Abstract
In this paper, we give a short introduction to machine learning and survey its applications in radiology. We focused on six categories of applications in radiology: medical image segmentation, registration, computer aided detection and diagnosis, brain function or activity analysis and neurological disease diagnosis from fMR images, content-based image retrieval systems for CT or MRI images, and text analysis of radiology reports using natural language processing (NLP) and natural language understanding (NLU). This survey shows that machine learning plays a key role in many radiology applications. Machine learning identifies complex patterns automatically and helps radiologists make intelligent decisions on radiology data such as conventional radiographs, CT, MRI, and PET images and radiology reports. In many applications, the performance of machine learning-based automatic detection and diagnosis systems has shown to be comparable to that of a well-trained and experienced radiologist. Technology development in machine learning and radiology will benefit from each other in the long run. Key contributions and common characteristics of machine learning techniques in radiology are discussed. We also discuss the problem of translating machine learning applications to the radiology clinical setting, including advantages and potential barriers.
Keywords: survey, radiology, machine learning, image registration, image segmentation, computer aided detection and diagnosis, functional MRI, content-based image retrieval, computed tomography, magnetic resonance imaging
1. Introduction
Radiologic imaging is of increasing importance in patient care. Both diagnostic and therapeutic indications for radiologic imaging are expanding rapidly (Bhargavan et al., 2009). The rapid expansion is a consequence of the need for more rapid, accurate, cost-effective, and less invasive treatment. Technologic advancements in radiologic imaging equipment have also fueled the utilization of imaging. Such technologic advancements include the capability to acquire higher and higher resolution images, enabling visualization of smaller anatomic structures and abnormalities. The higher resolution comes at the cost of an ever increasing average number of images per patient. Radiologists need to interpret these images and as the number of images increases, radiologists' workload increases as well. The increasing number and complexity of the images threatens to overwhelm radiologists' capacities to interpret them. In many real radiologic practices, automated and intelligent image analysis and understanding are becoming an essential part or procedure, such as image segmentation, registration, and computer-aided diagnosis and detection. In addition, in the area of cancer prognosis and treatment, automated and intelligent algorithms have a large market and are welcomed broadly, in areas such as radiation therapy planning or automatic identification of imaging biomarkers from radiological images of certain diseases, etc. Machine learning algorithms underpin the algorithms and software that make computer-aided diagnosis/prognosis/treatment possible.
Radiology is a branch of medical science which uses imaging technology and radiation to make diagnoses and treat disease. It has benefited greatly from the advances of physics, electronic engineering, and computer science. Based on different detection and imaging rationale, various modalities were developed in the past decades in the field of diagnostic radiology. Today, the mainstream modalities which are widely used in hospitals and medical centers include radiography, fluoroscopy, computed tomography (CT), ultrasound, magnetic resonance imaging (MRI), and positron emission tomography (PET).
In the daily practice of radiology, medical images from different modalities are read and interpreted by radiologists. Usually radiologists must analyze and evaluate these images comprehensively in a short time. But with the advances in modern medical technologies, the amount of imaging data is rapidly increasing. For example, CT examinations are being performed with thinner slices than in the past. The reading and interpretation time of radiologists will mount as the number of CT slices grows.
Machine learning provides an effective way to automate the analysis and diagnosis for medical images. It can potentially reduce the burden on radiologists in the practice of radiology. The applications of machine learning in radiology include medical image segmentation (e.g., brain, spine, lung, liver, kidney, colon); medical image registration (e.g., organ image registration from different modalities or time series); computer-aided detection and diagnosis systems for CT or MRI images (e.g., mammography, CT colonography, and CT lung nodule CAD); brain function or activity analysis and neurological disease diagnosis from fMR images; content based image retrieval systems for CT or MRI images; and text analysis of radiology reports using natural language processing (NLP) and natural language understanding (NLU).
Machine learning is the study of computer algorithms which can learn complex relationships or patterns from empirical data and make accurate decisions (Bishop, 2006; Duda et al., 2000; Mitchell, 1997). It is an interdisciplinary field that has close relationships with artificial intelligence, pattern recognition, data mining, statistics, probability theory, optimization, statistical physics, and theoretical computer science. Applications of machine learning include natural language processing, medical diagnosis, bioinformatics, video surveillance, and financial data analysis.
Machine learning algorithms can be organized into different categories based on different principles. For example, depending on the utilization of labels of training samples, they can be categorized into supervised learning, semi-supervised learning, and unsupervised learning algorithms.
In supervised learning, each sample contains two parts: one is input observations or features and the other is output observations or labels (Alpaydin, 2004; Hastie et al., 2009). Usually the input observations are causes and the output observations are effects. The purpose of supervised learning is to deduce a functional relationship from training data that generalizes well to testing data. The form of the relationship is a set of equations and numerical coefficients or weights. Examples of supervised learning include classification, regression, and reinforcement learning.
In unsupervised learning, we only have one set of observations and there is no label information for each sample (Hastie et al., 2009). Usually these observations or features are caused by a set of unobserved or latent variables. The main purpose of unsupervised learning is to discover relationships between samples or reveal the latent variables behind the observations. Examples of unsupervised learning include clustering, density estimation, and blind source separation.
Semi-supervised learning falls between supervised and unsupervised learning (Chapelle et al., 2006; Zhu, 2007). It utilizes both labeled data (usually a few) and unlabeled data (usually many) during the training process. Semi-supervised learning algorithms were developed mainly because the labeling of data is very expensive or impossible in some applications. Examples of semi-supervised learning include semi-supervised classification and information recommendation systems (Christakou et al., 2005).
Machine learning has many applications in real life. It is routinely used in banking (for detecting fraudulent transactions (Dorronsoro et al., 1997)), in finance (to predict stock prices (Huang et al., 2005a)), in marketing (to reveal patterns of consumer spending (Bose and Mahapatra, 2001)), and on the Internet (as part of search engines (Basili, 2003)). In biomedicine, MYCIN was proposed in the early 1970s at Stanford University. It is an expert system with about 600 rules designed to identify bacteria and recommend antibiotics (Swartout, 1985). Machine learning also showed capability in the field of drug design (Burbidge et al., 2001).You may not be aware of the existence of machine learning, but its applications are pervasive in our daily lives.
This review is structured as follows. In Sec. 2 we give a short introduction to machine learning and related algorithms. In Sec. 3 we describe six representative applications of machine learning in radiology. In Sec. 4 we discuss key contributions and common characteristics of machine learning techniques in radiology. In Sec. 5 we cover issues on translating machine learning techniques to clinical radiology practice. In Sec. 6 we review current research status and discuss future directions.
2. Overview of machine learning
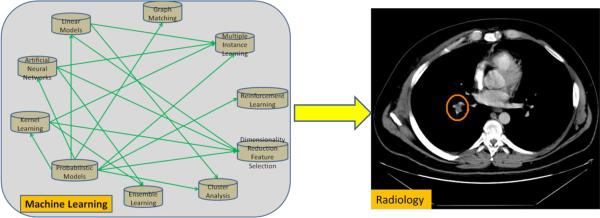
Because of the rapid development of machine learning, it is hard to introduce every aspect of machine learning in one article. So in this section we will give a concise introduction to the most important topics of machine learning (Bishop, 2006). These topics include linear models, learning with kernels, probabilistic models, clustering analysis and dimensionality reduction. Through this introduction, we hope readers may have a general idea about the content of machine learning research, what it is capable of, and what are the implications to other research areas and real applications. The topics that will be introduced and their inter-relationships are shown in Fig. 1.
Fig. 1.
Connections between different areas of machine learning.
In these topics of machine learning research, kernel learning and probabilistic models play key roles in machine learning-based radiology applications. Kernel learning usually provides the best classifier for computer-aided detection in radiology (El-Naqa et al., 2002; Malley et al., 2003); probabilistic models provide a theoretical framework for medical image analysis, such as image reconstruction (Levitan and Herman, 1987), segmentation (Zhang et al., 2001), and registration (Ashburner et al., 1997; Maintz and Viergever, 1998). Linear models, artificial neural networks, and ensemble learning provide other options for handling classification and regression problems in radiology besides kernel learning (Jerebko et al., 2003a; Yoshida and Nappi, 2001). Dimensionality reduction and feature selection is an essential part of computer-aided detection (CAD) systems in radiology (Wang et al., 2008a). Multiple instance learning addresses the common scenario in radiology CAD where a patient may have few positive instances of disease (e.g., lesions) and many false positives (Liang and Bi, 2007). Reinforcement learning is dedicated to accumulate domain experience in sequential learning (Sahba et al., 2006). Clustering analysis could be applied in medical images to identify similar lesions or meaningful findings (Chuang et al., 1999). Graph matching is employed to handle medical image registration problems (Wang et al., 2010a).
2.1 Linear models for classification and regression
Linear models assume that there is a linear relationship between the input of the model and the output of the model. Perhaps it is the simplest method for classification and regression. It has been widely used in computer-aided classification. For example, Chan et al. employed linear discriminant analysis (LDA) in texture feature space for classification of mammographic masses and normal tissue (Chan et al., 1995b). In the work of Preul et al. on accurate, noninvasive diagnosis of human brain tumors by proton magnetic resonance spectroscopy, they used LDA for classification in the “leave-one-out” test paradigm (Preul et al., 1996).
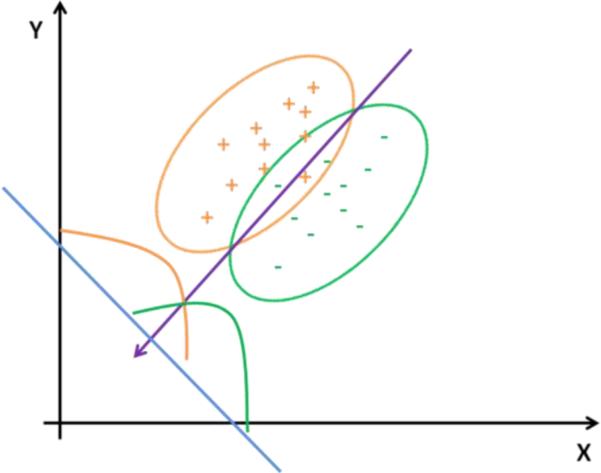
Given an input vector xεRd which describes features of objects we want to classify, a decision function in linear models usually is defined as f(x)= wTx + w0 where w is the weight vector and w0 is a constant and called threshold. Learning the optimal weight vector w and threshold w0 is a key problem in linear models. Once w is learned from training data, it can be applied to test cases and predicts the labels of them. For two-class classification problems, Fisher proposed the following criterion to locate the optimal parameters (Fisher, 1936): , where SB=(m1−m2)(m1+m2)T is called the “between” scatter matrix (mi is the mean of samples from class i,iε{1,2}), and Sw=S1+S2 is called the “within” scatter matrix (S1=ΣxεDi(x−m1)(x−m1)T, Di is the collection of samples from class i,iε{1,2}). This method is called linear discriminant analysis (LDA). The basic idea of LDA is to try to find an optimal projection w which can maximize the distances between samples from different classes and minimize the distances between samples from the same class. An illustration of LDA is shown in Fig. 2. Once the 2D data are projected to one dimensional line, threshold along the line will affect the classification error, as depicted by the 1-D distributions in Fig. 2. For multiple classes problems, the above scatter matrices can be extended to the following form: , where K is the number of classes, mi is the mean vector of class i, pi is the priori probability, is the overall mean (Loog et al., 2001).
Fig. 2.
Best projection direction (purple arrow) found by LDA. Two different classes of data with “Gaussian-like” distributions are shown in different markers and ellipses. 1-D distributions of the two-classes after projection are also shown along the line perpendicular to the projection direction.
Closely related to linear discriminant analysis, quadratic discriminant analysis tries to capture the quadratic relationship between the independent and dependent variables (Hastie et al., 2009). It provides more powerful discriminant ability compared with the linear separation interface of two classes learned by LDA.
2.2 Artificial neural networks
Artificial neural networks (ANNs) are techniques that were inspired by the brain and the way it learns and processes information. ANNs are frequently used to solve classification and regression problems in real world applications. Neural networks are composed of nodes and interconnections. Nodes usually have limited computation power. They simulate neurons by behaving like a switch, just as neurons will be activated only when sufficient neurotransmitter has accumulated. The density and complexity of the interconnections are the real source of a neural network's computational power.
Neural networks can be classified by their structures. In 1957 Rosenblatt proposed the first concrete neural network model, the perceptron (Rosenblatt, 1958). A perceptron has only one layer; in essence it is a linear classifier. In 1960, Bryson and Ho proposed the multiple neural network and introduced the fundamental backpropagation algorithm for training a neural network (Bryson and Ho, 1969). In theory, a three layer neural network can learn any complicated function. In 1982, the Hopfield network was proposed which has only one layer and all neurons are fully connected with each other (Hopfield, 1982). Boltzmann machines can be seen as the stochastic, generative version of Hopfield networks (Ackley et al., 1985). Boltzmann machines are able to solve difficult combinatorial problems and learn internal representations. The self-organizing map (SOM) was introduced around the same time (Kohonen, 1982). It is a unique network which conducts unsupervised learning. Since the final network topology learned by SOM can express certain characteristics of input signal, it was widely used for dimension reduction, visualization of high dimensional data and clustering. Cellular neural network (CNN) provides a parallel computing paradigm similar to human vision perception (Chua and Yang, 1988a, 1988b). In CNN, the communication is only allowed between neighboring nodes. Typical applications of CNN include image processing, analyzing 3D surface, modeling biological vision, etc. Besides these neural networks introduced above, other important neural networks include radial basis function (RBF) (Moody and Darken, 1989), probabilistic neural (Specht, 1990) and cascading neural networks (Fahlman and Lebiere, 1991).
Baker et al. showed that ANN could be used to categorize benign and malignant breast lesions based on the standardized lexicon of the Breast Imaging Recording and Data System (BIRADS) of the American College of Radiology (Baker et al., 1995). Tourassi et al. showed an application of ANN in acute pulmonary embolism detection (Tourassi et al., 1993). They found that the ANN significantly outperformed the physicians involved in this study.
2.3 Learning with kernels
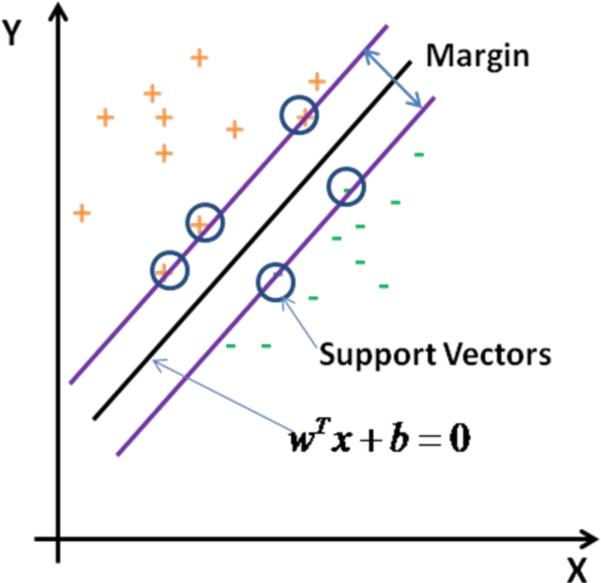
By applying traditional supervised and unsupervised learning methods in the feature space, kernel methods provide powerful tools for data analysis and have been found to be successful in a number of real applications. Support vector machines (SVMs) are a set of kernel-based supervised learning methods used for classification and regression (Burges, 1998). Here kernel means a matrix which encodes similarities between samples (evaluated by a certain kernel function which is a weighting function in the integral equation used to calculate similarities between samples). SVMs try to minimize the empirical classification error and maximize the geometric margin simultaneously on the training set which leads to high generalization ability on the new samples. For a two-class classification problem, given training samples {(x1, y1),…,(xn, yn)}, yi ε{−1,+1}, the optimization problem for learning a linear classifier in the feature space is defined as (hard margin): , subject to yi(〈w,Φ(xi〉)+b)≥1i=1,…,n,, where Φ is the mapping from original space to feature space and symbol <= means the inner product of two vectors. The matrix composed by inner products of samples in feature space (after linear or non-linear mapping) is called the kernel matrix which describes the similarities between samples and serves as evidence when we maximize the margin between two classes of samples. The above problem is a quadratic programming (QP) optimization problem and it is convex. The optimal (w*,b*) is a maximal margin classifier with geometric margin γ =1/||w*||2 if it exists. It can be applied to classify test samples once it is learned from the training set. The concept of the geometric margin learned by the SVM is shown in Fig. 3 in which samples on the margin are called support vectors.
Fig. 3.
Illustration of margin learned by SVM. Black line is the best hyperplane which can separate the two classes of data with maximum margin. Support vectors are shown in circles.
In many real clinical applications, we can extract many features with the help of modern medical test equipment, e.g., lab tests and radiology devices. Some features may not be relevant to the labels of subjects tested. If we feed all the features to SVMs, it may be affected by irrelevant and noisy features which can result in poor performance. In addition, with hundreds (or thousands or more) of features from test equipments doctors may also wonder which features are relevant or useful for the diagnosis of certain diseases so they can give better interpretations of the clinical findings. In the work of Weston et al., they proposed a method of feature selection for SVMs (Weston et al., 2001). The best features are identified by minimizing bounds on the leave-one-out error.
