Abstract
Histone modifications influence the interactions of transcriptional regulators with chromatin. Studies in embryos and embryonic stem (ES) cells have uncovered histone modification patterns that are diagnostic for different cell types and developmental stages. For example, bivalent domains consisting of regions of H3 lysine 27 trimethylation (H3K27me3) and H3 lysine 4 trimethylation (H3K4me3) mark lineage control genes in ES cells and zebrafish blastomeres. Such bivalent domains have garnered attention because the H3K27me3 mark might help repress lineage regulatory genes during pluripotency while the H3K4me3 mark could poise genes for activation upon differentiation. Despite the prominence of the bivalent domain concept, studies in other model organisms have questioned its universal nature and the function of bivalent domains has remained unclear. Histone marks are also associated with developmental regulatory genes in sperm. These observations have raised the possibility that specific histone modification patterns might persist from parent to offspring, but it is unclear whether histone marks are inherited or formed de novo. Here, we review the potential roles of H3K4me3 and H3K27me3 marks in embryos and ES cells and discuss how histone marks might be established, maintained and resolved during embryonic development.
Introduction
Histones are subject to various modifications, including methylation, acetylation, phosphorylation, ubiquitination and ribosylation [1]. These modifications alter protein-DNA and protein-protein interactions and regulate the interaction of transcriptional regulators with chromatin [2,3] (see Box 1 for more information about chromatin and specific histone modifications). Immunofluorescence studies have revealed that global patterns of histone modifications and chromatin architecture change during the early stages of development [4–8]. Genome-wide Chromatin Immuno Precipitation (ChIP) analyses have suggested that specific combinations of histone marks at promoters and enhancers correlate with the developmental potential and fate of cells [9–24]. For example, embryonic stem cells have a different histone modification landscape than cells with restricted fates [9–19,21–24]. The importance of these modifications in embryogenesis is highlighted by the severe phenotypes caused by mutations in histone modifying complexes (see Table 1 for a summary of mouse and ES cell phenotypes [25–74][75]). Here we review the potential roles of histone modifications during embryonic development with a focus on H3 lysine 27 trimethylation (H3K27me3) and H3 lysine 4 trimethylation (H3K4me3) marks at promoters in vertebrate embryos and embryonic stem cells.
Box 1. Chromatin at a glance.
Chromatin
Chromatin refers to DNA and its associated proteins. The basic subunit of chromatin is the nucleosome, an octamer of four core histone proteins; two copies each of H2A, H2B, H3 and H4, around which ~147 bp of DNA is wrapped [142]. Five major types of changes in chromatin structure that affect gene expression have been characterized: (i) DNA methylation. The methyl group that is added to the cytosine of CG dinucleotides (as well as cytosines in other contexts) is thought to alter chromatin density and accessibility of DNA, thereby modulating the transcriptional potential of the underlying DNA sequence [143]. (ii) Histone variants can replace canonical histones [144]. (iii) Histones can be modified post-translationally [1,144]. Histone variants and modifications can affect transcription either in cis (by sterically hindering DNA-protein interactions, by changing the charge of chromatin, or by changing the stability of the nucleosome) or in trans (by creating binding platforms for downstream effectors). (iv) Chromatin can be remodeled and compacted by ATP-dependent chromatin remodelers [145]. These chromatin remodelers can be recruited to specific locations in the genome by modified histones or by proteins with sequence specificity. (v) Long-range interactions can affect higher order chromatin structure and transcription by bridging distant sites in the genome [137,139]. This review focuses on two specific histone modifications, H3K4me3 and H3K27me3.
Histone modifications
Technological advances have allowed researchers to map histone modifications throughout the genome by combining chromatin immunoprecipitation (ChIP) with DNA microarray (ChIP-chip) or deep sequencing (ChIP-Seq). These studies have revealed that modifications can mark large chromatin domains or regulatory elements such as promoters or enhancers. They have also associated specific histone modifications with transcriptional output [20,81,146–148]. For example, histone acetylation increases the accessibility of DNA by weakening the interaction between histones and DNA and by binding chromatin-remodeling complexes that contain bromodomains. Acetylated lysines are generally associated with genes that are actively transcribed [1]. Histone methylation is more complex as lysines may be mono-, di- or trimethylated (me1, me2, me3). These modifications can provide binding sites for both positive and negative transcriptional regulators [1]. Lysine trimethylation (H3K4me3, laid down by Trithorax (Trx)/Mixed lineage leukemia (Mll) proteins) is often found at promoters. H3K4me3 binds chromatin remodelers that contain a chromodomain or a PHD finger [149,150]. H3K27me3 (laid down by Polycomb group proteins) is associated with genes that are repressed. Transcriptional repression by Polycomb group proteins is mediated by the action of two complexes: Polycomb Repressive Complex 1 and 2 (PRC1 and PRC2). Ezh2, a component of PRC2, catalyzes trimethylation of H3K27. A chromodomain protein in PRC1 specifically recognizes H3K27me3. Together, PRC1 and PRC2 repress transcription. While it was initially suggested that Polycomb-repressed chromatin restrains RNA polymerase II from entering the elongation phase via ubiquitination of H2A [128,129], it was recently shown that H3K27me3 marked genes have reduced levels of RNA polymerase II [125,126], perhaps due to the compaction of chromatin [127].
Table 1.
Representative examples that illustrate mouse and ES cell phenotypes associated with the loss of histone modifiers. Please note that the reported defects have not been causally linked to the loss of histone modifications and could thus be due to other functions of the histone modifiers. ko: knock-out, kd: knock-down. dko: double knockout.
