Abstract
It has been suggested that poor immunogenicity may explain the lack of vaccine efficacy in preventing or controlling HIV infection in the Step trial. To investigate this issue we vaccinated eight Indian rhesus macaques with a trivalent replication-incompetent adenovirus serotype 5 vaccine expressing SIV Gag, Pol, and Nef using a regimen similar to that employed in the Step trial. We detected broad vaccine-induced CD8+ (2–7 pool-specific responses) and CD4+ (5–19 pool-specific responses) T-cell responses in IFN-γ ELISPOT assays at one week post-boost using fresh PBMC. However, using cryopreserved cells at one and four weeks post-boost we observed a reduction in both the number and magnitude of most vaccine-induced responses. This demonstrates that the time points and conditions chosen to perform immune assays may influence the observed breadth and frequency of vaccine-induced T-cell responses. To evaluate protective efficacy, we challenged the immunized macaques, along with naïve controls, with repeated, limiting doses of the heterologous swarm isolate SIVsmE660. Vaccination did not significantly affect acquisition or control of virus replication in vaccinees compared to naïve controls. Post-infection we observed an average of only two anamnestic CD8+ T-cell responses per animal, which may not have been sufficiently broad to control heterologous virus replication. While the trivalent vaccine regimen induced relatively broad T-cell responses in rhesus macaques, it failed to protect against infection or control viral replication. Our results are consistent with those observed in the Step trial and indicate that SIV immunization and challenge studies in macaque models of HIV infection can be informative in assessing pre-clinical HIV vaccines.
Keywords: HIV vaccine, Adenovirus serotype 5, Simian Immunodeficiency Virus, CD8+ T cells, CD4+ T cells, Step trial
1. Introduction
The Step trial was a proof-of-efficacy human study of a replication-defective rAd51 expressing clade B HIV-1 gag, pol, and nef [1, 2]. It was initiated in 2004 with much anticipation, as it was the first large-scale study to assess the efficacy of a T-cell-based HIV vaccine. It targeted the induction of T-cell responses against viral proteins that are relatively well conserved across different HIV clades and recognized by many infected individuals. The vaccine was designed to induce potent cellular immune responses in the hope of preventing infection, or more realistically, controlling viral replication after infection, thereby, limiting pathogenesis and reducing secondary transmission [1, 3–7]. Early optimism, however, faded when the study was prematurely stopped after the first interim analysis revealed an inability of the vaccine regimen to prevent infection or lower viral replication in vaccinated individuals [1]. In the aftermath of the Step trial, questions arose as to why the vaccine regimen failed, the usefulness of cellular immune responses as HIV vaccine targets, and the ability of non-human primate models to predict HIV vaccine efficacy [8–11].
One of the hypotheses proposed to explain the lack of efficacy seen in the Step trial is the inability of the vaccine regimen to elicit sufficiently broad immune responses to handle the diversity of circulating strains of HIV-1 responsible for clinical infections [2]. Overall the vaccine regimen induced detectable cellular immune responses in 75% of participants [1, 2]. However, vaccinated individuals made a median of one CD8+ T-cell responses per protein using defined pools of vaccine-matched 9-mer peptides [12]. This rather narrow breadth of responses may have been insufficient to deal with the variability within circulating strains of HIV-1 [13, 14]. Evidence from the human trials also suggested that pre-existing immunity to the adenovirus can limit the magnitude and breadth as measured by cellular responses to the three antigen components [2, 15]. This has stimulated interest in developing novel viral vectors or adenoviral vectors with lower seroprevalence levels [16, 17].
Several primate challenge studies were conducted to examine the potential efficacy of the rAd5 HIV-1 vaccine approach used in the Step trial. An early pre-clinical study by Merck found that rAd5 encoding only Gag led to substantial and durable control of SHIV-89.6P [18]. Unfortunately, SHIV-89.6P has a different target cell tropism and pathogenesis than most SIV and HIV strains since it causes relatively fast CD4+ T-cell depletion than what is typically observed with human infections [19, 20]. A more rigorous tests of rAd5 encoding Gag using a single high-dose challenge of the highly pathogenic CCR5-tropic SIV molecular clone SIVmac239 yielded only transient viral control, which was limited to animals expressing the MHC class I molecule (Mamu2-A*01) previously associated with a measure of virological control [21]. Similar results were observed when the efficacy of trivalent rAd5 vaccine was tested in Mamu-A*01 positive macaques against a single challenge of SIVmac239 [22]. Additional pre-clinical challenge studies suggested that the inclusion of additional viral proteins and/or a DNA prime would be needed to engender protective responses [23–25].
The result of the Step trial raises several important questions on the role of pre-clinical challenge models in the field of HIV-1 vaccine discovery and development. Additionally, consideration must be taken in how the non-human primate model systems can be improved to better inform future vaccine candidate selection [8–11]. To partly address these questions, we examined the efficacy of the rAd5 vaccine expressing SIVmac239 gag, pol, and nef using macaques negative for the MHC class I molecules Mamu-A*01, -B*08, or -B*17 against a repeated, limiting dose challenge with the heterologous virus strain SIVsmE660. We did not include Mamu-A*01, -B*08, or -B*17 positive animals because macaques expressing these alleles have previously been associated with a measure of viral control and, particularly for Mamu-B*08 and -B*17, have similar traits to the human protective alleles HLA-B*27 and HLA-B*57 which were found in only a small fraction of the subjects in the human Step trial [26–30]. The repeated, limiting dose challenge method will help address the efficacy of this vaccine in terms of protection against acquisition and viral control of a heterologous virus. In principle, such a design might more effectively replicate natural human infection than previous models. In this study, we vaccinated a cohort of Indian rhesus macaques using a regimen designed to mimic as closely as possible that used in the human Step trial. We performed a comprehensive survey of the vaccine-induced immune responses. We also assessed the effectiveness of the vaccine regimen in rhesus macaques to protect against infection or control virus replication in comparison to naïve controls after a heterologous virus challenge.
2. Materials and Methods
2.1 Animals and Viruses
Indian rhesus m acaques (Macaca mulatta) from the Wisconsin National Primate Research Center were housed and cared for according to the regulations set forth in the Guide for the Care and Use of Laboratory Animals of the National Research Council (National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC), as approved by the University of Wisconsin Institutional Animal Care and Use Committee. All rhesus macaques used in the study were genotyped for their MHC class I alleles using PCR-SSP3 as previously described [31]. Animals expressing the MHC class I alleles Mamu-A*01, -B*08, and –B*17 were excluded from the study. We also genotyped the animals for TRIM5α based on the following three allelic classes: TRIM5CypA, TRIM5TFP, and TRIM5Q as previously described [32].
Eight macaques were inoculated intramuscularly with a 1:1:1 ratio of recombinant adenovirus serotype 5 (MRKAd5) encoding SIVmac239 gag, pol, or nef (1010 copies each) on weeks 0, 4, and 26 of the study. The vaccinated animals, along with eight naïve controls, were challenged approximately 15 weeks after the third immunization. All animals were challenged intrarectally with 6×106 SIV RNA copies (225 TCID50) of SIVsmE660 on a weekly basis. If an animal remained uninfected after the fifth challenge, the dose of SIVsmE660 was increased to 1.2×107 (450 TCID50). Challenges were stopped once an animal tested positive for SIVsmE660 infection as assessed by measurable plasma viremia by QRT-PCR.
2.2 Cryopreservation of PBMC
PBMC were cryopreserved within six hours of isolation by suspending the cells in 1.5 ml cryotubes (Nunc) at a concentration of between 5–20 million cells per ml of cold CryoStor CS5 (BioLife Solutions, Inc.). The cryotubes were immediately transferred to −80°C freezers in a 5100 Cryo 1% freezing container (Nalgene Labware). After at least 8 hours at −80°C the cells were transferred to liquid nitrogen freezers for long-term storage.
Cryopreserved cells were thawed by removing them from liquid nitrogen storage and placing them in a 37°C water bath until almost completely thawed. The cells were then added drop wise to pre-warmed RPMI 1640 containing 10% fetal bovine serum (R10). The cells were washed twice more in R10 to remove any residual freezing media. The viability of the recovered cells was tested by trypan blue exclusion.
2.3 IFN-γELISPOT assays
Fresh PBMC isolated from EDTA-anticoagulated blood were used in ELISPOT assays for the detection of IFN-γ secreting cells as previously described [33]. The assay was set up within four hours of isolation of the PBMC. Assays with cryopreserved cells were performed with 100,000 or 200,000 cells per well. Test wells were performed with two replicates and control wells were performed in replicates of 2 or 4 depending on the assay. Responses containing < 50 spot forming cells per 1×106 cells were considered negative and not tested statistically. Positive responses were determined using a one-tailed t-test and alpha = 0.05, where the null hypothesis (Ho): background level ≥ treatment level. If determined to be positive statistically, the reported values equal the average of the test wells minus the average of all negative control wells. We used the same premise to determine which anamnestic cellular immune responses significantly expanded above pre-challenge levels. Positive responses were again determined using a one-tailed t-test and alpha=0.05, but this time Ho: pre-challenge levels ≥ post-challenge levels. Peptides used in these assays were obtained through the AIDS Research and Reference reagent program, Division of AIDS, NIAID, and NIH.
We examined CD4+ T-cell responses by depleting PBMC of CD8+ cells using a CD8 Microbead Kit for non-human primates (Miltenyi Biotec Inc., Bergisch Gladbach, Germany) according to the manufacturer’s instructions.