In the basic optimization problems of SVMs introduced above, there is only one mapping function Φ which maps input vector to feature space. In real applications, we often have multiple information sources to describe the same object. A multiple kernel learning approach provides a feasible way to solve real applications which involve multiple, heterogeneous data sources (Chapelle et al., 2002; Lanckriet et al., 2004; Sonnenburg et al., 2006). This so-called “multiple kernel learning” problem usually can be solved by considering the convex combinations of K kernels, i.e., , with βk ≥ 0 and , where each kernel Kk uses a group of features from one information source and xi,xj are samples.
Typical applications of kernel-based learning methods in radiology are in CAD. SVMs perform well in detection of microcalcifications on mammography CAD (El-Naqa et al., 2002; Tang et al., 2009; Wei et al., 2005b). For computed tomographic colonography (CTC), colonic polyps were detected using statistical features extracted from polyp candidates and multiple kernel learning (Wang et al., 2010c).
2.4 Learning and inference in probabilistic models
Probabilistic models provide a concise representation of complicated real world phenomena and enable predictions of future events from present observations. For example, in radiology, dose control in clinical scanning is a critical issue. Giving a patient more dose than he/she needed may cause potential damage to the issue or induce cancer. Mohan et al. proposed a tumor control probability (TCP) model to predict the clinical consequences of different radiation dose distributions and optimize 3-D conformal treatment plans (Mohan et al., 1992). The model can be used to predict radiation effect on tissue for a given dose in simulation prior to exposing the patient. Martel et al. estimated TCP model parameters from 3-D dose distributions of non-small cell lung cancer patients (Martel et al., 1999).
The Naive Bayes classifier is a classifier based on probabilistic models with strong (naive) independence assumptions. In spite of its oversimplified assumptions, naive Bayes classifiers work well in many real life applications (Domingos and Pazzani, 1997). Assume C is a class variable depending on n input features: X1,X2,…,Xn. The prediction of C can be described by the following conditional model: p(C | X1,X2,…,Xn). By Bayes' theorem, , where p (C) is the prior probability of C, p(X1,X2,…,Xn | C is the conditional probability depending on C, and p(X1,X2,…,Xn) is the probability of input features. Assume that each feature Xi is conditionally dependent and observe that denominator p(X1,X2,…,Xn) does not depend on C which is actually a constant when features are given, the conditional probability over the class variable C can be expressed as where Z is a normalization constant. The above Naïve Bayes classifier can be trained based on the relative frequencies shown in the training set to get an estimation of the class priors and feature probability distributions. For a test sample, the decision rule will be picking the most probable hypothesis (value of C) which is known as the maximum a posteriori (MAP) decision rule using the above model.
As an example of Naïve Bayesian classifier in radiology application, Prasad et al. tackled the problem of lung parenchyma segmentation in the setting of pulmonary disease (Prasad et al., 2008). They used curvature of ribs for the segmentation and used Naïve Bayesian classifier to build the model to find the best-fitting lung boundary. The inputs of the Naïve Bayesian classifier are features of lung and rib curvature. Probability of a matched curvature can be obtained from the classifier.
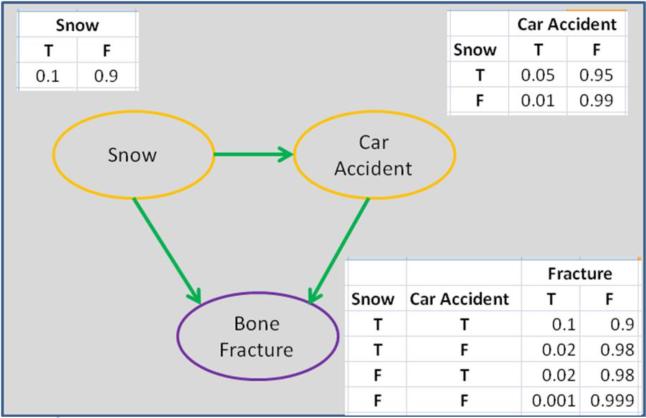
Graph models (Jordan, 1998; Jordan et al., 1999) are perhaps the most popular probabilistic models in which nodes represent random variables and links between nodes denote the conditional independence structure between random variables. Bayesian networks and Markov random fields are two typical graph models. Illustration of a Bayesian network on bone fracture modeling is shown in Fig. 4.
Fig. 4.
Modeling of bone fractures using a Bayesian network in which the bone fracture variable is caused by the states of the weather (e.g., snowing) and car accidents on the road. Each table in the figure shows the probabilities of the corresponding variables given states of father nodes (indentified by arrows). Snow is an independent variable and we show its a priori probabilities in the adjacent table.
The Bayesian network (Heckerman, 1996), also called a belief network or directed acyclic graphical model, represents conditional independencies via a directed acyclic graph (DAG). For example, in the area of medical diagnosis, the relationship between diseases and symptoms can be modeled by a Bayesian network. The probability of a specific disease can be calculated based on the presence of symptoms. Let G = (V,E) be a DAG (where G represents the graphical model, V a set of variables, and E an encoding of the casual relationships between variables) and X = (Xv)vεV be a set of random variables indexed by V. The joint probability density function of X can be factorized as follows: , where pa(v)are the parent nodes of v.
Learning of a Bayesian network includes parameter learning and structure learning. Structure learning is more challenging than parameter learning because the network structure is unknown and the solution space is much larger than that of parameter learning. Maximum likelihood (Lecam, 1990) and expectation-maximization (EM) (Dempster et al., 1977) are widely used in parameter learning of a Bayesian network and Markov chain Monte Carlo (MCMC) provides a global search method for the learning of structure of a Bayesian network.
A Markov random field (MRF), Markov network, or undirected graphical model, is a graphical model in which a set of random variables are connected by undirected links. These models have the so-called Markov property in which a random variable will only be affected by its direct neighbor variables. MRFs have wide applications in computer vision and image processing (Held et al., 1997; Li, 1994; Panjwani and Healey, 1995; Zhang et al., 2001). For example, Lei and Sewchand applied MRFs to CT image segmentation (Lei and Sewchand, 1992). They used an MRF to model CT images and segment them using a Bayesian classifier.
2.5 Ensemble learning
Learning by an ensemble of classifiers is a very effective learning mechanism and was paid much attention in recent years (Dietterich, 1997). Ensemble learning refers to a collection of methods that learn a target function by training a number of individual learners and combining their predictions together. The Bagging algorithm (Bootstrap aggregating) (Breiman, 1996) uses bootstrap samples to build base classifiers. Each bootstrap sample is formed by uniformly sampling from the training set with replacement. The accuracy can be improved through building multiple versions of the base classifier when unstable learning algorithms (e.g., neural networks, decision trees) are used. The AdaBoost algorithm (Freund and Schapire, 1995) calls a given base learning algorithm repeatedly and maintains a distribution of weights over the training set in a series of rounds t = 1, …, T . During the training process, the weights of incorrectly classified examples are increased so that the weak learner is forced to focus on the hard examples in the training set.
Exemplar applications of ensemble learning in medicine include lung cancer cell identification based on ANN ensembles (Zhou et al., 2002), colonic polyp detection using SVM ensembles (Jerebko et al., 2005), and automated classification of lung bronchovascular anatomy in CT using AdaBoost (Ochs et al., 2007).
2.6 Cluster analysis
Natural data usually show clustering properties: samples belonging to the same cluster are more similar , or have closer distance, under certain distance metrics than samples from different clusters. Analysis of the clustering properties of the data will help us understand the nature of the data and potential real applications. It has broad applications in radiology, such as medical image segmentation and diagnosis. Famous clustering algorithms include k -means clustering (Pena et al., 1999), hierarchical clustering (Hastie et al., 2009), DBSCAN (Ester et al., 1996), normalized cut (Shi and Malik, 2000), and mixtures of Gaussians (Bilmes, 1998).
The key idea of k -means clustering is assigning each sample or point to the cluster with the nearest center (also called centroid, the mean of all samples belonging to this cluster). The optimization of clustering is done by iteratively re-assigning labels (markers of clusters) and re-computing the centroids. k -means clustering has the advantage of simplicity and speed but is prone to local minima as there is no guarantee of a global minimum of intra-cluster variance. k -means clustering assigns a hard label to each sample. Fuzzy c-means clustering, a variant of k- means clustering, incorporates fuzzy logic to show the degree to which a sample belongs to each cluster.
Hierarchical clustering adopts a top-down or bottom-up strategy for clustering. It builds a tree structure (called a dendrogram) based on sample distance. For the top-down strategy, hierarchical clustering starts from one cluster as root. Then it splits the cluster successively till a desired number of clusters are derived. Agglomerative hierarchical clustering operates in the reverse direction by merging leaves or sub-clusters together step-by-step based on various similarity or distance measures. DBSCAN is a density-based clustering algorithm. It starts from a randomly selected and unlabeled point. Then it expands the cluster from the initial points based on the sample density around that point. All unvisited points near a point in the cluster which are density-reachable (i.e., density around the point is higher than a certain threshold) will be included in the cluster. DBSCAN has an overall runtime complexity of O(n log n) and usually is faster than k -means clustering (O(ndk+1 log n) where n is the number of samples, d is the dimension of samples and K is the number of clusters).
Gaussians mixture model (GMM) (Bilmes, 1998) is among the most statistically mature methods for clustering. Assume that we have a data set X composed of N samples generated by k components. Each component generates data from a d -dimension Gaussian distribution N (μi, Σi with mean μi and covariance Σi. The mixture model is expressed as follows: with parameters where x represents observed data and are the mixture coefficients. The incomplete-data log-likelihood expression for the mixture density from the data X is . Let us assume that a hidden variable z is attached to each observation which indicates from which component the sample was generated, . With the help of hidden variables, the incomplete-data log-likelihood shown above can be optimized by using the EM algorithm (Dempster et al., 1977) in which the latent variables or unobserved data z1,…, zN) can be estimated in the E-step and the parameters for each Gaussian distribution can be estimated in the M-step (Amari, 1995).
Clustering analysis has many applications in medical image segmentation. For example, Chen et al. proposed a robust algorithm for 3D image segmentation by combining adaptive k-means clustering and knowledge-based morphological operations together (Chen et al., 1998). They applied the proposed method to cardiac CT volumetric images to segment the volumes of the left ventricle chambers. Yao et al. applied fuzzy clustering and deformable models to colonic polyp segmentation in CT colonography (Yao et al., 2009).
2.7 Dimensionality reduction and feature selection
With the rapid development of modern measurement and detection instruments, we are able to sample more and more data (regarding both dimension and size of the sample) from real applications. For example, in radiology achievable image resolution has increased significantly compared with ten years ago. Higher resolution means more voxels in an image which corresponds to more input features if we feed a classifier with all voxels. The increase in dimensionality (number of voxels in radiology images or number of features in feature space that can be extracted from original images) is a significant obstacle to solving optimization problems. This, in turn, complicates machine learning because of the optimization tasks involved in the learning stage. Dimensionality reduction is dedicated to solve this problem by extracting or selecting useful information from the feature space. Classical techniques for dimensionality reduction, such as Principal Components Analysis (PCA), are designed for data whose submanifold is embedded linearly or almost linearly in the observation space (Jolliffe, 2002). Submanifold is a subset of a manifold in space and has its own structure. Because many data from real applications, such as visual perception (Tenenbaum et al., 2000), have nonlinear submanifold structures, there has been a surge in research on nonlinear dimensionality reduction (NLDR) in recent years. The representative methods of NLDR include local approaches such as Locally Linear Embedding (LLE) (Roweis and Saul, 2000) and Laplacian Eigenmaps (Belkin and Niyogi, 2003), and global approaches such as ISOMAP (Tenenbaum et al., 2000) and Diffusion Map (Coifman et al., 2005a, 2005b). In these nonlinear methods, local methods try to preserve the local geometry of the data in low-dimensional space; global approaches tend to give a more faithful representation of the data's global structure. Applications of NLDR in radiology include showing data structure and distribution in low dimensional space (which cannot be observed in the original high dimensional space), and classification (Wang et al., 2008b).
Similar to PCA, independent component analysis (ICA) looks for a linear transformation which can convert the original data to a new linear space (Comon, 1994). The difference is that for ICA the transformation matrix is designed to minimize the statistical dependence between its components; whereas in PCA the transformation matrix is to retain the components with maximal variance or energy.
By employing dimensionality reduction algorithms, we can extract useful information and build compact representations from the original data. Such benefits can also be obtained by feature selection. Feature selection is a machine learning technology which selects a subset of features based on various optimization criteria (Guyon and Elisseeff, 2003).
Feature selection methods can be classified into two types- filter and wrapper- depending on the integration of feature selection and the problem to be solved (Liu and Yu, 2005). For filter methods, the best features are selected according to a specific criterion (such as Pearson correlation and mutual information between features). The employed criterion is independent of the real problem. For wrapper methods, feature selection is embedded into the real problem to be solved and the optimal subset will be determined during iterative optimization. The optimization is based on a final criterion related to the real problem. For example, in a classification task, a wrapper method for feature selection tests a subset of the features on the classification problem. The subset of the features evolves depending on the classification results. Sequential forward selection and sequential backward selection are common wrapper methods for feature selection. In the sequential feature selection methods, an objective function or criterion will be defined first. Then a sequential search algorithm will be introduced to add or remove features from candidate subsets based on the evaluation results of the criterion.
In 2005, a method called minimal-redundancy-maximal-relevance (mRMR) feature selection was proposed by Peng et al. (Peng et al., 2005). In mRMR feature selection, the optimization criteria are affected by two factors: one is relevance between features and target classes and one is redundancy between features. Peng et al. proposed a heuristic framework to minimize redundancy and maximize relevance at the same time. mRMR showed better performance in many real applications compared with traditional feature selection methods. But as every learning algorithm has its own assumptions and conditions, depending on the specific feature selection problem to be solved, we might find other algorithms which are superior to the mRMR algorithm.
Applications of feature selection in radiology focus on selecting the best features for computer-aided detection and diagnosis systems. For example, Li et al. proposed an efficient feature selection algorithm based on piecewise linear network and orthonormal least square procedure for computer-aided polyp detection in CT colonography (Li et al., 2006). Mougiakakou et al. applied feature selection in conjunction with texture features and ensemble classifiers to the problem of differential diagnosis of CT focal liver lesions (Mougiakakou et al., 2007).
2.8 Reinforcement learning
Reinforcement learning studies how agents respond to the change of environment and maximize long-term reward (Barto and Sutton, 1999). It has broad applications in robot control (Schaal and Atkeson, 1994) and game playing (Schraudolph et al., 1994). In reinforcement learning the agent continuously updates her strategy through iterative interactions with the environment in order to develop an optimal strategy. Q-learning is a typical on-line, model-free reinforcement learning algorithm, where Q summarizes in a single number all the information needed by the agent in order to determine her discounted cumulative future reward (Watkins and Dayan, 1992). By using Q-learning, an agent can develop the optimal strategy in a Markovian decision process through a sequence of actions following a Boltzmann distribution strategy. It has been proven that Q-learning converges to the optimum action-values with a probability of 1 so long as all actions are repeatedly sampled in all states and the action-values are represented discretely.
Reinforcement learning has potential applications in radiology. For example, current medical CAD systems for radiology are typically trained on a fixed training set. An alternative approach is to incrementally enlarge the training set as new patient data becomes available. Reinforcement learning could be used to incorporate the knowledge gained from the new patients into the CAD systems. Sahba et al. have shown high potential of applying reinforcement learning in medical image segmentation (Sahba et al., 2006).
2.9 Multiple instance learning
All supervised machine learning methods introduced in previous sections are single-instance learning methods. In single-instance learning, each instance or sample has a label. But in reality, we may have single instances without labels but groups of instances with labels. Methods developed to handle such cases are called multiple-instance learning (MIL). Multiple-instance learning is currently a hot topic in machine learning (Dietterich et al., 1997; Maron and Lozano-Ṕerez, 1998). In multiple-instance learning, samples (also called instances) are wrapped in bags. A bag is defined as an ensemble of instances. The learner (computer learning algorithm) only knows the labels of bags and has no idea about the labels of instances. A bag is labeled negative if all the instances in it are negative; a bag is labeled positive if there is at least one instance in it which is positive. For example, in a radiology CAD application, a patient can be viewed as a bag and all the detections given by the CAD system can be treated as instances which include true lesions and false positives.
The axis-parallel rectangles (APR) method proposed by Dietterich is the first MIL algorithm (Dietterich et al., 1997). The idea behind APR is very simple: find an axis-parallel rectangle (APR) in the feature space to represent the target concept. APR should contain at least one instance from each positive bag and should not contain any instances from any negative bags. Experiments on drug activity prediction problems indicate that the APR method is superior to traditional supervised methods based on single instance learning such as backpropagation neural network and C4.5 decision tree.
