Summary
The spliceosome is a mega-Dalton ribonucleoprotein (RNP) assembly that processes primary RNA transcripts, producing functional mRNA. The electron microscopy structures of the native spliceosome and of several spliceosomal subcomplexes are available but the spatial arrangement of the latter within the native spliceosome is not known. We designed a new computational procedure to efficiently fit thousands of conformers into the spliceosome envelope. Despite the low resolution limitations, we obtained only one model that complies with the available biochemical data. Our model localizes the five small nuclear RNPs (snRNPs) mostly within the large subunit of the native spliceosome, requiring only minor conformation changes. The remaining free volume presumably accommodates additional spliceosomal components. The constituents of the active core of the spliceosome are juxtaposed, forming a continuous surface deep within the large spliceosomal cavity, which provides a sheltered environment for the splicing reaction.
Introduction
Most nuclear primary transcripts of RNA polymerase II are subject to a set of chemical transformations that make them ready for transport to the cytoplasm as functional mRNAs (Brow, 2002; Burge et al., 1999; Staley and Guthrie, 1998; Tycowski et al., 2006; Will and Luhrmann, 2010). These RNA processing steps include 5’-end capping, 3’-end polyadenylation, editing and splicing, all of which appear to occur within the supraspliceosome – a nuclear mega-Dalton ribonucleoprotein splicing-machine (Sperling et al., 2008). The major components of the spliceosome are five small nuclear ribonucleoprotein complexes (U1, U2, U4, U5 and U6 snRNPs) all of which are essential for functional spliceosomes (Newman and Nagai, 2010; Tycowski et al., 2006). Each of the spliceosomal U snRNPs is composed of the respective U-rich snRNA, the seven Sm proteins (except U6 snRNP which has the LSm proteins) and a set of specific proteins.
The spliceosome is a highly dynamic complex, thus changes in the networks of RNA-RNA, RNA-protein and protein-protein interactions accompany its assembly and catalytic activity. Of note are changes in the inter-snRNP base-pairing network, such as the disruption of U4/U6 pairing and formation of U2/U6 pairing, and changes in base-pairing interactions with the pre-mRNA, e.g. replacement of the pairing of U1 snRNA with the 5’ end of the intron by pairing with U6 snRNA. These changes in the interaction networks are likely to be accompanied by conformational changes; however the remodeling may be accounted for by local structural adjustments and does not necessarily require large-scale modulations in the overall shape and composition of the spliceosome (Brow, 2002; Burge et al., 1999; Staley and Guthrie, 1998; Tycowski et al., 2006).
Because of the huge mass, complexity and dynamic nature of the splicing machine, cryo-electron microscopy (cryo-EM) has been proven as the method of choice for the three-dimensional (3D) structural analyses of the spliceosomal U snRNPs and the complexes they form during the in vitro spliceosome assembly, as well as for determining the structure of the fully assembled native spliceosome (reviewed in Sperling et al., 2008; Stark and Luhrmann, 2006; Will and Luhrmann, 2010). The 3D structure of U1 snRNP was determined by cryo-EM single particle technique at 10 Å resolution (Stark et al., 2001). Further, X-ray crystallographic analysis of the functional core of U1 snRNP, combined with site specific labeling of individual proteins, produced a structure at 5.5 Å resolution (Pomeranz Krummel et al., 2009). The structure of splicing factor 3b (SF3b), a group of seven proteins within the U2 snRNP, was determined by cryo-EM single particle techniques at ~10 Å resolution (Golas et al., 2003). U2 snRNP and U11/U12 di-snRNP, which is part of the minor spliceosome, share the seven SF3b proteins. The Cryo-EM structure of U11/U12 at ~12 Å resolution revealed that SF3b has to undergo major conformational changes in order to be accommodate within U11/U12 di-snRNP (Golas et al., 2005). The U5 snRNP is the largest component of the spliceosome. Its structural analysis at 26–32 Å resolution, performed by cryo-negative staining, revealed a triangular shape (Sander et al., 2006). A similar overall shape was observed for U4/U6.U5 tri-snRNP, which contains the respective snRNAs and 29 distinct proteins. Its structure was determined by cryo-EM at 19–24 Å resolution (Sander et al., 2006). Structural analysis of the U4/U6 di-snRNP at ~40 Å resolution, showed two distinct globular domains connected by a bridge (Sander et al., 2006). The structure of U4 snRNP core, which consists of a heptameric Sm ring and the Sm site heptad (AUUUUUG), was solved by X-ray crystallography at 3.6 Å resolution (Leung et al., 2011).
The 3D structures of a number of complexes, identified as intermediates in the stepwise assembly of the spliceosome in vitro, have also been studied by cryo-EM (Staley and Guthrie, 1998; Will and Luhrmann, 2010). The structure of the BΔU1 complex was resolved at 40 Å resolution (Boehringer et al., 2004), and that of complex C at 30 Å resolution (Jurica et al., 2004). The structure of the yeast complex C was resolved by cryo-EM single particle techniques at a resolution of 29 Å (Ohi et al., 2007). Analysis of negatively stained spliceosomal A complex at 40–50-Å resolution was also performed (Behzadnia et al., 2007).