2.4 Intracellular cytokine staining (ICS)
ICS was performed using fresh or cryopreserved PBMC. Briefly, 5×105 to 1×106 PBMC were incubated in the presence of Brefeldin A (Sigma), anti-CD28 (BD Biosciences), anti-CD49d (BD Biosciences), and pools of peptides overnight at 37°C. Staphylococcal enterotoxin B (SEB) was used as a positive control and no peptide stimulation was used as the negative control. After incubation the cells were washed twice with PBS and stained for 30 minutes at 4°C with anti-CD3 FITC (BD Biosciences), anti-CD8α PerCP (BD Biosciences), anti-CD4 APC (Miltenyi Biotec Inc) and fixable yellow amine reactive dye (Invitrogen) for live/dead discrimination. After the incubation the cells were washed twice with PBS containing 10% fetal bovine serum, fixed with 2% paraformaldehyde, and stored at 4°C for at least 30 minutes. After the incubation the cells were washed twice with permeabilization buffer (PBS + 10% fetal bovine serum + 0.1% saponin) and stained with anti-IL-2 PE (BD Biosciences), anti-IFN-γ FITC (BD Biosciences), and anti-TNFα (BD Biosciences) for 50 minutes at room temperature. After washing the cells twice with permeabilization buffer the cells were fixed with 2% paraformaldehyde and stored at 4° C until being acquired on a BD LSR II Flow Cytometer (Becton Dickinson).
The data were analyzed using FlowJo software version 9.2 (Treestar). For each peptide pool, Boolean gates were created based on single-positive IFN-γ, TNF-α, or IL-2 for CD3+/CD4−/CD8+ cells (Supplemental Fig. 1). For each of the cytokine combinations positive responses were determined using 2×2 contingency tables whereby the four entries in the table were the number of cytokine positive cells and the number cytokine negative cells for both the test and negative control samples. A one-tailed chi-squared test with Yates’ correction was then applied to each table to test for a significant difference in the number of cytokine-producing cells in the test sample versus the negative control sample. Any cytokine subset with a p-value ≤ 0.05 was considered positive. Further data analysis was performed using PESTLE version 1.6.2 and SPICE version 5.2 provided by Mario Roederer, Vaccine Research Center, NIAID, NIH.
2.5 Viral Load Determination
Levels of circulating plasma virus were determined using a previously described QRT-PCR assay [34]. The threshold of sensitivity for this assay was 30 copy equivalents (Eq)/ml of plasma.
2.6 Statistics
Kaplan-Meier survival analysis was performed to test whether there was a difference in the number of challenges required to productively infect the vaccinated (n=8) and control (n=8) animals (Graphpad Prism, version 4.0c).
To test whether viral loads (log transformed) w ere significantly different between the vaccinated and control groups at 2, 3, 4, 6, 8, and 12 weeks post-infection, we used a repeated measures model with age and sex as covariates. We used a Toeplitz matrix because it was the model with the lowest Akaike Information Criterion (AIC) score. This model yielded residuals with a roughly normal distribution, suggesting that log transformation of the viral loads produced acceptable results. With a nominal alpha of 0.05 (2-tailed), we found neither sex nor age to be a significant predictor of outcome and these covariates were therefore dropped from the analysis. Sensitivity analysis was performed by conducting the nonparametric Wilcoxon-Mann-Whitney test at each time point for comparison to the mixed model’s conclusions. We also used the Wilcoxon-Mann-Whitney test to determine whether vaccinated and unvaccinated animals had significantly different maximum peak viral loads. The peak acute plasma viral load for each animal was determined to be the highest viral load occurring during the first three weeks of infection.
Analysis to determine the power of the study to detect a 75% difference in the rate of acquisition of SIV infection between the vaccine and control groups was performed as previously described using a two-sided alpha of 0.05 and the log rank test [35, 36].
Comparison of values between fresh and cryopreserved cells was performed using a paired sample t test. The p values are reported as the level of significance for a two-tailed analysis (Graphpad Prism).
3. Results
3.1 Vaccine-induced immune responses
We vaccinated eight Indian rhesus macaques mimicking as closely as possible the immunization protocol used in the Step trial [1]. We immunized the macaques intramuscularly with a 1:1:1 ratio of three separate rAd5 vectors expressing gag, pol, or nef derived from SIVmac239 at weeks 0, 4, and 26 of the study. The MRKAd5 vectors were formulated containing the same number of viral particles (1010 each) used in the Step trial. We excluded from this study any animal expressing MHC class I molecules previously associated with spontaneous control of SIVmac239 (Mamu-A*01, -B*08, and –B*17) [28–30].
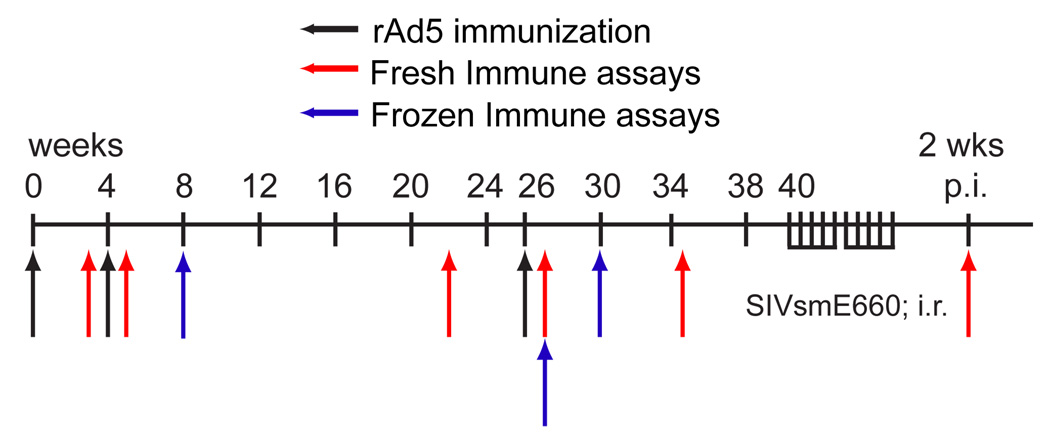
One of the advantages of the rhesus macaque/SIV model is the ability to efficiently monitor vaccine-induced immune responses at multiple time points post-immunization. We assessed vaccine-induced immune responses using a combination of time points employed in the Step trial (4 weeks after the 2nd and 3rd boosts) as well as those closer to the date of immunization (Fig. 1). To determine the breadth and magnitude of vaccine-induced T-cell responses we used IFN-γ ELISPOT assays with pools of ten peptides, 15 amino acids in length overlapping by 11 amino acids covering SIVmac239 Gag (13 pools), Pol (26 pools), and Nef (6 pools). To distinguish between CD8+ and CD4+ T-cell responses we used whole PBMC and PBMC depleted of CD8+ cells in the IFN-γ ELISPOT assays. Responses detected in whole PBMC but not PBMC depleted of CD8+ cells are classified as CD8+ T-cell responses. Conversely, responses detected in assays performed with PBMC depleted of CD8+ cells were classified as being mediated by CD4+ T cells. Responses were considered positive if they were detected at multiple time points or confirmed by intracellular cytokine staining (ICS).
Figure 1. Study overview.
Outline of the immunizations and immune assays performed in the study.
i.r.: intrarectal, p.i.: post-infection
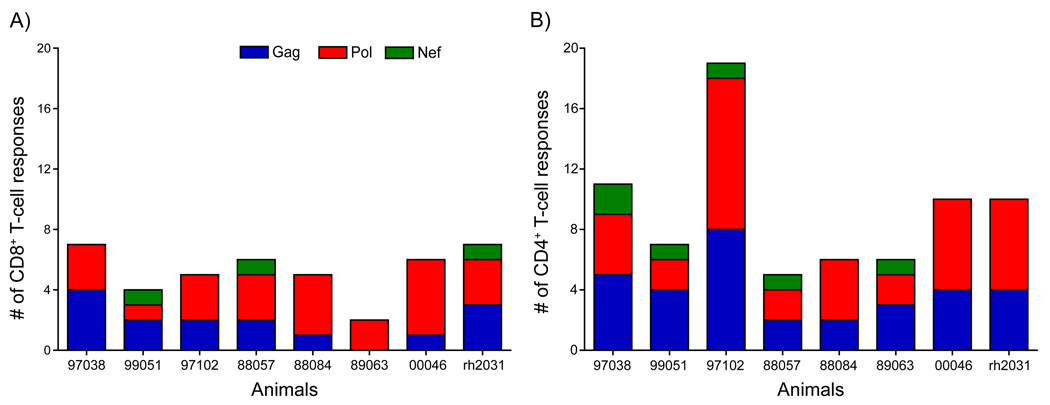
Given the limited cellular immune responses reported using the rAd5 vaccine in humans, we were surprised to detect broader cellular immune responses induced by this vaccine regimen in rhesus macaques. In this study, we detected an average of five distinct pool-specific CD8+ T-cell responses per animal during the course of the vaccine phase of the study, ranging between two and seven pool-specific responses (Fig 2A). Pol (a total of 24 different pool-specific CD8+ T-cell responses made by the eight animals in the study) was the most frequently targeted protein by CD8+ T cells, followed by Gag (15 responses) and Nef (3 responses). This trend roughly mirrors the size of the individual proteins with Pol at 1,066 amino acids (AA), Gag at 510 AA, and Nef at 165 AA. The CD8+ T-cell responses induced by vaccination were also of relatively high frequency. For example, at one week after the third rAd5 immunization all but one of the animals had at least one pool-specific response above 500 spot forming cells (SFC) per million PBMC (Fig. 3A). Thus, the rAd5 trial vaccine regimen induced high frequency CD8+ T-cell responses targeting multiple proteins in most of the animals in our study.
Figure 2. The number of distinct SIV peptide pools detected in freshly isolated lymphocytes during the vaccine phase of the study.