Maron and Lozano-Ṕerez (Maron and Lozano-Ṕerez, 1998) proposed another MIL algorithm called diversity density (DD) algorithm which tries to search for a point in the feature space with the maximum diverse density. Diverse density measures the intersection of positive bags, excluding the union of negative bags. This algorithm was improved by incorporating expectation maximization (EM) to estimate which instance(s) in a bag is responsible for the assigned class label (Zhang and Goldman, 2001). As a natural extension of the classical k-nearest neighbor (k-NN) classifier, citation-kNN was proposed by Wang and Zucker (Wang and Zucker, 2000), in which a Hausdorff distance is used to measure the distance between bags, and both “citers” and “references” are considered in calculating neighbors.
MIL has many potential applications in radiology, particularly when we are interested in patient-level diagnoses rather than lesion-level diagnoses. For example, in CT pulmonary angiography, we may wish to know whether the patient has pulmonary emboli. The number of pulmonary emboli may be of secondary priority. MIL has been successfully applied to CT pulmonary angiography to detect pulmonary emboli (Liang and Bi, 2007). MIL has also shown promising results in CT colonography to detect colonic polyps (Fung et al., 2007).
2.10 Graph Matching
Learning how to match two objects is a basic problem in computer vision and machine learning. Graph matching provides an elegant way to present and match objects. It can be applied to medical image registration.
In graph matching, an object usually is represented by a graph consisting of nodes and links that connect the nodes. The nodes represent key points of the object with obvious visual features or clues, i.e., anatomical landmarks. The links represent the spatial neighbor relationship between different nodes. Given two similar graphs, graph matching studies how to match nodes from one graph to the other graph accurately under various considerations and constraints. The graph matching problem has interested researchers for decades and many approaches have been developed. For example, Luo & Hancock treated matching matrix or assignment matrix which defines the correspondence between two vertex sets as hidden variable and proposed an integrative expectation-maximization (EM) algorithm to solve this problem (Luo and Hancock, 2001). Gold and Rangarajan proposed a graduated assignment graph matching algorithm (Gold and Rangarajan, 1996). A control parameter was introduced in the algorithm to gradually impose matching constraints during the iterative optimization process. Leordeanu and Hebert employed spectral technique to decompose the affinity matrix Q of all possible assignments (Leordeanu and Hebert, 2005). The principle eigenvector of Q is trimmed by imposing the mapping constraints required by the overall correspondence mapping (one-to-one or one-to-many). For more about graph matching, readers are referred to a review paper on this topic by Conte et al. (Conte et al., 2004).
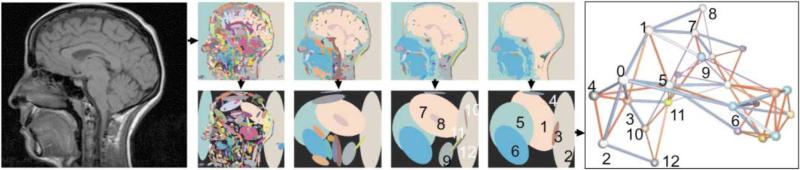
Graph matching has many applications in medical image diagnosis. For example, in content-based image retrieval in medical applications, Lehmann et al. employed graph matching to abstract medical images by hierarchical partitioning and corresponding blobs (Lehmann et al., 2004). Blobs are sub-regions of an image which usually are part of an anatomical structure. Fig. 5 shows an example of hierarchical blobs and graph representation. The authors used a database with 10,000 radiology images (CT, MRI) which were categorized by imaging modality, orientation, body region, and biological system. With the help of a graph representation, the distance or similarity of query image and database entry can be transformed into a graph matching problem. Wang et al. used graph matching to register supine and prone computed tomographic colonography scans (Wang et al., 2010a). After formulating 3D colon registration as a graph matching problem, the authors found an optimal solution by applying mean field theory to what was in essence a quadratic integer programming problem.
Fig. 5.
A hierarchical blob representation of a brain image. Right figure shows corresponding graph constructed from the blob image. Reproduced with permission from Ref. (Lehmann et al., 2004).
2.11 Training and testing of a learning algorithm
Training and testing play important roles in the evaluation of a machine learning algorithm. Usually a machine learning algorithm will be trained on a training set and tested on a test set. A good training strategy can help to find the optimal parameters for a computer-aided detection (CADe) or computer-aided diagnosis (CADx) system. Grid search is the simplest way to train an algorithm. In grid search, the parameter space of a learning algorithm will be divided into hyper-cubes of equal volume. The learning algorithm will be tested on each vertex of each hyper-cube. The vertex in the parameter space with the best performance (evaluated by the training set or validation set) will be selected as the optimal solution.
Grid search is computationally expensive and may be impossible for some large scale data. A recent approach to address this difficulty is called hyperparameter learning (Bengio, 2000; Duan et al., 2003). Hyperparameter learning tries to find the optimal parameters of a prior distribution based on different model selection criterion. Model selection criterion defines how to select the best parameters in a model. Typical strategies for hyperparameter learning include gradient-based optimization and maximum likelihood methods.
To evaluate machine learning algorithms on a particular dataset, one often partitions the dataset in different ways. Popular partition strategies include K-fold cross-validation, leave-one-out, and random sampling. In K-fold cross-validation, the whole data set is partitioned into K subsets. A learning algorithm is trained on K-1 of the subsets and tested on the remaining one. This procedure is conducted K times until all K subsets have been tested. Leave-one-out is similar to K-fold cross-validation but each subset consists of only a single sample from the dataset. Each time a sample will be held aside for testing and the learning algorithm is trained on the other samples. This process is repeated N times where N is number of samples in the dataset (each sample will be tested exactly once). For random sampling, the data subsets are formed by random sampling of the data without replacement as training samples and the remaining data serves as test data. Bootstrapping method (Efron and Tibshirani, 1986) can be viewed as a special case of random sampling with replacement during the test.
Different evaluating methods will affect the generalization ability of machine learning algorithms. An estimation method with low bias and low variance will be ideal to estimate the final accuracy of a classifier. Recent theoretical studies and experimental results on real-world datasets showed that ten-fold cross-validation is better than the more computationally expensive leave-one-out cross-validation for model selection (select a good classifier from a set of classifiers) (Kohavi, 1995). For random sampling, when the ratio between testing and training is high, it tends to give estimations with high bias and high variance due to insufficient training samples. The advantage of random sampling is that for small size datasets it can test a classifier's capability in a thorough way by varying the training set multiple times.
In many real radiology applications, detection and diagnostic decision-making play import roles in clinical practice. Receiver operating characteristic (ROC) analysis provides a practical tool for model selection. An ROC curve is a 1D curve which shows the trade-off in sensitivity versus false positive rate as the threshold is varied for the decision variable for a binary classifier (Zweig and Campbell, 1993). Cost/benefit factors which are import in diagnostic decision making can be directly and naturally embedded in the ROC analysis.
3. Applications of machine learning in radiology
In this section, we will introduce some typical applications of machine learning in radiology.
3.1 Medical image segmentation
Medical images contain many structures including normal structures such as organs, bones, muscles, fat, and abnormal structures such as tumors and fractures. Segmentation is the process of identifying structures, both normal and abnormal, in the images. It is fundamental to the interpretation of medical images. Learning how to segment anatomic structures is a critical part of medical image segmentation. Segmenting structures from medical images is not trivial due to the complexity and variability of the region-of-interest. Examples of problems that complicate medical image segmentation include normal anatomic variation, post-surgical anatomic variation, vague and incomplete boundaries, inadequate contrast, artifacts and noise.
The concept of the graph provides an elegant way to abstract image information that is useful for segmentation. Graph cuts are segmentation methods which are based on graphs and utilize flows between source and sink nodes on the graph (Greig et al., 1989). Shi and Malik proposed a graph partitioning method called “normalized cut” that performs image segmentation (Shi and Malik, 2000). In graph theoretic language, given a graph G = (V, E), where V represents a set of vertices and E a set of edges, a cut between two disjoint sets of G is defined as where w is the weight of the edge or link connecting two nodes in subsets A and B. The cut can be used to depict the dissimilarity between the two subsets. Minimizing the cut value usually will lead to the optimal bipartitioning of a graph. For real-world data, however, the minimum cut criterion favors cutting the graph into small sets of isolated nodes. To solve this problem, Shi and Malik proposed the measure called normalized cut (Ncut). Ncut penalizes partitions containing small isolated points and gives more balanced partitions compared with ordinary cut.
Graph cuts have many applications in medical image segmentation including interactive organ segmentation for 3D CT and MRI images (Boykov and Jolly, 2000), multiple sclerosis lesion segmentation in MRI (García-Lorenzo et al., 2009), segmentation of the left myocardium in four-dimensional (3D space + time) cardiac MRI data (Kedenburg et al., 2006), and lung segmentation from volumetric low-dose CT scans (Ali and Farag, 2008). Some of these applications are fully-automated.
As introduced in Sec. 2, Markov random fields (MRFs) have wide applications in medical image segmentation (Held et al., 1997; Towhidkhah et al., 2008; Zhang et al., 2001). The graph model of Markov random fields has a natural representation of the voxels in a 2D/3D medical image and their spatial relationship. Based on Bayes theory, given observed images I, the posterior probability of segmentation p(S|I) can be inferred from prior distribution p(S) of label S and conditional distribution p(I|S). The Markov model could be solved by using maximum a posterior (MAP) criterion. In MRF models, parameters controlling the spatial interactions have significant influence on the smoothness of segmentation results (Pham et al., 2000). In practice, usually we need to balance the smoothness and important structural details. In addition, MRF models usually are computationally hard to solve.
Although cluster analysis developed in machine learning and pattern recognition area was not designed for medical segmentation problems originally, many clustering algorithms can be applied directly to medical image segmentation problems because the objectives of clustering algorithms and image segmentation problems are highly overlapped. Examples include applying fuzzy c-means clustering algorithm to brain MR image segmentation (Chen et al., 2007), segmentation of thalamic nuclei from DTI using spectral clustering (Ziyan et al., 2006), and 3D cardiac CT data set segmentation using random walks (Grady, 2006).
In medical image segmentation, flexibility is often required to adapt the segmentation to the variability of biological structures over time and across different individuals (McInerney and Terzopoulos, 1996). Deformable models can sometimes help deal with such variability. Deformable models can be contours (known as snake or active contours) for 2D images and surfaces for 3D images. Deformable models combine elements from geometry, physics, approximation theory, and machine learning. Geometry provides a way to represent the object boundary. Constraints are imposed on the geometric representation of the object to limit the way it can evolve. The constraints often incorporate physical principles such as force and elasticity. Approximation theory and machine learning help fit the model to the data and learn the best parameters of the model and the deformation. For detailed information on how deformable models describe object shapes in a compact and analytical way, and incorporate anatomic constraints, readers are referred to a review paper on this topic (McInerney and Terzopoulos, 1996).
In many radiology images, objects to be segmented have irregular shapes and complicated topologies. Level set-based segmentation methods provide a natural and flexible way to handle those complicated objects (Cremers et al., 2007; Malladi et al., 1995). In level set, the boundary is defined as the zero value of a hypersurface. By casting the segmentation problem into a higher dimensional space, the motion of the hypersurface under the control of a speed function will cause the initial boundary to move. By utilizing image information, e.g., edges and grey value, the evolution of the hypersurface can be stopped at the object boundary (Malladi et al., 1995). Level set has been widely used in medical image segmentation, such as brain (Baillard et al., 2001; Ciofolo and Barillot, 2005; Yang et al., 2003), heart (Lin et al., 2003; Paragios, 2003; Yang et al., 2003), liver (Lee et al., 2007; Smeets et al., 2010), and colon (Franaszek et al., 2006; Konukoglu et al., 2007; Uitert and Summers, 2007).
For a machine learning algorithm, how to make full use of a training set is a key point of its success on the test set. In deformable models and level set-based segmentation methods, training information is incorporated into the segmentation method in an implicit way (through parameter learning). On the contrary, active shape models (ASMs) tries to utilize training shape information in a more explicit way by building a shape model from training images and adapting the model to a new test image through an alternative optimization way (Cootes et al., 1995). Later, Cootes et al. extended ASMs to active appearance models (AAMs) by incorporating appearance information of objects in an image (Cootes et al., 2001). Applications of AAMs and ASMs in radiology include cardiac MR and ultrasound images (Mitchell et al., 2002), prostate segmentation (Shen et al., 2003), and segmenting thrombus in abdominal aortic aneurysms (de Bruijne et al., 2003). More discussions on statistical shape models for 3D medical image segmentation can be found in a review paper (Heimann and Meinzer, 2009).
In recent years, joint categorization and segmentation (JCaS) has become a hot topic in computer vision (Ladicky et al., 2010; Singaraju and Vidal, 2011). In JCaS, interested objects in a 2D image are categorized and segmented simultaneously. Each pixel in the image is assigned an object category label. In JCaS, MRF was widely used to model the image and corresponding segmentation. To model the local properties of each node, and interactions among nodes in the MRF, we usually define some potential functions (known as “potentials”). Unary and pairwise potentials are typical choices used in 2D image segmentation. Given images, the MRF could be solved using maximum a-posteriori learning and expectation-maximization. Compared with traditional MRF for image segmentation, JCaS incorporates higher order potentials that encode the classification cost of statistical features extracted from objects in an image. Usually it is done by encoding the output of a classifier in a potential function which is able to capture global or higher order interactions of the objects-of-interest. Since in many radiological applications, segmentation and categorization of various organs are fundamental tasks, we expect that JCaS will have wide applications in radiology in the near future.
3.2 Medical image registration
Image registration is an application of machine learning. During a medical examination, a patient may be scanned by different imaging modalities (Studholme et al., 1996), or scanned by the same modality at different positions, times, or situations (with or without contrast agents). These images are usually complementary and in combination may lead to more accurate diagnosis. In order to integrate all the information, a first step is to align these images spatially, a procedure referred to as registration or matching (Hill et al., 2001; Lester and Arridge, 1999; Maintz and Viergever, 1998). Machine learning plays a key role in the medical image registration problem by learning the best registration or parameters under different matching criteria.
Mutual information provides a good measure on the interdependence of two images. Thus registration based on mutual information has drawn a lot of attention in recent years and served as the basis of many medical image registration methods (Pluim et al., 2003). Given two images (2D or 3D) to be registered, let us define one of the two image as the reference image u and the other one as the test image v. The registration problem can be formulated as follows: , where x are the coordinates of a voxel, T is a transformation from the coordinate system of reference image to that of the test image,I is the mutual information between u and v. I(u(x),v(T(x))) ≡ h(u(x)) + h(v(T(x))) − h(u(x),v(T(x))) where and is the joint probability of x and y which could be obtained using Parzen windowing (Pluim et al., 2003). The registration methods based on mutual information can be classified into different categories based on the transformation and optimization method employed. For example, the transformation can be used including rigid (Holden et al., 2000), affine (Radau et al., 2001), perspective, and curved (Chui and Rangarajan, 2003; Meyer et al., 1999) transformation. The optimization methods which are widely used include gradient-based (Maes et al., 1999) (steepest gradient descent, conjugate-gradient methods, quasi-Newton methods, least-squares methods) and non-gradient-based optimization methods (interpolation (Zhu and Cochoff, 2002), probability distribution estimation (pdf) estimation (Shekhar and Zagrodsky, 2002), optimization and acceleration (Studholme et al., 1997)). In addition, mutual information based on intensity information only may not be adequate. In the work of Papademetris et al., they combine point-feature and intensity information together for non-rigid registration (Papademetris et al., 2004).
Wang et al. proposed a graph matching method based on mean field theory for computed tomographic colonography (CTC) scan registration (Wang et al., 2010b). They first formulated colon registration as a graph matching problem. Then a matching algorithm was proposed based on mean field theory. During the iterative optimization process, one-to-one matching constraints were added to the system step by step. Prominent matching pairs found in previous iterations are used to guide subsequent mean field calculation. Graph also provides a concise and efficient representation of the medical objects.
The thin-plate spline (TPS) is an important tool for medical image registration (Bookstein, 1991). TPS can be considered to be a natural non-rigid extension of the affine map through minimizing a bending energy based on the second derivative of the spatial mapping (Bookstein, 1991). Chui et al. developed an algorithm which combines TPS and robust point matching (RPM) together where TPS provides the parameterization of the non-rigid spatial mapping and RPM solves the correspondence problem (Chui and Rangarajan, 2003). Combining mutual information and TPS is a very interesting topic. Interested readers can find such work in Meyer et al. (Meyer et al., 1997) by. Readers who are interested in brain registration problems could read reference (Klein et al., 2009) in which the authors evaluated and ranked 14 nonlinear deformation algorithms for human brain MRI registration.
Diffeomorphic Registration is also another type of widely used registration method in medical image analysis. It seeks an invertible function which is smooth and maps one differentiable manifold (image) to another. In ideal situation, the composition of the mapping function and its inverse should be close to the identity transform. Ashburner proposed a fast diffeomorphic registration framework called DARTEL which utilized Levenberg-Marquardt strategy to optimize the registration problem (Ashburner, 2007). Rueckert et al. used B-spines in diffeomorphic registration which serve as a way to parameterize a deformation field (Rueckert et al., 2006). Vercauteren et al. conducted research on non-parametric diffeomorphic image registration by adapting Thirion's demons algorithm to the space of diffeomorphic transformations (Vercauteren et al., 2007). Avants et al. tackled the problem from the viewpoint of cross-correlation within the space of diffeomorphic maps and developed a symmetric registration method (Avants et al., 2008).