When isolated from nuclei of mammalian cells, RNA pol II transcripts are found assembled in large 21 MDa RNP complexes - the supraspliceosome. The entire repertoire of nuclear pre-mRNAs, independent of their length or number of introns, appears to be individually assembled in supraspliceosomes (reviewed in Sperling et al., 2008). Supraspliceosomes harbor all five spliceosomal snRNPs, as well as non-snRNP proteins, including regulatory splicing factors such as hnRNP G (Heinrich et al., 2009), and all phosphorylated Ser/Arg-rich-proteins (Yitzhaki et al., 1996), which are essential for splicing and splicing regulation. Supraspliceosomes also harbor the editing enzymes ADAR1 and ADAR2 (Raitskin et al., 2001), cap binding proteins, and components of the 3'-end processing activity (Raitskin et al., 2002).
Structural studies revealed that the supraspliceosome is composed of four substructures that are held together by the pre-mRNA (Azubel et al., 2006; Azubel et al., 2004; Cohen-Krausz et al., 2007; Medalia et al., 2002; Müller et al., 1998; Sperling et al., 1997). Specific cleavage of the pre-mRNA, while keeping the U snRNA components of the supraspliceosome intact, enabled the isolation and purification of the monomeric subcomplexes of the supraspliceosome. These subcomplexes resemble in several parameters the spliceosomes assembled in vitro and were therefore termed native spliceosomes (Azubel et al., 2006; Azubel et al., 2004). Structural studies of the native spliceosome by cryo-EM single particle technique at 20 Å resolution (Azubel et al., 2004) revealed a globular particle made up of two distinct subunits, with the longest dimension of 28 nm. A tunnel is observed in between the two subunits, which is large enough to allow the pre-mRNA to pass through. The other side of the native spliceosome exposes a large cavity that can transiently store the pre-mRNA. It was proposed that the large subunit accommodates the five spliceosomal snRNPs, as the high density regions were confined to this subunit (Azubel et al., 2004).
The size, complexity and dynamic nature of the spliceosome hamper structural studies, and currently the location of the spliceosomal components within the native spliceosome is not known. We therefore used computational methods to fit the available structures of spliceosomal subcomplexes into the envelope of the native spliceosome. Rigid body matching with our previously described FitEM2EM tools (Frankenstein et al., 2008) did not produce acceptable solutions, therefore a new procedure was devised that allowed for conformational flexibility. Gross structural changes were represented by ensembles of normal modes analysis (NMA) conformers (Tama et al., 2002; Tirion, 1996). NMA identifies rigid domains and provides the trajectories of the normal modes; the low frequency trajectories describe the most facile deformations of the structure, which often follow functionally important deformations (Bahar and Rader, 2005; Chacon et al., 2003). Thousands of conformers were tested but only one model complied with the available biochemical data, a remarkable result in view of the low resolution of the structures. Our model shows that the five spliceosomal snRNPs can be simultaneously accommodated within the native spliceosome – mostly within its large subunit – leaving space for the U2-SF3a subcomplex whose structure is not available, and for other spliceosomal components. The free volume also grants tolerance toward local conformation changes, while keeping the integrity of the spliceosome. Moreover, the components proposed to be part of the catalytic core of the functional spliceosome form a continuous surface in the depth of the large spliceosomal cavity, which provides a sheltered environment for the enzymatic reaction.
Results
A model depicting the arrangement of spliceosomal snRNPs within the native spliceosome was constructed by fitting available EM structures of spliceosomal subcomplexes into the envelope of the native spliceosome. The model was constructed in steps; hence the larger subcomplexes, which would fill up most of the volume of the native spliceosome, were matched first, leaving a smaller search volume for the smaller subcomplexes. An efficient "conformer selection" procedure was devised to fit an ensemble of NMA conformers of a given particle into a larger entity that contains it, as described in Methods. Data regarding the subcomplexes used in this study are given in supplementary Table S1.
Fitting U5 snRNP into the U4/U6.U5 tri-snRNP: A test of the "conformer selection" procedure
The experimental EM maps of U4/U6.U5 and U5 snRNPs (Sander et al., 2006) were used to test of the conformer selection scheme. U4/U6.U5 tri-snRNP consists of a globular "head" domain attached to a larger "body" domain. U5 snRNP too consists of head and body domains, separated by a cleft. Initially, the whole U5 snRNP was matched into U4/U6.U5 tri-snRNP, using FitEM2EMIN (Frankenstein et al., 2008). This procedure incorporates thickening and smoothing of the surfaces of the matched objects, controlled by the parameter w, which depends on and can be calculated from the resolution of the EM maps. Matching of U5 into U4/U6.U5 tri-snRNP with w set to 2.4, as dictated by the resolutions of the two EM maps, produced negative scores for all tested poses (Table 1). This result persisted when w was gradually increased to 2.7, yet the scores increased consistently suggesting that a small modification of the overall shape of U5 snRNP may improve the fit into U4/U6.U5 tri-snRNP.
