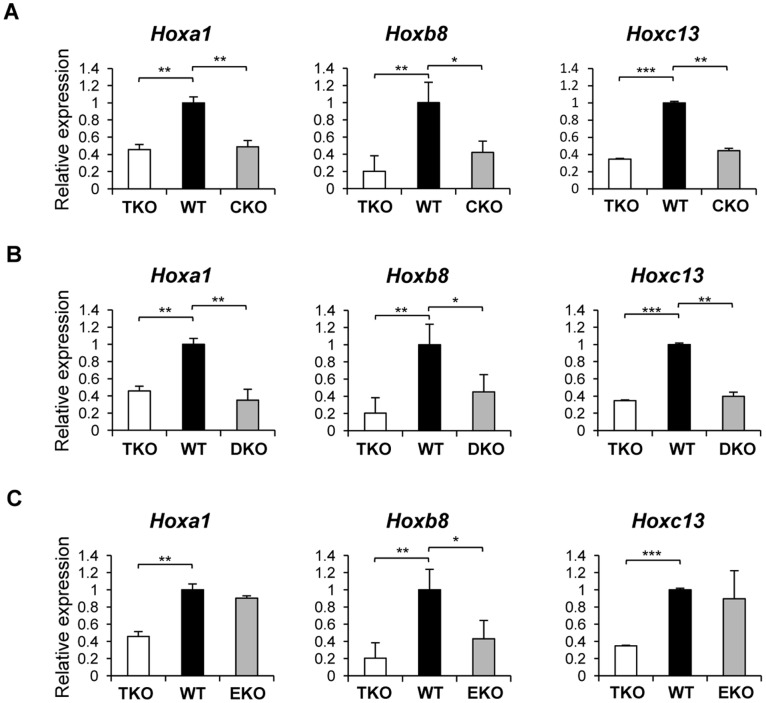
We focused our expression analysis in H1 single KO ESCs on the 6 Hox genes that displayed reduced expression in H1 TKO ESCs. Hoxb8 exhibited decreased expression in all three single-H1 KO ESCs, whereas Hoxa1 and Hoxc13 had reduced expression in H1c−/− and H1d−/−, but not in H1e−/− ESCs compared with WT (Figure 4), indicating that these Hox genes are differentially regulated by H1c, H1d and H1e. Interestingly, the expression levels of these Hox genes in single-H1 KO ESCs were similar to that in H1 TKO (Figure 4), suggesting that these genes may be especially sensitive to alterations of local chromatin structure or H1 to nucleosome stoichiometry. The other three Hox genes did not show consistent expression changes in any of the single-H1 null ESCs, indicating that their expression reduction in H1 TKO ESCs is likely due to the marked reduction of the total H1 levels in TKO cells.
Figure 4. The expression profiles of Hox genes in single-H1 KO ESCs.
Relative expression of Hoxa1, Hoxb8, and Hoxc13 in H1c−/− (A), H1d−/− (B), and H1e−/− (C) ESCs were shown. *: P<0.05, **: P<0.01, ***: P<0.001.
Dynamic Changes of H3K4me3 and H3K27me3 at Affected Hox Genes in H1 TKO ESCs
Trithorax group (TrxG) and polycomb group (PcG) proteins are known to regulate the expression of Hox genes [34], [35]. TrxG mediates H3K4 tri-methylation (H3K4me3), corresponding to transcriptional activation [36], [37], whereas PcG directs H3K27 tri-methylation (H3K27me3), correlating with transcriptional repression [22], [38], [39]. In ESCs, many developmental genes display both H3K4me3 and H3K27me3 marks, a “bivalent” chromatin signature for genes poised for expression and important for maintenance of ESC pluripotency [21], [40].