3.3 Computer-aided detection and diagnosis systems for CT or MRI images
To assist doctors in the interpretation of medical images, computer-aided detection (CADe) and computer-aided diagnosis (CADx) provide an effective way to reduce reading time, increase detection sensitivity, and improve diagnosis accuracy. CADe and CADx are young interdisciplinary technologies combining elements of digital image processing, machine learning, pattern recognition, and domain knowledge of medicine together (Doi, 2005, 2007; Kononenko, 2001; Sajda, 2006).
The top leading cause of cancer related deaths in men and women is lung cancer in the United States. According to cancer statistics, 221,130 new cases of lung cancer and 156,940 deaths were reported in the United States in 2011 (http://www.cancer.gov/cancertopics/types/lung). In current lung cancer diagnosis, computed tomography (CT) screening is a standard procedure which is superior to traditional chest radiography in the detection of lung nodules (potential lung cancers) (Kaneko et al., 1996; Swensen et al., 2002).
An essential initial component of image interpretation and diagnosis is the identification of normal anatomical structures in the image. Since the lung has very complicated structures, developing a fully automated approach to distinguish normal lung structures will improve the utility of a lung CAD system. Ochs et al. (Ochs et al., 2007) employed AdaBoost to train a set of ensemble classifiers. The training CT images were labeled by radiologists with the following categories: airways (trachea and bronchi to 6th generation), major and minor lobar fissures, nodules, vessels (hilum to peripheral), and normal lung parenchyma.
Usually a CAD system is designed and optimized for detecting a specific disease, i.e., single-task learning. However, different diseases may share similar characteristics (for example, lung nodule and ground glass opacity), hence training a classifier to do multiple tasks may improve its performance and utility. Multi-task machine learning has been proposed (Caruana, 1997). These related problems may share the same representation. A typical example of a multi-task CAD system can be found in the work of Bi et al. (Bi et al., 2008). The multi-task learning algorithms they proposed can eliminate irrelevant features and identify discriminative features for each sub-task. They showed promising results on predicting lung cancer prognosis and heart wall motion analysis.
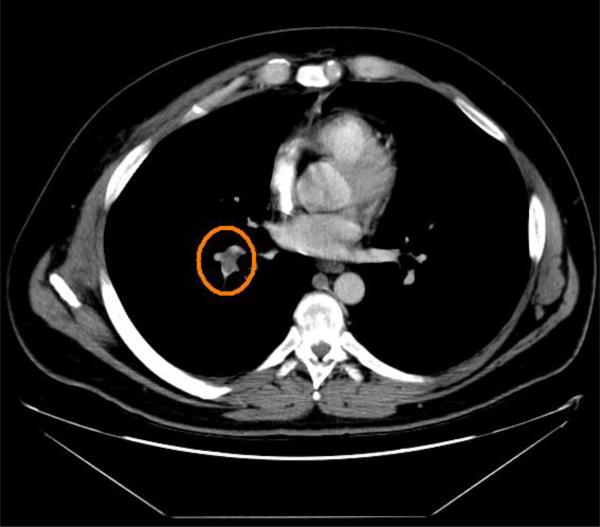
Pulmonary embolism (PE) is a blockage of the main artery of the lung or one of its branches by a substance (usually a blood clot) that has travelled to the lung through the bloodstream from another part of the body (Fig. 6). Since pulmonary embolism can be life-threatening, early diagnosis can improve survival rate. Computed tomography angiography (CTA) provides an accurate diagnostic tool for PE when it is combined with a CADx system (Schoepf and Costello, 2004; Schoepf et al., 2007). In (Liang and Bi, 2007), Liang et al. proposed a fast, effective approach for PE diagnosis. Segmentation of emboli in CTA is a very challenging task due to partial volume effects around the vessel boundaries that make PE voxels and vessel boundaries indistinguishable. Liang et al. proposed an algorithm called concentration-oriented tobogganing to solve this problem. They extracted 116 features from initial PE candidates. To reduce false positives, they applied multiple-instance classification to make the final diagnosis. This PE CAD system reported 80% sensitivity at 4 false positives per patient on a CTA dataset of 177 cases. Fung et al. also employed multiple-instance learning for diagnosis of PE (Fung et al., 2007). Their work focused on learning a convex hull representation of multiple instances. This representation enabled their algorithm to be significantly faster than existing multiple-instance learning algorithms. More information on PE CAD systems can be found in a review paper (Chan et al., 2008).
Fig. 6.
Pulmonary embolism (shown in yellow circle) in the artery of a 52-year old male patient.
Colon cancer is the second leading cause of cancer-related death in the United States. Computed tomographic colonography (CTC), also known as virtual colonoscopy (VC) when a fly-through viewing mode is used, provides a less-invasive alternative to optical colonoscopy in screening patients for colonic polyps. Computer-aided polyp detection software has improved rapidly and is highly accurate (Summers, 2010; Yoshida and Dachman, 2006).
CAD systems for detection of polyps on CTC have been under investigation over the past decade. Feature extraction and classification are two critical procedures in a successful CTC CAD system. CAD systems first extract multiple features from the images, such as curvature, shape index, curvedness, surface normal overlap, and texture. Summers et al. (Summers et al., 2005) employed curvature-related features to find polyp candidates. Yoshida and Nappi (Yoshida and Nappi, 2001) applied the shape index and curvedness measures to describe polyp candidates. Paik et al. (Paik et al., 2004) developed a method called surface normal overlap that can capture the shape of polyp candidates. Wang et al. (Wang et al., 2005) proposed a polyp detection method which employs geometrical, morphological, and textural features inside polyp candidates.
A well-functioning classifier is a critical component of a practical CTC CAD system. Example classifiers for CTC CAD systems include neural networks and binary classification trees (Jerebko et al., 2003b), committees of SVMs (Malley et al., 2003), quadratic discriminant analysis (Yoshida and Nappi, 2001), massive-training artificial neural networks (MTANNs) (Suzuki et al., 2008), and logistic regression (van Ravesteijn et al., 2010). To extract more useful information from noisy and high dimensional features, Wang et al. introduced dimensionality reduction and multiple kernel learning to CTC CAD and showed promising results (Wang et al., 2008a)(Wang et al., 2010c).
CADe and CADx have also been widely applied in breast tumor detection and diagnosis (Cheng et al., 2003). Breast cancer is the second leading cause of cancer death in U.S. women. In the United States, the lifetime risk for breast cancer is 12.5% with a 3% chance of death (http://www.cancer.org/Cancer/BreastCancer/index). The most widely used diagnostic and screening tool for breast cancer is mammography which uses low-dose X-rays for imaging the human breast (Kerlikowske et al., 1995). For practical breast tumor CADx system, differentiating benign tumor and normal tissue from malignant tumor is the top priority. Chan et al. analyzed texture features of microcalcifications (Chan et al., 1995a; Sahiner et al., 1998). El-Naqa et al. first showed that it is feasible to detect microcalcifications from digital mammograms using support vector machines (El-Naqa et al., 2002). Later, Wei et al. (Wei et al., 2005a) investigated several state-of-the-art machine learning methods for automated classification of clustered microcalcifications (MCs). The methods tested include support vector machine (SVM), kernel Fisher discriminant (KFD), relevance vector machine (RVM), and committee machines (ensemble averaging and AdaBoost).
3.4. Brain function or activity analysis and neurological disease diagnosis from fMR images
Brain function and activity analysis play important roles in research in cognition, psychology, and brain disease diagnosis. Functional magnetic resonance imaging (fMRI) provides a noninvasive and effective way to assess brain activity. Because of the complexity of the human brain and variations of brain activity, fMR images usually show complicated patterns and the interpretation of them usually requires significant computerized analysis. In recent years, machine learning algorithms have been used more and more to decode from fMRI the stimuli, mental states, behaviors, and other variables of interest (Pereira et al., 2009). After feature extraction, we can train a classifier, such as SVM, LDA, and neural network, to differentiate brain activity patterns. Developing automatic ways to learn the patterns from fMR images is challenging because the data are extremely high dimensional and noisy with small size (tens of training samples).
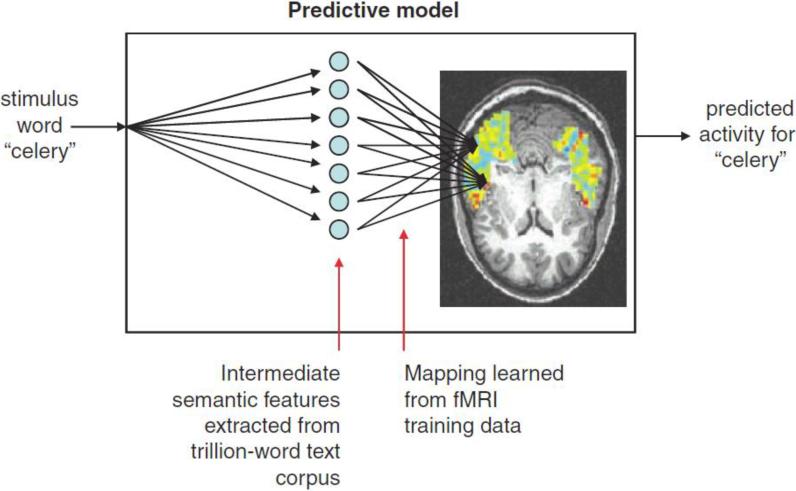
In early work, T. Mitchell et al., tried to decode cognitive states from brain images (Mitchell et al., 2004). They showed a human subject a picture or a sentence or asked the subject to read a word describing different concepts (food, people, building, etc.). The goal was to activate different patterns of brain activity that would be detected by fMRI. They explored several classifiers for analyzing the fMRI data including Gaussian naïve Bayes, SVMs, and k Nearest Neighbor (kNN). Their results showed that it was feasible to distinguish a variety of cognitive states of the brain by using machine learning algorithms. Then they conducted further research on human cognition to try to predict brain behavior under different stimulations (Mitchell et al., 2008). Previous research had shown that spatial patterns of neural activation are related with thinking about different objects and concepts. In this paper, they presented a computational model (Fig. 7) that is able to predict the neural patterns associated with words whose fMRI data have not been utilized by the training process. A striking finding was that their model could make accurate predictions for thousands of nouns in a text corpus based only on the fMRI data of 60 nouns.
Fig. 7.
Form of the model for predicting fMRI activation for arbitrary noun stimuli. fMRI activation is predicted in a two-step process. The first step encodes the meaning of the input stimulus word in terms of intermediate semantic features whose values are extracted from a large corpus of text exhibiting typical word use. The second step predicts the fMRI image as a linear combination of the fMRI signatures associated with each of these intermediate semantic features. Reproduced with permission from Ref. (Mitchell et al., 2008)
As a practical application of machine learning in spatial patterns of brain activity analysis, Davatzikos et al. proposed a new method for lie detection from fMR images (Davatzikos et al., 2005). After image acquisition and preprocessing, such as filtering and registration, they converted the original MR images into parameter estimate images (PEIs). Then, by subdividing the PEIs into 560 cubes (16mm × 16mm × 16mm), they extracted one feature from each cube. Average value of the PEI of each event was employed as the feature. In the last step, an SVM with Gaussian kernel was employed for classification. Experimental results showed that the proposed high-dimensional non-linear pattern recognition method can distinguish different brain activities associated with lying and truth-telling with high accuracy.
Alzheimer's disease (AD) is the most common form of dementia and will cause cognition disturbance gradually as it progresses. A study showed that it is feasible to predict whether persons are in the prodromal phase of AD using structural magnetic resonance imaging (Killiany et al., 2000). In brain MR images, mild cognitive impairment (MCI), which is a prodromal phase of AD, has certain patterns. Davatzikos et al. developed an automatic method to detect these patterns via high-dimensional image warping, robust feature extraction, and SVM (Davatzikos et al., 2008; Fan et al., 2005). Later, Kloppel et al. used linear SVM to classify pathologically proven AD patients and cognitively normal persons using T1-weighted MR scans from two centers and different scanners (Kloppel et al., 2008). They also showed that for dementia diagnosis, well-trained neuroradiologists were comparable to SVMs, which encourages deployment of computerized diagnostic methods in clinical practice (Stonnington et al., 2009). Other efforts on using machine learning algorithms for AD classification problems include linear programming boosting (Hinrichs et al., 2009a), multi-kernel learning (Hinrichs et al., 2009b), and relevance vector regression (Wang et al., 2010d).
Schizophrenia is another type of common neurological disease which affects about 1% of the general population (Shenton et al., 2001). MRI provides a good opportunity to evaluate brain abnormalities in schizophrenia. MRI structural findings in schizophrenia include ventricular enlargement, medial temporal lobe, superior temporal gyrus, parietal lobe, and subcortical brain region involvements (Shenton et al., 2001). Since schizophrenia MRI shows complex patterns and is hard to diagnose, machine learning based methods are preferred to identify psychiatric disorders from high dimensional imaging data. Caan et al. applied principal component analysis and linear discriminant analysis to diffusion tensor brain images of schizophrenia and controls (Caan et al., 2006). In (Demirci et al., 2008), Demirci et al. proposed a projection pursuit algorithm to classify schizophrenia using fMRI data. Their method also focuses on dimensionality reduction involving ICA and PCA in order to find a low dimensional embedding of the original data which was designed to classify schizophrenia and healthy control groups. Kim et al. proposed a hybrid machine learning framework for schizophrenia classification (Kim et al., 2008). They first used ICA to reduce the noise. Then a discrete dynamic Bayesian network was used to distinguish patients with schizophrenia from healthy controls.
In brain fMRI analysis, statistical parametric mapping (SPM) is a widely used statistical technique to test hypotheses on whether a certain region of the brain has some specific effects (Friston et al., 1995). SPM is based on the general linear model and the theory of Gaussian fields. The hypotheses are tested in a voxel-based way. The final parametric mapping image can be viewed as a compression image of original MR or PET image sequence across different time points or from different tasks which is very helpful to understanding brain activity. Applications of SPM in neurological disease diagnosis include Alzheimer's disease (Bookheimer et al., 2000; Scahill et al., 2002), schizophrenia (Sowell et al., 2000; Wilke et al., 2001; Wright et al., 1995), and obsessive-compulsive disorder (Kim et al., 2001; Saxena et al., 2001).
3.5 Content-based image retrieval systems for CT or MRI images
Content-based image retrieval (CBIR) aims to search digital images in large databases based on the contents of the image, such as colors, shapes, and textures, etc. CBIR can help radiologists in disease diagnosis by retrieving images with similar features or previously-confirmed cases with the same diagnosis. It can also help to train radiologists by creating teaching collections of similar images. CBIR is the application of computer vision to the image retrieval problem. In recent years, with the rapid development of machine learning, many machine learning algorithms have been embedded in CBIR systems to improve the query accuracy and efficiency. In the medical area, with the expanding role and quantity of digital imaging for diagnosis and therapy, there are many potential applications for CBIR systems (Tagare et al., 1997). For example, in the Radiology Department of the University Hospital of Geneva, 12,000 images were produced daily in 2002 (Muller et al., 2004). Searching target images in such a huge medical image database would be impossible without a CBIR system.
A practical and effective CBIR system has three key components: a feature extractor, a content comparator, and a query engine. For example, El-Naqa et al. (El-Naqa et al., 2004) studied content comparison for digital mammography CBIR. They proposed a two-stage supervised learning network to learn the similarity function which will assign a similarity coefficient (SC) to each pair of a query image and database entry. In the first stage, a linear classifier is employed to identify database entries which are sufficiently similar to the query image. In the second stage, they considered SVM regression and a general regression neural network to learn the optimal similarity function.
The idea of human interaction in CBIR has been explored by Brodley et al. (Brodley et al., 1999). For medical images, extraction of global information or feature to characterize the images usually will not help much for the CBIR because the clinically useful information is often localized. Brodley et al. proposed an approach called “Physician-in-the-Loop” in which a physician delineates the “pathology-bearing regions” (PBR). These PBRs are often difficult to segment using fully-automated techniques. The authors described their CBIR system as a synergy of human interaction, machine learning, and computer vision.
Global image descriptors based on color, texture, or shape often do not exhibit sufficient semantics for medical applications. Combining global and local features together is a key point for a successful CBIR system. Keysers et al. (Keysers et al., 2003) proposed a statistical framework for model-based image retrieval in medical applications. Their multi-step approach included image categorization based on global features, image registration (in geometry and contrast), feature extraction (using local features), feature selection, indexing (multiscale blob representation), identification (incorporating prior knowledge), and retrieval (on abstract blob level). They used global features such as image modality, body orientation, anatomic region, and biological system to classify and register images first. Then the query was done based on local features extracted from anatomic region.