The bars indicate the number of vaccine-induced CD8+ (A) and CD4+ (B) T-cell responses, measured as described in the Results. The number of responses are broken down by the proteins included in the vaccine.
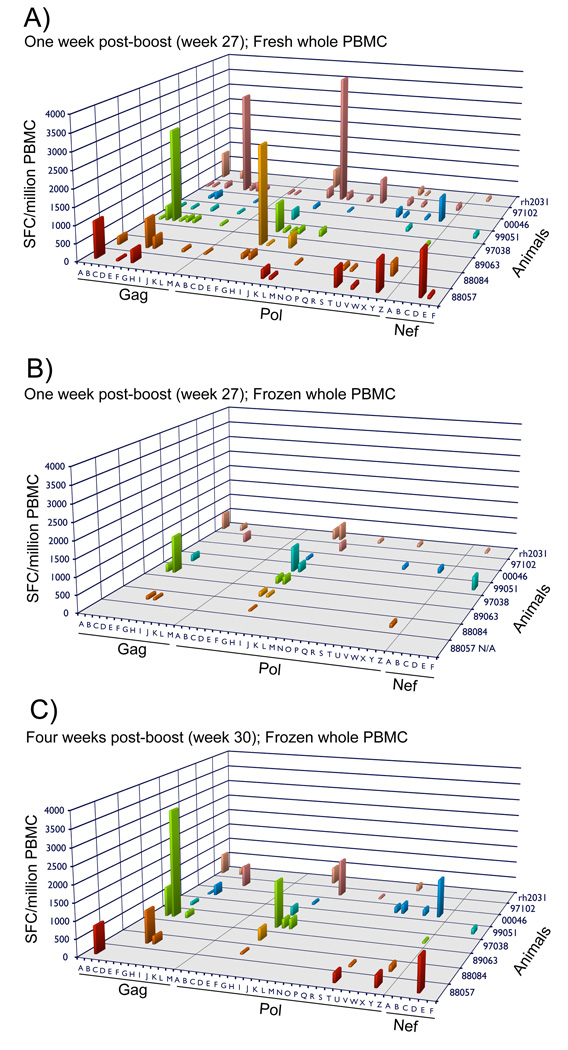
Figure 3. Cellular immune responses at one and four weeks post-rAd5 immunization using freshly isolated and cryopreserved PBMC in IFN-γ ELISPOT assays.
Pool-specific cellular immune responses at one (A, B) or four (C) weeks after the third rAd5 immunization using freshly isolated (A) or cryopreserved PBMC (B,C) in IFN-γ ELISPOT assays. Each response is presented as the mean spot forming cells (SFC) per million PBMC of the sample wells minus the mean of the SFC/million no peptide negative control wells. N/A: cryopreserved samples were not available from animal 88057 one week after the third rAd5 immunization.
Published reports from the Step trial indicate that CD4+ T-cell responses were detected in 41% of vaccine recipients [2, 37]. In contrast, we identified vaccine-induced CD4+ T-cell responses in all of our vaccinated macaques (Fig 2B). We detected an average of nine (range 5–19) distinct pool-specific CD4+ T-cell responses per animal during the course of the vaccine phase of the study. Just as with the vaccine-induced CD8+ T-cell responses, the predominant target of the vaccine-induced CD4+ T-cell responses was Pol (36 total pool-specific CD4+ T-cell responses in the eight macaques), followed closely by Gag (32 responses). However, only a few Nef-specific (6 responses) responses were detected. The magnitude of the vaccine-induced CD4+ T-cell responses was lower than observed for the CD8+ T-cell compartment, with vaccine-induced CD4+ T-cell responses ranging between 100 and 500 SFC/million CD8− PBMC (Supplemental Fig. 2). Although, it should be noted that the method used to detect vaccine-induced CD4+ T-cell responses differed between the human (ICS) and macaque (ELISPOT with subset depletion) studies. The relative sensitivity of these two assays for detecting CD4+ T-cell responses is unclear and may have contributed to the discrepancy in observed vaccine-induced CD4+ T-cell responses between the two studies. Additionally, to date, there have been no epitope-mapping analyses performed for CD4+ T-cell responses in rAd5 human recipients.
To compare our results to the published reports from the Step study we also collected PBMC at weeks 8 and 30 (4 weeks after the second and third rAd5 immunization). Using cryopreserved cells we demonstrate a significant decrease in the number of positive responses in both the whole PBMC (p=.005, paired t-test) and PBMC depleted of CD8+ cells (p=0.004, paired t-test) in comparison to responses identified at one week post-boost using fresh PBMC (Fig. 3A and C, Supplemental Fig. 2). For example, the average number of responses in freshly isolated whole PBMC dropped from 9.6 at week 27 to 4.3 at week 30 using cryopreserved cells. Furthermore, we detected an even larger decrease in the number of responses using PBMC depleted of CD8+ cells from an average of 8.6 responses in freshly isolated cells at week 27 to only 2.0 in cryopreserved cells at week 30 (Supplemental Fig. 2). When comparing responses in the whole PBMC at week 27 to those at week 30 we found that most of the missing responses corresponded to peptide pools containing CD4+ T-cell responses. For the CD4+ T-cell responses that remained we noted a significant decrease in the overall magnitude of the responses in the assays using cryopreserved PBMC depleted of CD8+ cells at week 30 in comparison to freshly isolated cells at week 27 (p=0.05, paired t-test)(Supplemental Fig. 2). The majority of the CD4+ responses detected at the later time point were barely measurable. We also observed the trend towards lower magnitude responses in the whole PBMC using cryopreserved cells at week 30 in comparison to freshly isolated cells at week 27, although the magnitudes were more comparable between the two time points than with the PBMC depleted of CD8+ cells and even a few responses were marginally higher at week 30 with the cryopreserved cells (Fig. 3A and C).
The decrease in the number and magnitude of responses between weeks 27 and 30 for most of the responses could be due to contraction of vaccine-induced immune responses over time or to the difference in using fresh versus cryopreserved cells. To address this issue we performed IFN-γ ELISPOT assays using whole PBMC cryopreserved at week 27 and pools of peptides eliciting positive responses with fresh PBMC at this time point. Unfortunately, cryopreserved samples were unavailable for animal 88057. Despite recovering cells with greater than 95% viability, we observed a significant decrease (p=0.02; paired t-test) in the number of responses using the cryopreserved cells in comparison to fresh PBMC. The overall magnitude of the responses with cryopreserved cells also trended lower than those detected with fresh PBMC but was not significantly different (p=0.07; paired t-test)(Fig. 3B). Once more the pools containing the CD4+ T-cell epitopes were most likely to be lost through the cryopreservation process. These results indicate that the time points and conditions chosen to perform immune assays can influence both the breadth and magnitude of T-cell responses.
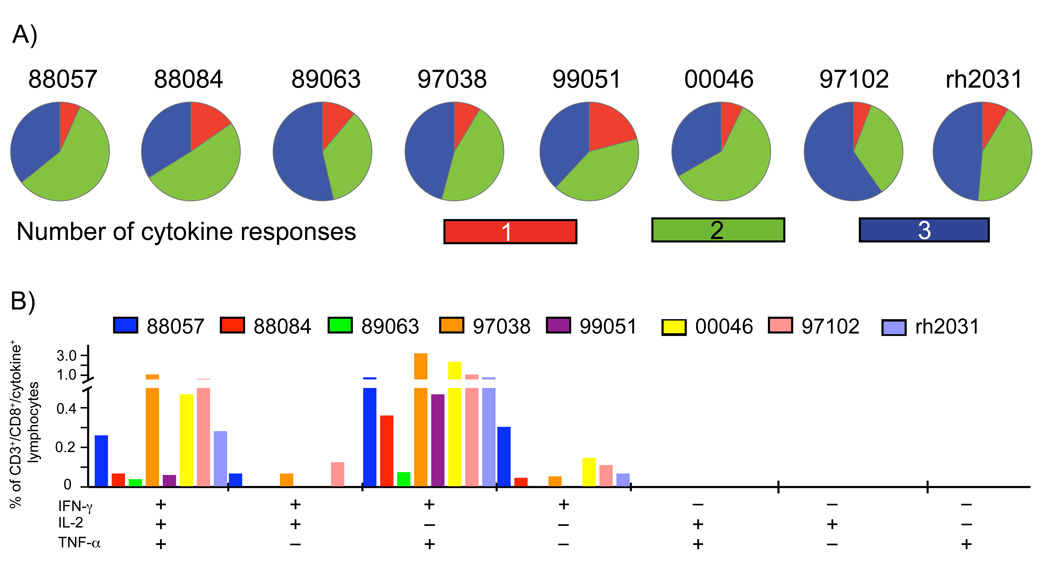
To evaluate the ability of vaccine-induced T cells to elaborate various cytokines we performed multiparameter ICS. For comparison to the Step trial we monitored expression of the same cytokines (IFN-γ, TNF-α, and IL-2) for both CD4+ and CD8+ T cells. We used one Gag pool, one Nef pool, and two Pol pools (consisting of 15-mer peptides overlapping by 11-AA) for stimulation and performed the assay with cryopreserved cells collected four weeks after the third rAd5 boost. The majority of pool-specific CD8+ T cells produced either two or three cytokines (Fig. 4), with cells producing both IFN-γ and TNF-α being the most frequent dual-producing populations. These results are consistent with those observed in the human trial where most vaccine-induced CD8+ T cells expressed two or three cytokines [2], although we detected a higher frequency of vaccine-induced CD8+ T-cells expressing all three cytokines and relatively few cells producing only a single cytokine. Unfortunately, the frequency of vaccine-specific CD4+ T cells was too low at this time point for a meaningful analysis of the cytokine profile of these cells. However, our results suggest that upon interaction with virally infected cells the vaccine-induced CD8+ T cells could react by secreting several cytokines.