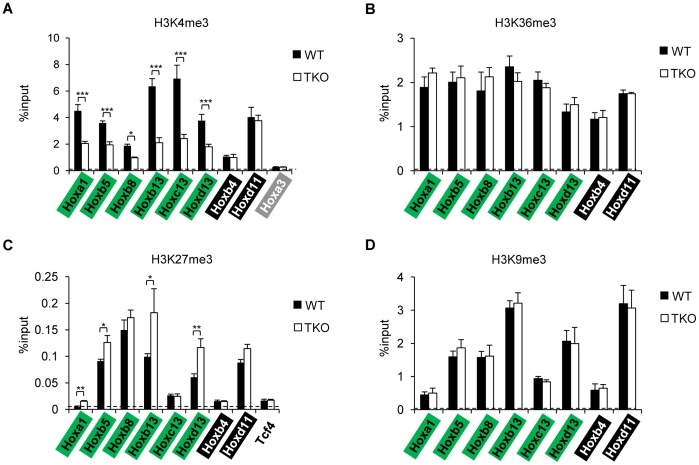
To investigate whether H1 depletion has an impact on bivalent chromatin marks on the 6 Hox genes (Hoxa1, Hoxb5, Hoxb8, Hoxb13, Hoxc13 and Hoxd13) affected in H1 TKO ESCs, we performed quantitative chromatin immunoprecipitation (qChIP) analysis on the promoter regions of these genes as well as two Hox genes (Hoxb4 and Hoxd11) whose expression levels were not altered by triple-H1 deletion. As expected, most Hox genes analyzed displayed the bivalent marks in WT ESCs, with higher levels of H3K4me3 and H3K27me3 compared with Hoxa3 and Tcf4 (Figure 5A&C), which have been shown to harbor minimum levels of respective histone marks [40]. The levels of H3K4me3 were decreased significantly at all six Hox genes affected in H1 TKO ESCs (Figure 5A), but not at Hoxb4 or Hoxd11 loci, suggesting that H1 depletion did not lead to a general reduction of H3K4me3 throughout the Hox gene clusters. The changes in H3K4me3 level at the promoters of the six Hox genes correlated with the reduction of gene expression in H1 TKO ESCs, indicating that the effects of H1 depletion on Hox genes may be mediated through regulating the establishment and/or maintenance of specific H3K4me3 patterns. Increased levels of H3K27me3 were observed at 4 of the 6 Hox genes affected in H1 TKO ESCs (Hoxa1, Hoxb5, Hoxb13, and Hoxd13) (Figure 5C), suggesting that an increase in the H3K27me3 level may also contribute to the reduced expression of these genes. In contrast, H3K36me3, which is enriched at gene bodies of active genes [41], and H3K9me3, which marks heterochromatin and associated with gene repression [42], remained unchanged at all sites after triple H1 depletion (Figure 5B&D), indicating that the effects of marked H1 reduction on H3K4me3 and H3K27me3 (to a less extent) are rather specific. qChIP analysis in single-KO ESCs indicated that H3K4me3 was decreased significantly at the promoters of the Hox genes with reduced expression in the respective H1 KO ESCs, but not at unaffected genes, such as Hoxd11 (Figure S2A). The level of H3K4me3 was not affected by single-H1 deletion at those genes which displayed reduced expression only in H1 TKO ESCs, such as Hoxb5 (Figure S2A). The increase of H3K27me3 occupancy was more restricted, detected only at Hoxa1 promoter in H1c- and H1d- KO ESCs with 2–3 fold over WT (Figure S2B). Taken together, our results demonstrate that H1 depletion leads to dynamic changes of the H3K4me3 and H3K27me3 marks, which may regulate Hox gene expression.
Figure 5. qChIP analysis of histone marks at Hox genes in WT and H1 TKO ESCs.
The levels of H3K4me3 (A), H3K36me3 (B), H3K27me3 (C), and H3K9me3 (D) were analyzed by qChIP. Promoter regions of the indicated Hox genes were assayed, except for (B), for which gene body regions were analyzed. Dashed lines denote the highest signal level of control IgG qChIP. *: P<0.05, **: P<0.01, ***: P<0.001.
Discussion
Hox genes encode a large family of transcription factors crucial for body patterning and positioning along the anterior-posterior axis during animal development [1], [43]. Multiple mechanisms have been shown to regulate the spatial and temporal collinearity of Hox genes, such as the antagonism between PcG and TrxG proteins [34], [35], local chromatin condensation and reorganization [10], [11], [13], spatial configuration or compartmentalization [14], targeting of miRNAs and long non-coding RNAs (lncRNAs) [44], [45]. Chromatin conformation and compaction appear to be key mediators for regulating the expression of Hox gene clusters [10], [11], [13], [14], however, whether changes in chromatin structure have a direct impact on Hox gene expression remains undetermined.
In this study, we have taken advantage of a number of mutants, null in one or several major somatic H1 subtypes, with different levels of reduction in total H1 proteins, to investigate the role of H1, a key component in promoting chromatin compaction, in regulating Hox gene clusters in mouse embryos and ESCs. We find that depletion of three H1 subtypes leads to the transcriptional reduction of a group of Hox genes in embryos and ESCs, and that the reduced expression levels correlate with dynamic changes in H3K4me3 and H3K27me3 marks. This is in contrast to the deletion of PRC1 or PRC2 repressive chromatin complexes, which causes upregulation of specific Hox genes in embryos [46]–[48] or ESCs [13], [22], [24].