| gene | domains | Phenotype in vitro (mESC) | Phenotype in vivo (mouse) | Key references |
|---|---|---|---|---|
| MLL | ||||
| mll | SET domain | Embryoid bodies display reduced hematopietic potential. Failure to activate hox genes. | Embryonic lethal (E11.5–14.5), impaired segmental identity, reduction in hematopoietic precursors. | [25, 31, 32] |
| mll2 | SET domain | ES cells pluripotent. Proliferation defects due to increased rates of apoptosis. Compromised timing and coordination of lineage commitment. | Conditional ko: genome-wide reduction in H3K4me3 in oocyte, oocyte death. Full ko: embryonic failure before E11.5. Slowed growth, increased apoptosis, retarded development, male sterility, failure to maintain hox gene expression. | [27–30] |
| dpy-30* | Self renewal unaffected but failure to properly differentiate. Lineage-associated genes are not properly activated. Global downregulation of H3K4me3. | [26] | ||
| wdr5* | WD repeats | Severe defects in ES cell maintenance, reduced expression of key pluripotency genes. Global downregulation of H3K4me3. | [57, 58] | |
| ash2l | SPRY domain | Failure to derive ES cells from mutant blastocyst. | Embryonic lethal early during gestation. | [56] |
| men1 | Embryonic lethal (E11.5–12.5). Heterozygotes develop endocrine tumors. | [53–55] | ||
| PRC2 | ||||
| ezh2 | SET domain | ES cells fail to undergo mesendoderm differentiation, but phenotype is less severe than eed null, because of partial redundancy with Ezh1. Decreased levels of bulk H3K27me2 and me3, not me1. Specifically at developmental genes, H3K27me3 and me1 are still significantly enriched, because of partial redundancy with Ezh1. | Depletion of maternal Ezh2: Eed localisation, H3K27me3 and H3K9me3 patterns affected. Severe growth retardation of neonates. Full ko: Early post-implantation lethality (E8.5). Gastrulation defects. | 51, 52, 74] |
| ezh1*;ezh2 | SET domain | Depletion of Ezh1 in Ezh2 null cells abolishes residual methylation on H3K27 and de-represses H3K27me3 target genes. | [51] | |
| eed | WD40 repeats | ES cells are pluripotent but fail to differentiate properly. Genome-wide decrease in H3K27me1, me2 and me3. Target genes are de-repressed. Decrease in Ezh2 protein levels. | Embryonic lethal (~E8.5). Failure to properly gastrulate and to produce embryonic mesoderm. Disrupted axial patterning. | [42, 44–48, 51] |
| suz12 | Zinc-finger domain | Failure to properly differentiate. Global loss of H3K27me2 and me3, de-repression of lineage-specific genes. Decrease in Ezh2 protein levels. | Early post-implantation lethality (~E7.5). Severe developmental (gastrulation) and proliferative defects. | [36, 40] |
| yy1 | Zinc-finger domain | Peri-implantation lethality. Developmental and proliferative defects. | [39] | |
| jarid2 | JmjC domain | Failure to properly differentiate. Global levels of H3K27me3 unaffected but H3K27me levels up on some target genes, down on others. | Embryonic lethal (E10.5–15.5, depending on genetic background). Neural, cardiac, liver and hematopoietic defects. | [35, 37, 38, 41, 49, 50] |
| pcl2/mtf2 | PHD-finger domain | Failure to properly differentiate. Global levels of H3K27me3 unaffected. Upregulated pluripotency factors. | Viable, but growth defects. Low penetrance posterior homeotic transformation. | [33, 34] |
| PRC1 | ||||
| ring1b (rnf2) | RING-finger domain | Embryoid body formation is abnormal. Global loss of H2Aub, not H3K27me3. Upregulation of target genes, e.g. lineage regulators. Decrease in Bmi1 levels. | Embryonic lethal by E10.5. Developmental arrest in early gastrulation, similar to PRC2 component mutants. | [43, 44, 71–73] |
| ring1a(ring1);ring1b(rnf2) | RING-finger domain | Loss of ES cell morphology. Developmental regulators are de-repressed. | Viable. Anterior transformation and other axial skeletal patterning abnormalities, both in heterozygous and homozygous mutants. | [69, 70] |
| bmi1 | RING-finger domain | Viable. Posterior homeotic transformations. Neurological abnormalities. Hematopoietic defects. Bmi1/Mel18 dko mice display strongly exacerbated phenotypes. | [65, 67, 68] | |
| mel18 | RING-finger domain | Mice die 4 weeks after birth exhibiting strong growth retardation. Posterior homeotic transformations. Bmi1/Mel18 dko mice display strongly exacerbated phenotypes. | [65, 66] | |
| m33 | Chromodomain | Most mice die between birth and 4 weeks of age. Severe growth defects. Homeotic transformations. | [64] | |
| rae28 | Zinc finger SPM domain | Perinatal lethality. Posterior skeletal transformations. Various defects in neural-crest related tissues. | [75] | |
| H3K9-methyltransferases | ||||
| glp/ehmt(euchromatin) | SET domain | Embryonic lethality (~E9.5). Severe growth retardation. Global loss of H3K9me1 and me2, H3K9me3 unaffected. | [62] | |
| g9a/ehmt2 (euchromatin) | SET domain | ES cells can be maintained in culture but display growth defects during differentiation. Decreased levels of bulk H3K9me2. | Embryonic lethality (E9.5–12.5). Severe growth retardation. Global loss of H3K9me2. | [63] |
| eset/setdb1 (euchromatin) | SET domain | Defects in ICM outgrowth, no derivation of mutant ES cells possible. H3K9me2 and me3 levels largely unaffected. RNAi kd results in loss of ES cell morphology and upregulation of differentiation markers. | Peri-implantation lethality (E3.5–5.5) | [60, 61] |
| suv39h1;suv39h2 (pericentric heterochromatin) | SET domain | Reduced viability after E12.5. Growth retardation. Increased risk of tumorigenesis. Chromosomal instability. Loss of H3K9me3 from heterochromatin. (single mutants are normally viable and do not exhibit apparent phenotypes) | [59] |
genes that have only been analyzed in RNAi knockdown studies.
Bivalent Promoters in Embryonic Stem Cells
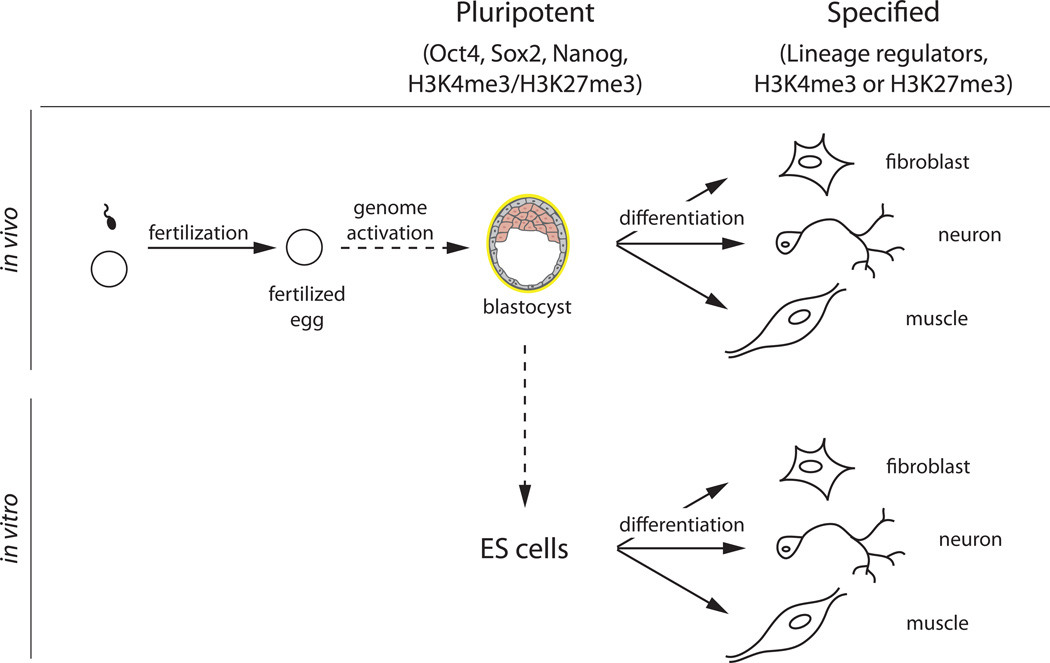
Pluripotent cells from the inner cell mass of mammalian blastocysts can generate embryonic stem (ES) cells [76]. These cells are self-renewing and can give rise to all lineages of the developing organism (Figure 1). Pluripotency is maintained by the activity of a set of transcriptional regulators that include Nanog, Oct4 and Sox2 [77]. In contrast, transcriptional regulators that determine specific cell lineages are not expressed at significant levels in pluripotent cells. During differentiation, these lineage regulators are activated and pluripotency genes are repressed (Figure 1).