Figure 4. Cytokine secretion profiles of vaccine-induced CD8+ T-cell responses four weeks after the third rAd5 immunization.
We monitored the capacity of the vaccine-induced CD8+ T-cell responses to secrete cytokines in an ICS assay by stimulating cryopreserved cells collected four weeks after the third rAd5 immunization with four pools of peptides: one Gag pool, one Nef pool, and two Pol pools. We assessed the expression of INF-γ, TNF-α, and IL-2. Displayed for each vaccinated animal are the proportions of CD8+ T cells making one, two, or three cytokines (A) and the frequency of CD8+ T cells making the different cytokine combinations (B) upon peptide stimulation. Data is presented as the sum of the positive responses for each animal.
3.2 SIV challenge
To assess the ability of the vaccine-induced immune responses to protect against infection or control viral replication we challenged the vaccinated animals, along with eight naïve controls, with repeated, limiting doses of the heterologous biological isolate SIVsmE660 approximately 15 weeks after the last rAd5 boost. The naïve control animals, like their vaccinated counterparts, were negative for MHC class I alleles (Mamu-A*01, -B*08, and –B*17) previously associated with control of SIVmac239 replication [28–30]. Additionally, since TRIM5 polymorphisms can affect SIV replication and acquisition of infection we also matched as closely as possible the TRIM5α genotype of the controls to those of the vaccinees (Supplemental Table 1)[32, 38–42]. Animals were inoculated intrarectally with SIVsmE660 once a week and challenges were stopped once an animal tested SIV positive. This study had a 70% power to detect a 75% reduction in the rate of acquisition between the vaccine and control groups.
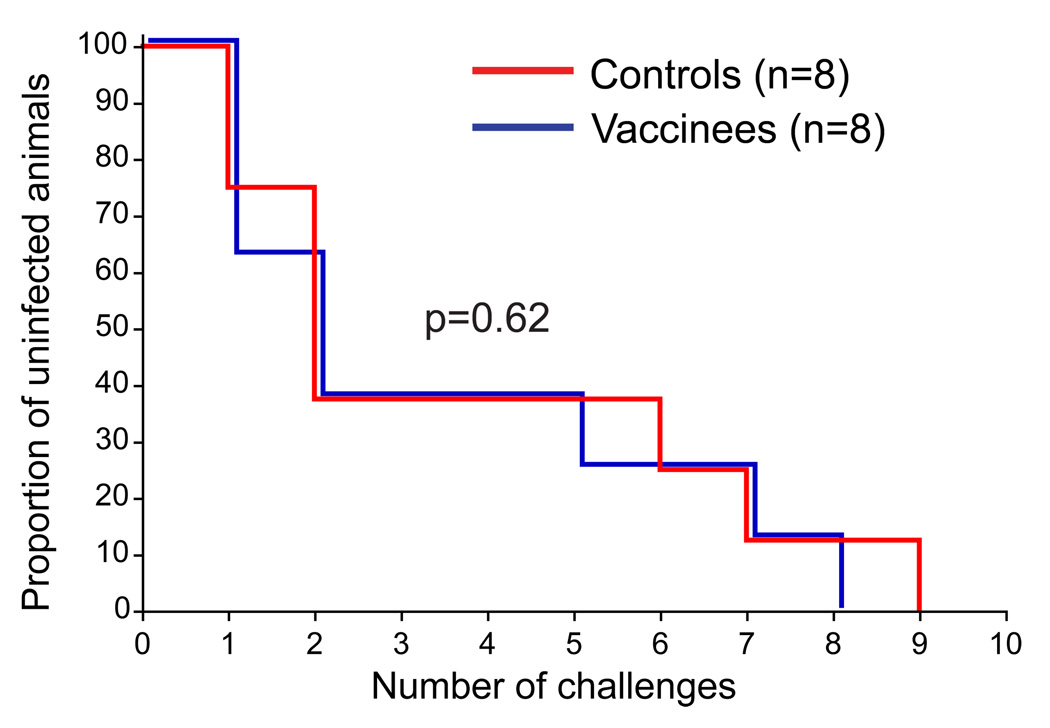
We found no difference in the rate of acquisition between the vaccinated and control animals (Fig. 5). Our results, thus, mirror those of the Step trial in which the vaccine regimen was not protective against infection [1]. The median number of challenges needed to infect both the vaccinees and controls was two. In both groups animals with the restrictive TRIM5TFP/CypA genotype were among the last to become productively infected, which is consistent with recent studies examining the effects of TRIM5 genotype on acquisition of SIV infection, but this did not influence outcome, as TRIM5 genotypes were balanced between the groups [39, 42, 43].
Figure 5. Kaplan-Meier analysis of the rate of infection after repeated, limiting dose inoculation with SIVsmE660.
The proportion of vaccinated or control animals remaining uninfected after each inoculation with SIVsmE660. The Log-rank test was used to assess for a significant difference between the two groups.
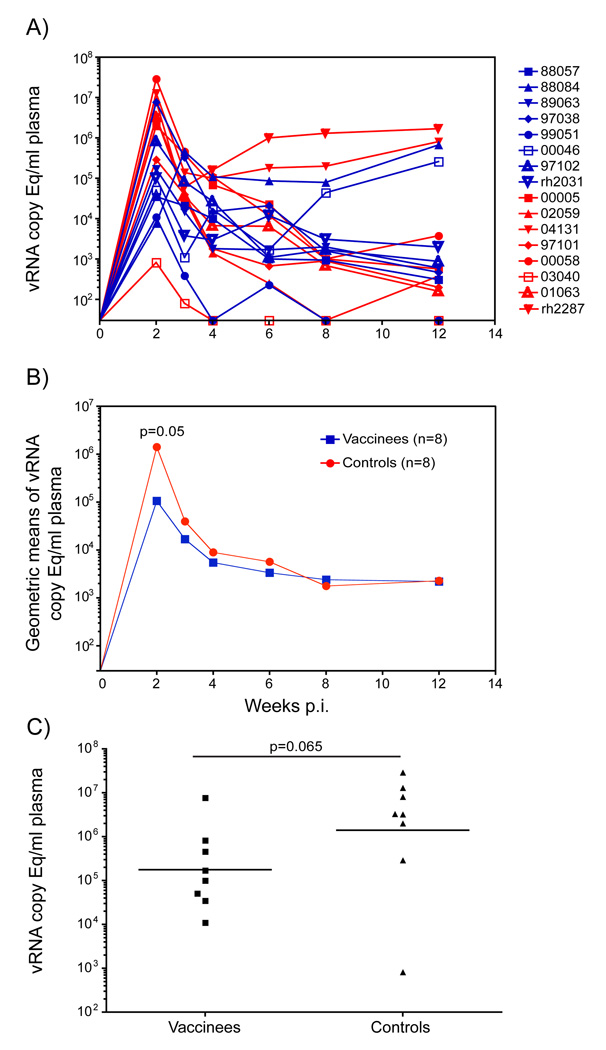
The Step trial was also designed to determine whether vaccine-induced T cells could reduce set-point viremia should infection occur. We therefore monitored the effect of vaccination on plasma virus replication for three months after infection with SIVsmE660. At two weeks post-detectable infection (the usual peak of virus replication) the vaccinated animals had marginally significantly lower plasma viral loads in comparison to the naïve controls (Fig. 6B, p=0.046; repeated measures mixed model). However, the vaccinated animal r88084 did not reach peak virus replication until week three post-infection. Comparison of the peak viral loads revealed no significant difference between the vaccinated and control groups (Fig. 6C, p=0.065; Wilcoxon-Mann-Whitney test). There was no significant difference in the plasma virus concentrations between the vaccinees and controls during the chronic phase of infection.
Figure 6. Plasma virus concentrations after detectable SIVsmE660 infection.
A) vRNA copies per ml of plasma for the individual vaccinated (blue) and control (red) animals in the 12 weeks following infection with SIVsmE660. The week prior to detectable virus replication is labeled as week 0. B) Geometric mean of the vRNA copy Eq/ml for the vaccinated (blue) and control (red) groups during the first 12 weeks post-infection. The difference between the geometric mean of the plasma virus concentrations between the vaccinees and controls is significant at week 2 by repeated measures mixed models. C) A scatter plot comparison of the peak plasma virus replication occurring at any point during the first three weeks of infection with SIVsmE660 for the vaccine and control groups. The horizontal line represents the geometric mean of each group. The difference between the vaccinated and control animals was not statistically significant by the Wilcoxon-Mann-Whitney test. The limit of detection for the QRT-PCR assay was 30 SIV RNA copy Eq/ml plasma therefore the X-axis crosses the Y-axis at this point in both A and B.
3.3 Anamnestic cellular immune responses
The inability of the vaccine-induced immune responses to adequately recognize the diverse infecting HIV strain(s) may have been responsible for the failure of the Step vaccine trial. Unfortunately, in human studies it is difficult to assess anamnestic immune responses in vaccinated individuals at the very earliest stages of infection. We, therefore, carried out these analyses in our vaccinated macaques. We measured anamnestic cellular immune responses at two weeks post-infection using IFN-γ ELISPOT assays containing pools of peptides corresponding to the SIVmac239 proteins in the vaccine. We previously detected expansion of anamnestic immune responses at this two-week time point prior to the development of de novo responses [33, 43, 44]. We compared these responses with assays performed approximately five weeks prior to the initiation of challenges to determine which of the vaccine-induced immune responses expanded significantly above pre-challenge levels, evaluating the presence, breadth, and magnitude of these responses.
