Abstract
Infection of the human host by Shigella species requires the coordinated production of specific Shigella virulence factors, a process mediated largely by the VirF/VirB regulatory cascade. VirF promotes the transcription of virB, a gene encoding the transcriptional activator of several virulence-associated genes. This study reveals that transcription of virB is also regulated by the small RNA RyhB, and importantly, that this regulation is not achieved indirectly via modulation of VirF activity. These data are the first to demonstrate that the regulation of virB transcription can be uncoupled from the master regulator VirF. It is also established that efficient RyhB-dependent regulation of transcription is facilitated by specific nucleic acid sequences within virB. This study not only reveals RyhB-dependent regulation of virB transcription as a novel point of control in the central regulatory circuit modulating Shigella virulence, but also highlights the versatility of RyhB in controlling bacterial gene expression.
Introduction
Shigellosis, a severe diarrheal disease caused by members of the Shigella genus (S. dysenteriae, S. flexneri, S. sonnei and S. boydii) remains a worldwide health concern with a conservative estimate of 165 million cases resulting in over one million deaths each year [1]. Following ingestion and transit through the gastrointestinal tract of the host, Shigella invade cells of the colonic epithelium, replicate within the cytoplasm of the infected cell and spread to neighboring epithelial cells via actin based motility [2], [3]. These virulence-associated processes are mediated by the coordinated production and activity of several Shigella virulence factors.
The complex and coordinated regulation of Shigella virulence gene expression is accomplished largely via the activity of two transcriptional activators, VirF and VirB [4], [5]. Activity of the VirF/VirB regulatory cascade, and thus Shigella virulence, is modulated in response to specific environmental signals such as changes in temperature, pH and osmolarity, as well as carbon source and iron availability [1], [6], [7], [8]. In response to precise environmental conditions, specifically those encountered within the host, VirF is produced and directly activates the transcription of two virulence-associated genes, icsA and virB. IcsA facilitates intracellular spread of Shigella by mediating actin-based motility [9], while VirB directly promotes the expression of multiple virulence-associated genes (Figure 1). The VirB regulon includes genes encoding components of the Type Three Secretion System (TTSS), icsP encoding the protease that modulates IcsA activity, and mxiE encoding a transcriptional activator of additional virulence associated genes [1], [3], [4], [10], [11]. As a central pathway controlling the coordinated expression of several virulence-associated genes, any factor that influences the production or activity of VirF or VirB consequently impacts Shigella virulence.
Figure 1. Schematic of the VirF/VirB cascade.
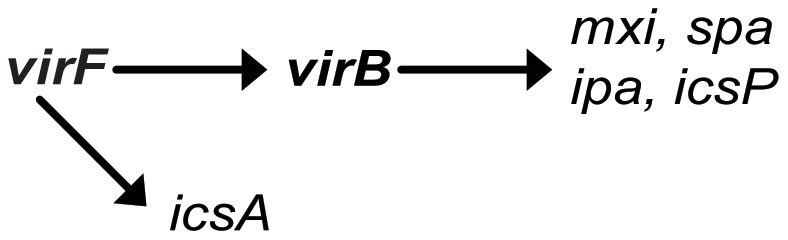
VirF is produced under permissive conditions and positively regulates the expression of icsA and virB. VirB in turn activates the expression of many virulence-associated genes including those encoding components of the Type Three Secretion System and associated effectors (mxi, spa and ipa) as well as IcsP, the protease specific for the actin based motility protein IcsA.
Expression of genes within the VirF and VirB regulons appears to be coupled: factors that impact the VirF regulon, also impact the VirB regulon. The coupled expression of genes within the VirF and VirB regulons results from the fact that both transcriptional and post-transcriptional regulation of virB is mediated by factors or environmental conditions that also modulate the expression of virF. Specifically, transcriptional regulation of virB is mediated by VirF itself, or by H-NS and IHF, proteins known to influence virF expression [5], [12], [13]. Post-transcriptional regulation of virB expression, via destabilization of the virB mRNA, is mediated in response to changes in environmental pH, osmolarity and temperature [14], [15], [16], conditions that also influence expression of virF [12], [17], [18], [19]. No study to date has identified a regulatory factor that controls the transcription of genes within the VirB regulon, such as icsP or mxiE, without also regulating icsA, a gene that lies solely within the VirF regulon (Figure 1).
Of the environmental conditions known to modulate Shigella virulence factors via regulation of the VirF/VirB cascade, the impact of iron-availability is the least well characterized to date. Under conditions of iron-limitation, the small RNA RyhB is produced and functions to inhibit virB expression, as evidenced by a reduction in the steady state level of virB mRNA [6], a decrease in the production of proteins within the VirB regulon [6], [20] and an inhibition of epithelial cell invasion by the bacterium [6]. While it is clear that RyhB functions to reduce the steady state level of virB mRNA and inhibit VirB activity [6], it remains unclear whether RyhB-dependent regulation of virB mRNA is achieved through the modulation of VirF activity, whether the regulation occurs in a VirF-independent manner at the level of virB transcription, or whether the virB message is simply targeted for accelerated degradation in the presence of RyhB. A greater understanding of the RyhB-dependent regulation of virB expression is needed to improve our knowledge of the VirF/VirB cascade, which controls virulence gene expression in Shigella.
Materials and Methods
Growth Conditions
Bacterial strains and plasmids used in this study are shown in Table 1. Escherichia coli was cultured in Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract and 1% NaCl) or on LB agar plates at 37°C. Shigella dysenteriae was cultured in LB broth or on tryptic soy broth (Becton, Dickenson and Company, Sparks, MD) agar plates containing 0.01% (wt/vol) Congo red dye (ISC Bioexpress, Kaysville, UT) at 37°C.
Table 1. Bacterial strains and plasmids.
| Strain or Plasmid | Description | Source |
| Escherichia coli strains | ||
| DH5α | Life Technologies | |
| DH5α(λpir) | [39] | |
| Shigella strains | ||
| O-4576S1 | Wild-type S. dysenteriae | [40] |
| ND100 (wild-type) | Spontaneous Streptomycin resistant mutant of O-4576S1 | [6] |
| ND100ryhB | ryhB deletion in ND100 | [6] |
| ND100ryhBaltvirB | Site-specific mutation within virB of ND100rhyB | This study |
| 2457T | Wild-type S. flexneri | [41] |
| Plasmids | ||
| pCVD442N2 | Suicide vector | [42] |
| pERM124 | Altered virB construct in pCVD442N2 | This study |
| pQE2 | Expression vector | QIAGEN |
| pryhB | ryhB in pQE-2 | [6] |
| pCompryhB | Compensatory ryhB in pQE-2 | This study |
| pRW50 | lacZ reporter plasmid | [24] |
| PicsA-lacZ | icsA promoter region transcriptionally fused to lacZ in pSRG12 | Gift from M. Goldberg.This study |
| PicsP-lacZ (previously pHJW20) | icsP promoter region transcriptionally fused to lacZ | [27] |
Oligonucleotide Primers and Probes
The name, nucleic acid sequence and function of each oligonucleotide primer and probe used in this study are presented in Table 2.