Table 1.
Matching spliceosomal components into the larger entities that contain them
| Spliceosomal components (small & large) |
wa | Top score |
Number of poses with positive scores |
|---|---|---|---|
| U5 & U4/U6.U5 |
2.4 2.5 2.6 2.7 |
−6847 −4415 −2459 −85 |
0 |
| U5 body & U4/U6.U5 | 2.4 | 5147 | 409 |
| U4/U6.U5 & native spliceosome |
2.6 2.7 2.8 2.9 |
−3869 −2669 −1581 −372 |
0 |
| U4/U6.U5 body & native spliceosome |
2.6 2.7 2.8 2.9 |
4022 4326 4742 5063 |
124 164 236 344 |
| BΔU1 (U2.[U4/U6.U5]) & native spliceosome |
1.9 2.0 2.1 2.2 |
−58605 −56093 −53101 −50365 |
0 |
| BΔU1 (U2.[U4/U6.U5]) body & native spliceosome |
1.9 2.0 2.1 2.2 |
−13208 −11704 −10392 −8856 |
0 |
| human C complex (U2/U6.U5) & native spliceosome |
2.2 2.3 2.4 2.5 |
−5699 −3980 −2188 −1011 |
0 |
| Yeast C complex (U2/U6.U5) & native spliceosome |
2.3 2.4 2.5 2.6 |
−54076 −49628 −44491 −39835 |
0 |
| A complex (U1/U2) & spliceosomeΔ[U4.U6/U5] |
1.9 2.0 2.1 2.2 |
−39503 −35103 −30925 −26298 |
0 |
| SF3b & spliceosomeΔ[U4.U6/U5] |
2.9 3.0 3.1 3.2 |
452 1604 2484 3028 |
1 4 8 25 |
| U1 & spliceosomeΔSF3b.[U4.U6/U5] | 2.8 | 3574 2662b |
16068 423b |
| Weighted matching: U1 & spliceosomeΔSF3b.[U4.U6/U5] | 2.8 | 3976 3784b |
16270 2b |
The w predicted from the EM maps resolution is indicated in bold.
Score and rank of the best pose that complies with experimental data (see text).
The conformational flexibility of U5 snRNP was explored using NMA. Snapshots along the trajectories depicted by the four lowest-frequency normal modes (the six rigid body modes were excluded) and their bimodal combinations were collected. This produced 560 conformers of which 181 had no significant clash between the body and head domains and only minor shape distortions within the domains. The, deformation along mode 1 (Figure 1A) generally resembled the experimentally observed flexibility of U5 snRNP (Sander et al., 2006). U5 snRNP was then divided into head and body domains. Matching of the larger U5 body domain into U4/U6.U5 tri-snRNP produced 409 poses with positive scores (Table 1). The 242 matching poses with score exceeding the mean score + 0.5σ (868 score units) were used in the next step. Thus, the body domain of each of the 181 U5 conformers was superposed on each of the 242 poses of the unperturbed U5 body within U4/U6.U5 tri-snRNP, and each of the 181×242 configurations was evaluated with FitEM2EMIN, determining the best translation and the matching score. The configurations with matching scores exceeding the threshold set for the U5 body fitting were optimized and their clustering produced 4 groups (Table 2a).
Figure 1.