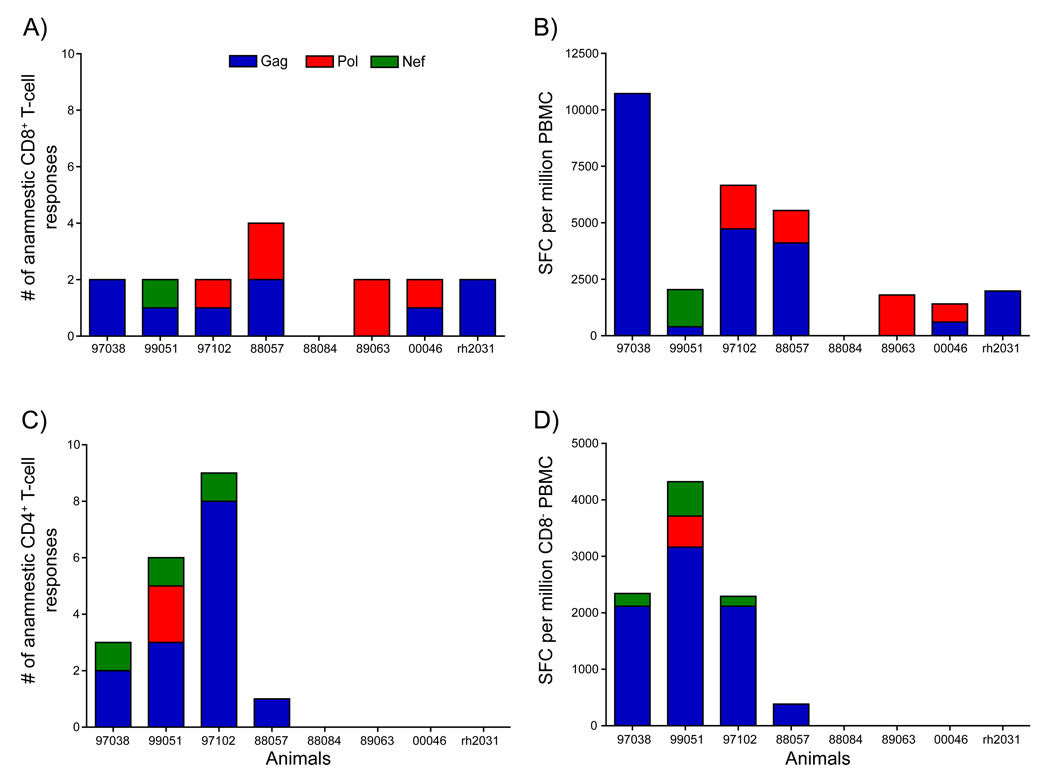
The breadth of recall responses may be especially important in controlling HIV strains that are likely to be substantially different from the vaccine sequences [45, 46]. We therefore examined the number of individual pool-specific responses that expanded post-infection. We detected expansion of anamnestic CD8+ T-cell responses above pre-challenge levels in seven of the eight vaccinated animals at week two post-infection (Fig. 7A). The absence of detectable anamnestic cellular immune responses in the remaining animal, r88084, was likely related to the relatively low level of virus replication at two weeks post-infection (7,600 vRNA copy Eq/ml plasma). This level of viral replication may not have been sufficient to drive expansion of the vaccine-induced immune responses at this time. While anamnestic responses were detected in seven of the eight vaccinated animals, the breadth of these responses was restricted. Six of eight of the vaccinated animals mounted only two distinct anamnestic CD8+ T-cell responses at two weeks post-infection. In the seventh animal, r88057, we observed anamnestic CD8+ T-cell responses against four regions of the virus. This suggests that the vaccine regimen induced only a limited number of CD8+ T cell responses capable of responding to the challenge virus. However, we did not break down individual pools into their constituent peptides, so it is formally possible that CD8+ T-cell responses against several epitopes were stimulated by some of the pools.
Figure 7. Expansion of anamnestic CD8+ and CD4+ T-cell responses at two weeks post-infection with SIVsmE660.
The number of anamnestic pool-specific CD8+ (A) and CD4+ (C) T-cell responses that significantly expanded above prechallenge levels at two weeks post-infection. The sum of the SFC per protein that significantly expanding above prechallenge levels for the anamnestic T-cell responses is shown in panels B (CD8+) and D (CD4+).
We then examined the magnitude of the anamnestic T-cell responses. For pool-specific responses containing CD8+ T-cell responses we observed expansions ranging from 1,500 to 11,000 SFC/million PBMC above pre-challenge levels (Fig. 7B). Despite the prevalence of numerous vaccine-induced Pol-specific CD8+ T-cell responses, Gag-specific responses dominated during acute infection. Only a single animal, r99051, had anamnestic CD8+ T cell responses directed against Nef. This was not surprising given that only three animals made vaccine-induced responses against Nef and there is an approximately 21% AA difference between the vaccine and SIVsmE660 sequences.
We also looked at the contribution of a namnestic CD4+ T cells to the antiviral immune response. We only detected expansion of anamnestic CD4+ T-cell responses in four of the eight vaccinated animals (Fig. 7C). However, in those four animals we detected between one and nine pool-specific responses, with animals r97102 and r99051 making eight and nine pool-specific responses respectively. Three of the four animals for which we observed anamnestic CD4+ T cell responses had an expansion of over 2,000 SFC/million CD8+ depleted PBMC above pre-challenge levels (Fig 7D). Impressively, animal r99051 had an expansion of anamnestic CD4+ T-cell responses exceeding 4,300 SFC/million CD8+ depleted PBMC. Interestingly, this animal also had the lowest viral load of 11,000 vRNA copies/ml plasma at peak. In the group of four vaccinated animals with detectable anamnestic CD4+ T cells, Gag was the primary target of these responses with only minor populations targeting Nef and Pol. Our study was too small to draw any firm conclusions regarding the utility of these CD4+ T cell responses but it is interesting to note that the animals making the highest frequency anamnestic CD4+ T-cell responses achieved the best control of viral replication among the vaccinated animals.
4. Discussion
Clinical trials designed to test the efficacy of HIV-1 vaccine candidates are both expensive and time-consuming. Hence, pre-clinical challenge studies that can help inform the selection of candidates for such clinical efficacy trials would provide a significant benefit to the field. Among the several advantages of pre-clinical studies, they provide the opportunity for careful analysis of vaccine-induced immune responses which otherwise can be challenging in human trials; such information potentially affords a detailed understanding of the mechanisms of the immune control against this virus. It is important to determine how well and what pre-clinical challenge system(s) could predict the clinical efficacy of a vaccine candidate. There are several parameters that define a challenge system, namely the viral challenge strain, the route of challenge, and frequency/dose of challenge virus (repeated, limiting-dose or single high-dose). Developing the appropriate model of HIV infection will not only depend on the transmission route of natural infection in question (vaginal intercourse, anal intercourse, intravenous drug use) but also on assimilating the lessons from both pre-clinical and clinical efficacy studies. Prior to the Step trial, several pre-clinical studies had been conducted in which components of the rAd5 vaccine were tested against single high-dose challenge of either SHIV or SIVmac239 viruses, with varying degrees of success [18, 21, 23–25, 47]. The trivalent rAd5 vaccine was later tested using Mamu-A*01+ macaques challenged with a single high-dose of the homologous SIVmac239 clone. These vaccinees did not control replication of this pathogenic virus [22]. We, therefore, retrospectively performed a parallel testing of the Step trial vaccine regimen in Mamu-A*01 negative rhesus macaques to elucidate the immune responses induced by this vaccination approach. We also used the mucosal heterologous repeated, limiting-dose SIVE660 challenge to more closely mimic exposure of human vaccinees to HIV. We, therefore, evaluated protective efficacy in this animal model by measuring prevention of infection and/or control of viral replication after infection.
In our study we detected relatively broader CD4+ and CD8+ T-cell responses using the Step trial vaccine regimen in macaques than the published immunogenicity results of the trivalent rAd5 vaccine in healthy human subjects [12]. There are several potential reasons for the different results observed in humans and monkeys. First, we performed immune assays at more time points in our monkey study compared to the Step trial in humans, including those closer to the immunizations (one week post-boost in our study versus four-weeks post-boost in the human study). Performing immune assays closer to when the rAd5 immunizations were given may have facilitated the detection of low-level responses that would otherwise have diminished by week four post-boost, particularly in the CD4+ T-cell compartment. It should be noted that at week 30 (four weeks after the final rAd5 boost), the responses seen in macaques are broadly comparable to those reported in humans at the level of the full antigen peptide pool; where 75% responded to Gag, 71% responded to Pol, and 70% responded to Nef for individuals with neutralizing Ad5 antibody titers below 200 [2]. Also, at this same time point, the median number of CD8+ T-cell responses was three in the clinical trial versus four in non-human primates. Second, most of our assays were performed with smaller pools of peptides than used in the Step trial. We used 45 pools (26 covering Pol, 13 covering Gag, and 6 covering Nef) in comparison to the four pools (entire Gag, entire Nef, and first and second half of Pol) used in the Step trial. These smaller pools of peptides allowed us to gain a better understanding of the breadth of the vaccine-induced immune responses. In our experience, smaller pools often result in higher magnitude responses than that observed using larger whole protein pools. Overall, we detected responses in IFN-γ ELISPOT assays using whole PBMC against Gag in seven out of eight animals, Pol eight out of eight animals, and Nef three out of eight animals. However, it should also be noted that there were methodological differences in identifying the human and monkey T-cell responses. These include the use of different sized peptides (9-mer for human samples versus 15-mer for the macaques), input cell number per well, the use or non-use of subset depletion and the nature of the reagents used, which may result in differences in the sensitivities of the assays. Finally, experience suggests that macaque responses to SIV vaccine immunogens are often higher than corresponding human responses to HIV vaccine immunogens, particularly for Gag [48].