Figure 1. Pluripotency and differentiation of embryonic cells.
The embryonic genome is initially transcriptionally inactive after fertilization. At the blastocyst stage, cells are pluripotent and transcriptionally active. Pluripotent cells from the inner cell mass of mammalian blastocysts can be used to generate embryonic stem (ES) cells. Pluripotent blastomeres and ES cells can give rise to all lineages of the developing organism. Pluripotency is characterized by the presence of the pluripotency factors (Oct4, Sox2 and Nanog). Transcriptional regulators that determine specific cell lineages are not expressed at significant levels in pluripotent cells, and they are often marked by bivalent chromatin domains (H3K4me3/H3K27me3). Recently, it has been shown that a subset of bivalent genes is also marked and repressed by the presence of H3K9me3 marks, adding another layer of repression to this subset of lineage regulators [60]. During differentiation, specific sets of lineage regulators are activated.
The analysis of histone modifications in embryonic stem cells has generated genome-wide location maps of H3K27me3 and H3K4me3 [9–14], catalyzed by Polycomb and Trithorax group proteins, respectively [78]. These studies indicate that many promoters are associated with both H3K4me3 and H3K27me3 [9–14]. The apparent co-localization of H3K4me3 and H3K27me3 might be due to population averaging and reflect heterogeneity within the ES cell population. In such cases, H3K4me3 marks occupy a given promoter in only a subset of cells, whereas H3K27me3 marks are present in a different subpopulation [24]. However, sequential Chromatin Immuno Precipitation (ChIP) has shown that H3K4me3 and H3K27me3 can co-occupy some promoters in ES cells [9,13]. Interestingly, these ‘bivalent’ chromatin domains often mark lineage regulatory genes.
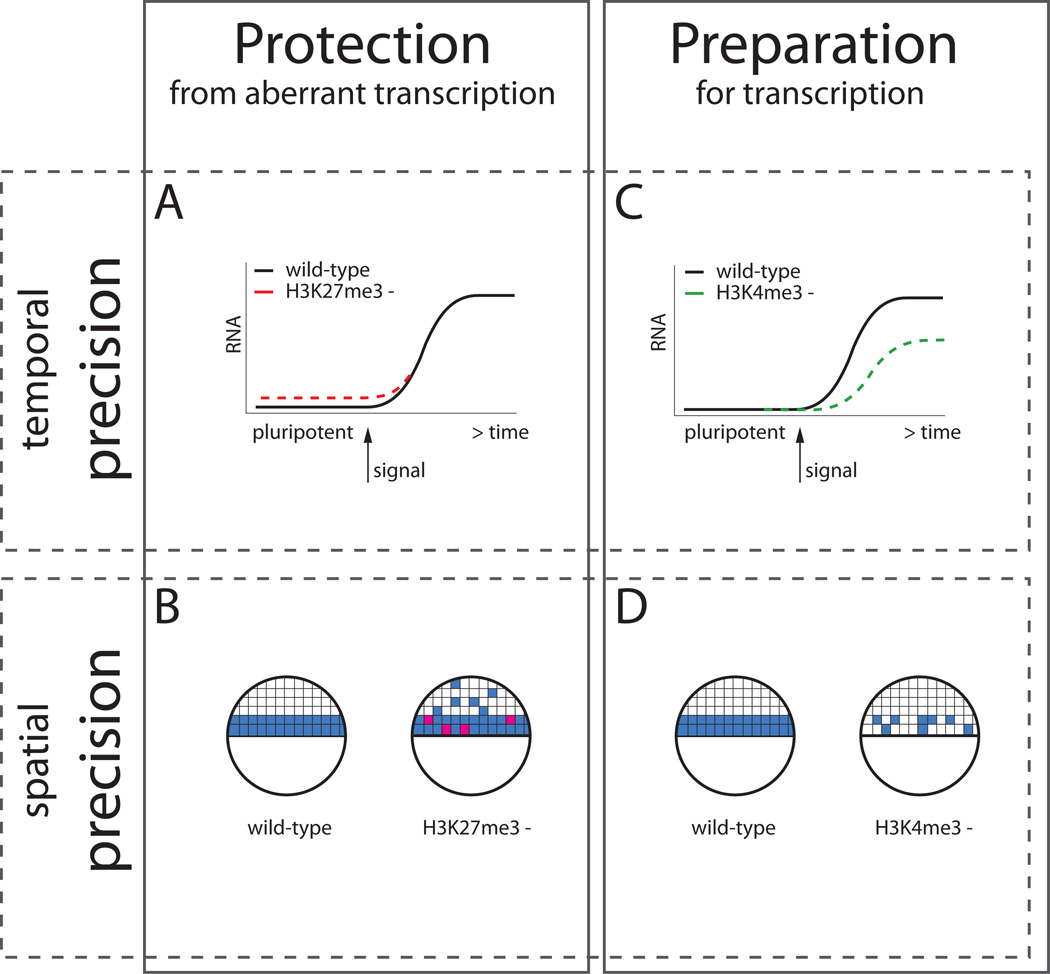
Bivalent domains have garnered wide attention, because they might contribute to the precise unfolding of gene expression programs during pluripotency and differentiation. In particular, it has been proposed that bivalent domains might repress lineage control genes (H3K27me3) during pluripotency while keeping them poised for activation upon differentiation (H3K4me3) (Figure 2). In this model, H3K27me3-mediated repression of developmental control genes might protect cells from the aberrant expression of lineage regulators and thus help maintain pluripotency (Figure 2A). During the differentiation into specific cell types, continued association with H3K27me3 might maintain the repression of the majority of developmental control genes while only a specific subset of regulators is activated in a given lineage (Figure 2B). Conversely, it has been proposed that H3K4me3 might poise developmental regulators for activation upon differentiation. In this scenario, H3K4me3 might make the induction of developmental genes more efficient (Figure 2C) or more synchronous [79] (Figure 2D). H3K4me3 might also protect genes from permanent silencing, for example by repelling transcriptional repressors or blocking DNA methylation [80]. Thus, it is possible that bivalent domains convey temporal and spatial precision to the expression of lineage control genes during pluripotency and differentiation. In the following sections we review the evidence for the postulated roles of bivalent domains in ES cells and their potential occurrence in embryonic cells in vivo.
Figure 2. Potential roles of H3K27me3 and H3K4me3 in developmental gene expression.