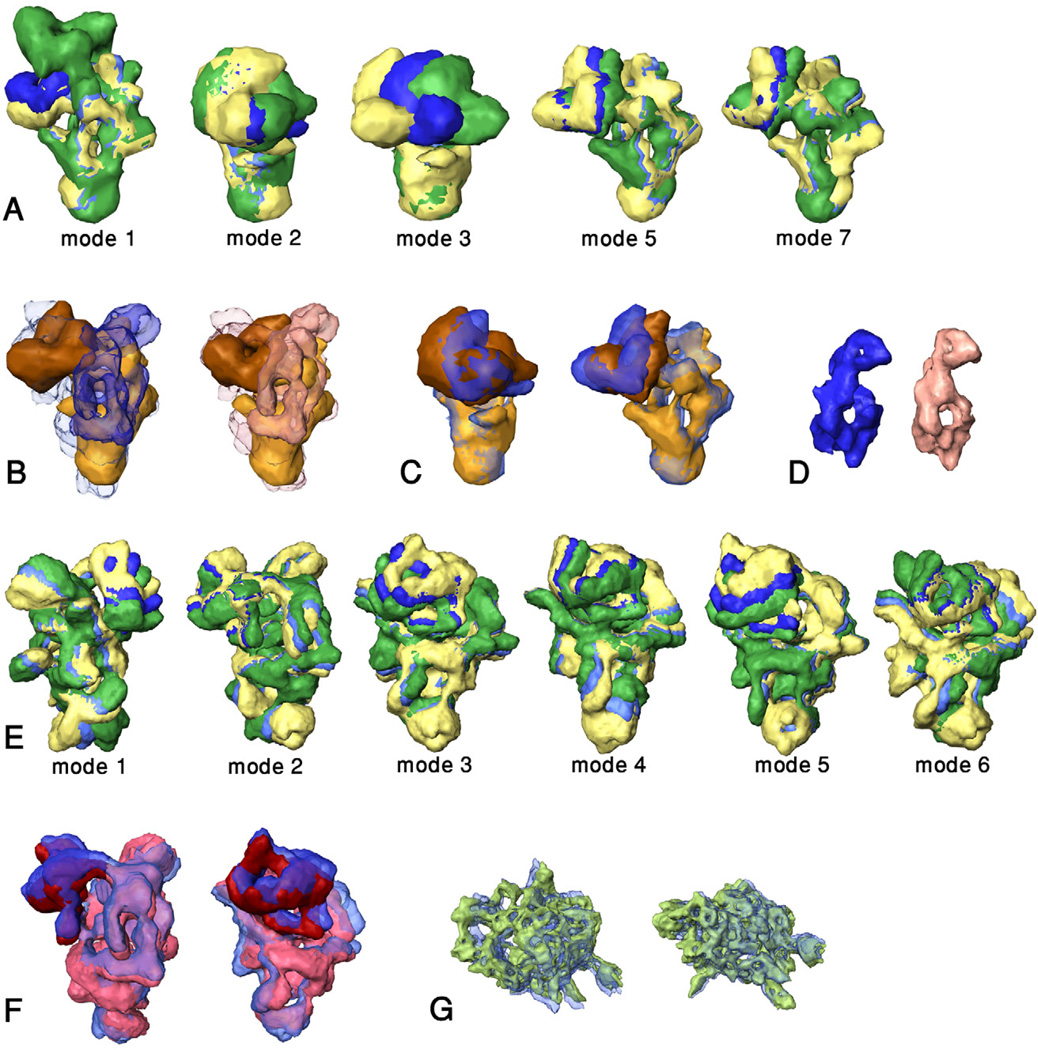
Matching ensembles of NMA conformers of spliceosomal subcomplexes into larger entities that contain them. Only normal modes that contribute to the best matching conformers are shown. The images are rotated to show the largest deformation. (A) Normal modes 1, 2, 3, 5 and 7 for U5 snRNP (the numbering excludes the six rigid body motion modes), that shape the conformers that match best within the free and “bound” U4/U6.U5 tri-snRNP. The unperturbed U5 snRNP is shown in blue. The yellow and green models depict the largest acceptable deformations (see Methods) in opposite directions. (B) The highest scoring match of U5 snRNP (gold body and brown head) within the unperturbed, “free” U4/U6.U5 tri-snRNP (blue; left image) and the “bound” U4/U6.U5 tri-snRNP (pink; right image). U4/U6.U5 tri-snRNP was made transparent in order to show the location of U5 snRNP. Note the empty blue or pink volume within the U4/U6.U5 tri-snRNP, which is attributed to the U4/U6 di-snRNP. (C) Left – comparison of the unperturbed U5 snRNP (transparent; blue with dark blue head) with the conformer that fits best within the free U4/U6.U5 tri-snRNP (gold with brown head). Right – similar comparison for the conformer that fits best within the “bound” U4/U6.U5 tri-snRNP. The structures were rotated to show the largest difference. (D) The U4/U6 di-snRNP density obtained by subtracting the density attributed to U5 snRNP from the density of the free or “bound” U4/U6.U5 tri-snRNP (blue and pink, respectively). (E) Normal modes 1 through 6 for U4/U6.U5 tri-snRNP; coloring as in A. (F) Two views comparing the unperturbed U4/U6.U5 tri-snRNP (transparent; dark blue head and blue body) with the best matching conformer of U4/U6.U5 tri-snRNP within the native spliceosome (dark red head and pink body). (G) Two views comparing the unperturbed SF3b complex (transparent; blue) with the conformer that fits best into native spliceosomeΔ[U4/U6.U5] (green). These images are not to scale with the other images.
Table 2.