We first systematically analyzed the impacts of H1 depletion on expression levels of all 39 Hox genes in mouse embryos. Consistent with previous findings [9], the posterior genes are not detected by qRT-PCR assays in E8.5 embryos. The 13 affected genes include many paralogous Hox gene members (Figure 1B), suggesting a broad effect of H1 on regulation of Hox genes. Hoxa2, expressed in hindbrain and crucial for trigeminal system development [49], [50], is drastically repressed in H1 TKO embryos. The remaining 12 of the 13 Hox genes with reduced expression in H1 TKO embryos are located within paralogous genes Hox3–10, a region important for axial morphology and patterning [1], [51]–[53]. H1 TKO embryos have significant reduction in total H1 levels and die during midgestation [19]. H1 depletion in vivo causes local reductions in chromatin compaction [19], [20]. The finding that all affected Hox genes are down-regulated in H1 TKO embryos is surprising because chromatin decompaction and progressive changes in 3D chromatin architecture coincide with activation of Hox genes during embryonic development [10]–[14] and thus one may expect that H1 depletion would result in up-regulation of certain Hox genes. We believe that the down-regulation of Hox genes is a direct effect due to H1 depletion, and contributes to, rather than merely reflects, the growth retardation observed in a fraction of H1 TKO embryos at a later stage [19]. The E8.5 H1 TKO embryos analyzed in this study did not exhibit obvious phenotypic difference compared with WT littermates. It is noteworthy that H1 depletion in embryos did not lead to changes in expression of any of the Hox genes on the entire Hoxb cluster, which harbors a large intergenic repeat-rich region with a different 3D chromatin structure compared with other Hox clusters [14]. Furthermore, similar to our findings from analyzing H1 TKO embryos, H1 depletion in ESCs does not lead to increased expression in any of the Hox genes, but causes further reduction in the expression of 6 Hox genes. The less prominent effects of H1 depletion on ESCs could be due to the following reasons: 1) ESCs have no or minimum expression of most Hox genes; 2) embryos consist of a more heterogeneous cell population which are likely to have very different bulk and/or local chromatin structure at Hox gene clusters compared with the undifferentiated ESCs. Indeed, embryos at midgestation have a H1/nuc of 0.74 [19], suggesting a more compact chromatin than ESCs with a H1/nuc of 0.45 [20]; and 3) triple-H1 deletion reduces H1/nuc by 0.34 (from 0.74 to 0.40) in embryos, a larger reduction in total H1 levels than the 0.20 (from 0.45 to 0.25) in ESCs [19], [20].
Importantly, we find that the levels of H3K4me3, a chromatin mark catalyzed by TrxG proteins, are decreased at promoters of all affected Hox genes, corresponding to the reduction in gene expression levels of these Hox genes in H1 TKO ESCs. Likewise, the correlation of changes in H3K4me3 and Hox gene expression extends to the single-H1 KO ESCs, suggesting that individual H1 subtypes may also contribute to epigenetic regulation of H3K4me3 at specific Hox genes. The effects of triple-H1 deletion on H3K27me3 are more limited, with mild increase at 4 of the 6 affected genes. We speculate that loss of H1 may lead to changes in occupancy of H3K4me3 methyltransferases/demethylases, and/or affect binding of polycomb complex components to the Hox genes [54], resulting in alterations in the histone H3K4 and H3K27 trimethyl marks. It is especially interesting to note that JARID proteins contain an AT-rich interacting domain (Arid) [55], [56] that preferentially binds to AT rich tracts [57] and the matrix attachment region (MAR) [58], a region that is involved in the regulation of Hox genes [59] and has a high affinity for H1 binding [60]. However, the levels of JARID1A and JARID1B, two H3K4me2/3 demethylases, do not appear to differ significantly in cellular protein amounts or at affected Hox genes in H1 TKO ESCs compared with WT (Cao, Zhang and Fan, unpublished observations). Similarly, H3K4 methyltransferase MLL1 [36] does not display consistent changes by H1 depletion in ESCs (Cao, Zhang and Fan, unpublished observations). Whether any other H3K4me3 methyltransferase(s)/demethylase(s) is responsible for H1 regulated H3K4me3 at Hox genes in ESCs remains to be determined. We also cannot exclude additional possible regulatory mechanisms mediated through changes in other epigenetic events upon H1 depletion. For instance, nucleosome positioning is thought to impact DNA accessibility and transcription [61], and H1 depletion leads to a reduction in nucleosome repeat length of bulk chromatin and at specific loci [19], [20]. Nucleosomes are found to be positioned at Hox gene clusters, preferentially at 3′ of the expressed Hox genes [62], thus the expression of Hox genes may be impaired by altered nucleosome positioning in H1 TKO embryos and ESCs. Alternatively, DNA methylation may be affected at Hox gene clusters by H1 depletion, which has been shown to affect specific DNA methylation patterns at specific imprinted genes and other loci [20], [63]–[65]. Furthermore, the distance between enhancers or regulatory regions for Hox clusters and individual Hox genes [66]–[68] may be altered by H1 loss, which in turn reduces Hox gene expression.