3.6 Text analysis of radiology reports using NLP/NLU
Another application of machine learning in radiology is the processing of radiology text reports. The accumulated reports from daily radiology practice fill huge text databases. Exploiting these radiology report databases by using modern information processing technologies may improve report search and retrieval and help radiologists in diagnosis. Compared with search reports using keywords, natural language processing (NLP) and natural language understanding (NLU) provide a more efficient way to organize and retrieve relevant information hidden in the radiology reports. An advantage of NLP/NLU is that they can handle large scale data and extract meaningful information in a way that is not feasible by human readers. Natural language processing and natural language understanding are important subsets of machine learning and provide a feasible way for text analysis of radiology report databases (Dreyer et al., 2005; Friedman and Hripcsak, 1999). NLP/NLU can extract useful information from human language and organize them into more formal representations such as parse trees or first-order logic (Bates, 1995). These formal representations enable more efficient computer processing compared with handling human language directly.
The MedLEE system is the first NLP system deployed in patient care (Bakken et al., 2004; Friedman et al., 1994; Friedman and Hripcsak, 1998, 1999). The main purpose of MedLEE is to identify clinical information in narrative reports and build structured representation about the diagnosis and analysis. There are three major steps in the system. The first step parses the text report based on grammar and identifies the main structures of sentences in the text. The second step regularizes the phrases; similar expressions (regarding semantics) are transformed into standardized forms reducing stylistic variations in natural language. The last step is the encoding phase which maps extracted and standardized items from the text to concepts in a controlled vocabulary. Experiments on a collection of 230 radiology reports showed high precision and recall in text retrieval (Friedman et al., 1994). Interested readers can try a demo of MedLEE at http://zellig.cpmc.columbia.edu/medlee/demo/ which shows examples of structured output for mammography reports. Huang et al. (Huang et al., 2005b) studied the improvement of noun phrase identification in radiology reports. They first employed maximum entropy modeling for sentence boundary detection. Then they used the Stanford parser (Klein and Manning, 2002) to parse each sentence. The last module is noun phrase identification. A Unified Medical Language System (UMLS) (Humphreys et al., 1998) specialist lexicon was integrated into the parser to improve the noun phrase identification performance.
In addition to text retrieval, text classification has important applications in radiologic diagnosis. In (Chapman et al., 2003), Chapman et al. proposed an automatic detection system which was applied to radiology reports to identify patients with mediastinal findings consistent with inhalational anthrax. The Identify Patient Sets model (Cooper et al., 1998) was utilized to create a classifier based on keywords extracted from clinical chest radiograph reports.
Dang et al. employed NLP/NLU technologies to find trends in a large radiology practice (Dang et al., 2009). Experimental results showed that NLP/NLU technologies could help to analyze yield of different radiology exams from a large radiology report database based on presence of positive/negative radiology findings.
4. Key contributions and common characteristics of machine learning techniques in radiology
In the previous section, we showed that machine learning has many applications in radiology. These applications vary from each other and may have very different forms regarding problems to be solved, input data, output data, anatomical constraints, prior knowledge, and hidden variables. Also machine learning methods utilized to solve these problems seem complicated and many of them have strict assumptions. At first glance, it seems difficult for a researcher in radiology to decide whether to employ machine learning techniques and which ones to use for a real radiology problem. In this section, we identify key contributions and common characteristics of machine learning techniques in radiology area.
As we introduced above, machine learning studies the design and development of algorithms which make computers recognize complex patterns or make intelligent decisions based on empirical data. The most significant contribution of machine learning to radiology is that it provides an automatic way to generalize (human) knowledge obtained from training data to future unknown test data. For example, in a mammography CAD system, we could teach a computer to describe (feature extraction) and make decisions (classification) on masses and/or microcalcifications of the human breast. In this way, we are actually transferring the knowledge of radiologists on mammography diagnosis to computers. As general guidance, given empirical data from a radiology application, first we need to identify the input data, objective, and related constraints. For example, for medical image segmentation, the input data would be original CT/MRI/PET/US images, the objective would be segmentation of an object-of-interest in the image (e.g., organ, bone, vessels), the related constraints would be anatomical structure of the patient and prior knowledge (e.g., shape, density, texture) of the object-of-interest. The second step would be extracting useful features, identifying or designing an appropriate objective function, and solution of the objective function. For example, if we can extract some anatomical key points from original medical images and extract related features from these key points, then probably we could utilize graph cut-based method and use cut between two clusters as the optimization objective. Meanwhile different constraints could be added to the objective function in order to fit the anatomical structures of the target. The last step would be to train the algorithm and find the best parameters for the graph cut model. The trained machine learning segmentation algorithm would then be applied to new scans.
In the machine learning techniques introduced in Sec. 2, classification techniques are the most widely used machine learning techniques in radiology. That is because in clinical practice, many applications of radiology can be formulated as a classification problem. Especially for many CADx applications in radiology, such as lung, breast, and colon CAD, intelligent classification of lesions is a central task for these computerized systems (Doi, 2005, 2007; Giger et al., 2001). Classification techniques employed in these applications range from simple classification techniques, such as linear classifier, LDA, and neural networks to more complicated and modern techniques, e.g., SVMs, multiple-instance learning and ensemble learning.
For radiology applications focusing on segmentation and statistical analysis of image content, probabilistic models play an import role in solving these problems (Bromiley et al., 2003). Typical probability modeling includes following procedures: independent and dependent variables identification, probability density function design or assignment between variables, and integration or marginalization of complete probability model to get distributions on target variable or objective function. For example, Zhang et al. utilized a hidden Markov random field model and the expectation-maximization algorithm to solve the segmentation problem of brain MR images (Zhang et al., 2001). In their work, they used hidden Markov random field to model the relationship between observations and unknown cluster labels under spatial constraints. The emission probability of latent variable is designed as a Gaussian distribution. Expectation-maximization algorithm was used to solve the MRF - MAP estimation problem.
Given a probability model, maximum likelihood (ML) and MAP were widely used to obtain a point estimation in parametric space (Bishop, 2006). Suppose we have empirical data composed by N independent and identically distributed (IID) observations. These data were sampled from an unknown distribution p(x∣θ) parameterized by θ, and that the likelihood of θ is defined as . The ML estimation will be . MAP is also designed for point estimation of unobserved variable based on empirical data. It is close to ML estimation. where g(θ) is a prior distribution over θ. MAP estimation could be viewed as a regularization of ML estimation where the prior distribution g(θ) serves as a regularization term.
To solve ML and MAP estimation problems, gradient-based methods were most widely used to find optimal solutions. In real applications, when the probabilistic models are too complicated and there are also unobserved latent variables involved, it may be very hard to use gradient-based methods due to the complexity of gradient calculation and they may fail to find the optimal solution. For such complicated statistical models, expectation-maximization (EM) algorithm is widely used to find ML or MAP estimations of parameters (Dempster et al., 1977). EM is an iterative method which performs an expectation (E) step and a maximization (M) step alternatively to get the estimations of latent variables and model parameters, respectively.
Besides probability models, optimization is also an important aspect of machine learning. Many medical image processing problems can be formulated as optimization problems. Chan et al. (Chan et al., 2006) showed how to convert some non-convex minimization problems from image processing and computer vision into convex ones. After the conversion it is then feasible to find global minimizers for image segmentation models such as the Mumford-Shah model using standard convex minimization methods. Optimization based on regularization of variables to be optimized is also a widely used technology. Yan et al. proposed a new regularization model based on the boundary element method to recover the left ventricular deformation (Yan et al., 2007).
5. The translation from machine learning to clinical practice
In the previous section we illustrated six representative domains in radiology where machine learning could contribute. In this section, we will discuss advantages of utilizing machine learning in radiology and potential barriers which could hinder the deployment of machine learning in clinical practice.
The major advantages of applying machine learning in radiology will be labor saving and accurate diagnostic results. Many radiology practices are very time consuming for human labor. For example, in medical image segmentation, e.g., brain white matter and liver segmentation, it requires a long time and high attention for radiologists and technologists to delineate the boundary of the object in 3D images. Such segmentation tasks could be done quickly by computers once a segmentation algorithm is well designed and trained. For another example, in CTC practice, it would be time consuming to identify all potential colonic polyp candidates. Radiologists need to go through the whole CT scan and check suspicious spots segment by segment (in 3D fly-through mode) or slice-by-slice (on original 2D CT slices). In CTC CAD, an initial filtering task could be done rapidly by calculating curvatures of the inner colon surface in a brute force way (voxel-by-voxel). With the help of machine learning algorithms that deal with low-level and time-consuming image analysis problems such as these, radiologists could focus on high-level diagnostic issues.
The second major advantage of machine learning technologies is comparable performance to humans. In many radiology applications, e.g. mammography and colon CAD, computerized CADx systems have shown comparable, or even higher, performance compared with well-trained and experienced radiologists and technologists (Cheng et al., 2003; Doi, 2005, 2007; Giger et al., 2001). In addition, a good machine learning predictor usually will give predictions with low bias and variance at any time and any place. On the contrary, radiologists' performance may be affected by various factors: fatigue, emotion, reading time, and environment, etc. In principle, machine learning-based computer systems will perform more consistently than human beings.
Although machine learning has shown its power in many radiology applications, there are still some barriers to the translation of machine learning techniques to radiology clinical practice. The first issue is about data size. In the radiology, many studies were done using relatively small data sets. The proposed machine learning methods to solve radiology problems may not generalize well from small data sets to large data sets. For example, the distribution of features in a large data set may differ from that of a small data set. So a machine learning algorithm trained based on a small data set may have lower performance on a large data set (Raudys and Jain, 1991). To solve the problem, re-training the algorithm will be necessary, but it requires intervention of knowledgeable experts which hinders the deployment of machine learning based systems in hospitals or medical centers. One possible solution would be utilizing incremental learning and adjust the computerized systems in an automatic way (Li et al., 2007). In addition, increased large scale data may bring computational issues to radiology applications. Machine learning techniques employed in these applications may not scale well as training data increases. For example, many machine learning techniques are based on quadratic optimization. It is well known that the training time for quadratic optimization problems will increase exponentially as input variables and samples increase. Converting the batch learning mode to online learning mode would solve this problem to some extent but may result in performance degeneration.
The second issue is about complexity which arises from both sides (machine learning and radiology) (Abu-Mostafa, 1989; Bialek et al., 2001; Kearns, 1990; Madabhushi et al., 2005). Some machine learning methods are too complicated to apply to real radiology problems. They may have strict assumptions on the problems to be solved and many variables which make it hard to apply to radiology problems directly. So currently simple and robust machine learning techniques, e.q. linear classifier, PCA, and SVMs, are widely used in clinical practice.
From the viewpoint of radiology application, there are also issues about complexity. Some applications are so complicated that there are no known machine learning methods that could solve them. For example, in interventional radiology, human-guided operation is still the mainstream way. A fully automatic interventional procedure, which requires knowledge of human anatomy, real-time tracking of needles or catheters through blood vessels, and treatment of lesions is still too complicated for current machine learning techniques. In addition, in real radiology applications, there may be many complex variables involved that may change over time, e.g., scanning protocols. So machine learning algorithms trained on previous data sets may not adapt well to new or evolving situations.
The third issue is about psychology. Although machine learning showed many advantages in many areas, many people in clinical practice still believe in human decisions or diagnostic results by radiologists. They doubt the helpfulness or accuracy of the computerized medical systems. In other words, people usually place more trust in human decisions than machine decisions. Such psychological issues make it hard to fully benefit from machine learning in daily radiology practice. So in current clinical practice, computerized medical systems are mainly viewed or treated as an aid or helper in a secondary position, not a problem solver or a necessary part of the whole medical system.
6. Discussion and conclusion
In this paper, we have presented a short introduction to machine learning and surveyed its applications in radiology. We focused on six applications in radiology: image segmentation, image registration, computer-aided detection and diagnosis, brain function or activity analysis and neurological disease diagnosis from fMR images, content-based image retrieval, and text analysis of radiology reports using NLP/NLU. This survey shows that machine learning plays a key role in many radiology applications. Machine learning helps the computer identify complex patterns in diverse types of radiology data.
In many applications, the performance of the machine learning-based systems is comparable to that of experienced radiologists. The application of machine learning may benefit patients either by reducing costs, improving accuracy, or disseminating expertise that is in short supply.
Statistical approaches are playing an expanding role in machine learning. In many radiology applications, data are in high dimensional spaces. The data may be corrupted by noise and variables generating the data may have complicated relationships. Statistical methods based on probability models provide practical tools to model these complicated relationships. Probability inferences based on these statistical models can usually provide the optimal solution under specific settings. Research on statistical approaches will be a major direction in the future of machine learning in radiology.
The use of machine learning in radiology is still evolving. As machine learning research progresses, we expect there to be more applications to radiology. Machine learning will be a critical component of advanced software systems for radiology and is likely to have wider and wider application in the near future.
Highlights
Mainstream machine learning techniques relevant for radiology are introduced.
Six major applications of machine learning in radiology are surveyed.
Central themes of machine learning research in radiology are described.
Factors impacting translation of machine learning to radiology are discussed.