Table 2. Primers and Probes.
| Sequence | Use in study | |
| Primers | ||
| virB2-for | TCCAATCGCGTCAGAACTTAACT | Real-time primer-Taqman |
| virB2-rev | CCTTTAATATTGGTAGTGTAGAACTAAGAGATTC | Real-time primer-Taqman |
| sodB1-for | CTGGAAAAGTCGCTGAAGCTATC | Real-time primer-Taqman |
| sodB1-rev | CGCTTTGAAATCGGCAAAG | Real-time primer-Taqman |
| rrsA-for | CACGATTACTAGCGATTCCGACTT | Real-time primer-Taqman |
| rrsA-rev | CGTCGTAGTCCGGATTGGA | Real-time primer-Taqman |
| virB 3′-for | GCGCGAAAGTCACTCGTC | Real-time primer-SYBR |
| virB 3′-rev | GAGATTCATTAGCCTTTTCAAGTCC | Real-time primer-SYBR |
| virB 5′-for | AACAGAAGAATCATTAGCCGATA | Real-time primer-SYBR |
| virB 5′-rev | TACGAGTGCCATCCAGAA | Real-time primer-SYBR |
| sodB-for | TCCGCTGCTGACCGTTGATG | Real-time primer-SYBR |
| sodB-rev | CGCCCAGAAGTGCTCCAGATAG | Real-time primer-SYBR |
| rrsA-for | AACGTCAATGAGCAAAGGTATTAA | Real-time primer-SYBR |
| rrsA-rev | TACGGGAGGCAGCAGTGG | Real-time primer-SYBR |
| MBG608 | TCCAGAATTCAGTTGATTTGAC | Construction of pSRG12 |
| MBG607 | ACAAAAGCTTGAACATTGGGTCATATTACA | Construction of pSRG12 |
| virB5 | GCTCTAGAGCAAAAGATGTAACCACTTCCC | Construction of altered virB |
| virB6 | GCTCTAGAGAACGAAGTCTGTTAATGGTGG | Construction of altered virB |
| virB12 | GCATTCGTGAGTTGGGCATCGGCCTTAATTTTCTGAAAGTATCAGGG | Construction of altered virB |
| virB13 | TAAGGCCGATGCCCAACTCACGAATGCTATGCTC | Construction of altered virB |
| sodB probe for | CTACTGGAACTGCCTGG | Amplification of northern blot probe |
| sodB probe rev | GTGCTCCAGATAGCCAG | Amplification of northern blot probe |
| Probes | ||
| virB2 | 6FAMAGGACTTGAAAAGGCTMGBNFQ | Real-time probe-Taqman |
| sodB1 | 6FAMCCGCATCTTTTGGCMGBNFQ | Real-time probe-Taqman |
| rrsA | 6FAMATGGAGTCGAGTTGCAGMGBNFQ | Real-time probe-Taqman |
| ssvirB-sense | TGCTCCTGCATATATTGCAGATGCTCTTCTACGAGTGCCAATTTCAATTCTACC ATCAATCTCCCTTCCTATTACAGGGAATTGTTGTAGC | Northern blot |
| ssvirB-antisense | TAGCATCCGAGAACTTGGTATTGGTCTTAATTTTCTGAAAGTATCAGGGATGTCCT ATAAAGACATAGCCAAAAAAGAGAATCTGTCTCGCGCGAAAGTC | Northern blot |
Site-directed Mutagenesis of virB
Specific nucleic acid modifications were introduced into virB by allelic exchange [21]. Briefly, splice overlap polymerase chain reaction (PCR) was used to amplify virB containing the desired nucleic acid changes and 500 bp of flanking sequence on either side [21]. Initial upstream and downstream products were amplified using oligonucleotides virB-5/virB-12 and virB-6/virB-13 respectively. PCR conditions: denaturation for 30 seconds at 95°C, annealing for 45 seconds at 50°C, and extension for 60 seconds at 72°C for 30 cycles in a Peltier thermal cycler (MJ Research, Watertown, MA). The final 2 kb product containing the desired nucleic acid changes, including an additional HaeIII restriction endonuclease recognition site, was amplified with oligonucleotides virB-5 and virB-6. The resulting product was digested with XbaI and cloned into the XbaI site of pCVD442N2 to generate pERM124. The insert of pERM124 was sequenced to confirm that only the desired nucleic acid changes were incorporated. Osmotic shock was used to transform DH5α(λpir) with pERM124, and the plasmid was subsequently moved by conjugation into S. dysenteriae O-4576S1-G. Primary integrants were selected by growth in the presence of 200 µg/ml streptomycin and 250 µg/ml carbenicillin. Primary integrants were analyzed by PCR using oligonucleotide sets virB-1/virB-10 and virB-4/virB-9, followed by the purification of each product and digestion with the HaeIII restriction endonuclease. Once confirmed, the selected primary integrant was cultured to mid-logarithmic phase in LB supplemented with 0.1% sucrose at 30°C. The culture was then serially diluted and plated onto LB agar plates containing 5% sucrose. The plates were incubated overnight at 37°C. Resulting colonies were screened for sensitivity to 250 µg/ml carbenicillin. Colonies sensitive to carbenicillin (8 of 120) were screened for the presence of the altered virB nucleic acid sequence by PCR using virB-4/virB-9 and virB-1/virB-10 oligonucleotide sets followed by digestion of each purified product with the HaeIII restriction endonuclease. Sequence analysis was used to verify that only the desired nucleic acid changes were present in the resulting altered virB open reading frame.
Real-time Polymerase Chain Reaction
RNA was isolated using RNeasy® Midi Kit (QIAGEN, Valencia, CA) per the product directions, from bacteria cultured for approximately 20 hours at 37°C on TSB agar plates supplemented with 0.1% Congo red, 250 µg/ml carbenicillin and 200 µM IPTG. Each RNA sample was then treated with 16 units of amplification grade DNaseI (Invitrogen, Carlsbad, CA), ethanol precipitated and dried. The RNA pellet was resuspended in DEPC-treated water, and the nucleic acid concentration was measured using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). No more than 10 µg of total RNA was used to generate cDNA with the High Capacity cDNA Archival Kit (Applied Biosystems, Foster City, CA) per the product directions. Each cDNA sample was diluted 10 fold in water, and 5 µl was used as the template for each amplification reaction. All Taqman based reactions utilized TaqMan Universal Master Mix (Applied Biosystems, Foster City CA). Primers and FAM labeled minor grove binding probes were designed using Primer Express software (Applied Biosystems, Foster City, CA). All SYBR based reactions utilized iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) and primers were designed using Beacon Designer 7 software (Premier Biosoft, Palo Alto, CA). Regardless of the chemistry, rrsA was used as the normalizer for each sample, and all values were calibrated to the value obtained from the given parental strain carrying the vector control plasmid using the ΔΔCt method [22]. All primer pairs and amplification conditions were validated by the inclusion of a standard curve on each reaction plate, from which efficiency was calculated. All reactions were run in a 7300 Real-Time PCR system (Applied Biosystems, Foster City, CA) under standard reaction conditions.
Tissue Culture and in vivo Plaque Assays
Henle cell monolayers were cultured in 6 well polystyrene tissue culture plates (Corning Inc. Costar, Corning, NY) in Gibco Minimum Essential Medium (MEM) (Invitrogen Corp. Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine and a 1 X final concentration of non-essential amino acids (Lonza, Switzerland). Plates were incubated at 37°C in an atmosphere of 5% CO2.