Matching NMA conformers
| a) Four groups of conformers of U5 snRNP within U4/U6.U5 tri-snRNP. | ||||
|---|---|---|---|---|
| Conformer information | Body domain pose |
|||
| Mode A (amplitude)a | Mode B (amplitude)a | Matching scoreb | Rank | Score |
| 2 (−0.62) | 3 (+0.12) | 3776 | ||
| 2 (−0.62) | 3 (+0.41) | 3410 | 67 | 2660 |
| 2 (−1.00) | 1 (−0.13) | 2561 | ||
| 2 (−0.62) | 4 (+1.00) | 3479 | ||
| 2 (−0.23) | 4 (+0.29) | 2628 | 1 | 5147 |
| 2 (−0.62) | 4 (+0.29) | 2476 | ||
| 1 (+0.58) | 2 (−0.23) | 2318 | 183 | 1325 |
| 1 (+0.58) | - | 1043 | ||
| 2 (−0.23) | 1 (+0.17) | 2054 | 1 | 5147 |
| 2 (−0.23) | 3 (+0.12) | 1428 | ||
| 1 (+0.58) | 4 (+0.29) | 1158 | 183 | 1325 |
| b) Conformers of U4/U6.U5 tri-snRNP within the native spliceosomec | ||||
|---|---|---|---|---|
| Conformer information | Body domain pose |
|||
| Mode A (amplitude)a | Mode B (amplitude)a | Matching scoreb | Rank | Score |
| 6 (−1.00) | 3 (−0.57) | 3193 | 193 | 1313 |
| 5 (−0.57) | 3143 | |||
| 3 (−0.35) | 3119 | |||
| 5 (−0.35) | 3081 | |||
| c) Conformers of U5 snRNP within “bound” U4.U6/U5 tri-snRNP | ||||
|---|---|---|---|---|
| Conformer information | Body domain pose |
|||
| Mode A (amplitude)a | Mode B (amplitude)a | Matching scoreb | Rank | Score |
| 5 (−1.00) | 7 (−0.38) | 5465 | 67 | 2660 |
| 3 (+0.41) | 3398 | |||
| d) Conformers of U2-SF3b within spliceosomeΔ[U4/U6.U5]d | ||||
|---|---|---|---|---|
| Conformer Information | Unperturbed particle pose |
|||
| Mode A (amplitude)a | Mode B (amplitude)a | Matching scoreb | Rank | Score |
| 1 (+0.13) | 3 (+0.13) | 4270 | 1 | 3674 |
| 4 (−0.25) | 3 (−0.15) | 2611 | 9 | 1017 |
The normal modes numbering excludes the 6 rigid body modes. The amplitude is given as a fraction of the maximal amplitude determined by the number of clashes and level of distortions (see Methods). The direction of the perturbation from the starting geometry is given by + or −.
w was set to the calculated value + 0.3.
The cluster includes 63 configurations. We listed the four configurations with scores exceeding 1185, the limit set for the U4/U6.U5 tri-snRNP body matching.
Only the two configurations that comply with experimental contact data (see text) are listed.
U5 snRNP in the highest scoring configuration fits well into the U4/U6.U5 tri-snRNP envelope (Figure 1B, left). The body domain of U5 is located within the body domain of U4/U6.U5 in all four groups but only for the top-ranking group the head of U5 fits into the head of U4/U6.U5. Notably, a similar position of U5 snRNP within U4/U6.U5 tri-snRNP was obtained in the rigid-body matching of the whole U5 snRNP; it was ranked 3 and scored −1610. The improved match of the best configuration (score 3776) was achieved by a minor modification of the shape of U5 snRNP in which the head rotates with respect to the body (Figure 1C, left). Thus, U5 snRNP is deformed along mode 2 with a small adjustment along mode 3 suggesting that the assembly of U5 snRNP with U4/U6 di-snRNP to form the tri-snRNP does not utilize the experimentally observed deformation for "free" U5 snRNP.
The conformer-selection procedure successfully determined the position and the conformation of U5 snRNP within U4/U6.U5 tri-snRNP. The model is consistent with the previously proposed positioning of U5 snRNP within the tri-snRNP (Sander et al., 2006). The remaining density depicts an elongated particle (Figures 1B, 1D; left) that generally fits the shape and volume of the unbound U4/U6 di-snRNP (Sander et al., 2006).
Applying the conformer selection procedure to position U4/U6.U5 tri-snRNP within the native spliceosome
We first attempted to match the large spliceosomal subcomplexes into the native spliceosome because their large volumes strongly restrict the number of possible matches. This included the U4/U6.U5 tri-snRNP, the in vitro assembled human C-complex U5.U2/U6, the yeast C-complex, and the in vitro assembled BΔU1 (U2.[U4/U6.U5]) (Table S1). Matching any of these particles and even the BΔU1 body domain into the native spliceosome produced negative scores for all tested poses (Table 1). Significantly less negative scores were obtained for the U4/U6.U5 tri-snRNP and the human C-complex when w was increased by 0.3 units, raising the possibility that small shape modifications would improve the fit. However, the use of NMA to estimate the shape deformations of the human C-complex was hindered by the low resolution (30Å) of this structure (Tama et al., 2002).