Acknowledgments
We thank Andrew Dwyer, MD, for critical review of the manuscript. This manuscript was support by the Intramural Research Program of the National Institutes of Health Clinical Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu-Mostafa YS. The Vapnik-Chervonenkis dimension: Information versus complexity in learning. Neural Computation. 1989;1:312–317. [Google Scholar]
- Ackley DH, Hinton GE, Sejnowski TJ. A Learning Algorithm for Boltzmann Machines. Cognitive Science. 1985;9:147–169. [Google Scholar]
- Ali AM, Farag AA. Automatic Lung Segmentation of Volumetric Low-Dose CT Scans Using Graph Cuts. Advances in Visual Computing. 2008 [Google Scholar]
- Alpaydin E. Introduction to Machine Learning. The MIT Press; 2004. [Google Scholar]
- Amari SI. Information geometry of the EM and em algorithms for neural networks. Neural Networks. 1995;8:1379–1408. [Google Scholar]
- Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Neelin P, Collins DL, Evans A, Friston K. Incorporating prior knowledge into image registration. Neuroimage. 1997;6:344–352. doi: 10.1006/nimg.1997.0299. [DOI] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillard C, Hellier P, Barillot C. Segmentation of brain 3D MR images using level sets and dense registration. Medical Image Analysis. 2001;5:185–194. doi: 10.1016/s1361-8415(01)00039-1. [DOI] [PubMed] [Google Scholar]
- Baker JA, Kornguth PJ, Lo JY, Williford ME, Floyd CE. Breast-Cancer - Prediction with Artificial Neural-Network-Based on Bi-Rads Standardized Lexicon. Radiology. 1995;196:817–822. doi: 10.1148/radiology.196.3.7644649. [DOI] [PubMed] [Google Scholar]
- Bakken S, Hyun S, Friedman C, Johnson S. A Comparison of Semantic Categories of the ISO Reference Terminology Models for Nursing and the MedLEE Natural Language Processing System. Medinfo. 2004:472–476. [PubMed] [Google Scholar]
- Barto A, Sutton R. Reinforcement learning: an introduction. Cambridge University Press; 1999. [Google Scholar]
- Basili R. Learning to classify text using support vector machines: Methods, theory, and algorithms. Comput Linguist. 2003;29:655–661. [Google Scholar]
- Bates M. Models of Natural-Language Understanding. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:9977–9982. doi: 10.1073/pnas.92.22.9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkin M, Niyogi P. Laplacian eigenmaps for dimensionality reduction and data representation. Neural Computation. 2003;15:1373–1396. [Google Scholar]
- Bengio Y. Gradient-based optimization of hyperparameters. Neural Computation. 2000;12:1889–1900. doi: 10.1162/089976600300015187. [DOI] [PubMed] [Google Scholar]
- Bhargavan M, Kaye AH, Forman HP, Sunshine JH. Workload of Radiologists in United States in 2006–2007 and Trends Since 1991–1992. Radiology. 2009;252:458–467. doi: 10.1148/radiol.2522081895. [DOI] [PubMed] [Google Scholar]
- Bi J, Xiong T, Yu S, Dundar M, Rao RB. An Improved Multi-task Learning Approach with Applications in Medical Diagnosis, Machine Learning and Knowledge Discovery in Databases. Springer Berlin / Heidelberg; 2008. pp. 117–132. [Google Scholar]
- Bialek W, Nemenman I, Tishby N. Predictability, complexity, and learning. Neural Computation. 2001;13:2409–2463. doi: 10.1162/089976601753195969. [DOI] [PubMed] [Google Scholar]
- Bilmes J. A gentle tutorial of the EM algorithm and its application to parameter estimation for Gaussian mixture and hidden Markov models. UC Berkeley; 1998. [Google Scholar]
- Bishop CM. Pattern Recognition and Machine Learning. Springer; 2006. [Google Scholar]
- Bookheimer SY, Strojwas MH, Cohen MS, Saunders AM, Pericak-Vance MA, Mazziotta JC, Small GW. Patterns of brain activation in people at risk for Alzheimer's disease. New Engl J Med. 2000;343:450–456. doi: 10.1056/NEJM200008173430701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookstein FL. Thin-Plate Splines and the Atlas Problem for Biomedical Images. Lecture Notes in Computer Science. 1991;511:326–342. [Google Scholar]
- Bose I, Mahapatra RK. Business data mining - a machine learning perspective. Inform Manage. 2001;39:211–225. [Google Scholar]
- Boykov Y, Jolly MP. Interactive organ segmentation using graph cuts. Medical Image Computing and Computer-Assisted Intervention - Miccai 2000. 2000;1935:276–286. [Google Scholar]
- Breiman L. Bagging predictors. Mach Learn. 1996;24:123–140. [Google Scholar]
- Brodley C, Kak A, Shyu C, Dy J, Broderick L, Aisen A. Content-based retrieval from medical image databases: A synergy of human interaction, machine learning and computer vision, AAAI Conference on Artificial Intelligence.1999. [Google Scholar]
- Bromiley PA, Thacker NA, Scott MLJ, Pokric M, Lacey AJ, Cootes TF. Bayesian and non-Bayesian probabilistic models for medical image analysis. Image Vision Comput. 2003;21:851–864. [Google Scholar]
- Bryson AE, Ho Y-C. Applied optimal control: optimization, estimation, and control. Blaisdell Publishing Company or Xerox College Publishing. 1969;481 [Google Scholar]
- Burbidge R, Trotter M, Buxton B, Holden S. Drug design by machine learning: support vector machines for pharmaceutical data analysis. Comput Chem. 2001;26:5–14. doi: 10.1016/s0097-8485(01)00094-8. [DOI] [PubMed] [Google Scholar]
- Burges CJC. A tutorial on Support Vector Machines for pattern recognition. Data Min Knowl Disc. 1998;2:121–167. [Google Scholar]
- Caan MWA, Vermeer KA, van Vliet LJ, Majoie CBLM, Peters BD, den Heeten GJ, Vos FM. Shaving diffusion tensor images in discriminant analysis: A study into schizophrenia. Medical Image Analysis. 2006;10:841–849. doi: 10.1016/j.media.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Caruana R. Multitask learning. Mach Learn. 1997;28:41–75. [Google Scholar]
- Chan H-P, Hadjiiski L, Zhou C, Sahiner B. Computer-Aided Diagnosis of Lung Cancer and Pulmonary Embolism in Computed Tomography—A Review. Academic Radiology. 2008;15:535–555. doi: 10.1016/j.acra.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HP, Lo SCB, Sahiner B, Lam KL, Helvie MA. Computer-Aided Detection of Mammographic Microcalcifications - Pattern-Recognition with an Artificial Neural-Network. Medical Physics. 1995a;22:1555–1567. doi: 10.1118/1.597428. [DOI] [PubMed] [Google Scholar]
- Chan HP, Wei DT, Helvie MA, Sahiner B, Adler DD, Goodsitt MM, Petrick N. Computer-Aided Classification of Mammographic Masses and Normal Tissue - Linear Discriminant-Analysis in Texture Feature Space. Physics in Medicine and Biology. 1995b;40:857–876. doi: 10.1088/0031-9155/40/5/010. [DOI] [PubMed] [Google Scholar]
- Chan TF, Esedoglu S, Nikolova M. Algorithms for finding global minimizers of image segmentation and denoising models. Siam Journal on Applied Mathematics. 2006;66:1632–1648. [Google Scholar]
- Chapelle O, Schölkopf B, Zien A. Semi-supervised learning. MIT Press; Cambridge, MA, USA: 2006. [Google Scholar]
- Chapelle O, Vapnik V, Bousquet O, Mukherjee S. Choosing multiple parameters for support vector machines. Mach Learn. 2002;46:131–159. [Google Scholar]
- Chapman WW, Cooper GF, Hanbury P, Chapman BE, Harrison LH, Wagner MM. Creating a text classifier to detect radiology reports describing mediastinal findings associated with inhalational anthrax and other disorders. Journal of the American Medical Informatics Association. 2003;10:494–503. doi: 10.1197/jamia.M1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CW, Luo JB, Parker KJ. Image segmentation via adaptive K-mean clustering and knowledge-based morphological operations with biomedical applications. Ieee T Image Process. 1998;7:1673–1683. doi: 10.1109/83.730379. [DOI] [PubMed] [Google Scholar]
- Chen SC, Cai WL, Zhang DQ. Fast and robust fuzzy c-means clustering algorithms incorporating local information for image segmentation. Pattern Recognition. 2007;40:825–838. [Google Scholar]
- Cheng HD, Cai XP, Chen XW, Hu LM, Lou XL. Computer-aided detection and classification of microcalcifications in mammograms: a survey. Pattern Recognition. 2003;36:2967–2991. [Google Scholar]
- Christakou C, Lefakis L, Vrettos S, Stafylopatis A. A Movie Recommender System Based on Semi-supervised Clustering, International Conference on Computational Intelligence for Modelling, Control and Automation and International Conference on Intelligent Agents, Web Technologies and Internet Commerce; Vienna, Austria. 2005. [Google Scholar]
- Chua LO, Yang L. Cellular Neural Networks - Applications. Ieee Transactions on Circuits and Systems. 1988a;35:1273–1290. [Google Scholar]
- Chua LO, Yang L. Cellular Neural Networks - Theory. Ieee Transactions on Circuits and Systems. 1988b;35:1257–1272. [Google Scholar]
- Chuang KH, Chiu MJ, Lin CC, Chen JH. Model-free functional MRI analysis using Kohonen clustering neural network and fuzzy c-means. Ieee Transactions on Medical Imaging. 1999;18:1117–1128. doi: 10.1109/42.819322. [DOI] [PubMed] [Google Scholar]
- Chui HL, Rangarajan A. A new point matching algorithm for non-rigid registration. Computer Vision and Image Understanding. 2003;89:114–141. [Google Scholar]
- Ciofolo C, Barillot C. Brain segmentation with competitive level sets and fuzzy control. Information Processing in Medical Imaging, Proceedings. 2005;3565:333–344. doi: 10.1007/11505730_28. [DOI] [PubMed] [Google Scholar]
- Coifman RR, Lafon S, Lee AB, Maggioni M, Nadler B, Warner F, Zucker SW. Geometric diffusions as a tool for harmonic analysis and structure definition of data: Diffusion maps. Proceedings of the National Academy of Sciences of the United States of America. 2005a;102:7426–7431. doi: 10.1073/pnas.0500334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coifman RR, Lafon S, Lee AB, Maggioni M, Nadler B, Warner F, Zucker SW. Geometric diffusions as a tool for harmonic analysis and structure definition of data: Multiscale methods. Proceedings of the National Academy of Sciences of the United States of America. 2005b;102:7432–7437. doi: 10.1073/pnas.0500896102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comon P. Independent Component Analysis, a New Concept. Signal Process. 1994;36:287–314. [Google Scholar]
- Conte D, Foggia P, Sansone C, Vento M. Thirty years of graph matching in pattern recognition. International Journal of Pattern Recognition and Artificial Intelligence. 2004;18:265–298. [Google Scholar]
- Cooper GF, Buchanan BG, Kayaalp M, Saul M, Vries JK. Using computer modeling to help identify patient subgroups in clinical data repositories. Journal of the American Medical Informatics Association. 1998:180–184. [PMC free article] [PubMed] [Google Scholar]
- Cootes TF, Edwards GJ, Taylor CJ. Active appearance models. Ieee Transactions on Pattern Analysis and Machine Intelligence. 2001;23:681–685. [Google Scholar]
- Cootes TF, Taylor CJ, Cooper DH, Graham J. Active Shape Models - Their Training and Application. Computer Vision and Image Understanding. 1995;61:38–59. [Google Scholar]
- Cremers D, Rousson M, Deriche R. A review of statistical approaches to level set segmentation: Integrating color, texture, motion and shape. Int J Comput Vision. 2007;72:195–215. [Google Scholar]
- Dang PA, Kalra MK, Blake MA, Schultz TJ, Stout M, Halpern EF, Dreyer KJ. Use of Radcube for Extraction of Finding Trends in a Large Radiology Practice. J Digit Imaging. 2009;22:629–640. doi: 10.1007/s10278-008-9128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C, Fan Y, Wu X, Shen D, Resnick SM. Detection of prodromal Alzheimer's disease via pattern classification of magnetic resonance imaging. Neurobiology of Aging. 2008;29:514–523. doi: 10.1016/j.neurobiolaging.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C, Ruparel K, Fan Y, Shen DG, Acharyya M, Loughead JW, Gur RC, Langleben DD. Classifying spatial patterns of brain activity with machine learning methods: Application to lie detection. Neuroimage. 2005;28:663–668. doi: 10.1016/j.neuroimage.2005.08.009. [DOI] [PubMed] [Google Scholar]
- de Bruijne M, van Ginneken B, Viergever MA, Niessen WJ. Adapting active shape models for 3D segmentation of tubular structures in medical images. Information Processing in Medical Imaging, Proceedings. 2003;2732:136–147. doi: 10.1007/978-3-540-45087-0_12. [DOI] [PubMed] [Google Scholar]
- Demirci O, Clark VP, Calhoun VD. A projection pursuit algorithm to classify individuals using fMRI data: Application to schizophrenia. Neuroimage. 2008;39:1774–1782. doi: 10.1016/j.neuroimage.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum Likelihood from Incomplete Data Via Em Algorithm. Journal of the Royal Statistical Society Series B-Methodological. 1977;39:1–38. [Google Scholar]
- Dietterich TG. Machine-learning research - Four current directions. Ai Magazine. 1997;18:97–136. [Google Scholar]
- Dietterich TG, Lathrop RH, LozanoPerez T. Solving the multiple instance problem with axis-parallel rectangles. Artificial Intelligence. 1997;89:31–71. [Google Scholar]
- Doi K. Current status and future potential of computer-aided diagnosis in medical imaging. British Journal of Radiology. 2005;78:S3–S19. doi: 10.1259/bjr/82933343. [DOI] [PubMed] [Google Scholar]
- Doi K. Computer-aided diagnosis in medical imaging: Historical review, current status and future potential. Computerized Medical Imaging and Graphics. 2007;31:198–211. doi: 10.1016/j.compmedimag.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingos P, Pazzani M. On the optimality of the simple Bayesian classifier under zero-one loss. Mach Learn. 1997;29:103–130. [Google Scholar]
- Dorronsoro JR, Ginel F, Sanchez C, Cruz CS. Neural fraud detection in credit card operations. Ieee T Neural Networ. 1997;8:827–834. doi: 10.1109/72.595879. [DOI] [PubMed] [Google Scholar]
- Dreyer KJ, Kalra MK, Maher MM, Hurier AM, Asfaw BA, Schultz T, Halpern EF, Thrall JH. Application of recently developed computer algorithm for automatic classification of unstructured radiology reports: Validation study. Radiology. 2005;234:323–329. doi: 10.1148/radiol.2341040049. [DOI] [PubMed] [Google Scholar]
- Duan K, Keerthi SS, Poo AN. Evaluation of simple performance measures for tuning SVM hyperparameters. Neurocomputing. 2003;51:41–59. [Google Scholar]
- Duda RO, Hart PE, Stork DG. Pattern Classification. 2nd Edition Wiley-Interscience; 2000. [Google Scholar]
- Efron B, Tibshirani R. Bootstrap Methods for Standard Errors, Confidence Intervals, and Other Measures of Statistical Accuracy. Statistical Science. 1986;1:54–75. [Google Scholar]
- El-Naqa I, Yang YY, Galatsanos NP, Nishikawa RM, Wernick MN. A similarity learning approach to content-based image retrieval: Application to digital mammography. Ieee Transactions on Medical Imaging. 2004;23:1233–1244. doi: 10.1109/TMI.2004.834601. [DOI] [PubMed] [Google Scholar]
- El-Naqa I, Yang YY, Wernick MN, Galatsanos NP, Nishikawa RM. A support vector machine approach for detection of microcalcifications. Ieee Transactions on Medical Imaging. 2002;21:1552–1563. doi: 10.1109/TMI.2002.806569. [DOI] [PubMed] [Google Scholar]
- Ester M, Kriegel H-P, Sander J, Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise, the Second International Conference on Knowledge Discovery and Data Mining (KDD-96) AAAI Press. 1996:226–231. [Google Scholar]
- Fahlman S, Lebiere C. created for National Science Foundation, Contract Number EET-8716324, and Defense Advanced Research Projects Agency (DOD), ARPA Order No. 4976 under Contract F33615-87-C-1499. 1991. The Cascade-Correlation Learning Architecture. [Google Scholar]
- Fan Y, Shen DG, Davatzikos C. Classification of structural images via high-dimensional image warping, robust feature extraction, and SVM. Lect Notes Comput Sc. 2005;3749:1–8. doi: 10.1007/11566465_1. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The Use of Multiple Measurements in Taxonomic Problems. Annals of Eugenics. 1936;7:179–188. [Google Scholar]
- Franaszek M, Summers RM, Pickhardt PJ, Choi JR. Hybrid segmentation of colon filled with air and opacified fluid for CT colonography. Ieee Transactions on Medical Imaging. 2006;25:358–368. doi: 10.1109/TMI.2005.863836. [DOI] [PubMed] [Google Scholar]
- Freund Y, Schapire RE. A desicion-theoretic generalization of on-line learning and an application to boosting Computational Learning Theory. Springer Berlin / Heidelberg; 1995. pp. 23–37. [Google Scholar]
- Friedman C, Alderson PO, Austin JHM, Cimino JJ, Johnson SB. A General Natural-Language Text Processor for Clinical Radiology. Journal of the American Medical Informatics Association. 1994;1:161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman C, Hripcsak G. Evaluating natural language processors in the clinical domain. Methods of Information in Medicine. 1998;37:334–344. [PubMed] [Google Scholar]
- Friedman C, Hripcsak G. Natural language processing and its future in medicine. Academic Medicine. 1999;74:890–895. doi: 10.1097/00001888-199908000-00012. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Holmes AP, Worsley KJ, Poline JB, Frith CD, Frackowiak RSJ. Statistical parametric maps in functional imaging: a general linear approach. Human Brain Mapping. 1995;2:189–210. [Google Scholar]