In order to determine if any of the three deleted H1 subtypes is responsible for the reduction of Hox genes identified in H1 TKO ESCs, we derived single-H1 KO ESCs that are null for H1c, or H1d, or H1e. Surprisingly, unlike adult tissues of the single-H1 knockout mice [29], which display no changes in the total H1 levels, single-H1 KO ESCs established in this study exhibit a moderate reduction in the total H1 levels, and a lack of significant compensation for the deleted H1s by the remaining H1 subtypes. Interestingly, the analysis of the 6 Hox genes whose expression levels were significantly reduced in H1 TKO ESCs shows that loss of H1c or H1d has similar effects on Hoxa1, Hoxb8, and Hoxc13 as triple-H1 deletions. On the other hand, 5 of these 6 Hox genes show no expression change in H1e−/− ESCs (Figure 4C). This differential role of the individual H1 subtypes in activating expression of specific genes is reminiscent of the effects of loss of H1a on the expression of non-variegating transgenes in mice [69] and the activation of MMTV promoter by overexpression of H10 and H1c [70]. Hoxb5, Hoxb13 and Hoxd13 are not changed in single-H1 null ESCs, suggesting that the expression reduction of these genes in H1 TKO ESCs may be due to additive effects of deficiency of all three H1 subtypes. It is interesting to note that the levels of H3K4me3 are differentially affected at several Hox genes, suggesting potential roles of individual H1 subtypes in contributing to the patterns of this histone mark at specific Hox genes.
Taken together, the results in this study establish a novel link between histone H1 and Hox gene regulation. Furthermore, the reduction of Hox gene expression by marked H1 depletion correlates with dynamic patterns of H3K4me3 and H3K27me3 marks. The single-H1 KO ESCs established in this study should be useful cell resources for studying specificity of the individual H1 subtypes in regulating gene expression and epigenetic events.
Materials and Methods
Establishment of Mouse Single-H1 KO ESCs and Formation of Embryoid Bodies
Mouse ESCs deficient in histone H1c, or H1d, or H1e were derived from outgrowth of the respective H1c−/−, H1d−/−, and H1e−/− blastocysts (E3.5) as described previously [20]. Two ESC lines were established for each single KO. Genotyping analysis of WT and KO alleles of H1c, H1d, and H1e loci was carried out as reported [19]. Animal breeding and experimental procedures were approved by Georgia Tech Institutional Animal Care and Use Committee. Embryoid bodies were formed by seeding 1×106 ESCs in a 10-cm ultra-low attachment culture dish (Corning) and cultured for 10 days in media containing Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies) with 15% fetal bovine serum (Gemini), 0.1 mM MEM Non-essential amino acids (Life Technologies), 55 µM 2-mercaptoethanol (Life Technologies) and 100 U/ml penicillin/100 µg/ml streptomycin (Life Technologies).
RNA Extraction and Quantitative Reverse Transcription PCR (qRT-PCR)
Total RNAs from ESCs were extracted with Trizol reagent (Life Technologies) according to the manufacturers’ instructions. Total RNAs from embryos were prepared using Allprep DNA/RNA Micro kit (Qiagen). Reverse transcription was carried out using a SuperScript III First-Strand cDNA Synthesis kit (Life Technologies). cDNAs were subsequently analyzed with real-time quantitative PCR (qPCR) using iQ SYBR Green Supermix (Bio-Rad) with a MyIQ Single Color real-time PCR Detection System (Bio-Rad). Hox gene specific primers used for qRT-PCR are listed in Table 1.
Table 1. Primers for qRT-PCR analysis.