Plaque assays were performed as published with minor modifications [23]. Briefly, bacterial strains were grown overnight at 37°C on TSB agar plates containing the appropriate antibiotic. Ten colonies were used to inoculate a 3 ml LB culture containing 250 µg/ml carbenicillin, 0.1% DOC, and 200 µM IPTG. Cultures were grown to mid-logarithmic phase at 37°C. 104 bacteria, diluted in phosphate buffered saline (PBS), were used to infect the Henle cell monolayer in each tissue culture well containing 2 ml MEM supplemented with 250 µg/ml carbenicillin and 200 µM IPTG. Plates were centrifuged for 10 minutes at 600 x g in a Centra GP8 (International Equipment Company, Needham Heights, MA). Plates were incubated at 37°C for 1.5 hours, washed with 2 ml PBS, then covered with a 2 ml overlay composed of MEM supplemented with 0.3% glucose, 250 µg/ml carbenicillin, 200 µM IPTG, and 20 µg/ml gentamicin to kill any bacteria that had not invaded into a eukaryotic cell. Following incubation for 72 hours at 37°C, the plates were washed with 2 ml PBS, stained with Wright-Giemsa stain (Camco, Ft. Lauderdale, FL), washed with distilled water, and air dried.
Construction of the PicsA-lacZ Reporter Plasmid pSRG12
The PicsA-lacZ reporter plasmid pSRG12 is derived from the low-copy number, broad host range, lac expression vector pRW50 [24] and carries the S. flexneri icsA promoter region (597 bp upstream of the beginning of the icsA gene) on an EcoRI/Hind III restriction fragment. This places the lacZ gene directly under the control of the icsA promoter.
mRNA Stability Assay
Bacterial strains were grown on TSB agar plates containing 0.01% Congo red and 250 µg/ml carbenicillin. Following overnight incubation at 37°C, a single colony of each strain was used to inoculate a 3 ml culture of LB containing carbenicillin. Each culture was incubated overnight at 30°C in a shaking incubator. 250 µl of each overnight culture was used to inoculate a 25 ml LB culture containing carbenicillin and 0.1% DOC which was incubated at 37°C in a shaking incubator to an optical density (OD650) of 0.7. Next, 200 µM IPTG was added to each culture followed two minutes later by the addition of 250 µg/ml rifampicin. The time of rifampicin addition was designated t=0. At each time point, 1 ml of culture was removed and mixed with 250 µl of RNAlater (Ambion, Austin, TX) to preserve the mRNA profile of the sample. RNA isolation and Real-time PCR was performed using SYBR technology as detailed above.
Northern Blot Analysis
Northern blots were performed using the Ambion Northern Max kit (Ambion, Austin, TX) per kit directions. No more than 4 µg of total RNA was loaded into the wells of a 0.8% agarose gel and BrightStar Biotinylated RNA Millenium Markers (Ambion, Austin, TX) were used as size standards. Blots were hybridized overnight with 10 pM of the indicated probe at 46°C or 42°C with either single stranded, biotin labeled DNA probe (IDT, Coralville, IA) or double stranded DNA probe labeled using the BrightStar Psoralen-Biotin Nonisotopic Labelling Kit (Ambion, Austin, TX), respectively. Blots were visualized using the BrightStar Biodetect Kit (Ambion, Austin, TX) per the directions.
β-Galactosidase Assays
β-Galactosidase assays were performed as detailed previously [21]. Briefly, strains were grown overnight at 37°C on TSBA with Congo red containing 250 µg/ml ampicillin and 20 µg/ml tetracycline. Single colonies were selected and cultured in 3 ml LB with appropriate antibiotics in a shaking incubator overnight at 30°C. 130 µl of each culture was used to inoculate a 3 ml subculture in LB with antibiotics and 200 µM IPTG. Each subculture was incubated in a 37°C shaking incubator and grown to an OD600 between 0.3 and 0.6. The bacteria present in 1 ml of each culture were pelleted and resuspended in 1 ml Z buffer. 400 µl of the resuspended bacteria were then diluted 1∶1 in Z buffer. Bacteria were then permeabilized by the addition of 50 µl 0.1% sodium dodecyl sulfate (SDS) and 100 µl chloroform and mixed by vortexing on high for 10 sec. The permeabilized bacteria were incubated at 30°C for 15 min prior to the addition of 160 µl of ortho-Nitrophenyl-β-galactopyranoside (ONPG), diluted to 4 mg/ml in Z buffer. Each tube was mixed by vortexing on high for 5 seconds and incubated again at 30°C until a color change was observed. The time required for each reaction to change color was noted. Each reaction was stopped by the addition of 400 µl of 1 M Na2CO3. Tubes were centrifuged at maximum speed for 2 minutes and the optical density at both 550 nM (OD550) and 420 nM (OD420) was measured. A reaction containing all components except the bacterial culture was used as the negative control in the experiment.
Results
RyhB does not Influence VirF Activity
Previous studies have demonstrated that the production of S. dysenteriae RyhB results in a significant decrease in the steady state level of virB mRNA, but does not influence the steady state level of virF mRNA [6]. These findings indicate that RyhB-dependent regulation of virB expression is not mediated indirectly via inhibition of virF transcription or by the destabilization of the virF message [6], but do not rule out the possibility that RyhB-dependent regulation of virB expression is mediated indirectly via decreasing translational efficiency of the virF message, decreasing VirF protein levels or decreasing VirF activity. Since all three of these possibilities are likely to lower the activity of VirF in the cell, a necessary first step in characterizing the full impact of RyhB on the VirB/VirF regulon was to determine if RyhB influences the ability of VirF to function as a transcriptional activator.
VirF increases the activity of the icsA promoter [25], [26] (Figure 1), therefore to test the impact of RyhB production on the activity of VirF, the activity of the icsA promoter was measured in the presence or absence of increased RyhB production. Wild-type S. dysenteriae and S. flexneri were transformed with PicsA-lacZ, a reporter plasmid carrying the icsA promoter fused to an otherwise promoter-less lacZ gene [27], and the ryhB expressing plasmid pryhB [6] or pQE, the empty vector control, were subsequently introduced. To ensure that RyhB production was being up-regulated from pryhB in the presence of the inducer, a PicsP-lacZ fusion plasmid [20] was used as a control since RyhB has been shown to negatively regulate this promoter via its modulation of virB [6], [20].
As seen previously in S. flexneri [20], production of RyhB from the pryhB plasmid in S. dysenteriae resulted in a significant reduction in β-galactosidase activity produced from the PicsP-lacZ reporter (Figure 2A), indicating that the plasmid-based induction of ryhB was working as expected. In contrast, the activity of the PicsA-lacZ reporter in either S. dysenteriae or S. flexneri (Figure 2B) was not affected by the production of RyhB from the pryhB plasmid. These data indicate that RyhB production does not influence icsA promoter activity. These findings strongly suggest that RyhB does not decrease the activity of VirF, the positive regulator of icsA promoter activity.
Figure 2. RyhB does not influence the activity of VirF.