Another possible contributing factor to our enhanced detection of vaccine-induced immune responses may have been the use of freshly isolated PBMC in some of our IFN-γ ELISPOT assays. In this study we observed a reduction in the number and magnitude of most responses when using cryopreserved cells, especially for vaccine-induced CD4+ T cells. This agrees with a previous study reporting a similar phenomenon in specimens from HIV-1 infected individuals [49]. Conversely, additional studies comparing fresh versus cryopreserved cells in IFN-γ ELISPOT assays have shown minimal loss of antigen-specific specificity through the cryopreservation process [50–52]. The differences between our results and those showing minimal effects of cryopreservation on antigen-specific cells may lie in the nature of the cells we are trying to retrieve. In our study we recovered cells with overall viability of 95% or greater when measured immediately after thawing. So, it does not appear that the recovery of sufficient numbers of viable cells alone accounts for a loss of pool-specific responses in our study. It is possible that recently activated antigen-specific cells are particularly sensitive to the cryopreservation process. The cells used in our study were cryopreserved within a month of being activated by rAd5 immunizations and thawed for use in immune assays 6–12 months later. Despite the potential draw back of using cryopreserved cells it is impractical to use fresh PBMC in immune assays in large-scale HIV vaccine studies. Therefore, cryopreserved cells will remain an essential re source for analyzing vaccine-induced immune responses in these studies.
Despite the overall lack of efficacy observed in the Step trial, recent studies have provided evidence that the vaccine regimen influenced the nature of infection in some vaccinated individuals. In a follow-up of 87 male participants in the Step study, a subset of vaccinated individuals expressing HLA alleles previously associated with slow progression to disease (HLA-B*27, -B*57, and –B*5801) had significantly lower viral loads than individuals expressing these alleles that received the placebo [26]. This suggests that a T-cell-based vaccine can generate biologically functional immune responses. Additionally, a study of 68 HIV-infected participants in the Step study by Rolland et al. found evidence of a “sieving effect” whereby the Step vaccine regimen influenced which viral variants established infection [53]. They found that the sequence of the infecting virus(es) in the vaccinees was more disparate from the vaccine sequence than those arising in the placebo group, but only for the proteins included in the vaccine. Interestingly, they also observed evidence of a greater breadth of vaccine-induced cytotoxic T-lymphocyte responses than initially described, indicating that the Step vaccine regimen may have been more immunogenic than previously appreciated. This corresponds with recent data published from the Phambili study, the companion to the Step trial conducted in South Africa that was prematurely terminated after the Step trial interim analysis. In the Phambili cohort, more vaccinated individuals made vaccine-induced responses compared to the frequency of responders in the Step trial, with significant cross-reactivity against Clade C peptides [54]. Together these findings suggest that CD8+ T cells directed against a broad array of HIV determinants that have some ability to inhibit HIV replication can impact viral replication.
The ability of vaccine-induced responses to recognize the infecting strain(s) of virus is likely a key factor in determining whether an HIV vaccine is effective. The leading hypothesis for the lack of efficacy seen in the Step trial is the limited breadth of the vaccine-induced immune responses and their inability to deal with the variability of circulating strains of the virus [2]. To address this issue we used the sequence of the molecular clone SIVmac239 as the basis for our immunogens and SIVsmE660 as our challenge virus. SIVmac239 and SIVsmE660 are derived from different SIV lineages and the genetic distance between these viruses approximates the estimated differences between consensus clade-based immunogens and circulating strains within that clade, which are similar to the conditions for the Step trial [55, 56]. Gag and Pol are relatively conserved between SIVmac239 and SIVsmE660 and differ at only 8.0% and 8.3% of their amino acids respectively [44]. Nef is more variable between the two strains and differs at 21.3% of its amino acids. Our vaccine regimen induced an average of five pool-specific CD8+ T-cell responses per animal. However, after infection we observed an anamnestic expansion of only two of these vaccine-induced CD8+ T-cell responses in six of the seven animals with detectable responses post-infection. This suggests that the majority of animals only had two vaccine-induced CD8+ T-cell responses that were able to recognize SIV-infected cells. It may be beneficial for future vaccine modalities to incorporate more HIV proteins and/or additional conserved regions of the virus to increase the chances of inducing broadly reactive CD8+ T-cell responses.
Unexpectedly, a subgroup of the Step trial volunteers experienced an increased rate of infection [1]. We did not, however, detect an enhancement of acquisition of infection in our vaccinated animals. There are several potential reasons for this. First, the increase in infection detected in the Step trial was among uncircumcised men. Since we challenged our animals intrarectally our study may have failed to recapitulate relevant features of virus exposure associated with enhancement of infection as was recently observed in a penile transmission model [57]. Secondly, the animals used in our study had no pre-existing Ad5 immunity, which was a key risk factor for the increased risk of infection for vaccinated, uncircumcised men.
There is still considerable debate as to which viral proteins should be included in vaccines [58–60]. For the Step trial vaccine gag, pol, and nef genes were selected for inclusion because these proteins are commonly recognized during natural infection and are relatively conserved across different HIV-1 clades [1]. During the vaccination phase of our study the majority of CD8+ T-cell responses targeted Pol, followed by Gag with only a few responses against Nef. The hierarchy of the number of protein-specific responses roughly mirrored the size of each protein. After infection we observed high frequency anamnestic CD8+ T cell responses directed against several epitopes in Gag. Low frequency Pol-specific responses, by contrast, were directed against only a few epitopes in Pol. The switch from targeting Pol after vaccination to targeting Gag after infection might have been due to several factors. First, the amount of protein produced by the rAd5 immunization differs from that produced by the virus during natural infection. Once the rAd5 vectors deliver the genes to target cells they are designed to efficiently produce abundant levels of the SIV proteins, providing ample stimulation to engender CD8+ T-cell responses against all of the proteins included in the vaccine [61]. During natural SIV infection, however, due to ribosomal frameshifting, the structural proteins of gag are produced in greater quantities than are the enzymatic proteins of pol [62]. This difference in antigen abundance may result in the preferential expansion of the Gag-specific instead of Pol-specific T-cell responses. Alternatively, it is also possible that differences between the Pol sequences of the vaccine and the infecting SIVsmE660 virus(es) prevented their expansion post-infection. Since we did not map the CD8+ T-cell responses to their minimal optimal epitopes it is, therefore, formally possible (but unlikely) that sequence differences between SIVmac239 and SIVsmE660 were largely found in regions of Pol containing CD8+ T-cell epitopes. The inclusion of env in HIV vaccine design may be critical to inhibiting the acquisition of infection [39]. The RV144 HIV vaccine trial in Thailand reported a modest level of protection against infection that early correlate analysis has associated with the level of antibodies binding the V1/V2 loops of HIV Env [63, 64]. Furthermore, a recent study in non-human primates demonstrates that the addition of Env to vaccine modalities was essential for delaying acquisition of SIV infection[65].
Chronic phase viral loads in both the vaccinees and controls were unusually low. This may have been a result of the variability in pathogenicity associated with mucosal inoculation of SIVsmE660. One of the advantages of using SIVsmE660 in pre-clinical vaccine studies is that it provides a heterologous challenge to SIVmac239-based immunogens in a manner analogous to the predicted variation of consensus clade-based vaccines and circulating strains of those clades. SIVsmE660 is also similar to HIV in that it is derived from a biological isolate and contains many quasispecies within the inoculum. These quasispecies, however, may have varying degrees of replicative fitness. Studies have shown that repeated, limiting dose mucosal challenges in rhesus macaques typically results in one to three viruses establishing infection [66, 67]. Under these conditions the nature of the virus(es) that cross the mucosal barrier to establish infection may influence how effectively the virus replicates. Investigation of the sensitivity of transmitted viruses to neutralizing antibody activity after repeated, limiting dose SIVsmE660 infection shows that certain clones are more sensitive to neutralization than others [68]. This suggests that a subset of quasispecies present in SIVsmE660 stocks may be sensitive to neutralizing antibody activity and this may affect viral set point, regardless of the immunization status of the macaque. Although it should also be noted that despite the advancement in developing limiting dose SIV challenges to more effectively mimic human exposure to HIV these “limiting” doses still result in a greater risk of infection for the macaques than is typically encountered during human sexual exposure. TRIM5α genotype may also affect virus acquisition and replication. Recent studies have shown that polymorphisms at the TRIM5α locus can have differential effects on peak and chronic virus replication in animals infected with SIVsmE543, a closely related molecular clone to SIVsmE660, and SIVmac251 [38, 41]. Control of viral replication does not appear to be associated with TRIM5α genotype in our study. Of the four animals (two vaccinees and two naïve controls) with plasma virus concentrations above 1×105 vRNA copy Eq/ml in the chronic phase of infection, three of them had restrictive TRIM5α genotypes. Additionally, others and we have recently reported that these polymorphisms at the TRIM5α locus can affect the acquisition of SIVsmE660 after limiting dose mucosal exposure [ 32, 39, 42]. For these reasons we balanced the TRIM5α genotypes of the animals in the vaccine and control groups.
5. Conclusions
In this study we retrospectively assessed the ability of the Step trial vaccine regimen to induce cellular immune responses in rhesus macaques and protect them from infection or control virus replication after repeated, limited dose challenge with a heterologous virus. The vaccine regimen induced relatively broad CD8+ and CD4+ T-cell responses, primarily targeting Gag and Pol. These responses were similar to but broader and of a higher frequency than detected in samples from human recipients of the rAd5 vaccine trial. This may have been due to a combination of differences in the methods used to detect vaccine-induced responses. However, these vaccine-induced immune responses were unable to prevent infection or significantly reduce viral set-point in comparison to naïve controls. Our efficacy results of this non-human primate study are consistent with those observed in the Step trial and suggest that pre-clinical trials in macaques, depending on the model design, may help inform the immunogenicity and effectiveness of vaccines in human trials.