| Name | Forward | Reverse |
| Homeobox A1 | tggccacgtataataactcc | aagtggaactccttctccag |
| Homeobox A2 | agtatccctggatgaaggag | aagctgagtgttggtgtacg |
| Homeobox A3 | aacaaatctttccctggatg | cataggtagcggttgaagtg |
| Homeobox A4 | cctggatgaagaagatccac | tctgaaaccagatcttgacc |
| Homeobox A6 | agcagcagtacaaacctgac | agtggaattccttctcaagc |
| Homeobox A7 | tcctacgaccaaaacatcc | aattccttctccagttccag |
| Homeobox A9 | ttgtccctgactgactatgc | aactccttctccagttccag |
| Homeobox A10 | cccttcagaaaacagtaaagc | ttcacttgtctgtccgtgag |
| Homeobox A11 | gacccgagagcagcag | gacgcttctctttgttgatg |
| Homeobox A13 | aaatgtactgccccaaagag | gatatcctcctccgtttgtc |
| Homeobox B1 | acctcctctctgaggacaag | aaatgaaatcccttctccag |
| Homeobox B2 | aagaaatccaccaagaaacc | aagtggaactccttctccag |
| Homeobox B3 | atgaaagagtcgaggcaaac | aagtggaactccttctccag |
| Homeobox B4 | aaagagcccgtcgtctac | ggtagcgattgtagtgaaactc |
| Homeobox B5 | cagatattcccctggatgag | aaccagattttgatctgacg |
| Homeobox B6 | aagagcgtgttcggagag | tgaaattccttctccagctc |
| Homeobox C6 | tcaatcgctcaggattttag | aattccttctccagttccag |
| Homeobox B8 | cagctctttccctggatg | cacttcattctccgattctg |
| Homeobox B9 | taatcaaagagctggctacg | ccctggtgaggtacatattg |
| Homeobox B13 | atgtgttgccaaggtgaac | aacttgttggctgcatactc |
| Homeobox C4 | aagcaacccatagtctaccc | gtcaggtagcggttgtaatg |
| Homeobox C8 | aggacaaggccacttaaatc | tggaaccaaatcttcacttg |
| Homeobox C9 | cgcagctacccggactac | aactccttctccagttccag |
| Homeobox C10 | gtccagacacctcggataac | aatggtcttgctaatctccag |
| Homeobox C11 | aggaggagaacacgaatcc | ttttcacttgtcggtctgtc |
| Homeobox C12 | actccagttcgtccctactc | tgaactcgttgaccagaaac |
| Homeobox C13 | gtcaggtgtactgctccaag | ccttctctagctccttcagc |
| Homeobox D3 | ctacccttggatgaagaagg | aagaggagcaggaagatgag |
| Homeobox D9 | gaaggaggaggagaagcag | tggaaccagattttgacttg |
| Homeobox D10 | gaagtgcaggagaaggaaag | tgaaaccaaatcttgacctg |
| Homeobox D11 | cagtccctgcgccaag | cgagagagttggagtcttttc |
| Homeobox D12 | cttcaaggaagacaccaaag | tgaggttcagcctgttagac |
| Homeobox D13 | gaacagccaggtgtactgtg | gagctgcagtttggtgtaag |
Statistical Analysis
Statistical analyses and P-values were calculated by the Student T two-tailed test. A P-value of less than 0.05 was considered to be statistically significant.
Preparation and HPLC/MS Analysis of Histones
Total histones were extracted from ES cells as described previously [30], [31]. Briefly, the cells were washed with PBS and harvested. The cell pellet was resuspended in Sucrose Buffer (0.3 M Sucrose, 15 mM NaCl, 10 mM HEPES [pH 7.9], 2 mM EDTA, 0.5 mM PMSF, protease inhibitor) with 0.5% NP-40 and homogenized with a dounce homogenizer (Wheaton). 0.2 N H2SO4 was used to extract histones from chromatin pellet. HPLC and mass spectrometry analysis of histone proteins were carried out as described previously [30], [31], [65]. Approximately 50 µg histone proteins were injected to a C18 reverse phase column (Vydac) on an Äktapurifier UPC 900 instrument (GE Healthcare). The effluent was monitored at 214 nm (A214), and the profiles were recorded and analyzed with UNICORN 5.11 software (GE Healthcare). The values of all peaks were adjusted according to the peptide bonds present in respective proteins. Percentage of total H1 for individual H1 subtypes was determined by the ratio of A214 values of individual H1 subtype to that of all H1 peaks. H1 to nucleosome ratio was determined by the ratio of A214 values of individual H1 subtype to that of half of the H2B peak.
Karyotyping
Exponentially growing ESCs were treated with colcemid (Life Technologies) at 37°C for 60 minutes, trypsinized, and harvested. Cells were subsequently resuspended with pre-warmed hypotonic solution (75 mM KCl) and incubated at 37°C for 6 minutes, and fixed as described previously [65]. Fixed cells were concentrated and dropped onto an angled, humidified microscope slide, dried and stained with Hoechst dye for 60 minutes in the dark. Images were collected at a 60x objective on an Olympus Fluorescence Microscope.