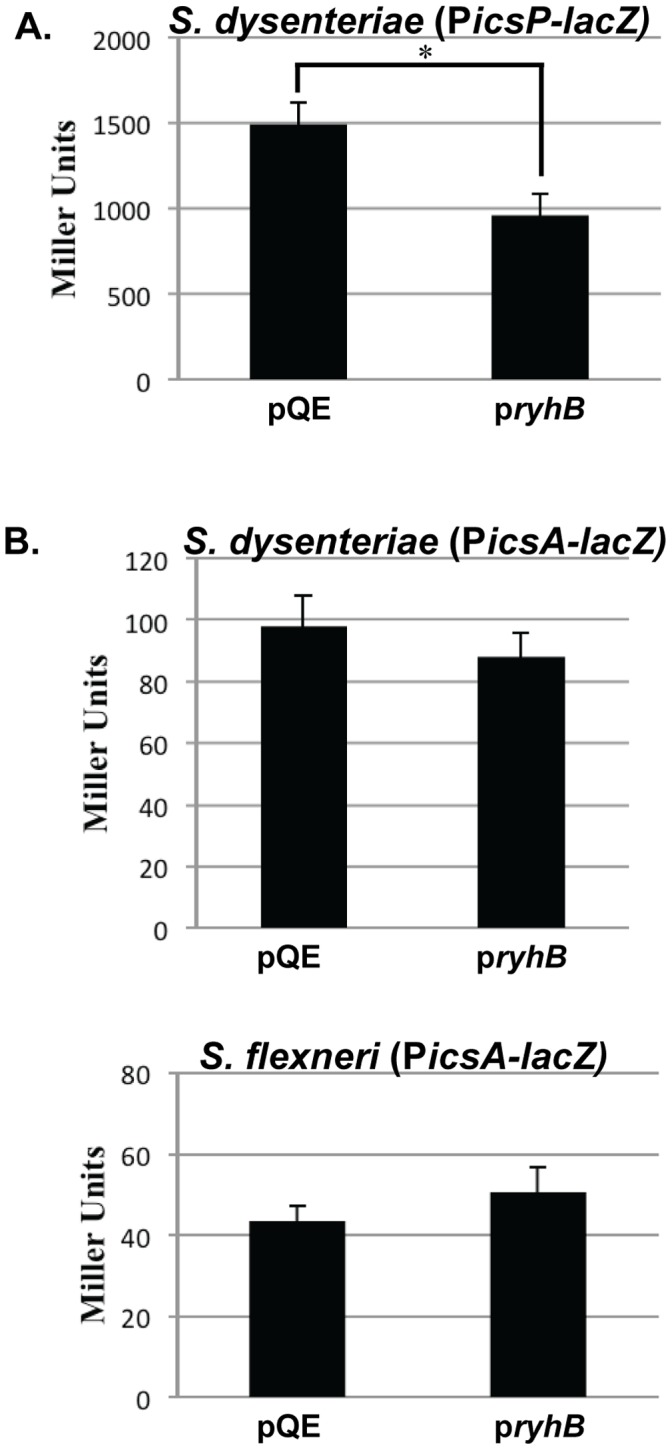
β-galactosidase activity measured in wild-type S. dysenteriae carrying PicsP-lacZ (A), and S. dysenteriae or S. flexneri carrying PicsA-lacZ (B), in both the presence (pryhB) and absence (pQE2) of increased production of RyhB from the pryhB plasmid. All cultures were grown in the presence of 200 µM IPTG to induce expression of ryhB from pryhB when present. The data is the average of three independent experiments and error bars represent one standard deviation. *represents a significant difference from the activity of the strain carrying the pQE vector control (p≤0.01).
RyhB Represses virB Transcription
The RyhB-dependent reduction of virB mRNA levels could result from lower levels of virB transcription, or from post-transcriptional regulation leading to the destabilization of the virB mRNA molecule. RyhB is known to down-regulate specific gene targets by facilitating destabilization of target mRNA molecules [28], [29], [30]. Furthermore, by a regulatory mechanism that is not yet fully characterized, stability of the S. sonnei virB mRNA molecule is altered in response to changes in environmental temperature, osmolarity and pH [14], [15], [16]. Based on these observations we next chose to determine whether or not RyhB production leads to destabilization of the virB mRNA molecule using in vivo mRNA stability assays and northern blot analyses. sodB mRNA was used as a control in each assay, as it is well established that RyhB functions to destabilize the sodB mRNA molecule [28], [31].
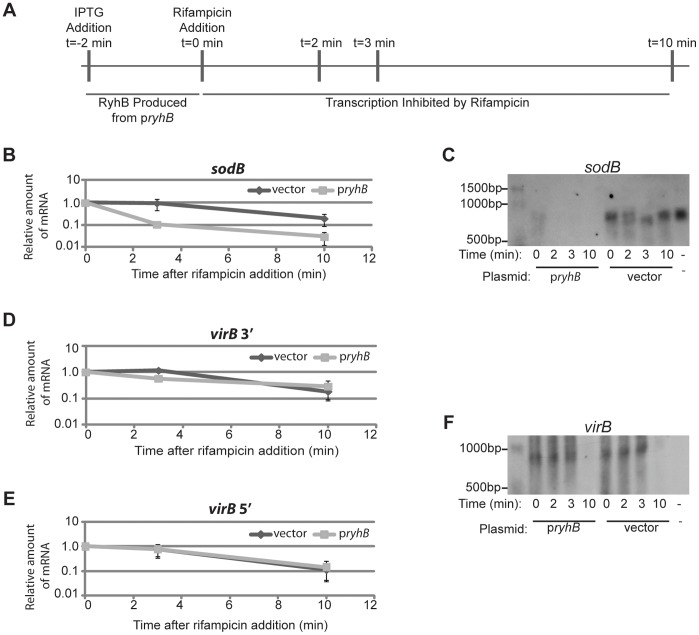
To investigate the effect of RyhB on the stability of virB mRNA and sodB mRNA, an mRNA decay assay was performed (Figure 3A). Briefly, wild-type S. dysenteriae carrying either pryhB or the vector control was cultured to the mid-logarithmic phase of growth and IPTG added to induce production of RyhB from pryhB. Two minutes after the addition of IPTG, transcription was halted by the addition of rifampicin. Samples were collected, and RNA isolated at the time of rifampicin addition (t=0) as well as 2, 3, and 10 minutes after rifampicin addition (t=2, 3 or 10). Relative amounts of virB mRNA and sodB mRNA were quantified by Real-time PCR. Analysis of sodB mRNA levels in the presence and absence of RyhB production demonstrated that the stability of S. dysenteriae sodB mRNA, like that of E. coli sodB mRNA [31], is dramatically reduced in the presence of RyhB (Figure 3B). These findings were confirmed by northern blot analysis, demonstrating that the production of RyhB from pryhB dramatically accelerates the rate of sodB mRNA degradation as compared to the rate of sodB mRNA degradation in the absence of increased RyhB production (Figure 3C). The obvious reduction in the amount of sodB mRNA seen at t=0 in the strain carrying pryhB as compared to that seen at the same time point in the strain carrying the vector control is to be expected given that RyhB causes the rapid degradation of sodB message, and that RyhB production was initiated two minutes prior to the collection of the sample (Figure 3A). These data confirm RyhB-dependent destabilization of the S. dysenteriae sodB mRNA and indicate that the in vivo assay is working as expected. Surprisingly, production of RyhB from the pryhB plasmid had no effect on the stability of virB mRNA as determined by measuring the relative abundance of both the 3′ end (Figure 3D) and 5′ end (Figure 3E) of the message using quantitative Real-time PCR analysis. As above, these findings were confirmed by northern blot analysis (Figure 3F). Northern blot analysis of virB mRNA levels was performed using a single stranded DNA probe to eliminate the possibility that a transcript encoded anti-sense to virB was being detected (see Discussion). Together, these data clearly demonstrate that, unlike RyhB-dependent regulation of sodB mRNA, RyhB-dependent regulation of virB expression is not mediated by an increase in the rate of mRNA degradation.