Supplementary Material
Highlights.
We modeled the Step trial HIV vaccine regimen in rhesus macaques
We observed broad vaccine-induced T-cell responses in freshly isolated PBMC
We detected fewer vaccine-induced T-cell responses with cryopreserved cells
The vaccine regimen failed to protect against infection or control SIV replication
Our results in macaques are consistent with those observed in Step trial in humans
Acknowledgements
We would like to thank the veterinary staff at the Wisconsin National Primate Research Center (WNPRC) for their assistance.
This research was supported by funds from the International AIDS Vaccine Initiative, National Institutes of Health (NIH) grants RO1 AI049120, R24 RR016038, R24 RR015371, and contract HHSN266200400088C to D.I.W., P51 RR000167 to the WNPRC from the National Center for Research Resources (NCRR), a component of the NIH, and in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. Additionally, this research was conducted at a facility constructed with support from Research Facilities Improvement Program grant numbers RR15459-01 and RR020141-01. This publication's contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. Employees of the International AIDS Vaccine Initiative contributed to the study design and preparation of the manuscript.
Drs. Bett, Casimiro, and Shiver are employees of Merck & Co., Inc. and own stock in Merck. This may be considered a potential conflict of interest for this work.
Dr. Allison has, anticipates, or has had financial interests with the following entities: the Frontiers Foundation; Vivus, Inc; Kraft Foods; University of Wisconsin; University of Arizona; Paul, Weiss, Wharton & Garrison LLP; and Sage Publications.
All authors reviewed and approved the final version of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations
recombinant Adenovirus serotype 5
Macaca mulatta
PCR amplification with sequence-specific primers
quantitative reverse transcription-PCR
Contributor Information
Matthew R. Reynolds, Email: mrreynol@wisc.edu.
Andrea M. Weiler, Email: amweiler@wisc.edu.
Shari M. Piaskowski, Email: sharip@primate.wisc.edu.
Michael Piatak, Jr., Email: Michael.Piatak2@nih.gov.
Henry T. Robertson, Email: HRobertson@ms.soph.uab.edu.
David B. Allison, Email: DAllison@ms.soph.uab.edu.
Andrew J. Bett, Email: andrew_bett@merck.com.
Danilo R. Casimiro, Email: danilo_casimiro@merck.com.
John W. Shiver, Email: john_shiver@merck.com.
Nancy A. Wilson, Email: nwilson@primate.wisc.edu.
Jeffrey D. Lifson, Email: Jeffrey.Lifson@nih.gov.
Wayne C. Koff, Email: WKoff@iavi.org.
David I. Watkins, Email: DWatkins@med.miami.edu.
References
- 1.Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McElrath MJ, De Rosa SC, Moodie Z, Dubey S, Kierstead L, Janes H, et al. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet. 2008;372:1894–1905. doi: 10.1016/S0140-6736(08)61592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langford SE, Ananworanich J, Cooper DA. Predictors of disease progression in HIV infection: a review. AIDS Res Ther. 2007;4:11. doi: 10.1186/1742-6405-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mellors JW, Margolick JB, Phair JP, Rinaldo CR, Detels R, Jacobson LP, et al. Prognostic value of HIV-1 RNA CD4 cell count, and CD4 Cell count slope for progression to AIDS and death in untreated HIV-1 infection. JAMA. 2007;297:2349–2350. doi: 10.1001/jama.297.21.2349. [DOI] [PubMed] [Google Scholar]
- 5.Mellors JW, Rinaldo CRJ, Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 6.Gray RH, Wawer MJ, Brookmeyer R, Sewankambo NK, Serwadda D, Wabwire-Mangen F, et al. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet. 2001;357:1149–1153. doi: 10.1016/S0140-6736(00)04331-2. [DOI] [PubMed] [Google Scholar]
- 7.Quinn TC, Wawer MJ, Sewankambo N, Serwadda D, Li C, Wabwire-Mangen F, et al. Viral load and heterosexual transmission of human immunodeficiency virus type-1. Rakai Project Study Group. N Engl J Med. 2000;342:921–929. doi: 10.1056/NEJM200003303421303. [DOI] [PubMed] [Google Scholar]
- 8.Cohen J. AIDS research. Did Merck's failed HIV vaccine cause harm? Science. 2007;318:1048–1049. doi: 10.1126/science.318.5853.1048. [DOI] [PubMed] [Google Scholar]
- 9.Fauci AS, Johnston MI, Dieffenbach CW, Burton DR, Hammer SM, Hoxie JA, et al. HIV vaccine research: the way forward. Science. 2008;321:530–532. doi: 10.1126/science.1161000. [DOI] [PubMed] [Google Scholar]
- 10.Sekaly RP. The failed HIV Merck vaccine study: a step back or a launching point for future vaccine development? J Exp Med. 2008;205:7–12. doi: 10.1084/jem.20072681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watkins DI, Burton DR, Kallas EG, Moore JP, Koff WC. Nonhuman primate models and the failure of the Merck HIV-1 vaccine in humans. Nat Med. 2008;14:617–621. doi: 10.1038/nm.f.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li F, Finnefrock AC, Dubey SA, Korber BT, Szinger J, Cole S, et al. Mapping HIV-1 vaccine induced T-cell responses: bias towards less-conserved regions and potential impact on vaccine efficacy in the Step study. PLoS One. 2011;6:e20479. doi: 10.1371/journal.pone.0020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corey L, McElrath MJ, Kublin JG. Post-step modifications for research on HIV vaccines. AIDS. 2009;23:3–8. doi: 10.1097/QAD.0b013e32830e6d6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li F, McKenney DM, Malhotra U, Crimi C, Nolin J, Corey L, et al. Towards prediction of degenerate CTL epitope recognition. Hum Vaccin. 2008;4:115–121. doi: 10.4161/hv.4.2.5215. [DOI] [PubMed] [Google Scholar]
- 15.Priddy FH, Brown D, Kublin J, Monahan K, Wright DP, Lalezari J, et al. Safety and immunogenicity of a replication-incompetent adenovirus type 5 HIV-1 clade B gag/pol/nef vaccine in healthy adults. Clin Infect Dis. 2008;46:1769–1781. doi: 10.1086/587993. [DOI] [PubMed] [Google Scholar]
- 16.Abbink P, Lemckert AA, Ewald BA, Lynch DM, Denholtz M, Smits S, et al. Comparative seroprevalence and immunogenicity of six rare serotype recombinant adenovirus vaccine vectors from subgroups B and D. J Virol. 2007;81:4654–4663. doi: 10.1128/JVI.02696-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barouch DH. Novel adenovirus vector-based vaccines for HIV-1. Curr Opin HIV AIDS. 2010;5:386–390. doi: 10.1097/COH.0b013e32833cfe4c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiver JW, Fu TM, Chen L, Casimiro DR, Davies ME, Evans RK, et al. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature. 2002;415:331–335. doi: 10.1038/415331a. [DOI] [PubMed] [Google Scholar]
- 19.Feinberg MB, Moore JP. AIDS vaccine models: challenging challenge viruses. Nat Med. 2002;8:207–210. doi: 10.1038/nm0302-207. [DOI] [PubMed] [Google Scholar]
- 20.Lifson JD, Martin MA. One step forwards, one step back. Nature. 2002;415:272–273. doi: 10.1038/415272b. [DOI] [PubMed] [Google Scholar]
- 21.Casimiro DR, Wang F, Schleif WA, Liang X, Zhang ZQ, Tobery TW, et al. Attenuation of simian immunodeficiency virus SIVmac239 infection by prophylactic immunization with dna and recombinant adenoviral vaccine vectors expressing Gag. J Virol. 2005;79:15547–15555. doi: 10.1128/JVI.79.24.15547-15555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casimiro DR, Cox K, Tang A, Sykes KJ, Feng M, Wang F, et al. Efficacy of multivalent adenovirus-based vaccine against simian immunodeficiency virus challenge. J Virol. 2010;84:2996–3003. doi: 10.1128/JVI.00969-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letvin NL, Mascola JR, Sun Y, Gorgone DA, Buzby AP, Xu L, et al. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science. 2006;312:1530–1533. doi: 10.1126/science.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattapallil JJ, Douek DC, Buckler-White A, Montefiori D, Letvin NL, Nabel GJ, et al. Vaccination preserves CD4 memory T cells during acute simian immunodeficiency virus challenge. J Exp Med. 2006;203:1533–1541. doi: 10.1084/jem.20060657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson NA, Reed J, Napoe GS, Piaskowski S, Szymanski A, Furlott J, et al. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J Virol. 2006;80:5875–5885. doi: 10.1128/JVI.00171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fitzgerald DW, Janes H, Robertson M, Coombs R, Frank I, Gilbert P, et al. An Ad5-vectored HIV-1 vaccine elicits cell-mediated immunity but does not affect disease progression in HIV-1-infected male subjects: results from a randomized placebo controlled trial (the Step study) J Infect Dis. 2011;203:765–772. doi: 10.1093/infdis/jiq114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaslow RA, Carrington M, Apple R, Park L, Munoz A, Saah AJ, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 28.Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, et al. Mamu-B*08-positive Macaques Control Simian Immunodeficiency Virus Replication. J Virol. 2007 doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mothe BR, Weinfurter J, Wang C, Rehrauer W, Wilson N, Allen TM, et al. Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2003;77:2736–2740. doi: 10.1128/JVI.77.4.2736-2740.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, et al. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2006;80:5074–5077. doi: 10.1128/JVI.80.10.5074-5077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaizu M, Borchardt GJ, Glidden CE, Fisk DL, Loffredo JT, Watkins DI, et al. Molecular typing of major histocompatibility complex class I alleles in the Indian rhesus macaque which restrict SIV CD8+ T cell epitopes. Immunogenetics. 2007;59:693–703. doi: 10.1007/s00251-007-0233-7. [DOI] [PubMed] [Google Scholar]