The ensemble of NMA conformers of U4/U6.U5 tri-snRNP along the 8 lowest frequency normal modes trajectories and their bimodal combinations included 2311 conformations. The deformations depicted by modes 1, 3 and 5 (Figure 1E) show rotation of the head domain away from the body and thus they generally resemble the low-resolution deformation observed for U4/U6.U5 tri-snRNP (Sander et al., 2006). Next, U4/U6.U5 tri-snRNP was divided into head and body domains and the larger body domain was fitted into the native spliceosome, producing 205 matches with scores above the mean score + 0.5σ (1185 score units). Superposition of the body domain in each of the U4/U6.U5 tri-snRNP conformers on each of its body poses within the native spliceosome produced 2311×205 configurations that were re-evaluated by FitEM2EMIN. The score exceeded zero for 63 configurations, which formed a single cluster. All 63 configurations used the same starting pose of the U4/U6.U5 tri-snRNP body within the native spliceosome; the highest scoring ones were deformed along mode 6 with adjustments along mode 3 or 5 (Table 2b).
The U4/U6.U5 tri-snRNP configuration that fitted best within the native spliceosome was only slightly deformed compared to the unperturbed particle, showing small rotation of the head toward the body and minor deformations within the body domain (Figure 1F). A very similar position was obtained in the matching scan for the whole U4/U6.U5 tri-snRNP, where it was ranked 62 and scored −4951. The minor shape modifications shown in Figure 1F facilitated the better match, which scored 3193.
The conformer selection procedure was also used to match U5 snRNP into the fitted U4/U6.U5 tri-snRNP ("bound" U4/U6.U5). The 8 lowest frequency normal modes and their bimodal combinations produced 593 structurally acceptable conformers of U5 snRNP. These conformers were superposed on the 242 rigid body poses of the U5 body within the "free" U4/U6.U5 (the body domains of the free and bound U4/U6.U5 tri snRNPs are very similar). The U5 snRNP configurations that matched well into the bound U4/U6.U5 were deformed along mode 5 with minor adjustment along mode 3 or 7 (Table 2c) and formed a single cluster. The score of the best configuration was significantly higher than the score of the best match within the free U4/U6.U5 tri-snRNP, 5465 versus 3776, possibly because only four normal modes were considered in the aforementioned conformer selection test.
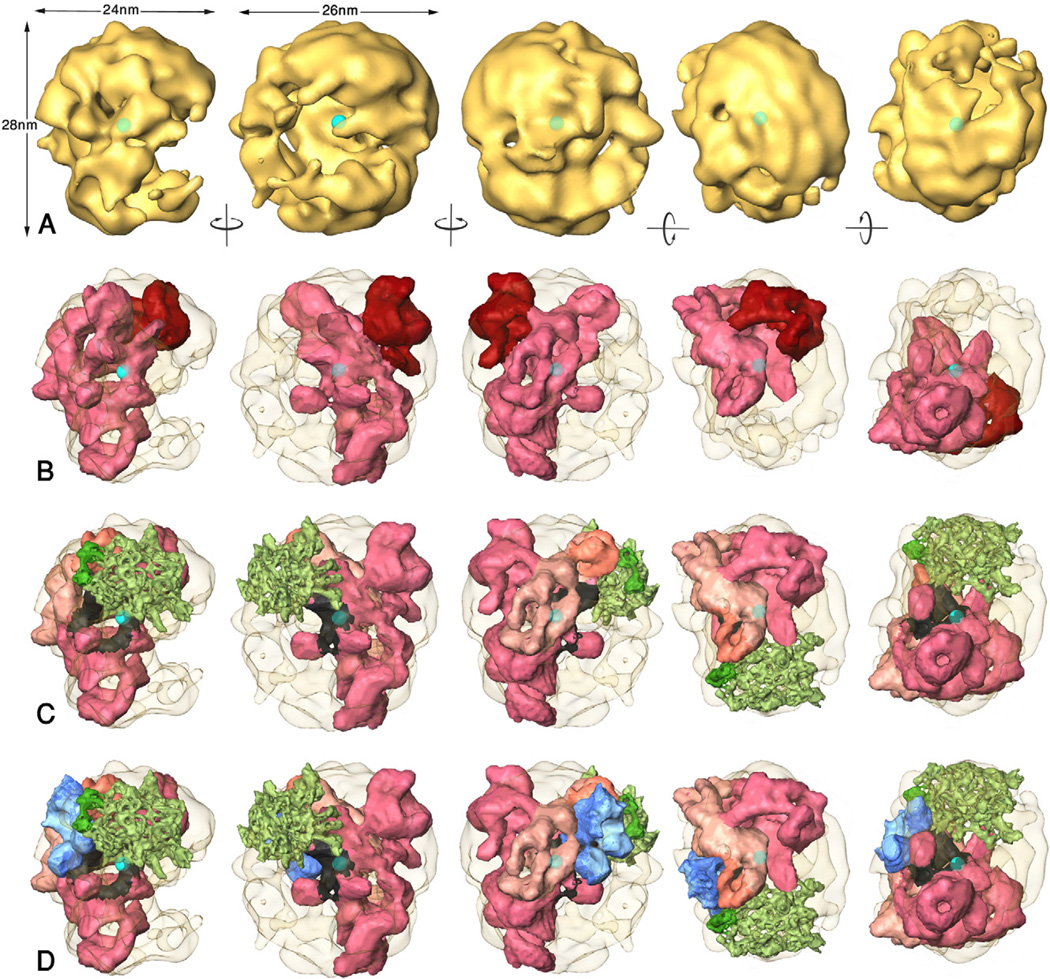
Figure 2A shows several views of the native spliceosome (Azubel et al., 2004). The predicted position of the U4/U6.U5 tri-snRNP within the native spliceosome is shown in Figure 2B, in which the tri-snRNP is colored by domains, and in Figure 2C, in which functional regions of the tri-snRNP are indicated. While the U5 head domain and the U4/U6 units are located in the large subunit of the spliceosome, adjusted to the outer surface, the U5 body domain penetrates into the small subunit and adjusts to the surface of the large cavity. Combining the predicted positions of U4/U6.U5 tri-snRNP within the native spliceosome and of U5 snRNP within the bound U4/U6.U5, with the previously proposed position of U5 snRNA loop I (Sander et al., 2006), we find that this loop is located at the surface of the deepest pocket of the native spliceosome cavity (Figure 2C). Since loop I of the U5 snRNA aligns the exons to be spliced (McConnell and Steitz, 2001), its location marks the site of the functional core of the spliceosome.