(A) Loss of H3K27me3 might result in the derepression of developmental genes that are normally not expressed in pluripotent cells. (B) During lineage specification, the loss of H3K27me3 might result in both the derepression of developmental control genes (ectopic blue cells) and a failure to properly activate genes (pink cells), perhaps due to the misexpression of pluripotency genes or other lineage genes. (C) Loss of H3K4me3 might result in a less efficient induction of gene expression during differentiation. (D) Loss of H3K4me3 might result in loss or stochastic activation of gene expression.
Bivalent Promoters in Embryonic Cells?
The identification of bivalent domains in permanently pluripotent ES cells (and potentially in differentiated cell types [11,13,81–83]) raises the question how relevant these findings are to transiently pluripotent cells in the embryo. Direct evidence for bivalent domains in vivo comes from studies in zebrafish: sequential ChIP has established H3K4me3/H3K27me3 co-occupancy of promoters in zebrafish blastomeres [84]. A study of mouse epiblast cells has also found putative bivalent domains but did not assess the simultaneous association of both chromatin marks in the same cell [85]. As in ES cells, H3K4me3 and H3K27me3 marks are enriched at the promoters of lineage regulators in mouse and zebrafish [84–86]. Surprisingly, however, H3K27me3 and bivalent domains have not been found in Xenopus blastomeres [87,88], and studies in Drosophila embryos have been unable to identify bivalent domains [89]. These observations do not exclude the possibility that these domains arise later during development [11,13,18,81–83], but it remains unclear how universal bivalent domains are across species.
The Function of H3K27me3 in Bivalent Chromatin Domains – Repression
It has been postulated that bivalently marked lineage-specific genes in ES cells are kept transcriptionally inactive by H3K27me3 [9,10] (Figure 2A). Indeed, loss of components of Polycomb Repressive Complex 2 (PRC2) results in a loss of H3K27me3 and a partial derepression of genes that are normally bivalent and repressed [36,45,46,51]. It was initially proposed that H3K27me3-mediated repression of lineage regulators was essential for maintenance of ES cell pluripotency [45,90]. However, despite the ectopic expression of transcription factors involved in lineage specification and a higher propensity to differentiate, ES cells can be derived from PRC2-deficient blastocysts and maintained in culture for many generations [36,44–46,51,69]. During differentiation, however, mutant cells display multiple phenotypes. While mutant ES cells can differentiate into ectoderm, mesoderm and endoderm [36,44,46,51], lineage regulators are not properly activated [36,51] (Figure 2B). This defect might seem paradoxical, because lineage regulators are prematurely activated in PRC2-deficient ES cells and the loss of PRC2 and H3K27me3 should promote gene activation. It is possible that the ectopic activation of genes from alternative lineages interferes with the execution of the proper developmental programs (Figure 2B). Furthermore, the failure to extinguish the expression of pluripotency genes may also affect proper differentiation and the activation of lineage specific gene expression programs [36].
Deficiencies in subunits of PRC2 also cause severe developmental defects in vivo (Table 1). In agreement with the differentiation problems observed in vitro, mutant mouse embryos form all three germ layers, but display severe gastrulation and patterning defects and die around implantation [33,37–40,47,48,52]. Similarly, interfering with PRC2 activity in C. elegans and Xenopus results in the prolonged activity of early-expressed genes [6] and the reduced activation of differentiation genes [6,49]. Together, these studies are consistent with the idea that H3K27me3 in bivalent chromatin domains is important for the repression of developmental genes and suggest that H3K27me3 is essential for lineage specification in vivo.
The Function of H3K4me3 in Bivalent Chromatin Domains –Poising?
It has been postulated that bivalently marked lineage-specific genes in ES cells are kept transcriptionally poised by H3K4me3 i.e. the association of H3K4me3 with an inactive gene facilitates the future activation of that gene [9,10] (Figure 2C). This putative function of H3K4me3 might extend beyond bivalent domains. For example, in Xenopus and zebrafish embryos and in ES cells, many inactive genes are marked with H3K4me3 in the absence of H3K27me3 [84,86,87].
Despite its prominence, evidence for the poising model is sparse, and the function of H3K4me3 is complex, as exemplified by two recent studies in ES cells [26,58]. Jiang et al. found that depletion of Dpy-30, a core subunit of MLL histone methyltransferase complexes, results in a partial reduction of H3K4me3. Consistent with the poising model, some lineage-associated genes are not properly activated upon differentiation [26]. In contrast, ES cell specific genes are expressed normally. These results suggest that the function of H3K4me3 in ES cells is to allow for the proper activation of lineage regulators upon differentiation. However, it is also conceivable that Dpy-30 and normal levels of H3K4me3 associated with lineage-regulatory genes are only required upon differentiation and not in ES cells.
In apparent contradiction to Jiang et al., a related study found that reduction of H3K4me3 levels upon depletion of Wdr5, another subunit of MLL histone methyltransferase complexes, results in severe defects in ES cell maintenance [58]. For example, Wdr5 depletion reduces the expression levels of key pluripotency genes [58]. The early effects of Wdr5-depleted ES cells precluded the detailed analysis of differentiation and suggest an earlier role for H3K4me3 than found in Dpy-30-depleted cells. The observed differences in these two studies may be due to different levels of H3K4me3 depletion and / or pleiotropic functions of Wdr5 and Dpy-30. Both studies establish essential roles of H3K4me3 in the regulation of developmental control genes (pluripotency factors and lineage regulators, respectively), but it remains unclear whether H3K4me3 has a function in poising the expression of embryonic genes.
Mutant and knockdown studies support an in vivo role for H3K4me3 in transcription regulation and lineage specification (Table 1) but have not addressed the poising model. For example, knockdown of Wdr5 in Xenopus results in a reduction of H3K4me3 levels and Hox gene expression [57]. Furthermore, deletion of Mll1, one of the H3K4 methyltransferases in mammals, results in an absence of Hox gene expression in mouse embryos [25,32]. Mll2 mutant mouse embryos display several developmental defects and embryonic lethality [29,30], and Mll2 mutant oocytes give rise to embryos that may be impaired in the activation of zygotic transcription [28]. While these studies are in agreement with a role for H3K4me3 in transcription regulation and lineage specification, further studies are required to determine precisely when H3K4me3 is required and whether H3K4me3 poises genes for activation during embryogenesis.
Establishing H3K4me3 and H3K27me3 marks
How is the positioning of H3K4me3 and H3K27me3 marks directed? Several mechanisms could guide the de novo methylation of histones. For example, long noncoding RNAs can provide sequence specificity to Polycomb and Trithorax proteins [91–95], or DNA binding proteins can recruit methyltransferases to specific sequence elements. Such elements might include Polycomb and Trithorax response elements [96–98] or CpG islands (genomic regions that contain a high frequency of mostly unmethylated CpG sites) [9,11,14,99,100].