Figure 3. RyhB does not alter the stability of the virB mRNA molecule.
Schematic of the procedure used to induce RyhB production, inhibit transcription, and collect RNA samples for Real-time PCR and northern blot based investigations into the impact of RyhB on the stability of virB and sodB mRNA molecules (A). The relative amount of sodB mRNA (B) and virB mRNA (D and E) was quantified by Real-time PCR in the presence (light lines) and absence (dark lines) of increased RyhB production. At each time point the amount of mRNA present is expressed relative to the level of mRNA at the time of transcriptional inhibition by the addition of rifampicin (t=0) and is normalized to the amount of rrsA measured in the sample. The data is an average of three independent experiments and error bars represent one standard deviation. The same set of RNA samples was used to measure levels of both sodB and virB mRNA. Northern blot analysis was used to detect full-length sodB and virB mRNA in each RNA sample collected (C and F, respectively). The negative control lane in each northern blot contains RNA isolated from wild-type S. dysenteriae carrying the pQE vector grown at 30°C, as virB expression is inhibited at this temperature. Each northern blot shown is representative of analysis performed with three biological replicates.
Unlike that observed for sodB mRNA, virB mRNA levels were not dramatically different in the S. dysenteriae carrying the vector control as compared to the strain carrying the pryhB plasmid at the point of transcriptional inhibition (t=0, Figure 3C and Figure 3F). This finding indicates that virB mRNA levels are not altered in the short time between the induction of RyhB production and the inhibition of transcription, further supporting the conclusion that RyhB does not function to destabilize virB mRNA.
RyhB-dependent reduction in the steady state level of virB mRNA, as seen in previous studies [6], together with the finding that this reduction is not mediated by destabilization of the transcript are consistent with RyhB regulating virB mRNA levels at the level of transcription.
RyhB-dependent Regulation of virB Transcription is Facilitated by Nucleic acid Sequences within the virB Open Reading Frame
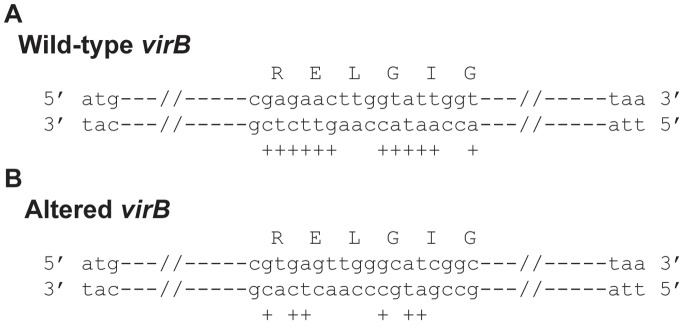
Each mechanism of direct RyhB-dependent regulation characterized to date has been shown to be mediated, at least in part, by nucleic acid complementarity between RyhB and the target mRNA molecule [28], [30], [32], [33]. The nucleic acid sequence of virB mRNA was examined for complementarity to RyhB, however no such complementarity was found. Instead, nucleic acid complementarity was observed between RyhB and the template DNA strand within the virB gene. Specifically, within the 930 nucleotide long virB coding sequence, twelve of eighteen nucleotides between base numbers 403 and 420 on the template DNA strand share perfect nucleic acid complementarity to nucleic acid sequences within RyhB (Figure 4A). Furthermore, five of the seven nucleotides between base numbers 414 and 420 of the virB gene share nucleic acid complementarity to five identically spaced nucleotides within the region of RyhB previously shown to mediate repression of virB expression [6] (Figure 4A).
Figure 4. Nucleic acid complementarity exists between RyhB and the template DNA strand within virB.
(A) A comparison of nucleic acid sequences within the wild-type virB gene to that within RyhB. The double stranded DNA sequence between base numbers 403 and 420 of 930 total bases in the wild-type virB gene is shown. The encoded amino acids are indicated using the single letter code. ";+"; indicates the location of nucleic acid sequence complementarity between the template strand DNA in the virB open reading frame and RyhB. (B) A comparison of nucleic acid sequences within the Altered virB gene to that within RyhB. The double stranded DNA sequence between base numbers 403 and 420 of 930 total bases in the Altered virB gene is shown. The encoded amino acids are indicated using the single letter code. Underlined sequences denote those that were mutated to construct Altered virB. ";+"; indicates the location of nucleic acid sequence complementarity between the template strand DNA in the Altered virB open reading frame and RyhB.
The role of nucleic acid complementarity between RyhB and the DNA sequence of the template strand within the virB gene in mediating RyhB-dependent regulation of virB transcription was investigated by site-directed mutagenesis of virB. The nucleic acid mutations introduced into the virB gene, designated Altered virB, conserve the amino acid sequence of the encoded protein, but reduced the nucleic acid complementarity between virB and RyhB to six of the original twelve bases (Figure 4B). The impact of the site-directed mutagenesis on RyhB-mediated repression of virB expression was investigated using quantitative Real-time PCR. The relative amount of either wild-type or Altered virB was measured in the presence (pryhB) or absence (vector) of increased production of RyhB. It is important to note that prior to normalization the level of Altered virB message was not significantly different than that of wild-type virB in each strain carry the vector control (data not shown). As seen previously [6] production of RyhB from the pryhB plasmid resulted in a significant decrease in the steady state levels of wild-type virB mRNA, however production of RyhB had no significant effect on the steady state level of Altered virB mRNA (Figure 5). These data indicate that the identified nucleic acid sequence within the virB gene directly or indirectly facilitate RyhB-dependent regulation of virB expression.
Figure 5. Alteration of nucleic acid sequences within virB reduces the efficiency of RyhB-mediated repression.
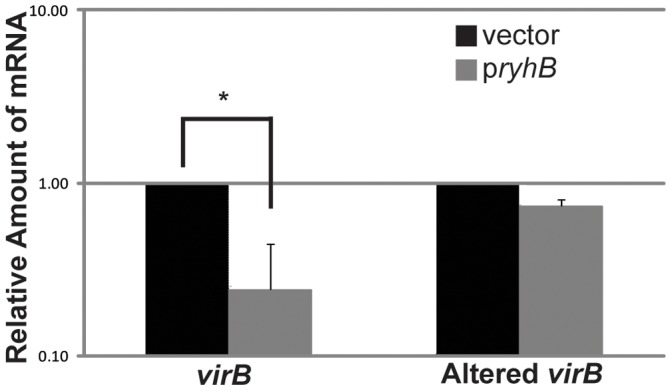
Real-time PCR analysis of the amount of wild-type virB mRNA and Altered virB mRNA in the absence (vector) and presence (pryhB) of ryhB expressed from an inducible plasmid promoter. The amount of mRNA is reported relative to the level of each in the strain carrying the vector control and is normalized to the amount of rrsA mRNA present in each sample. Each set of data is an average of three independent experiments. * denotes the existence of a significant difference between the indicated data points (p≤0.05).