Figure 2.
Stepwise modeling of the arrangement of spliceosomal subcomplexes within the native spliceosome. The indicated rotations are of ~90° with respect to the leftmost view. The five views are, from left to right: Side view that depicts the large and small subunits and the large spliceosomal cavity, view into the cavity, “back view” of the spliceosome, “top view” at the large subunit and “bottom view” at the small subunit. The centroid of the spliceosome is indicated by the cyan sphere. (A) The native spliceosome. (B) Positioning of U4/U6.U5 tri-snRNP within the native spliceosome. The native spliceosome is transparent. The U4/U6.U5 tri-snRNP is colored by domains, dark red head and pink body. (C) Positioning of U4/U6.U5 tri-snRNP and U2-SF3b within the native spliceosome. The U4/U6.U5 tri-snRNP is colored by functional regions: The U5 snRNP is shown in pink with the region attributed to loop I (Sander et al., 2006) emphasized in black; the density attributed to U4/U6 di-snRNP is shown in beige with the region attributed to the 3’end of the U4-snRNA emphasized in orange (Hacker et al., 2008). SF3b is shown in green with the region attributed to SF3b49 indicated in dark green. (D) Positioning the five U snRNPs within the native spliceosome. The coloring of U4/U6.U5 tri-snRNP and SF3b is as in C. U1 snRNP is shown in blue with the region that is found near U2 (Donmez et al., 2007), including the U1 snRNA 5’-end, indicated in darker blue.
Positioning the U2-SF3b subcomplex within the native spliceosome
The remaining volume of the native spliceosome, represented by the difference map between the native spliceosome and the bound U4/U6.U5 tri-snRNP (spliceosomeΔ[U4/U6.U5]), is expected to accommodate the U1 and U2 snRNPs and additional splicing factors and RNA processing components (Sperling et al., 2008). EM structures are available for the very small U1 snRNP, the significantly larger SF3b subcomplex of U2 snRNP and the A-complex, harboring U1/U2 di-snRNP and additional proteins (Table S1). Matching the low resolution (40–50 Å) structure of the A complex into the spliceosomeΔ[U4/U6.U5] density, employing several settings of w, produced large negative scores for all tested poses (Table 1). Hence, the A complex could not be fitted into the native spliceosome together with the U4/U6.U5 tri-snRNP.
The SF3b subcomplex matched well into the spliceosomeΔ[U4/U6.U5] density. The 25 matches with positive scores were optimized and clustered, producing 10 groups. In every case SF3b was located within the large subunit of the native spliceosome in approximately the same location; however, in only two groups (ranked 1 and 9) the orientation complied with the previously described contact information from human and yeast studies, according to which the 5’ end of the U2 snRNA, near protein SF3b49 (Dybkov et al., 2006), should point towards the U4/U6.U5 tri-snRNP (Igel et al., 1998; Madhani and Guthrie, 1992; Sun and Manley, 1995). The structure of SF3b is of relatively high resolution and quite rigid (Golas et al., 2003; Kramer et al., 1999) yet some structural changes were detected in raw 2D images (Kramer et al., 1999). We therefore checked if introducing conformational flexibility would improve the match to the spliceosomeΔ[U4/U6.U5] density. The 4 lowest frequency normal modes, in which the central body of the particle was not deformed, and their bimodal combinations were used to produce an ensemble of 1938 conformers. Each of these conformers was superposed on either of the two biologically relevant rigid body matches and re-scored. The best matching configurations were only slightly deformed along modes 3 and 1 or 3 and 4 (Table 2d and Figure 1G). Final optimization of the positions of the emerging U2-SF3b.[U4/U6.U5] complexes within the native spliceosome produced scores of 5022 and 5505, for the initial SF3b rigid-body poses 1 and 9, respectively.