- Fung G, Dundar M, Krishnapuram B, Rao RB. Multiple Instance Learning for Computer Aided Diagnosis, ADVANCES IN NEURAL INFORMATION PROCESSING SYSTEMS. 2007:425–432. [Google Scholar]
- García-Lorenzo D, Lecoeur J, Arnold DL, Collins DL, Barillot C. Multiple Sclerosis Lesion Segmentation Using an Automatic Multimodal Graph Cuts Medical Image Computing and Computer-Assisted Intervention – MICCAI 2009. 2009 doi: 10.1007/978-3-642-04271-3_71. [DOI] [PubMed] [Google Scholar]
- Giger ML, Karssemeijer N, Armato SG. Computer-aided diagnosis in medical imaging. Ieee Transactions on Medical Imaging. 2001;20:1205–1208. doi: 10.1109/tmi.2001.974915. [DOI] [PubMed] [Google Scholar]
- Gold S, Rangarajan A. A graduated assignment algorithm for graph matching. Ieee Transactions on Pattern Analysis and Machine Intelligence. 1996;18:377–388. [Google Scholar]
- Grady L. Random walks for image segmentation. Ieee Transactions on Pattern Analysis and Machine Intelligence. 2006;28:1768–1783. doi: 10.1109/TPAMI.2006.233. [DOI] [PubMed] [Google Scholar]
- Greig DM, Porteous BT, Seheult AH. Exact Maximum a-Posteriori Estimation for Binary Images. Journal of the Royal Statistical Society Series B-Methodological. 1989;51:271–279. [Google Scholar]
- Guyon I, Elisseeff A.e. An Introduction to Variable and Feature Selection. Journal of Machine Learning Research. 2003;3:1157–1182. [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. The Elements of. Statistical Learning: Data Mining, Inference, and Prediction. Second ed. Springer; 2009. [Google Scholar]
- Heckerman D. A tutorial on learning with Bayesian networks. 1996 [Google Scholar]
- Heimann T, Meinzer HP. Statistical shape models for 3D medical image segmentation: A review. Medical Image Analysis. 2009;13:543–563. doi: 10.1016/j.media.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Held K, Kops ER, Krause BJ, Wells WM, Kikinis R, Muller-Gartner HW. Markov random field segmentation of brain MR images. Ieee Transactions on Medical Imaging. 1997;16:878–886. doi: 10.1109/42.650883. [DOI] [PubMed] [Google Scholar]
- Hill DLG, Batchelor PG, Holden M, Hawkes DJ. Medical image registration. Physics in Medicine and Biology. 2001;46:R1–R45. doi: 10.1088/0031-9155/46/3/201. [DOI] [PubMed] [Google Scholar]
- Hinrichs C, Singh V, Mukherjee L, Xu GF, Chung MK, Johnson SC. Spatially augmented LPboosting for AD classification with evaluations on the ADNI dataset. Neuroimage. 2009a;48:138–149. doi: 10.1016/j.neuroimage.2009.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs C, Singh V, Xu G, Johnson S. MKL for Robust Multi-modality AD Classification, Medical Image Computing and Computer-Assisted Intervention. 2009b:786–794. doi: 10.1007/978-3-642-04271-3_95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden M, Hill DLG, Denton ERE, Jarosz JM, Cox TCS, Rohlfing T, Goodey J, Hawkes DJ. Voxel similarity measures for 3-D serial MR brain image registration. Ieee Transactions on Medical Imaging. 2000;19:94–102. doi: 10.1109/42.836369. [DOI] [PubMed] [Google Scholar]
- Hopfield JJ. Neural Networks and Physical Systems with Emergent Collective Computational Abilities. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences; 1982. pp. 2554–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Nakamori Y, Wang SY. Forecasting stock market movement direction with support vector machine. Comput Oper Res. 2005a;32:2513–2522. [Google Scholar]
- Huang Y, Lowe HJ, Klein D, Cucina RJ. Improved identification of noun phrases in clinical radiology reports using a high-performance statistical natural language parser augmented with the UMLS Specialist Lexicon. Journal of the American Medical Informatics Association. 2005b;12:275–285. doi: 10.1197/jamia.M1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys BL, Lindberg DAB, Schoolman HM, Barnett GO. The Unified Medical Language System: An informatics research collaboration. Journal of the American Medical Informatics Association. 1998;5:1–11. doi: 10.1136/jamia.1998.0050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerebko AK, Malley JD, Franaszek M, Summers RM. Multi neural network classification scheme for detection of colonic polyps in CT colonography data sets. Acad Radiol. 2003a;10:154–160. doi: 10.1016/s1076-6332(03)80039-9. [DOI] [PubMed] [Google Scholar]
- Jerebko AK, Malley JD, Franaszek M, Summers RM. Support vector machines committee classification method for computer-aided polyp detection in CT colonography. Acad. Radiol. 2005;12:479–486. doi: 10.1016/j.acra.2004.04.024. [DOI] [PubMed] [Google Scholar]
- Jerebko AK, Summers RM, Malley JD, Franaszek M, Johnson CD. Computer-assisted detection of colonic polyps with CT colonography using neural networks and binary classification trees. Medical Physics. 2003b;30:52–60. doi: 10.1118/1.1528178. [DOI] [PubMed] [Google Scholar]
- Jolliffe IT. Principal Component Analysis. second ed. Springer-Verlag; 2002. [Google Scholar]
- Jordan MI. Learning in graphical models. Kluwer Acdemic Publishers; 1998. [Google Scholar]
- Jordan MI, Ghahramani Z, Jaakkola TS, Saul LK. An introduction to variational methods for graphical models. Mach Learn. 1999;37:183–233. [Google Scholar]
- Kaneko M, Eguchi K, Ohmatsu H, Kakinuma R, Naruke T, Suemasu K, Moriyama N. Peripheral lung cancer: Screening and detection with low-dose spiral CT versus radiography. Radiology. 1996;201:798–802. doi: 10.1148/radiology.201.3.8939234. [DOI] [PubMed] [Google Scholar]
- Kearns JJ. The computational complexity of machine learning. The MIT Press; 1990. [Google Scholar]
- Kedenburg G, Cocosco CA, Köthe U, Niessen WJ, Vonken E.-j.P.A., Viergever MA. Automatic cardiac MRI myocardium segmentation using graphcut, SPIE Medical Imaging 2006: Image Processing. 2006 [Google Scholar]
- Kerlikowske K, Grady D, Rubin SM, Sandrock C, Ernster VL. Efficacy of Screening Mammography - a Metaanalysis. Jama-J Am Med Assoc. 1995;273:149–154. [PubMed] [Google Scholar]
- Keysers D, Dahmen J, Ney H, Wein BB, Lehmann TM. Statistical framework for model-based image retrieval in medical applications. Journal of Electronic Imaging. 2003;12:59–68. [Google Scholar]
- Killiany RJ, Gomez-Isla T, Moss M, Kikinis R, Sandor T, Jolesz F, Tanzi R, Jones K, Hyman BT, Albert MS. Use of structural magnetic resonance imaging to predict who will get Alzheimer's disease. Ann Neurol. 2000;47:430–439. [PubMed] [Google Scholar]
- Kim D, Burge J, Lane T, Pearlson GD, Kiehl KA, Calhoun VD. Hybrid ICA-Bayesian network approach reveals distinct effective connectivity differences in schizophrenia. Neuroimage. 2008;42:1560–1568. doi: 10.1016/j.neuroimage.2008.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Lee MC, Kim J, Kim IY, Kim SI, Han MH, Chang KH, Kwon JS. Grey matter abnormalities in obsessive-compulsive disorder - Statistical parametric mapping of segmented magnetic resonance images. Brit J Psychiat. 2001;179:330–334. doi: 10.1192/bjp.179.4.330. [DOI] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D, Manning CD. Fast exact inference with a factored model for natural language parsing, Advances in Neural Information Processing Systems. 2002;15 [Google Scholar]
- Kloppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR, Ashburner J, Frackowiak RSJ. Automatic classification of MR scans in Alzheimers disease. Brain. 2008;131:681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection, International joint Conference on artificial intelligence.1995. [Google Scholar]
- Kohonen T. Self-Organized Formation of Topologically Correct Feature Maps. Biological Cybernetics. 1982;43:59–69. [Google Scholar]
- Kononenko I. Machine learning for medical diagnosis: history, state of the art and perspective. Artificial Intelligence in Medicine. 2001;23:89–109. doi: 10.1016/s0933-3657(01)00077-x. [DOI] [PubMed] [Google Scholar]
- Konukoglu E, Acar B, Paik DS, Beaulieu CF, Rosenberg J, Napel S. Polyp enhancing level set evolution of colon wall: Method and pilot study. Ieee Transactions on Medical Imaging. 2007;26:1649–1656. doi: 10.1109/tmi.2007.901429. [DOI] [PubMed] [Google Scholar]
- Ladicky L, Sturgess P, Alahari K, Russell C, Torr PHS. What,Where & How Many?. Combining Object Detectors and CRFs, the 11th European Conference on Computer Vision.2010. [Google Scholar]
- Lanckriet GRG, Cristianini N, Bartlett P, El Ghaoui L, Jordan MI. Learning the kernel matrix with semidefinite programming. Journal of Machine Learning Research. 2004;5:27–72. [Google Scholar]
- Lecam L. Maximum-Likelihood - an Introduction. International Statistical Review. 1990;58:153–171. [Google Scholar]
- Lee J, Kim N, Lee H, Seo JB, Won HJ, Shin YM, Shin YG, Kim SH. Efficient liver segmentation using a level-set method with optimal detection of the initial liver boundary from level-set speed images. Comput Meth Prog Bio. 2007;88:26–38. doi: 10.1016/j.cmpb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Lehmann TM, Guld MO, Thies O, Fisher B, Spitzer K, Keysers D, Ney H, Kohnen M, Schubert H, Wein BB. Content-based image retrieval in medical applications. Methods of Information in Medicine. 2004;43:354–361. [PubMed] [Google Scholar]
- Lei TH, Sewchand W. Statistical Approach to X-Ray Ct Imaging and Its Applications in Image-Analysis .2. A New Stochastic Model-Based Image Segmentation Technique for X-Ray Ct Image. Ieee Transactions on Medical Imaging. 1992;11:62–69. doi: 10.1109/42.126911. [DOI] [PubMed] [Google Scholar]
- Leordeanu M, Hebert M. A Spectral Technique for Correspondence Problems Using Pairwise Constraints, Tenth IEEE International Conference on Computer Vision.2005. pp. 1482–1489. [Google Scholar]
- Lester H, Arridge SR. A survey of hierarchical non-linear medical image registration. Pattern Recognition. 1999;32:129–149. [Google Scholar]
- Levitan E, Herman GT. A Maximum a Posteriori Probability Expectation Maximization Algorithm for Image-Reconstruction in Emission Tomography. Ieee Transactions on Medical Imaging. 1987;6:185–192. doi: 10.1109/TMI.1987.4307826. [DOI] [PubMed] [Google Scholar]
- Li FF, Fergus R, Perona P. Learning generative visual models from few training examples: An incremental Bayesian approach tested on 101 object categories. Computer Vision and Image Understanding. 2007;106:59–70. [Google Scholar]
- Li J, Yao J, Summers RM, Petrick N, Hara A. An efficient feature selection algorithm for computer-aided polyp detection. International Journal on Artificial Intelligence Tools. 2006;15:893–915. [Google Scholar]
- Li SZ. Markov Random Field Models in Computer Vision, The European Conference on Computer Vision.1994. [Google Scholar]
- Liang J, Bi J. Computer Aided Detection of Pulmonary Embolism with Tobogganing and Mutiple Instance Classification in CT Pulmonary Angiography, Information Processing in Medical Imaging. Springer; 2007. pp. 630–641. [DOI] [PubMed] [Google Scholar]
- Lin N, Yu WC, Duncan JS. Combinative multi-scale level set framework for echocardiographic image segmentation. Medical Image Analysis. 2003;7:529–537. doi: 10.1016/s1361-8415(03)00035-5. [DOI] [PubMed] [Google Scholar]
- Liu H, Yu L. Toward Integrating Feature Selection Algorithms for Classification and Clustering. IEEE Transactions on knowledge and data engineering. 2005;17:491–502. [Google Scholar]
- Loog M, Duin RPW, Haeb-Umbach R. Multiclass linear dimension reduction by weighted pairwise Fisher criteria. Ieee Transactions on Pattern Analysis and Machine Intelligence. 2001;23:762–766. [Google Scholar]
- Luo B, Hancock ER. Structural graph matching using the EM algorithm and singular value decomposition. Ieee Transactions on Pattern Analysis and Machine Intelligence. 2001;23:1120–1136. [Google Scholar]
- Madabhushi A, Feldman MD, Metaxas DN, Tomaszeweski J, Chute D. Automated detection of prostatic adenocarcinoma from high-resolution ex vivo MRI. IEEE Trans Med Imaging. 2005;24:1611–1625. doi: 10.1109/TMI.2005.859208. [DOI] [PubMed] [Google Scholar]
- Maes F, Vandermeulen D, Suetens P. Comparative evaluation of multiresolution optimization strategies for multimodality image registration by maximization of mutual information. Medical Image Analysis. 1999;3:373–386. doi: 10.1016/s1361-8415(99)80030-9. [DOI] [PubMed] [Google Scholar]
- Maintz JBA, Viergever MA. A survey of medical image registration. Medical Image Analysis. 1998;2:1–36. doi: 10.1016/s1361-8415(01)80026-8. [DOI] [PubMed] [Google Scholar]
- Malladi R, Sethian JA, Vemuri BC. Shape Modeling with Front Propagation - a Level Set Approach. Ieee Transactions on Pattern Analysis and Machine Intelligence. 1995;17:158–175. [Google Scholar]
- Malley JD, Jerebko AK, Summers RM. Committee of support vector machines for detection of colonic polyps from CT scans, Medical Imaging 2003: Physiology and Function from Multidimensional Images. 2003 [Google Scholar]
- Maron O, Lozano-P'erez T.a. A Framework for Multiple-Instance Learning, Advances in neural information processing. 1998:570–576. [Google Scholar]
- Martel MK, Ten Haken RK, Hazuka MB, Kessler ML, Strawderman M, Turrisi AT, Lawrence TS, Fraass BA, Lichter AS. Estimation of tumor control probability model parameters from 3-D dose distributions of non-small cell lung cancer patients. Lung Cancer-J Iaslc. 1999;24:31–37. doi: 10.1016/s0169-5002(99)00019-7. [DOI] [PubMed] [Google Scholar]
- McInerney T, Terzopoulos D. Deformable models in medical image analysis: a survey. Medical Image Analysis. 1996;1:91–108. doi: 10.1016/s1361-8415(96)80007-7. [DOI] [PubMed] [Google Scholar]
- Meyer CR, Boes JL, Kim B, Bland PH, Lecarpentier GL, Fowlkes JB, Roubidoux NA, Carson PL. Semiautomatic registration of volumetric ultrasound scans. Ultrasound in Medicine and Biology. 1999;25:339–347. doi: 10.1016/s0301-5629(98)00148-3. [DOI] [PubMed] [Google Scholar]
- Meyer CR, Boes JL, Kim B, Bland PH, Zasadny KR, Kison PV, Koral K, Frey KA, Wahla RL. Demonstration of accuracy and clinical versatility of mutual information for automatic multimodality image fusion using affine and thin-plate spline warped geometric deformations. Medical Image Analysis. 1997;1:195–206. doi: 10.1016/s1361-8415(97)85010-4. [DOI] [PubMed] [Google Scholar]
- Mitchell SC, Bosch JG, Lelieveldt BPF, van der Geest RJ, Reiber JHC, Sonka M. 3-D active appearance models: Segmentation of cardiac MR and ultrasound images. Ieee Transactions on Medical Imaging. 2002;21:1167–1178. doi: 10.1109/TMI.2002.804425. [DOI] [PubMed] [Google Scholar]
- Mitchell TM. Machine Learning. McGraw-Hill Science/Engineering/Math. 1997 [Google Scholar]
- Mitchell TM, Hutchinson R, Niculescu RS, Pereira F, Wang XR, Just M, Newman S. Learning to decode cognitive states from brain images. Machine Learning. 2004;57:145–175. [Google Scholar]
- Mitchell TM, Shinkareva SV, Carlson A, Chang KM, Malave VL, Mason RA, Just MA. Predicting human brain activity associated with the meanings of nouns. Science. 2008;320:1191–1195. doi: 10.1126/science.1152876. [DOI] [PubMed] [Google Scholar]
- Mohan R, Mageras GS, Baldwin B, Brewster LJ, Kutcher GJ, Leibel S, Burman CM, Ling CC, Fuks Z. Clinically Relevant Optimization of 3-D Conformal Treatments. Medical Physics. 1992;19:933–944. doi: 10.1118/1.596781. [DOI] [PubMed] [Google Scholar]
- Moody J, Darken CJ. Fast learning in networks of locally-tuned processing units. Neural Computation. 1989;1:281–294. [Google Scholar]
- Mougiakakou SG, Valavanis IK, Nikita A, Nikita KS. Differential diagnosis of CT focal liver lesions using texture features, feature selection and ensemble driven classifiers. Artificial Intelligence in Medicine. 2007;41:25–37. doi: 10.1016/j.artmed.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Muller H, Michoux N, Bandon D, Geissbuhler A. A review of content-based image retrieval systems in medical applications - clinical benefits and future directions. International Journal of Medical Informatics. 2004;73:1–23. doi: 10.1016/j.ijmedinf.2003.11.024. [DOI] [PubMed] [Google Scholar]
- Ochs RA, Goldin JG, Abtin F, Kim HJ, Brown K, Batra P, Roback D, McNitt-Gray MF, Brown MS. Automated classification of lung bronchovascular anatomy in CT using AdaBoost. Medical Image Analysis. 2007;11:315–324. doi: 10.1016/j.media.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik DS, Beaulieu CF, Rubin GD, Acar B, Jeffrey RB, Yee J, Dey J, Napel S. Surface normal overlap: A computer-aided detection algorithm, with application to colonic polyps and lung nodules in helical CT. IEEE Trans. Med. Imaging. 2004;23:661–675. doi: 10.1109/tmi.2004.826362. [DOI] [PubMed] [Google Scholar]
- Panjwani DK, Healey G. Markov Random-Field Models for Unsupervised Segmentation of Textured Color Images. Ieee Transactions on Pattern Analysis and Machine Intelligence. 1995;17:939–954. [Google Scholar]