The Altered virB Mutant Displays Virulence-associated Phenotypes in S. dysenteriae, but the Phenotypes are not Affected by the Production of RyhB
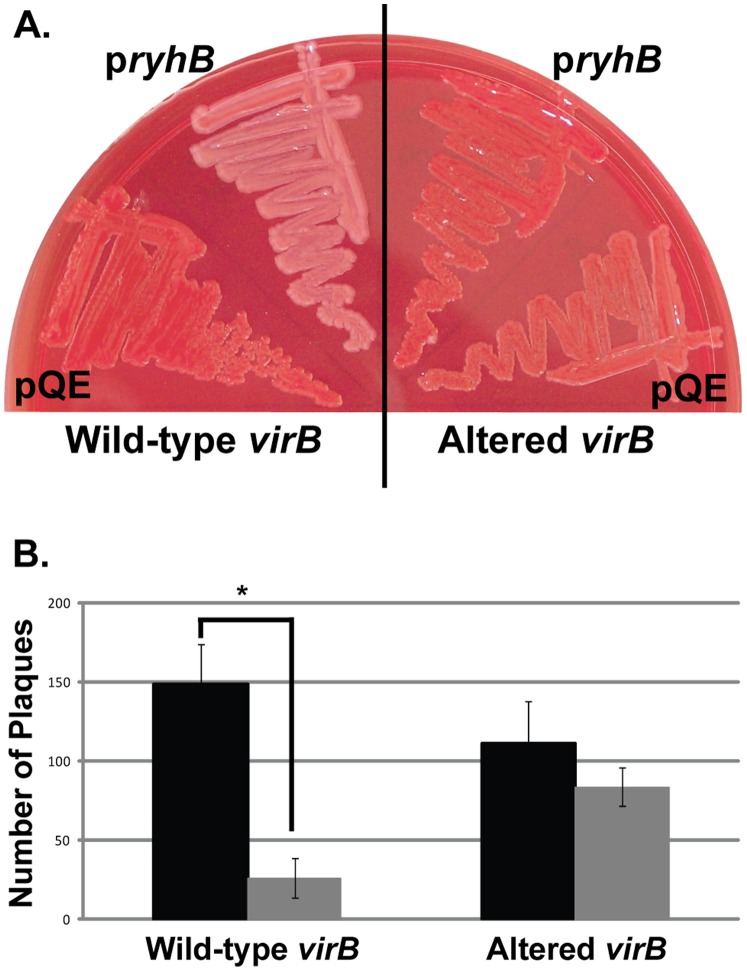
Shigella virulence is positively correlated with the ability of the bacterium to bind Congo red from an agar medium [34] and this phenotype has been used previously to determine the effects of RyhB on production of VirB-dependent gene products [6]. Therefore we next tested the ability of RyhB to suppress Congo red binding by wild-type S. dysenteriae or an Altered virB mutant. Each strain carrying the pQE vector control, displayed a positive Congo red phenotype, strongly suggesting that the mutations introduced into the Altered virB mutant do not disrupt function of the encoded protein (Figure 6A). Strikingly, unlike Congo red binding by wild-type S. dysenteriae, which was dramatically reduced following production of RyhB from the pryhB plasmid, Congo red binding by the Altered virB mutant was only slightly lower in the presence of increased RyhB production (Figure 6A).
Figure 6. RyhB has no significant affect on VirB-dependent phenotypes in S. dysenteriae expressing Altered virB.
A. Congo red binding by wild-type and Altered virB in S. dysenteriae in the presence (pryhB) and absence (pQE2) of increased production of RyhB. All strains were cultured in the presence of 200 µM IPTG to induce expression of ryhB from pryhB. B. Quantification of plaques formed by S. dysenteriae expressing wild-type (dark bar) or Altered virB (light bar) in the presence of ryhB expressed from the inducible plasmid promoter of pryhB and that measured in each strain carrying the vector control. All strains were cultured in the presence of 200 µM IPTG to induce expression of ryhB from pryhB. Data represents the average of four independent experiments. *represents a significant difference from the number of plaques formed by the strain carrying the pQE vector control (p≤0.01).
The ability of Shigella to form plaques within a Henle cell monolayer is a quantifiable virulence-associated phenotype that is dependent on the activity of VirB [23]. As a means to evaluate the ability of RyhB to inhibit VirB activity, the impact of RyhB production on plaque formation by wild-type S. dysenteriae and the Altered virB mutant was measured. As demonstrated by the number of plaques formed by wild-type S. dysenteriae carrying the vector control (149+/−25) and the Altered virB mutant carrying the vector control (111+/−27), the nucleic acid changes introduced into the mutant did not significantly affect the ability of this strain to form plaques as compared to wild-type S. dysenteriae (p=0.149) (Figure 6B). Consistent with previous data [6], production of RyhB from the pryhB plasmid significantly repressed plaque formation by wild-type S. dysenteriae as compared to the strain carrying the pQE vector control (p=0.002) (Figure 6B). In contrast, in the Altered virB mutant, increased production of RyhB had no significant effect on plaque formation, as compared to the strain carrying the pQE vector control (p=0.188).
Together, these data clearly demonstrate that alteration of nucleic acid sequences within virB that reduce the level of nucleic acid complementarity between RyhB and the DNA sequence of the template strand within virB decrease the ability of RyhB to inhibit each VirB-dependent virulence-associated phenotype investigated.
Discussion
The VirF/VirB regulatory cascade plays a central role in Shigella virulence, coordinately regulating the production of several virulence factors in response to environmental conditions encountered by the pathogen during the course of a natural infection. The hierarchical regulation of the VirF and VirB regulons [1] and the observation that factors affecting the post-transcriptional regulation of virB mRNA also affect virF expression [1], [14], [15], [16] has suggested that the expression of the VirF and VirB regulons is linked. This study reveals that RyhB modulates virB transcription and that this regulation is independent of VirF, providing the first experimental evidence of differential regulation of genes within the VirF and VirB regulons.
So far, three mechanisms of RyhB-dependent regulation have been characterized in E. coli. These mechanisms are exemplified by the regulation of sodB, the isc operon (not to be confused with the Shigella ics genes, also discussed in this work) and shiA [28], [29], [30], [31], [32], [33]. RyhB controls the expression of E. coli sodB, encoding superoxide dismutase, by binding to and ultimately destabilizing the sodB mRNA molecule [28], [29], [30]. Additionally, RyhB can facilitate differential gene expression from polycistronic mRNA molecules by altering the stability of a portion of the message, as has been demonstrated for the isc operon [33]. Finally, RyhB is predicted to promote the translational efficiency of E. coli shiA by binding to the 5′ untranslated region of the message and altering the structure such that an inhibitory element is eliminated [32]. Although the molecular mechanisms vary, common to each identified mechanism of RyhB-dependent regulation is the fact that regulation occurs following transcription and is dependent upon nucleic acid complementarity between RyhB and the target mRNA molecule.
As demonstrated in this study, several features of RyhB-mediated regulation of virB differentiate it from RyhB-dependent regulation of previously characterized gene targets. Unlike other targets of RyhB-mediated regulation that are chromosomally located, virB is encoded on a large virulence plasmid [4]. The regulation of a horizontally acquired, genus specific, virulence factor, by a conserved, chromosomally located, regulatory RNA, provides further evidence of the versatility of RyhB in controlling bacterial gene expression. The lack of nucleic acid complementarity between RyhB and the virB mRNA, as well as the presence of nucleic acid complementarity between RyhB and the template DNA strand within the virB gene distinguishes virB regulation from that of previously characterized RyhB targets.