Studies in ES cells suggest that pluripotency factors might play a role in the positioning of H3K4me3 and H3K27me3 [9,58,90,101]: bivalent chromatin domains and the position of core subunits of MLL and PRC2 histone methyltransferase complexes often coincide with the binding sites of pluripotency transcription factors [9,58,90]. Moreover, Oct4 has been shown to interact with components of MLL and PRC protein complexes [101]. While these observations are correlative, a recent study revealed that depletion of Oct4 in ES cells results in a reduction of H3K4me3 levels on selected genes, providing evidence for a causal relationship between the pluripotency network and H3K4me3 levels [58]. It remains unclear, however, whether Oct4 and other pluripotency factors are required for the establishment or maintenance of bivalent and monovalent chromatin domains and what other factors play a role.
Inheritance from Sperm?
In embryos, it is not only unclear how H3K4me3 and H3K27me3 marks are established but also controversial when they first appear. Studies in human, mouse and zebrafish have shown that some developmental regulatory genes are already marked by H3K4me3 and H3K27me3 in sperm [102–104]. It has been proposed that some of these marks are inherited after fertilization [86,102,103], but other studies have suggested that H3K4me3 and H3K27me3 marks are erased after fertilization and re-established during early embryogenesis [84,86]. For example, studies in zebrafish indicate that the majority of bivalent and monovalent marks are established when the embryo transitions from a stage when the genome is inactive to a stage when pluripotent blastomeres are transcriptionally active [84,86]. Interestingly, a small subset of genes (e.g. Hox genes) have H3K4me3 and H3K27me3 marks both in sperm and in early embryos, but it is not yet clear if these marks are permanently associated with specific genomic regions through cleavage stages or established de novo after fertilization [86,102,103]. Doubts about the inheritance of histone marks are also raised by studies in Xenopus embryos where H3K4me3 marks are established only during genome activation and H3K27me3 marks appear even later [87,88]. Similarly, studies in Drosophila embryos identified H3K27me3 later during development than H3K4me3 [105]. It thus remains unclear whether histone marks are established de novo or are inherited from sperm (or oocytes; their chromatin landscape has not yet been analyzed due to technical challenges).
How might histone marks that are established in the parent be transmitted to offspring? During replication, parental histones re-associate locally with newly synthesized DNA [106]. Histone modifications could thus be re-established by complexes that recognize a specific modification on an inherited parental histone and catalyze the same type of modification on adjacent, newly deposited nucleosomes. For example, H3K27me3 might recruit PRC2 to maintain the mark through replication [107,108]. Similarly, the histone methyltransferase MES-4 might recognize and maintain H3K36me3 domains from the parental germ line to offspring in C. elegans [109]. Notably, however, a mechanism by which histone modifications alone are sufficient to direct their own inheritance has not been established unequivocally in any system. Rather, specificity factors such as sequence elements or RNA scaffolds are thought to cooperatively contribute to the reestablishment of the parental chromatin state [110]. Functional analyses of non-coding RNAs, sequence-specific transcription factors, and histone marks during early embryonic stages might help to determine if chromatin states are inherited or re-established after fertilization [84,86–88,105,111–113].
Activation of Lineage Specific Genes
How do lineage regulators transition from an inactive state in ES cells to an active state during differentiation? In ES cells, many lineage regulators are inactive, associated with bivalent domains [9–14] and occupied by pluripotency factors [9,114–118]. It is thought that these factors recruit signal transducers [119], which then overcome H3K27me3-mediated repression and activate lineage regulatory genes [9–11,113,120–124]. For example, upon Nodal signaling, Smad2 binds to its target sites and recruits the histone demethylase Jmjd3, resulting in demethylation of H3K27me3 and gene activation [121]. Interestingly, H3K27me3-mediated repression can also be overcome without demethylating H3K27. One study reported that phosphorylation of Serine 28 in the tail of Histone 3 (the neighbor of Lysine 27) in response to stress signaling results in the displacement of PRC2, relieving transcriptional repression [122]. This mechanism might allow for the transient activation of PRC2-regulated genes until dephosphorylation of S28 reestablishes PRC2 binding and repression. While these in vitro studies have started to reveal how signaling pathways can overcome H3K27me3-mediated repression, the interaction between developmental signaling and chromatin during the transition from pluripotency to cell fate specification remains unclear.
Perspectives
There have been impressive advances in the genome-wide mapping of histone modifications and the phenotypic analysis of mutants that affect histone modifications. Novel concepts such as the bivalent poising of lineage regulators and the epigenetic inheritance from sperm have garnered wide attention. However, it remains poorly understood whether bivalency is a universally conserved principle across species, whether H3K4me3 truly poises genes for activation, and how parental histone marks can be transmitted to offspring.
It also remains largely unclear how embryonic histone marks act at smaller scales and higher-order dimensions i.e. how histone marks regulate the assembly of the transcriptional machinery and affect genome folding, respectively. For example, how does H3K27me3 repress transcription at the molecular level? Polycomb repressed chromatin can prevent RNA polymerase from accumulating at promoters [125,126], potentially by compacting chromatin and rendering it inaccessible for RNA polymerase II [127]. It has also been suggested that H3K27me3 and Polycomb group proteins can prevent the release of paused polymerases into the elongation phase of transcription via the ubiquitination of H2A [128,129]. The poising model predicts that H3K4me3 positively influences the recruitment or activity of RNA polymerase II. Although it has been assumed that H3K4 trimethylation follows the binding of RNA polymerase II [130–132], recent work has suggested that H3K4me3 marks can be established independently of RNA polymerase II association [84,100,126] and that H3K4me3 may facilitate RNA polymerase II recruitment [133,134]. Understanding the molecular function of bivalent domains in the regulation of transcription will be essential to understand their role during embryogenesis.
In the broader context of transcription regulation, it is important to note that the concept of bivalency has recently been extended from promoters to enhancers. Analogously to H3K4me3/H3K27me3 bivalent promoters, H3K4me1/H3K27me3 bivalent enhancers are thought to be associated with repressed but poised genes [19,21–23]. It will be interesting to determine the roles of bivalent marks on enhancers and to uncover the relationship between bivalent promoters and bivalent enhancers.
Finally, we also need to consider histone modifications within the larger context of chromatin structure and nuclear organization. For example, PRC2 has been shown to promote the compaction of chromatin and repress gene expression during differentiation in C. elegans [6]. Furthermore, Polycomb-repressed domains interact with each other over long distances in PcG bodies, stabilizing their silencing [135,136]. It will be achallenge for the future to integrate the role of histone modifications with long-range chromatin interactions [137,138], higher order chromatin structures [139], and the spatial organization of genes in the nucleus [140,141].