Quantitative Chromatin Immunoprecipitation (qChIP)
qChIP assays were performed as described previously [20] with modifications. The following antibodies were used: anti-H3K4me3 (Millipore 07–473), anti-H3K9me3 (Abcam 8898), anti-H3K27me3 (Millipore 07–449), anti-H3K36me3 (abcam 9050), anti-JARID1A (abcam 65769), anti-JARID1B (abcam 50958), anti-MLL1 (Bethyl Lab A300–086A) and rabbit IgG (Millipore 12–370). Briefly, crosslinked chromatin was sheared by sonication. Pre-blocked Protein G Dynabeads (Life Technologies) were incubated with the antibody and 40 µg of soluble chromatin overnight in 4°C, and subsequently washed with Washing Buffer (50 mM HEPES pH 7.6, 1 mM EDTA pH 8.0, 500 mM LiCl, 0.7% Sodium Deoxycholate, 1% NP-40). Immunoprecipated protein-DNA complexes were eluted and reverse-crosslinked at 65°C, and DNA was purified with a Qiagen DNA Isolation column (Qiagen). The amount of each specific immunoprecipitated DNA fragment was determined by real-time PCR. All samples were analyzed in triplicate in two independent experiments. The percentage of input was calculated by dividing the amount of each specific DNA fragment in the immunoprecipitates by the amount of DNA present in input DNA. qChIP primers are listed in Table 2.
Table 2. Primers for qChIP analysis.
| Name | Forward | Reverse |
| Homeobox A1 | gggaatccaacagacaccac | tcctcccagtcaatcctctg |
| Homeobox A1–2 | ggcaccctacaccactcact | gaaaccctcccaaaacaggt |
| Homeobox A3 | aattacctccctgcatctcaaa | ttatcagagcagacccacaatg |
| Homeobox B4 | atttccttatccgggaatcg | gtttccgaaagccctcctac |
| Homeobox B4–2 | gtgggcaattcccagaaa | gctggaagccgctctctc |
| Homeobox B5 | taacgaccacgatccacaaa | agagctgccactgccataat |
| Homeobox B5 –2 | cctccaaaatcacccaaatg | gctgagatccatcccattgt |
| Homeobox B8 | gctccgttccaaacacctac | cctccttcaaaggaagcaaa |
| Homeobox B8 –2 | taagcaaggactccctcgtc | gaattacggcgtgaataggc |
| Homeobox B13 | ccctctctttttccaccaca | ttgcgcctcttgtccttagt |
| Homeobox B13 –2 | gagggggtcggaatctagtc | cgcctccaaagtagccataa |
| Homeobox C13 | agctggagcagatcatgtca | gcgctgtcctcatagacgta |
| Homeobox C13 –2 | tgctgaccctgctcactgta | aattctgagcttccctccag |
| Homeobox D11 | tgaacgactttgacgagtgc | ggttggaggagtaggggaaa |
| Homeobox D11 –2 | cctagctcagtggccagagt | agcatccgagagagttggag |
| Homeobox D13 | agctcgaggagccaaagag | gacccaggagttgactttgc |
| Homeobox D13 –2 | gaaaagggtgccttacacca | tgtccttcacccttcgattc |
| Tcf4 | cggatgtgaatggattacaatg | attgttcttcggtcttgttggt |
Supporting Information
Characterization of the single-H1 KO ESCs and EBs. (A, B) Karyotypes (A) and phase images (B) of the single-H1 KO ESCs. Scale bar: 50 µm. (C) Characterization of EBs. (i) hematoxylin and eosin staining images of single-H1 KO EBs. Scale bar: 50 µm. (ii) Western blotting analysis of OCT4 in single-H1 KO ESCs and EBs. GAPDH expression levels indicate equal loading of cell lysates. (iii) qRT-PCR analysis of differentiation markers in single-H1 KO ESCs and EBs.
(TIF)
qChIP analysis of H3K4me3 in single -H1 KO ESCs. qChIP signals of H3K4me3 (A) and H3K27me3 (B) at indicated Hox genes in single-H1 KO ESCs were normalized to input controls and represented as fold changes over that of WT ESCs. *: P<0.05, **: P<0.01.