The presence of nucleic acid complementarity between RyhB and template strand DNA within virB raises the possibility that RyhB is complementary to an mRNA or sRNA encoded anti-sense to virB. If present, the anti-sense transcript may play an intermediary role in facilitating RyhB-dependent regulation of virB transcription. In this study, the presence of such a transcript was investigated by northern blot using a single-stranded DNA probe with nucleic acid identity to virB. This analysis failed to detect a transcript anti-sense to virB in the region of the message with sequence similarity to RyhB (data not shown). These findings strongly suggest that RyhB affects virB mRNA levels via its complementarity with the template stand of virB.
While disruption of the nucleic acid complementarity between RyhB and the template strand of virB ( [6], Figure 4), reduces the efficiency of both RyhB-dependent regulation of virB expression (Figure 5) and RyhB-dependent modulation of virulence phenotypes (Figure 6), the precise role of these sequences in facilitating regulation remains unknown.
Finally, in contrast to previously characterized mechanisms of RyhB-dependent gene regulation, we have demonstrated that RyhB-dependent regulation of virB expression is mediated, directly or indirectly, at the level of virB transcription. Aside from RyhB and VirF, all previously identified factors shown to regulate virB transcription also influence virF expression. Thus, if RyhB-dependent regulation of virB transcription is indirect, it is mediated via the regulation of an as-of-yet unidentified transcriptional regulator. Future in vitro assays will explore the molecular mechanism underlying RyhB-dependent regulation of virB transcription.
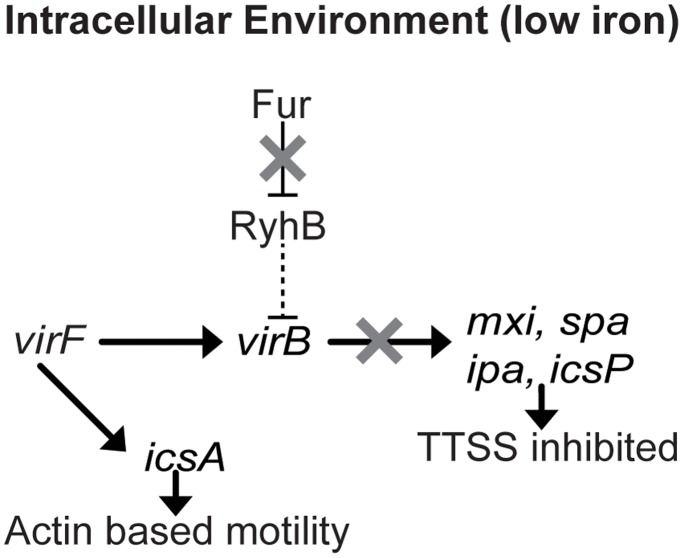
In Shigella species, the RyhB-dependent regulation of VirB activity may allow the VirB regulon (genes encoding the TTSS) to be regulated, without affecting the expression of genes in the VirF regulon (icsA, which is required for actin-based motility). Independent regulation of the VirF and VirB regulons may be advantageous to the bacterium at specific times during an infection within the human host. Specifically, once within the intracellular environment Shigella must repress production of the TTSS in order to prevent premature lysis of the eukaryotic cell, while allowing the production of IcsA in order to facilitate intracellular spread. As transcription of ryhB is under control of the iron-responsive regulator Fur [6], [28], [35], and gene expression analysis has demonstrated that several Fur-regulated genes are expressed within the intracellular environment of epithelial cells [36], [37], [38], it is reasonable to expect that RyhB will be produced when S. dysenteriae is within this intracellular environment, as discussed in Africa et al [20]. Under these conditions, RyhB may function to inhibit virB transcription, thus reducing production of TTSS while simultaneously allowing VirF-activated transcription of icsA to proceed (Figure 7).
Figure 7. Working model of RyhB-dependent repression of virB transcription under iron poor conditions encountered within the intracellular environment.
Under iron poor conditions, mimicking those likely encountered within the eukaryotic cell, Fur does not inhibit RyhB production and the RyhB functions to, directly or indirectly, inhibit virB transcription. Under such conditions RyhB-dependent VirF-independent regulation of virB expression facilitates the differential modulation of TTSS activity and actin based motility.
In conclusion, this study demonstrates that virB transcription can be uncoupled from the activity of VirF by RyhB, revealing a unique mechanism by which genes within the VirF and VirB regulons can be differentially regulated. These findings not only expand the current understanding of the regulatory circuit controlling Shigella virulence gene expression, but also highlight the versatility of RyhB in regulating bacterial gene expression.
Acknowledgments
The authors would like to thank Dr. Marcia B. Goldberg for providing the PicsA-lacZ reporter plasmid. The authors would also like to thank Dr. Peter Coschigano and members of the Murphy laboratory for critical reading of this manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded in part by Ruth L. Kirschstein National Research Service Award National Institutes of Health (NIH) 8-FGM069312A, a grant from the Ohio University Research Committee and by the Ohio University Heritage College of Osteopathic Medicine (to ERM), as well as by NIH grants AI-16935 (to SMP) and 5P20RR016464-10 (to HJW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jennison AV, Verma NK. Shigella flexneri infection: pathogenesis and vaccine development. FEMS Microbiol Rev. 2004;28:43–58. doi: 10.1016/j.femsre.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Clerc P, Baudry B, Sansonetti PJ. Molecular mechanisms of entry, intracellular multiplication and killing of host cells by shigellae. Curr Top Microbiol Immunol. 1988;138:3–13. [PubMed] [Google Scholar]
- 3.Dorman CJ, Porter ME. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol Microbiol. 1998;29:677–684. doi: 10.1046/j.1365-2958.1998.00902.x. [DOI] [PubMed] [Google Scholar]
- 4.Adler B, Sasakawa C, Tobe T, Makino S, Komatsu K, et al. A dual transcriptional activation system for the 230 kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol Microbiol. 1989;3:627–635. doi: 10.1111/j.1365-2958.1989.tb00210.x. [DOI] [PubMed] [Google Scholar]
- 5.Tobe T, Yoshikawa M, Mizuno T, Sasakawa C. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by VirF and repression by H-NS. J Bacteriol. 1993;175:6142–6149. doi: 10.1128/jb.175.19.6142-6149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy ER, Payne SM. RyhB, an Iron-Responsive Small RNA Molecule, Regulates Shigella dysenteriae Virulence. Infect Immun. 2007;75:3470–3477. doi: 10.1128/IAI.00112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gore AL, Payne SM. CsrA and Cra influence Shigella flexneri pathogenesis. Infection And Immunity. 2010;78:4674–4682. doi: 10.1128/IAI.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tobe T, Nagai S, Okada N, Adler B, Yoshikawa M, et al. Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Molecular Microbiology. 1991;5:887–893. doi: 10.1111/j.1365-2958.1991.tb00762.x. [DOI] [PubMed] [Google Scholar]
- 9.Goldberg MB, Theriot JA, Sansonetti PJ. Regulation of surface presentation of IcsA, a Shigella protein essential to intracellular movement and spread, is growth phase dependent. Infection And Immunity. 1994;62:5664–5668. doi: 10.1128/iai.62.12.5664-5668.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wing HJ, Yan AW, Goldman SR, Goldberg MB. Regulation of IcsP, the outer membrane protease of the Shigella actin tail assembly protein IcsA, by virulence plasmid regulators VirF and VirB. Journal Of Bacteriology. 2004;186:699–705. doi: 10.1128/JB.186.3.699-705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Gall T, Mavris M, Martino MC, Bernardini ML, Denamur E, et al. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiology (Reading, England) 2005;151:951–962. doi: 10.1099/mic.0.27639-0. [DOI] [PubMed] [Google Scholar]