Acknowledgements
We thank Vincenzo Pirrotta and Leonie Ringrose for discussions and Brad Bernstein, Shelby Blythe, Brad Cairns, James Gagnon, Susan Mango, Andrea Pauli, John Rinn, Will Talbot, and Joanna Wysocka for helpful comments on the manuscript. NLV and AFS are supported by NIH grants 1K99HD067220-01 and 5RO1 GM056211, respectively. We apologize to colleagues whose work could not be discussed due to space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 4.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One. 2010;5:e10531. doi: 10.1371/journal.pone.0010531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yuzyuk T, Fakhouri T, Kiefer J, Mango S. The polycomb complex protein mes-2/E(z) promotes the transition from developmental plasticity to differentiation in C. elegans embryos. Dev Cell. 2009;16:699–710. doi: 10.1016/j.devcel.2009.03.008.. > This paper shows that the PRC2 protein MES-2 orchestrates large-scale changes in chromatin organization and gene expression to promote the transition from developmental plasticity to differentiation.
- 7.Wongtawan T, Taylor JE, Lawson KA, Wilmut I, Pennings S. Histone H4K20me3 and HP1alpha are late heterochromatin markers in development, but present in undifferentiated embryonic stem cells. J Cell Sci. 2011;124:1878–1890. doi: 10.1242/jcs.080721. [DOI] [PubMed] [Google Scholar]
- 8.Burton A, Torres-Padilla ME. Epigenetic reprogramming and development: a unique heterochromatin organization in the preimplantation mouse embryo. Brief Funct Genomics. 2010;9:444–454. doi: 10.1093/bfgp/elq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein B, Mikkelsen T, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 10.Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 11.Mikkelsen T, Ku M, Jaffe D, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T, Koche R, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao XD, Han X, Chew JL, Liu J, Chiu K, Choo A, Orlov YL, Sung W, Shahab A, Kuznetsov VA, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Ku M, Koche R, Rheinbay E, Mendenhall E, Endoh M, Mikkelsen T, Presser A, Nusbaum C, Xie X, Chi A, et al. Genomewide Analysis of PRC1 and PRC2 Occupancy Identifies Two Classes of Bivalent Domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krejci J, Uhlirova R, Galiova G, Kozubek S, Smigova J, Bartova E. Genome-wide reduction in H3K9 acetylation during human embryonic stem cell differentiation. J Cell Physiol. 2009;219:677–687. doi: 10.1002/jcp.21714. [DOI] [PubMed] [Google Scholar]
- 18.Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Heintzman N, Hon GC, Hawkins R, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart R, Ching C, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heintzman N, Stuart R, Hon G, Fu Y, Ching C, Hawkins R, Barrera L, Van Calcar S, Qu C, Ching K, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 21. Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692.. > Together with Creyghton et al., this paper identifies H3K27ac as a histone mark that distinguishes active from poised enhancers in pluripotent cells. In addition, this paper shows that poised enhancers are often marked with H3K27me3. When a gene is activated during differentiation, H3K27me3 at a poised enhancer is exchanged for H3K27ac.
- 22. Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107.. > Together with Rada-Iglesias et al., this paper identifies H3K27ac as a histone mark that distinguishes active from poised enhancers in pluripotent cells.
- 23.Hawkins RD, Hon GC, Yang C, Antosiewicz-Bourget JE, Lee LK, Ngo QM, Klugman S, Ching KA, Edsall LE, Ye Z, et al. Dynamic chromatin states in human ES cells reveal potential regulatory sequences and genes involved in pluripotency. Cell Res. 2011;21:1393–1409. doi: 10.1038/cr.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong SH, Rampalli S, Lee JB, McNicol J, Collins T, Draper JS, Bhatia M. Cell fate potential of human pluripotent stem cells is encoded by histone modifications. Cell Stem Cell. 2011;9:24–36. doi: 10.1016/j.stem.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 26. Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2011;144:513–525. doi: 10.1016/j.cell.2011.01.020.. > This paper shows that Dpy-30, a core member of MLL complexes, plays an important role during differentiation but not in ES cell self-renewal.
- 27.Lubitz S, Glaser S, Schaft J, Stewart AF, Anastassiadis K. Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Mol Biol Cell. 2007;18:2356–2366. doi: 10.1091/mbc.E06-11-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andreu-Vieyra CV, Chen R, Agno JE, Glaser S, Anastassiadis K, Stewart AF, Matzuk MM. MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glaser S, Lubitz S, Loveland KL, Ohbo K, Robb L, Schwenk F, Seibler J, Roellig D, Kranz A, Anastassiadis K, et al. The histone 3 lysine 4 methyltransferase, Mll2, is only required briefly in development and spermatogenesis. Epigenetics Chromatin. 2009;2:5. doi: 10.1186/1756-8935-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, Tufteland KR, Aasland R, Anastassiadis K, Ang SL, Stewart AF. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133:1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- 31.Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr Biol. 2004;14:2063–2069. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 32.Yagi H, Deguchi K, Aono A, Tani Y, Kishimoto T, Komori T. Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood. 1998;92:108–117. [PubMed] [Google Scholar]
- 33.Wang S, He F, Xiong W, Gu S, Liu H, Zhang T, Yu X, Chen Y. Polycomblike-2-deficient mice exhibit normal left-right asymmetry. Dev Dyn. 2007;236:853–861. doi: 10.1002/dvdy.21070. [DOI] [PubMed] [Google Scholar]
- 34.Walker E, Chang WY, Hunkapiller J, Cagney G, Garcha K, Torchia J, Krogan NJ, Reiter JF, Stanford WL. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2010;6:153–166. doi: 10.1016/j.stem.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasini D, Bracken A, Hansen J, Capillo M, Helin K. The Polycomb Group Protein Suz12 Is Required for Embryonic Stem Cell Differentiation. Molecular and Cellular Biology. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Motoyama J, Kitajima K, Kojima M, Kondo S, Takeuchi T. Organogenesis of the liver, thymus and spleen is affected in jumonji mutant mice. Mech Dev. 1997;66:27–37. doi: 10.1016/s0925-4773(97)00082-8. [DOI] [PubMed] [Google Scholar]
- 38.Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, Motoyama J, Higashinakagawa T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995;9:1211–1222. doi: 10.1101/gad.9.10.1211. [DOI] [PubMed] [Google Scholar]
- 39.Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pasini D, Bracken A, Jensen M, Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. doi: 10.1038/sj.emboj.7600402. In EMBO J. Edited by; 2004:4061–4071. vol 23.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 42.Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 43.Leeb M, Wutz A. Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J Cell Biol. 2007;178:219–229. doi: 10.1083/jcb.200612127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 2010;24:265–276. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boyer L, Plath K, Zeitlinger J, Brambrink T, Medeiros L, Lee T, Levine S, Wernig M, Tajonar A, Ray M, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733.. > This study shows that Eed null ES cells remain pluripotent even though expression of developmental regulators is increased.