(TIF)
Acknowledgments
We thank colleagues and lab members for critical reading of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work is supported by a Georgia Cancer Coalition Distinguished Scholar award (to YF), National Institutes of Health (United States) grant GM085261, and Georgia Tech. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mallo M, Wellik DM, Deschamps J. Hox genes and regional patterning of the vertebrate body plan. Dev Biol. 2010;344:15. doi: 10.1016/j.ydbio.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leucht P, Kim JB, Amasha R, James AW, Girod S. Embryonic origin and Hox status determine progenitor cell fate during adult bone regeneration. Development. 2008;135:2854. doi: 10.1242/dev.023788. [DOI] [PubMed] [Google Scholar]
- 3.Gouti M, Gavalas A. Hoxb1 controls cell fate specification and proliferative capacity of neural stem and progenitor cells. Stem Cells. 2008;26:1997. doi: 10.1634/stemcells.2008-0182. [DOI] [PubMed] [Google Scholar]
- 4.Goodman FR. Limb malformations and the human HOX genes. Am J Med Genet. 2002;112:265. doi: 10.1002/ajmg.10776. [DOI] [PubMed] [Google Scholar]
- 5.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 6.Graham A, Papalopulu N, Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989;57:378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- 7.Ruddle FH, Bartels JL, Bentley KL, Kappen C, Murtha MT. Evolution of Hox genes. Annu Rev Genet. 1994;28:442. doi: 10.1146/annurev.ge.28.120194.002231. [DOI] [PubMed] [Google Scholar]
- 8.Wagner GP, Amemiya C, Ruddle F. Hox cluster duplications and the opportunity for evolutionary novelties. Proc Natl Acad Sci U S A. 2003;100:14606. doi: 10.1073/pnas.2536656100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soshnikova N, Duboule D. Epigenetic temporal control of mouse Hox genes in vivo. Science. 2009;324:1323. doi: 10.1126/science.1171468. [DOI] [PubMed] [Google Scholar]
- 10.Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chambeyron S, Da Silva NR, Lawson KA, Bickmore WA. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development. 2005;132:2223. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- 12.Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134:919. doi: 10.1242/dev.02779. [DOI] [PubMed] [Google Scholar]
- 13.Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W. The dynamic architecture of Hox gene clusters. Science. 2011;334:225. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- 15.Izpisua-Belmonte JC, Falkenstein H, Dolle P, Renucci A, Duboule D. Murine genes related to the Drosophila AbdB homeotic genes are sequentially expressed during development of the posterior part of the body. EMBO J. 1991;10:2289. doi: 10.1002/j.1460-2075.1991.tb07764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci U S A. 1998;95:14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431:12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol. 2003;23:4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R. Histone h1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 21.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 22.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 23.Jorgensen HF, Giadrossi S, Casanova M, Endoh M, Koseki H. Stem cells primed for action: polycomb repressive complexes restrain the expression of lineage-specific regulators in embryonic stem cells. Cell Cycle. 2006;5:1414. doi: 10.4161/cc.5.13.2927. [DOI] [PubMed] [Google Scholar]
- 24.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 26.Endoh M, Endo TA, Endoh T, Fujimura Y, Ohara O. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- 27.Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan Y, Sirotkin A, Russell RG, Ayala J, Skoultchi AI. Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype. Mol Cell Biol. 2001;21:7943. doi: 10.1128/MCB.21.23.7933-7943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medrzycki M, Zhang Y, Cao K, Fan Y. Expression Analysis of Mammalian Linker-histone Subtypes. J Vis Exp. 2012;61:e3577. doi: 10.3791/3577. DOI :3510.3791/3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan Y, Skoultchi AI. Genetic analysis of H1 linker histone subtypes and their functions in mice. Methods Enzymol. 2004;377:107. doi: 10.1016/S0076-6879(03)77005-0. [DOI] [PubMed] [Google Scholar]
- 32.Panyim S, Chalkley R. A new histone found only in mammalian tissues with little cell division. Biochem Biophys Res Commun. 1969;37:1049. doi: 10.1016/0006-291x(69)90237-x. [DOI] [PubMed] [Google Scholar]
- 33.Pehrson J, Cole RD. Histone H10 accumulates in growth-inhibited cultured cells. Nature. 1980;285:44. doi: 10.1038/285043a0. [DOI] [PubMed] [Google Scholar]
- 34.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 35.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 36.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 37.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 39.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 41.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 43.Alexander T, Nolte C, Krumlauf R. Hox genes and segmentation of the hindbrain and axial skeleton. Annu Rev Cell Dev Biol. 2009;25:456. doi: 10.1146/annurev.cellbio.042308.113423. [DOI] [PubMed] [Google Scholar]