- 32.Reynolds MR, Sacha JB, Weiler AM, Borchardt GJ, Glidden CE, Sheppard NC, et al. The TRIM5{alpha} Genotype of Rhesus Macaques Affects Acquisition of Simian Immunodeficiency Virus SIVsmE660 Infection after Repeated Limiting-Dose Intrarectal Challenge. J Virol. 2011;85:9637–9640. doi: 10.1128/JVI.05074-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reynolds MR, Weiler AM, Piaskowski SM, Kolar HL, Hessell AJ, Weiker M, et al. Macaques vaccinated with simian immunodeficiency virus SIVmac239Delta nef delay acquisition and control replication after repeated low-dose heterologous SIV challenge. J Virol. 2010;84:9190–9199. doi: 10.1128/JVI.00041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cline AN, Bess JW, Piatak M, Jr, Lifson JD. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J Med Primatol. 2005;34:303–312. doi: 10.1111/j.1600-0684.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 35.Hudgens MG, Gilbert PB. Assessing vaccine effects in repeated low-dose challenge experiments. Biometrics. 2009;65:1223–1232. doi: 10.1111/j.1541-0420.2009.01189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hudgens MG, Gilbert PB, Mascola JR, Wu CD, Barouch DH, Self SG. Power to detect the effects of HIV vaccination in repeated low-dose challenge experiments. J Infect Dis. 2009;200:609–613. doi: 10.1086/600891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fellay J, Frahm N, Shianna KV, Cirulli ET, Casimiro DR, Robertson MN, et al. Host genetic determinants of T cell responses to the MRKAd5 HIV-1 gag/pol/nef vaccine in the step trial. J Infect Dis. 2011;203:773–779. doi: 10.1093/infdis/jiq125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirmaier A, Wu F, Newman RM, Hall LR, Morgan JS, O'Connor S, et al. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Letvin NL, Rao SS, Montefiori DC, Seaman MS, Sun Y, Lim SY, et al. Immune and Genetic Correlates of Vaccine Protection Against Mucosal Infection by SIV in Monkeys. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002351. 81ra36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim SY, Chan T, Gelman RS, Whitney JB, O'Brien KL, Barouch DH, et al. Contributions of Mamu-A*01 status and TRIM5 allele expression, but not CCL3L copy number variation, to the control of SIVmac251 replication in Indian-origin rhesus monkeys. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000997. e1000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim SY, Rogers T, Chan T, Whitney JB, Kim J, Sodroski J, et al. TRIM5alpha Modulates Immunodeficiency Virus Control in Rhesus Monkeys. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000738. e1000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeh WW, Rao SS, Lim SY, Zhang J, Hraber PT, Brassard LM, et al. The TRIM5 Gene Modulates Penile Mucosal Acquisition of Simian Immunodeficiency Virus in Rhesus Monkeys. J Virol. 2011;85:10389–10398. doi: 10.1128/JVI.00854-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reynolds MR, Rakasz E, Skinner PJ, White C, Abel K, Ma ZM, et al. CD8+ T-lymphocyte response to major immunodominant epitopes after vaginal exposure to simian immunodeficiency virus: too late and too little. J Virol. 2005;79:9228–9235. doi: 10.1128/JVI.79.14.9228-9235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reynolds MR, Weiler AM, Weisgrau KL, Piaskowski SM, Furlott JR, Weinfurter JT, et al. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J Exp Med. 2008;205:2537–2550. doi: 10.1084/jem.20081524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 46.Liu J, O'Brien KL, Lynch DM, Simmons NL, La Porte A, Riggs AM, et al. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature. 2009;457:87–91. doi: 10.1038/nature07469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang X, Casimiro DR, Schleif WA, Wang F, Davies ME, Zhang ZQ, et al. Vectored Gag and Env but not Tat show efficacy against simian-human immunodeficiency virus 89.6P challenge in Mamu-A*01-negative rhesus monkeys. J Virol. 2005;79:12321–12331. doi: 10.1128/JVI.79.19.12321-12331.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bett AJ, Dubey SA, Mehrotra DV, Guan L, Long R, Anderson K, et al. Comparison of T cell immune responses induced by vectored HIV vaccines in non-human primates and humans. Vaccine. 2010;28:7881–7889. doi: 10.1016/j.vaccine.2010.09.079. [DOI] [PubMed] [Google Scholar]
- 49.Owen RE, Sinclair E, Emu B, Heitman JW, Hirschkorn DF, Epling CL, et al. Loss of T cell responses following long-term cryopreservation. J Immunol Methods. 2007;326:93–115. doi: 10.1016/j.jim.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kreher CR, Dittrich MT, Guerkov R, Boehm BO, Tary-Lehmann M. CD4+ and CD8+ cells in cryopreserved human PBMC maintain full functionality in cytokine ELISPOT assays. J Immunol Methods. 2003;278:79–93. doi: 10.1016/s0022-1759(03)00226-6. [DOI] [PubMed] [Google Scholar]
- 51.Maecker HT, Moon J, Bhatia S, Ghanekar SA, Maino VC, Payne JK, et al. Impact of cryopreservation on tetramer, cytokine flow cytometry, and ELISPOT. BMC Immunol. 2005;6:17. doi: 10.1186/1471-2172-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kierstead LS, Dubey S, Meyer B, Tobery TW, Mogg R, Fernandez VR, et al. Enhanced rates and magnitude of immune responses detected against an HIV vaccine: effect of using an optimized process for isolating PBMC. AIDS Res Hum Retroviruses. 2007;23:86–92. doi: 10.1089/aid.2006.0129. [DOI] [PubMed] [Google Scholar]
- 53.Rolland M, Tovanabutra S, deCamp AC, Frahm N, Gilbert PB, Sanders-Buell E, et al. Genetic impact of vaccination on breakthrough HIV-1 sequences from the STEP trial. Nat Med. 2011;17:366–371. doi: 10.1038/nm.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gray GE, Allen M, Moodie Z, Churchyard G, Bekker LG, Nchabeleng M, et al. Safety and efficacy of the HVTN 503/Phambili study of a clade-B-based HIV-1vaccine in South Africa: a double-blind, randomised, placebo-controlled test-of concept phase 2b study. Lancet Infect Dis. 2011;11:507–515. doi: 10.1016/S1473-3099(11)70098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Apetrei C, Kaur A, Lerche NW, Metzger M, Pandrea I, Hardcastle J, et al. Molecular epidemiology of simian immunodeficiency virus SIVsm in U. S. primate centers unravels the origin of SIVmac and SIVstm. J Virol. 2005;79:8991–9005. doi: 10.1128/JVI.79.14.8991-9005.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 57.Qureshi H, Ma ZM, Huang Y, Hodge G, Thomas MA, Dipasquale J, et al. Low dose penile SIVmac251 exposure of rhesus macaques infected with adenovirus 5 then immunized with a replication defective Ad5-based SIV gag/pol/nef vaccine recapitulates the results of the Phase IIb Step trial of a similar HIV-1 vaccine. J Virol. 2011 doi: 10.1128/JVI.06175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fischer W, Perkins S, Theiler J, Bhattacharya T, Yusim K, Funkhouser R, et al. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med. 2007;13:100–106. doi: 10.1038/nm1461. [DOI] [PubMed] [Google Scholar]
- 59.Koff WC. Accelerating HIV vaccine development. Nature. 2010;464:161–162. doi: 10.1038/464161a. [DOI] [PubMed] [Google Scholar]
- 60.Nickle DC, Rolland M, Jensen MA, Pond SL, Deng W, Seligman M, et al. Coping with viral diversity in HIV vaccine design. PLoS Comput Biol. 2007;3:e75. doi: 10.1371/journal.pcbi.0030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shiver JW, Emini EA. Recent advances in the development of HIV-1 vaccines using replication-incompetent adenovirus vectors. Annu Rev Med. 2004;55:355–372. doi: 10.1146/annurev.med.55.091902.104344. [DOI] [PubMed] [Google Scholar]
- 62.Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 63.Haynes B. Case control study of the RV144 trial for immune correlates: the analysis and way forward. AIDS Vaccine Conference. 2011;2011 [Google Scholar]
- 64.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, et al. Vaccination with ALVAC and AIDSVAX to Prevent HIV-1 Infection in Thailand. N Engl J Med. 2009 doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 65.Barouch DH, Liu J, Li H, Maxfield LF, Abbink P, Lynch DM, et al. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature. 2012 doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Keele BF, Li H, Learn GH, Hraber P, Giorgi EE, Grayson T, et al. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J Exp Med. 2009;206:1117–1134. doi: 10.1084/jem.20082831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilson NA, Keele BF, Reed JS, Piaskowski SM, MacNair CE, Bett AJ, et al. Vaccine-induced cellular responses control simian immunodeficiency virus replication after heterologous challenge. J Virol. 2009;83:6508–6521. doi: 10.1128/JVI.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bar KJ, Easlick J, Sun C, Decker J, Sterret S, Li H, et al. Neutralization Sensitivity of SIVsmE660 and SIVmac251 Challenge Stocks and Transmitted/Founder Viruses. 2010 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.