- Papademetris X, Jackowski AP, Schultz RT, Staib LH, Duncan JS. Integrated Intensity and Point-Feature Nonrigid Registration Medical Image Computing and Computer-Assisted Intervention. 2004 doi: 10.1901/jaba.2001.3216-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paragios N. A level set approach for shape-driven segmentation and tracking of the left ventricle. Ieee Transactions on Medical Imaging. 2003;22:773–776. doi: 10.1109/TMI.2003.814785. [DOI] [PubMed] [Google Scholar]
- Pena JM, Lozano JA, Larranaga P. An empirical comparison of four initialization methods for the K-Means algorithm. Pattern Recognition Letters. 1999;20:1027–1040. [Google Scholar]
- Peng HC, Long FH, Ding C. Feature selection based on mutual information: Criteria of max-dependency, max-relevance, and min-redundancy. Ieee Transactions on Pattern Analysis and Machine Intelligence. 2005;27:1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- Pereira F, Mitchell T, Botvinick M. Machine learning classifiers and fMRI: A tutorial overview. Neuroimage. 2009;45:S199–S209. doi: 10.1016/j.neuroimage.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham DL, Xu CY, Prince JL. Current methods in medical image segmentation. Annual Review of Biomedical Engineering. 2000;2:315–+. doi: 10.1146/annurev.bioeng.2.1.315. [DOI] [PubMed] [Google Scholar]
- Pluim JPW, Maintz JBA, Viergever MA. Mutual-information-based registration of medical images: A survey. Ieee Transactions on Medical Imaging. 2003;22:986–1004. doi: 10.1109/TMI.2003.815867. [DOI] [PubMed] [Google Scholar]
- Prasad MN, Brown MS, Ahmad S, Abtin F, Allen J, da Costa I, Kim HJ, McNitt-Gray MF, Goldin JG. Automatic segmentation of lung parenchyma in the presence of diseases based on curvature of ribs. Academic Radiology. 2008;15:1173–1180. doi: 10.1016/j.acra.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Preul MC, Caramanos Z, Collins DL, Villemure JG, Leblanc R, Olivier A, Pokrupa R, Arnold DL. Accurate, noninvasive diagnosis of human brain tumors by using proton magnetic resonance spectroscopy. Nat Med. 1996;2:323–325. doi: 10.1038/nm0396-323. [DOI] [PubMed] [Google Scholar]
- Radau PE, Slomka PJ, Julin P, Svensson L, Wahlund LO. Evaluation of linear registration algorithms for brain SPECT and the errors due to hypoperfusion lesions. Medical Physics. 2001;28:1660–1668. doi: 10.1118/1.1388894. [DOI] [PubMed] [Google Scholar]
- Raudys SJ, Jain AK. Small sample size effects in statistical pattern recognition: recommendations for practitioners and open problems. Ieee Transactions on Pattern Analysis and Machine Intelligence. 1991;13:252–264. [Google Scholar]
- Rosenblatt F. The Perceptron - a Probabilistic Model for Information-Storage and Organization in the Brain. Psychological Review. 1958;65:386–408. doi: 10.1037/h0042519. [DOI] [PubMed] [Google Scholar]
- Roweis ST, Saul LK. Nonlinear dimensionality reduction by locally linear embedding. Science. 2000;290:2323–+. doi: 10.1126/science.290.5500.2323. [DOI] [PubMed] [Google Scholar]
- Rueckert D, Aljabar P, Heckemann RA, Hajnal JV, Hammers A. Diffeomorphic registration using B-splines. Medical Image Computing and Computer-Assisted Intervention - Miccai 2006. 2006;(Pt 2):702–709. doi: 10.1007/11866763_86. 4191. [DOI] [PubMed] [Google Scholar]
- Sahba F, Tizhoosh HR, Salama MMA. A reinforcement learning framework for medical image segmentation. International Joint Conference on Neural Networks. 2006:511–517. [Google Scholar]
- Sahiner B, Chan HP, Petrick N, Helvie MA, Goodsitt MM. Computerized characterization of masses on mammograms: The rubber band straightening transform and texture analysis. Medical Physics. 1998;25:516–526. doi: 10.1118/1.598228. [DOI] [PubMed] [Google Scholar]
- Sajda P. Machine learning for detection and diagnosis of disease. Annual Review of Biomedical Engineering. 2006;8:537–565. doi: 10.1146/annurev.bioeng.8.061505.095802. [DOI] [PubMed] [Google Scholar]
- Saxena S, Brody AL, Ho ML, Alborzian S, Ho MK, Maidment KM, Huang SC, Wu HM, Au SC, Baxter LR. Cerebral metabolism in major depression and obsessive-compulsive disorder occurring separately and concurrently. Biol Psychiat. 2001;50:159–170. doi: 10.1016/s0006-3223(01)01123-4. [DOI] [PubMed] [Google Scholar]
- Scahill RI, Schott JM, Stevens JM, Rossor MN, Fox NC. Mapping the evolution of regional atrophy in Alzheimer's disease: Unbiased analysis of fluid-registered serial MRI. P Natl Acad Sci USA. 2002;99:4703–4707. doi: 10.1073/pnas.052587399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaal S, Atkeson CG. Robot Juggling - Implementation of Memory-Based Learning. Ieee Control Systems Magazine. 1994;14:57–71. [Google Scholar]
- Schoepf UJ, Costello P. CT angiography for diagnosis of pulmonary embolism: State of the art. Radiology. 2004;230:329–337. doi: 10.1148/radiol.2302021489. [DOI] [PubMed] [Google Scholar]
- Schoepf UJ, Schneider AC, Das M, Wood SA, Cheema JL, Costello P. Pulmonary embolism: Computer-aided detection at multidetector row spiral computed tomography. Journal of Thoracic Imaging. 2007;22:319–323. doi: 10.1097/RTI.0b013e31815842a9. [DOI] [PubMed] [Google Scholar]
- Schraudolph NN, Dayan P, Sejnowski TJ. Temporal difference learning of position evaluation in the game of Go, Advances in Neural Information Processing Systems. Morgan Kaufmann. 1994:817–824. [Google Scholar]
- Shekhar R, Zagrodsky V. Mutual information-based rigid and nonrigid registration of ultrasound volumes. Ieee Transactions on Medical Imaging. 2002;21:9–22. doi: 10.1109/42.981230. [DOI] [PubMed] [Google Scholar]
- Shen DG, Zhan YQ, Davatzikos C. Segmentation of prostate boundaries from ultrasound images using statistical shape model. Ieee Transactions on Medical Imaging. 2003;22:539–551. doi: 10.1109/TMI.2003.809057. [DOI] [PubMed] [Google Scholar]
- Shenton ME, Dickey CC, Frumin M, McCarley RW. A review of MRI findings in schizophrenia. Schizophr Res. 2001;49:1–52. doi: 10.1016/s0920-9964(01)00163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JB, Malik J. Normalized cuts and image segmentation. Ieee Transactions on Pattern Analysis and Machine Intelligence. 2000;22:888–905. [Google Scholar]
- Singaraju D, Vidal R.e. Using Global Bag of Features Models in Random Fields for Joint Categorization and Segmentation of Objects, 24th IEEE Conference on Computer Vision and Pattern Recognition.2011. [Google Scholar]
- Smeets D, Loeckx D, Stijnen B, De Dobbelaer B, Vandermeulen D, Suetens P. Semi-automatic level set segmentation of liver tumors combining a spiral-scanning technique with supervised fuzzy pixel classification. Medical Image Analysis. 2010;14:13–20. doi: 10.1016/j.media.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Sonnenburg S, Ratsch G, Schafer C, Scholkopf B. Large scale multiple kernel learning. Journal of Machine Learning Research. 2006;7:1531–1565. [Google Scholar]
- Sowell ER, Levitt J, Thompson PM, Holmes CJ, Blanton RE, Kornsand DS, Caplan R, McCracken J, Asarnow R, Toga AW. Brain abnormalities in early-onset schizophrenia spectrum disorder observed with statistical parametric mapping of structural magnetic resonance images. Am J Psychiat. 2000;157:1475–1484. doi: 10.1176/appi.ajp.157.9.1475. [DOI] [PubMed] [Google Scholar]
- Specht DF. Probabilistic Neural Networks. Neural Networks. 1990;3:109–118. doi: 10.1109/72.80210. [DOI] [PubMed] [Google Scholar]
- Stonnington CM, Kloppel S, Barnes J, Chen F, Chu C, Good CD, Mader I, Mitchell LA, Patel AC, Roberts CC, Fox NC, Jack CR, Ashburner J, Frackowiak RSJ. Accuracy of Dementia Diagnosis: A Direct Comparison of Radiologists and a Computerized Method. Neurology. 2009;72:A151–a151. doi: 10.1093/brain/awn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studholme C, Hill D, Hawkes D. Automated 3-D registration of MR and CT images of the head. Medical image analysis. 1996;1:163–175. doi: 10.1016/s1361-8415(96)80011-9. [DOI] [PubMed] [Google Scholar]
- Studholme C, Hill DLG, Hawkes DJ. Automated three-dimensional registration of magnetic resonance and positron emission tomography brain images by multiresolution optimization of voxel similarity measures. Medical Physics. 1997;24:25–35. doi: 10.1118/1.598130. [DOI] [PubMed] [Google Scholar]
- Summers RM. Improving the Accuracy of CT Colonography Interpretation: Computer-Aided Diagnosis. Gastrointestinal Endoscopy Clinics of North America. 2010;20:245–257. doi: 10.1016/j.giec.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers RM, Yao J, Pickhardt PJ, Franaszek M, Bitter I, Brickman D, Krishna V, Choi JR. Computed tomographic virtual colonoscopy computer-aided polyp detection in a screening population. Gastroenterology. 2005;129:1832–1844. doi: 10.1053/j.gastro.2005.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Yoshida H, Nappi J, Armato SG, Dachman AH. Mixture of expert 3D massive-training ANNs for reduction of multiple types of false positives in CAD for detection of polyps in CT colonography. Med. Phys. 2008;35:694–703. doi: 10.1118/1.2829870. [DOI] [PubMed] [Google Scholar]
- Swartout WR. Rule-Based Expert Systems - the Mycin Experiments of the Stanford Heuristic Programming Project - Buchanan,Bg, Shortliffe,Eh. Artificial Intelligence. 1985;26:364–366. [Google Scholar]
- Swensen SJ, Jett JR, Sloan JA, Midthun DE, Hartman TE, Sykes AM, Aughenbaugh GL, Zink FE, Hillman SL, Noetzel GR, Marks RS, Clayton AC, Pairolero PC. Screening for lung cancer with low-dose spiral computed tomography. Am J Resp Crit Care. 2002;165:508–513. doi: 10.1164/ajrccm.165.4.2107006. [DOI] [PubMed] [Google Scholar]
- Tagare HD, Jaffe CC, Duncan J. Medical image databases: A content-based retrieval approach. Journal of the American Medical Informatics Association. 1997;4:184–198. doi: 10.1136/jamia.1997.0040184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JS, Rangayyan RM, Xu J, El Naqa I, Yang YY. Computer-Aided Detection and Diagnosis of Breast Cancer With Mammography: Recent Advances. Ieee T Inf Technol B. 2009;13:236–251. doi: 10.1109/TITB.2008.2009441. [DOI] [PubMed] [Google Scholar]
- Tenenbaum JB, de Silva V, Langford JC. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;290:2319–+. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- Tourassi GD, Floyd CE, Sostman HD, Coleman RE. Acute Pulmonary-Embolism - Artificial Neural-Network Approach for Diagnosis. Radiology. 1993;189:555–558. doi: 10.1148/radiology.189.2.8210389. [DOI] [PubMed] [Google Scholar]
- Towhidkhah F, Khayati R, Vafadust M, Nabavi SM. Fully automatic segmentation of multiple sclerosis lesions in brain MR FLAIR images using adaptive mixtures method and markov random field model. Computers in Biology and Medicine. 2008;38:379–390. doi: 10.1016/j.compbiomed.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Uitert RL, Summers RM. Automatic correction of level set based subvoxel precise centerlines for virtual colonoscopy using the colon outer wall. Ieee Transactions on Medical Imaging. 2007;26:1069–1078. doi: 10.1109/TMI.2007.896927. [DOI] [PubMed] [Google Scholar]
- van Ravesteijn VF, van Wijk C, Vos FM, Truyen R, Peters JF, Stoker J, van Vliet LJ. Computer-Aided Detection of Polyps in CT Colonography Using Logistic Regression. IEEE Trans. Med. Imaging. 2010;29:120–131. doi: 10.1109/TMI.2009.2028576. [DOI] [PubMed] [Google Scholar]
- Vercauteren T, Pennec X, Perchant A, Ayache N. Non-parametric Diffeomorphic Image Registration with the Demons Algorithm Medical Image Computing and Computer-Assisted Intervention. 2007:319–326. doi: 10.1007/978-3-540-75759-7_39. [DOI] [PubMed] [Google Scholar]
- Wang J, Zucker J. Solving the multi-instance problem: A lazy learning approach, Proc. 17th International Conf. on Machine Learning.2000. pp. 1119–1125. [Google Scholar]
- Wang S, Petrick N, Uitert RLV, Periaswamy S, Summers RM. Graph Matching Based on Mean Field Theory, the International Conference on Image Processing.2010a. [Google Scholar]
- Wang S, Petrick N, Uitert RLV, Periaswamy S, Summers RM. Graph Matching Based on Mean Field Theory, the International Conference on Image Processing.2010b. [Google Scholar]
- Wang S, Yao J, Petrick N, Summers RM. Combining Statistical and Geometric Features for Colonic Polyp Detection in CTC Based on Multiple Kernel Learning. International Journal of Computational Intelligence and Applications. 2010c;9:1–15. doi: 10.1142/S1469026810002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Yao J, Summers RM. Improved classifier for computer-aided polyp detection in CT colonography by nonlinear dimensionality reduction. Med Phys. 2008a;35:1377–1386. doi: 10.1118/1.2870218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SJ, Yao JH, Summers RM. Improved classifier for computer-aided polyp detection in CT Colonography by nonlinear dimensionality reduction. Med. Phys. 2008b;35:1377–1386. doi: 10.1118/1.2870218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fan Y, Bhatt P, Davatzikos C. High-dimensional pattern regression using machine learning: From medical images to continuous clinical variables. Neuroimage. 2010d;50:1519–1535. doi: 10.1016/j.neuroimage.2009.12.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Liang Z, Li L, Li X, Li B, Anderson J, Harrington D. Reduction of false positives by internal features for polyp detection in CT-based virtual colonoscopy. Med Phys. 2005;32:3602–3616. doi: 10.1118/1.2122447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins CJCH, Dayan P. Q-Learning. Mach Learn. 1992;8:279–292. [Google Scholar]
- Wei LY, Yang YY, Nishikawa RM, Jiang YL. A study on several machine-learning methods for classification of malignant and benign clustered microcalcifications. Ieee Transactions on Medical Imaging. 2005a;24:371–380. doi: 10.1109/tmi.2004.842457. [DOI] [PubMed] [Google Scholar]
- Wei LY, Yang YY, Nishikawa RM, Wernick MN, Edwards A. Relevance vector machine for automatic detection of clustered microcalcifications. Ieee Transactions on Medical Imaging. 2005b;24:1278–1285. doi: 10.1109/TMI.2005.855435. [DOI] [PubMed] [Google Scholar]
- Weston J, Mukherjee S, Chapelle O, Pontil M, Poggio T, Vapnik V. Feature selection for SVMs, Advances in Neural Information Processing Systems. 2001 [Google Scholar]
- Wilke M, Kaufmann C, Grabner A, Putz B, Wetter TC, Auer DP. Gray matter-changes and correlates of disease severity in schizophrenia: A statistical parametric mapping study. Neuroimage. 2001;13:814–824. doi: 10.1006/nimg.2001.0751. [DOI] [PubMed] [Google Scholar]
- Wright IC, McGuire PK, Poline JB, Travere JM, Murray RM, Frith CD, Frackowiak RSJ, Friston KJ. A voxel-based method for the statistical analysis of gray and white matter density applied to schizophrenia. Neuroimage. 1995;2:244–252. doi: 10.1006/nimg.1995.1032. [DOI] [PubMed] [Google Scholar]
- Yan P, Sinusas A, Duncan JS. Boundary element method-based regularization for recovering of LV deformation. Medical Image Analysis. 2007;11:540–554. doi: 10.1016/j.media.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Yang J, Staib LH, Duncan JS. Neighbor-constrained segmentation with 3D deformable models. Information Processing in Medical Imaging, Proceedings. 2003;2732:198–209. doi: 10.1007/978-3-540-45087-0_17. [DOI] [PubMed] [Google Scholar]
- Yao JH, Li J, Summers RM. Employing topographical height map in colonic polyp measurement and false positive reduction. Pattern Recognition. 2009;42:1029–1040. doi: 10.1016/j.patcog.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Dachman AH. CAD techniques, challenges, andcontroversies in computed tomographic colonography. Abdominal Imaging. 2006;30:26–41. doi: 10.1007/s00261-004-0244-x. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Nappi J. Three-dimensional computer-aided diagnosis scheme for detection of colonic polyps. IEEE Trans. Med. Imaging. 2001;20:1261–1274. doi: 10.1109/42.974921. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Goldman S. EM-DD: An improved multiple-instance learning technique, Advances in neural information processing systems. 2001 [Google Scholar]
- Zhang YY, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. Ieee Transactions on Medical Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Jiang Y, Yang YB, Chen SF. Lung cancer cell identification based on artificial neural network ensembles. Artificial Intelligence in Medicine. 2002;24:25–36. doi: 10.1016/s0933-3657(01)00094-x. [DOI] [PubMed] [Google Scholar]
- Zhu X. Semi-supervised learning literature survey. University of Wisconsin; Madison: 2007. [Google Scholar]
- Zhu YM, Cochoff SM. Influence of implementation parameters on registration of MR and SPECT brain images by maximization of mutual information. Journal of Nuclear Medicine. 2002;43:160–166. [PubMed] [Google Scholar]
- Ziyan U, Tuch D, Westin CR. Segmentation of thalamic nuclei from DTI using spectral clustering. Medical Image Computing and Computer-Assisted Intervention - Miccai 2006. 2006;(Pt 2):807–814. doi: 10.1007/11866763_99. 4191. [DOI] [PubMed] [Google Scholar]
- Zweig MH, Campbell G. Receiver-Operating Characteristic (Roc) Plots - a Fundamental Evaluation Tool in Clinical Medicine. Clin Chem. 1993;39:561–577. [PubMed] [Google Scholar]