- 44.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 45.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akasaka T, van Lohuizen M, van der Lugt N, Mizutani-Koseki Y, Kanno M. Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development. 2001;128:1597. doi: 10.1242/dev.128.9.1587. [DOI] [PubMed] [Google Scholar]
- 47.van der Lugt NM, Alkema M, Berns A, Deschamps J. The Polycomb-group homolog Bmi-1 is a regulator of murine Hox gene expression. Mech Dev. 1996;58:164. doi: 10.1016/s0925-4773(96)00570-9. [DOI] [PubMed] [Google Scholar]
- 48.van Lohuizen M. Functional analysis of mouse Polycomb group genes. Cell Mol Life Sci. 1998;54:79. doi: 10.1007/s000180050126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erzurumlu RS, Murakami Y, Rijli FM. Mapping the face in the somatosensory brainstem. Nat Rev Neurosci. 2010;11:263. doi: 10.1038/nrn2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oury F, Murakami Y, Renaud JS, Pasqualetti M, Charnay P. Hoxa2- and rhombomere-dependent development of the mouse facial somatosensory map. Science. 2006;313:1413. doi: 10.1126/science.1130042. [DOI] [PubMed] [Google Scholar]
- 51.Wellik DM. Hox patterning of the vertebrate axial skeleton. Dev Dyn. 2007;236:2463. doi: 10.1002/dvdy.21286. [DOI] [PubMed] [Google Scholar]
- 52.McIntyre DC, Rakshit S, Yallowitz AR, Loken L, Jeannotte L. Hox patterning of the vertebrate rib cage. Development. 2007;134:2989. doi: 10.1242/dev.007567. [DOI] [PubMed] [Google Scholar]
- 53.Vinagre T, Moncaut N, Carapuco M, Novoa A, Bom J. Evidence for a myotomal Hox/Myf cascade governing nonautonomous control of rib specification within global vertebral domains. Dev Cell. 2010;18:661. doi: 10.1016/j.devcel.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 54.Christensen J, Agger K, Cloos PA, Pasini D, Rose S. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 55.Kortschak RD, Tucker PW, Saint R. ARID proteins come in from the desert. Trends Biochem Sci. 2000;25:299. doi: 10.1016/s0968-0004(00)01597-8. [DOI] [PubMed] [Google Scholar]
- 56.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang TH, Oka T, Asai T, Okada T, Merrills BW. Repression by a differentiation-specific factor of the human cytomegalovirus enhancer. Nucleic Acids Res. 1996;24:1701. doi: 10.1093/nar/24.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Herrscher RF, Kaplan MH, Lelsz DL, Das C, Scheuermann R. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev. 1995;9:3082. doi: 10.1101/gad.9.24.3067. [DOI] [PubMed] [Google Scholar]
- 59.Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 60.Izaurralde E, Kas E, Laemmli UK. Highly preferential nucleation of histone H1 assembly on scaffold-associated regions. J Mol Biol. 1989;210:585. doi: 10.1016/0022-2836(89)90133-2. [DOI] [PubMed] [Google Scholar]
- 61.Bai L, Morozov AV. Gene regulation by nucleosome positioning. Trends Genet. 2010;26:483. doi: 10.1016/j.tig.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 62.Kharchenko PV, Woo CJ, Tolstorukov MY, Kingston RE, Park PJ. Nucleosome positioning in human HOX gene clusters. Genome Res. 2008;18:1561. doi: 10.1101/gr.075952.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giambra V, Volpi S, Emelyanov AV, Pflugh D, Bothwell AL. Pax5 and linker histone H1 coordinate DNA methylation and histone modifications in the 3' regulatory region of the immunoglobulin heavy chain locus. Mol Cell Biol. 2008;28:6133. doi: 10.1128/MCB.00233-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maclean JA, Bettegowda A, Kim BJ, Lou CH, Yang SM. The rhox homeobox gene cluster is imprinted and selectively targeted for regulation by histone h1 and DNA methylation. Mol Cell Biol. 2011;31:1287. doi: 10.1128/MCB.00734-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Cooke M, Panjwani S, Cao K, Krauth B. Histone h1 depletion impairs embryonic stem cell differentiation. PLoS Genet. 2012;8:e1002691. doi: 10.1371/journal.pgen.1002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- 67.Deschamps J. Ancestral and recently recruited global control of the Hox genes in development. Curr Opin Genet Dev. 2007;17:427. doi: 10.1016/j.gde.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 68.Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 69.Alami R, Fan Y, Pack S, Sonbuchner TM, Besse A. Mammalian linker-histone subtypes differentially affect gene expression in vivo. Proc Natl Acad Sci U S A. 2003;100:5925. doi: 10.1073/pnas.0736105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gunjan A, Brown DT. Overproduction of histone H1 variants in vivo increases basal and induced activity of the mouse mammary tumor virus promoter. Nucleic Acids Res. 1999;27:3363. doi: 10.1093/nar/27.16.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of the single-H1 KO ESCs and EBs. (A, B) Karyotypes (A) and phase images (B) of the single-H1 KO ESCs. Scale bar: 50 µm. (C) Characterization of EBs. (i) hematoxylin and eosin staining images of single-H1 KO EBs. Scale bar: 50 µm. (ii) Western blotting analysis of OCT4 in single-H1 KO ESCs and EBs. GAPDH expression levels indicate equal loading of cell lysates. (iii) qRT-PCR analysis of differentiation markers in single-H1 KO ESCs and EBs.
(TIF)
qChIP analysis of H3K4me3 in single -H1 KO ESCs. qChIP signals of H3K4me3 (A) and H3K27me3 (B) at indicated Hox genes in single-H1 KO ESCs were normalized to input controls and represented as fold changes over that of WT ESCs. *: P<0.05, **: P<0.01.
(TIF)