- 12.Prosseda G, Fradiani PA, Di Lorenzo M, Falconi M, Micheli G, et al. A role for H-NS in the regulation of the virF gene of Shigella and enteroinvasive Escherichia coli. . Research In Microbiology. 1998;149:15–25. doi: 10.1016/s0923-2508(97)83619-4. [DOI] [PubMed] [Google Scholar]
- 13.Porter ME, Dorman CJ. Positive regulation of Shigella flexneri virulence genes by integration host factor. J Bacteriol. 1997;179:6537–6550. doi: 10.1128/jb.179.21.6537-6550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitobe J, Arakawa E, Watanabe H. A sensor of the two-component system CpxA affects expression of the type III secretion system through posttranscriptional processing of InvE. Journal Of Bacteriology. 2005;187:107–113. doi: 10.1128/JB.187.1.107-113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitobe J, Morita-Ishihara T, Ishihama A, Watanabe H. Involvement of RNA-binding protein Hfq in the post-transcriptional regulation of invE gene expression in Shigella sonnei. The Journal Of Biological Chemistry. 2008;283:5738–5747. doi: 10.1074/jbc.M710108200. [DOI] [PubMed] [Google Scholar]
- 16.Mitobe J, Morita-Ishihara T, Ishihama A, Watanabe H. Involvement of RNA-binding protein Hfq in the osmotic-response regulation of invE gene expression in Shigella sonnei. BMC Microbiology. 2009;9:110–110. doi: 10.1186/1471-2180-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakayama S, Watanabe H. Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J Bacteriol. 1995;177:5062–5069. doi: 10.1128/jb.177.17.5062-5069.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falconi M, Colonna B, Prosseda G, Micheli G, Gualerzi CO. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. The EMBO Journal. 1998;17:7033–7043. doi: 10.1093/emboj/17.23.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falconi M, Prosseda G, Giangrossi M, Beghetto E, Colonna B. Involvement of FIS in the H-NS-mediated regulation of virF gene of Shigella and enteroinvasive Escherichia coli. Molecular Microbiology. 2001;42:439–452. doi: 10.1046/j.1365-2958.2001.02646.x. [DOI] [PubMed] [Google Scholar]
- 20.Africa LAA, Murphy ER, Egan NR, Wigley AF, Wing HJ. The Iron-Responsive Fur/RyhB Regulatory Cascade Modulates the Shigella Outer Membrane Protease IcsP. Infection And Immunity. 2011;79:4543–4549. doi: 10.1128/IAI.05340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual; Argentine J, editor. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. 2001.
- 22.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2∧-ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Oaks EV, Wingfield ME, Formal SB. Plaque formation by virulent Shigella flexneri. Infect Immun. 1985;48:124–129. doi: 10.1128/iai.48.1.124-129.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lodge J, Fear J, Busby S, Gunasekaran P, Kamini NR. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiology Letters. 1992;74:271–276. doi: 10.1016/0378-1097(92)90441-p. [DOI] [PubMed] [Google Scholar]
- 25.Porter ME, Dorman CJ. Differential regulation of the plasmid-encoded genes in the Shigella flexneri virulence regulon. Mol Gen Genet. 1997;256:93–103. doi: 10.1007/s004380050550. [DOI] [PubMed] [Google Scholar]
- 26.Sakai T, Sasakawa C, Yoshikawa M. Expression of four virulence antigens of Shigella flexneri is positively regulated at the transcriptional level by the 30 kiloDalton VirF protein. Mol Microbiol. 1988;2:589–597. doi: 10.1111/j.1365-2958.1988.tb00067.x. [DOI] [PubMed] [Google Scholar]
- 27.Castellanos MI, Harrison DJ, Smith JM, Labahn SK, Levy KM, et al. VirB alleviates H-NS repression of the icsP promoter in Shigella flexneri from sites more than one kilobase upstream of the transcription start site. Journal Of Bacteriology. 2009;191:4047–4050. doi: 10.1128/JB.00313-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Afonyushkin T, Vecerek B, Moll I, Blasi U, Kaberdin VR. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005;33:1678–1689. doi: 10.1093/nar/gki313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vecerek B, Moll I, Afonyushkin T, Kaberdin V, Blasi U. Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol Microbiol. 2003;50:897–909. doi: 10.1046/j.1365-2958.2003.03727.x. [DOI] [PubMed] [Google Scholar]
- 31.Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. . Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prevost K, Salvail H, Desnoyers G, Jacques J-Fo, Phaneuf E, et al. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Molecular Microbiology. 2007;64:1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- 33.Desnoyers G, Morissette A, Prevost K, Masse E. Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. The EMBO Journal. 2009;28:1551–1561. doi: 10.1038/emboj.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Payne SM, Finkelstein RA. Detection and differentiation of iron-responsive avirulent mutants on Congo red agar. Infect Immun. 1977;18:94–98. doi: 10.1128/iai.18.1.94-98.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oglesby AG, Murphy ER, Iyer VR, Payne SM. Fur regulates acid resistance in Shigella flexneri via RyhB and ydeP. Mol Microbiol. 2005;58:1354–1367. doi: 10.1111/j.1365-2958.2005.04920.x. [DOI] [PubMed] [Google Scholar]
- 36.Payne SM, Wyckoff EE, Murphy ER, Oglesby AG, Boulette ML, et al. Iron and pathogenesis of Shigella: iron acquisition in the intracellular environment. Biometals: An International Journal On The Role Of Metal Ions In Biology, Biochemistry, And Medicine. 2006;19:173–180. doi: 10.1007/s10534-005-4577-x. [DOI] [PubMed] [Google Scholar]
- 37.Lucchini S, Liu H, Jin Q, Hinton JC, Yu J. Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect Immun. 2005;73:88–102. doi: 10.1128/IAI.73.1.88-102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Runyen-Janecky LJ, Payne SM. Identification of chromosomal Shigella flexneri genes induced by the eukaryotic intracellular environment. Infect Immun. 2002;70:4379–4388. doi: 10.1128/IAI.70.8.4379-4388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. Journal Of Bacteriology. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mills M, Payne SM. Identification of shuA, the gene encoding the heme receptor of Shigella dysenteriae, and analysis of invasion and intracellular multiplication of a shuA mutant. Infect Immun. 1997;65:5358–5363. doi: 10.1128/iai.65.12.5358-5363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Formal SB, Dammin GJ, Labrec EH, Schneider H. Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. Journal Of Bacteriology. 1958;75:604–610. doi: 10.1128/jb.75.5.604-610.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wyckoff EE, Mey AR, Leimbach A, Fisher CF, Payne SM. Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. Journal Of Bacteriology. 2006;188:6515–6523. doi: 10.1128/JB.00626-06. [DOI] [PMC free article] [PubMed] [Google Scholar]