- 46.Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faust C, Schumacher A, Holdener B, Magnuson T. The eed mutation disrupts anterior mesoderm production in mice. doi: 10.1242/dev.121.2.273. In Development. Edited by; 1995:273–285. vol 121.] [DOI] [PubMed] [Google Scholar]
- 48.Schumacher A, Faust C, Magnuson T. Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature. 1996;384:648. doi: 10.1038/384648a0. [DOI] [PubMed] [Google Scholar]
- 49.Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biondi CA, Gartside MG, Waring P, Loffler KA, Stark MS, Magnuson MA, Kay GF, Hayward NK. Conditional inactivation of the MEN1 gene leads to pancreatic and pituitary tumorigenesis but does not affect normal development of these tissues. Mol Cell Biol. 2004;24:3125–3131. doi: 10.1128/MCB.24.8.3125-3131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bertolino P, Tong WM, Galendo D, Wang ZQ, Zhang CX. Heterozygous Men1 mutant mice develop a range of endocrine tumors mimicking multiple endocrine neoplasia type 1. Mol Endocrinol. 2003;17:1880–1892. doi: 10.1210/me.2003-0154. [DOI] [PubMed] [Google Scholar]
- 55.Crabtree JS, Scacheri PC, Ward JM, Garrett-Beal L, Emmert-Buck MR, Edgemon KA, Lorang D, Libutti SK, Chandrasekharappa SC, Marx SJ, et al. A mouse model of multiple endocrine neoplasia, type 1, develops multiple endocrine tumors. Proc Natl Acad Sci U S A. 2001;98:1118–1123. doi: 10.1073/pnas.98.3.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stoller JZ, Huang L, Tan CC, Huang F, Zhou DD, Yang J, Gelb BD, Epstein JA. Ash2l interacts with Tbx1 and is required during early embryogenesis. Exp Biol Med (Maywood) 2010;235:569–576. doi: 10.1258/ebm.2010.009318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036.. > This study shows that Wdr5, a core component of MLL complexes, interacts with pluripotency factor Oct4. Wdr5 and Oct4 co-occupy many genes and Wdr5 is required for ES cell self-renewal.
- 58.Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 60.Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol. 2004;24:2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Core N, Bel S, Gaunt SJ, Aurrand-Lions M, Pearce J, Fisher A, Djabali M. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development. 1997;124:721–729. doi: 10.1242/dev.124.3.721. [DOI] [PubMed] [Google Scholar]
- 65.Akasaka T, van Lohuizen M, van der Lugt N, Mizutani-Koseki Y, Kanno M, Taniguchi M, Vidal M, Alkema M, Berns A, Koseki H. Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development. 2001;128:1587–1597. doi: 10.1242/dev.128.9.1587. [DOI] [PubMed] [Google Scholar]
- 66.Akasaka T, Kanno M, Balling R, Mieza MA, Taniguchi M, Koseki H. A role for mel-18, a Polycomb group-related vertebrate gene, during theanteroposterior specification of the axial skeleton. Development. 1996;122:1513–1522. doi: 10.1242/dev.122.5.1513. [DOI] [PubMed] [Google Scholar]
- 67.van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, van der Valk M, Deschamps J, Sofroniew M, van Lohuizen M, et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 protooncogene. Genes Dev. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 68.van der Lugt NM, Alkema M, Berns A, Deschamps J. The Polycomb-group homolog Bmi-1 is a regulator of murine Hox gene expression. Mech Dev. 1996;58:153–164. doi: 10.1016/s0925-4773(96)00570-9. [DOI] [PubMed] [Google Scholar]
- 69.Endoh M, Endo TA, Endoh T, Fujimura Y, Ohara O, Toyoda T, Otte AP, Okano M, Brockdorff N, Vidal M, et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- 70.del Mar Lorente M, Marcos-Gutierrez C, Perez C, Schoorlemmer J, Ramirez A, Magin T, Vidal M. Loss-and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development. 2000;127:5093–5100. doi: 10.1242/dev.127.23.5093. [DOI] [PubMed] [Google Scholar]
- 71.van der Stoop P, Boutsma EA, Hulsman D, Noback S, Heimerikx M, Kerkhoven RM, Voncken JW, Wessels LF, van Lohuizen M. Ubiquitin E3 ligase Ring1b/Rnf2 of polycomb repressive complex 1 contributes to stable maintenance of mouse embryonic stem cells. PLoS One. 2008;3:e2235. doi: 10.1371/journal.pone.0002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 73.Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, Deschamps J, van Lohuizen M. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci U S A. 2003;100:2468–2473. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Erhardt S, Su IH, Schneider R, Barton S, Bannister AJ, Perez-Burgos L, Jenuwein T, Kouzarides T, Tarakhovsky A, Surani MA. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development. 2003;130:4235–4248. doi: 10.1242/dev.00625. [DOI] [PubMed] [Google Scholar]
- 75.Takihara Y, Tomotsune D, Shirai M, Katoh-Fukui Y, Nishii K, Motaleb MA, Nomura M, Tsuchiya R, Fujita Y, Shibata Y, et al. Targeted disruption of the mouse homologue of the Drosophila polyhomeotic gene leads to altered anteroposterior patterning and neural crest defects. Development. 1997;124:3673–3682. doi: 10.1242/dev.124.19.3673. [DOI] [PubMed] [Google Scholar]
- 76.Evans M. Discovering pluripotency: 30 years of mouse embryonic stem cells. Nat Rev Mol Cell Biol. 2011;12:680–686. doi: 10.1038/nrm3190. [DOI] [PubMed] [Google Scholar]
- 77.Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 79.Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. doi: 10.1016/j.stem.2007.12.011. In Cell Stem Cell. Edited by; 2008:160–169. vol 2.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 82.Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci U S A. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103.. > Using sequential ChIP, this paper provides direct evidence for the existence of bivalent chromatin domains (H3K4me3/H3K27me3) in pluripotent blastomeres in vivo. This study also suggests that most H3K4me3 and H3K27me3 marks appear during genome activation and that H3K4me3 can mark genes in the absence of gene-specific transcription factors and stable association with RNA polymerase II.
- 84. Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464:922–926. doi: 10.1038/nature08866.. > This paper shows that H3K4me3 and H3K27me3 associate with the promoters of developmental regulatory genes in mouse epiblast cells.
- 85. Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc Natl Acad Sci U S A. 2010;107:10783–10790. doi: 10.1073/pnas.0914507107.. > This study suggests that some genes are marked with H3K4me3 prior to the activation of the zygotic genome.
- 86. Lindeman LC, Andersen IS, Reiner AH, Li N, Aanes H, Ostrup O, Winata C, Mathavan S, Muller F, Alestrom P, et al. Prepatterning of Developmental Gene Expression by Modified Histones before Zygotic Genome Activation. Dev Cell. 2011 doi: 10.1016/j.devcel.2011.10.008.. > This paper suggests that bivalent (co-occupancy by H3K4me3/ H3K27me3) domains might be rare or non-existent in Xenopus embryos. This finding contrasts with results in zebrafish that provided evidence for the existence of bivalent domains in embryonic blastomeres (Vastenhouw et al., 2010).
- 87.Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Veenstra GJ. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell. 2009;17:425–434. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schneider TD, Arteaga-Salas JM, Mentele E, David R, Nicetto D, Imhof A, Rupp RA. Stage-specific histone modification profiles reveal global transitions in the Xenopus embryonic epigenome. PLoS One. 2011;6:e22548. doi: 10.1371/journal.pone.0022548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 2009;7:e13. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee T, Jenner RG, Boyer L, Guenther MG, Levine S, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rinn J, Kertesz M, Wang J, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham P, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819.. > This paper is one of the first to show that long non-coding RNAs can function as a scaffold for histone modification complexes. Specifically, HOTAIR is shown to interact with PRC2 and play a role in its targeting to specific sites in the genome.
- 94.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43:1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 96.Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. doi: 10.1242/dev.02723. In Development. Edited by; 2007:223–232. vol 134.] [DOI] [PubMed] [Google Scholar]
- 97.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 98.Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE. GC-Rich Sequence Elements Recruit PRC2 in Mammalian ES Cells. PLoS Genet. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244.. > This paper shows that non-methylated CG dense sequences are sufficient to establish H3K4me3 domains in mouse ES cells. RNA polymerase II is not required.
- 100.Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ding J, Xu H, Faiola F, Ma'ayan A, Wang J. Oct4 links multiple epigenetic pathways to the pluripotency network. Cell Res. 2012;22:155–167. doi: 10.1038/cr.2011.179.. > While most nucleosomes are replaced by protamine in human sperm, retained nucleosomes are enriched at developmental regulators. H3K4me3 and H3K27me3 are often found at these loci, leading to the hypothesis that modified histones in sperm prepare genes for development.
- 102.Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009 doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu SF, Zhang H, Cairns BR. Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 2011;21:578–589. doi: 10.1101/gr.113167.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AH. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- 105.Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Radman-Livaja M, Verzijlbergen KF, Weiner A, van Welsem T, Friedman N, Rando OJ, van Leeuwen F. Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLoS Biol. 2011;9:e1001075. doi: 10.1371/journal.pbio.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 108. Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398.. > This paper shows that MES-4 is a maintenance methyltransferase required to transmit the H3K36me3 mark through the germline.
- 109. Rechtsteiner A, Ercan S, Takasaki T, Phippen TM, Egelhofer TA, Wang W, Kimura H, Lieb JD, Strome S. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001091.. > This review discusses how chromatin states can be inherited through replication. It is proposed that silencing elements or RNA scaffolds act cooperatively with parentally inherited histones to re-establish and maintain chromatin states.
- 110.Moazed D. Mechanisms for the inheritance of chromatin states. Cell. 2011;146:510–518. doi: 10.1016/j.cell.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, et al. Systematic identification of long non-coding RNAs expressed during zebrafish embryogenesis. Genome Res. 2011 doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved Function of lincRNAs in Vertebrate Embryonic Development despite Rapid Sequence Evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055.. > This study shows that β-catenin establishes a poised chromatin structure at target gene promoters before the onset of zygotic transcription and might mark genes for later expression.
- 113.Blythe SA, Cha SW, Tadjuidje E, Heasman J, Klein PS. beta-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2. Dev Cell. 2010;19:220–231. doi: 10.1016/j.devcel.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 117.Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mathur D, Danford TW, Boyer LA, Young RA, Gifford DK, Jaenisch R. Analysis of the mouse embryonic stem cell regulatory networks obtained by ChIP-chip and ChIP-PET. Genome Biol. 2008;9:R126. doi: 10.1186/gb-2008-9-8-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042.. > This paper describes a mechanism by which Nodal signaling modulates chromatin to activate gene expression: the intracellular signal transducer Smad2 recruits the histone demethylase Jmjd3, resulting in demethylation of H3K27me3 and activation of transcription.
- 121. Dahle O, Kumar A, Kuehn MR. Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci Signal. 2010;3:ra48. doi: 10.1126/scisignal.2000841.. > This paper suggests that transcription can be activated in the presence of H3 Lysine 27 trimethylation. Phosphorylation of Serine 28 in the tail of Histone 3 results in the displacement of PRC2, relieving transcriptional repression.
- 122.Gehani SS, Agrawal-Singh S, Dietrich N, Christophersen NS, Helin K, Hansen K. Polycomb Group Protein Displacement and Gene Activation through MSK-Dependent H3K27me3S28 Phosphorylation. Mol Cell. 2010;39:886–900. doi: 10.1016/j.molcel.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 123.Akizu N, Estaras C, Guerrero L, Marti E, Martinez-Balbas MA. H3K27me3 regulates BMP activity in developing spinal cord. Development. 2010;137:2915–2925. doi: 10.1242/dev.049395. [DOI] [PubMed] [Google Scholar]
- 124.Xi Q, Wang Z, Zaromytidou AI, Zhang XH, Chow-Tsang LF, Liu JX, Kim H, Barlas A, Manova-Todorova K, Kaartinen V, et al. A Poised Chromatin Platform for TGF-beta Access to Master Regulators. Cell. 2011;147:1511–1524. doi: 10.1016/j.cell.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chopra VS, Hendrix DA, Core LJ, Tsui C, Lis JT, Levine M. The polycomb group mutant esc leads to augmented levels of paused Pol II in the Drosophila embryo. Mol Cell. 2011;42:837–844. doi: 10.1016/j.molcel.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stock J, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher A, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 129.Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 131.Lee J, Skalnik D. Wdr82 Is a C-Terminal Domain-Binding Protein That Recruits the Setd1A Histone H3-Lys4 Methyltransferase Complex to Transcription Start Sites of Transcribed Human Genes. doi: 10.1128/MCB.01356-07. In Molecular and Cellular Biology. Edited by; 2008:609–618. vol 28.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase. II. doi: 10.1016/j.cell.2006.04.029. In Cell. Edited by; 2006:703–717. vol 125.] [DOI] [PubMed] [Google Scholar]
- 133.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 134.Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase. II. Mol Cell Biol. 2009;29:6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 136.Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–226. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 137.Miele A, Dekker J. Long-range chromosomal interactions and gene regulation. Mol Biosyst. 2008;4:1046–1057. doi: 10.1039/b803580f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev. 2011;21:175–186. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Arib G, Akhtar A. Multiple facets of nuclear periphery in gene expression control. Curr Opin Cell Biol. 2011;23:346–353. doi: 10.1016/j.ceb.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 141.Meister P, Mango SE, Gasser SM. Locking the genome: nuclear organization and cell fate. Curr Opin Genet Dev. 2011;21:167–174. doi: 10.1016/j.gde.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 143.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 144.Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 145.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schübeler D, Macalpine D, Scalzo D, Wirbelauer C, Kooperberg C, Van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. doi: 10.1101/gad.1198204. In Genes & Development. Edited by; 2004:1263–1271. vol 18.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Guenther MG, Levine S, Boyer L, Jaenisch R, Young R. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. doi: 10.1016/j.molcel.2006.12.014. In Molecular Cell. Edited by; 2007:15–30. vol 25.] [DOI] [PubMed] [Google Scholar]
- 150.Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ, et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]