A high scoring match of SF3b within the spliceosomeΔ[U4/U6.U5] density does not ascertain good shape complementarity with U4/U6.U5 tri-snRNP; it may reflect good shape complementarity in other regions. Therefore the surface complementarity between SF3b and the bound U4/U6.U5 conformer was tested by docking the two structures with FitEM2EMOUT (w was set to 3.0), starting from the matching models 1 and 9. The docking test clearly distinguished between the two interfaces: while the surface complementarity for model 1 was weak and SF3b shifted away from this starting position, the surface complementarity for model 9 was good with a score of 1980. Notably, refinement of the position of model 9 complex within the native spliceosome produced a score of 5505, larger than the score for U4/U6.U5 tri-snRNP alone, indicating that SF3b contributes to the formation of the surface of the spliceosome. According to this model SF3b is located within the large subunit, adjusted to the surface of the main cavity (Figure 2C), and protein SF3b49 (dark green), which resides next to the 5' end of U2 snRNA, makes contact with the upper portion of U4/U6.
Positioning the U1 snRNP within the native spliceosome
The EM structure of the complete U1 snRNP (Stark et al., 2001) reveals a ring-shaped main body with several protruding density elements. Although a higher resolution crystal structure of U1 snRNP is available (Pomeranz Krummel et al., 2009), we chose to use the EM structure in this study, because the crystal structure lacks the apical region of stem-loop 2 of the U1 snRNA and the U1 A protein, which form part of the U1 surface that is near the U2 snRNP (Donmez et al., 2007).
Matching U1 snRNP into the difference map obtained by subtracting the model of U2-SF3b.[U4/U6.U5] from the native spliceosome (spliceosomeΔU2-SF3b.[U4/U6.U5]) produced over 16000 matches with positive scores (Table 1). This result is expected in view of the small size of U1 snRNP and the numerous other components that are still missing. Therefore, only in this case, we used a weighted matching scan, which is biased according to external data (Ben-Zeev and Eisenstein, 2003). Thus, contacts via surface regions of U2-SF3b.[U4/U6.U5] implicated in U1 snRNP binding were rewarded. This included the surface region attributed to the U5 snRNA loop I and the region attributed to SF3b49, which were shown to be near the 5' end of the U1 snRNA(Ast and Weiner, 1997; Donmez et al., 2007; Malca et al., 2003) (indicated in Figure 2D). Similar contacts were also detected in yeast (McGrail and O'Keefe, 2008), and in view of the sequence conservation with humans (Jurica and Moore, 2003) they provide additional substantiation of the contact data. The weighted matching scan produced only one group of 3 matches that complied with the contact information; the best match was ranked 2. The same match was detected in the non-weighted scan ranking 423, hence the weighting only improved the ranking of the good match, facilitating its detection. The model was further examined by evaluating the surface complementarity between U1 snRNP and the U2-SF3b.[U4/U6.U5] complex using the FitEM2EMOUT tool. A good docking model was obtained, ranking 110 with score 2508.
The matched U1 snRNP is located in the large subunit of the native spliceosome, adjusted to the surface of the large cavity. The face that exposes the 5' end of the U1 snRNA is located near SF3b49, U5 snRNA loop I, and the U4/U6 di-snRNP. Importantly, a region of unaccounted for density is found within the large subunit, near the SF3b complex, which can be attributed to U2ΔSF3b; it can also represent large conformational changes in the structure of SF3b.
Discussion
The currently available structural data pertaining the RNA splicing machine include X-ray structures of a number of spliceosomal proteins and small subcomplexes, and EM structures of the native spliceosome, spliceosomal snRNPs, and of several in vitro assembled spliceosomal subcomplexes (reviewed in Newman and Nagai, 2010; Sperling et al., 2008; Will and Luhrmann, 2010). The 3D structures of U4/U6.U5 tri-snRNP, U2-SF3b and U1 snRNP, resolved at resolutions of 10–24 Å, combined with biochemical data regarding the interactions between them were used in this study to model the spatial arrangement of the snRNPs within the native spliceosome. The stepwise construction of our model allowed for gross deformations of the subcomplexes and involved testing of almost a million of different configurations. Despite the low resolutions of the structures used, we found that only one model complies with the biochemical data. Moreover, only minor shape modifications were necessary in order to position all five spliceosomal snRNPs within the native spliceosome. This result can be attributed to restrictions imposed by the shape of the native spliceosome, e.g. the occurrence of a large inter-subunit cavity.
In our model, the five spliceosomal snRNPs mostly occupy the large subunit of the native spliceosome; only the body domain of U5 snRNP penetrates into the small subunit. This result is in accordance with the observation that the highest-density region, attributed to RNA, is located in the large subunit (Azubel et al., 2004). Yet, density data were not considered in our matching procedure, which relies only on the shapes of the objects.
The estimated mass of the five spliceosomal snRNPs amounts to ~70% of the measured mass (4.8 MDa) of the native spliceosome (Müller et al., 1998). In our model, the collective volume of the U4/U6.U5 tri-snRNP, U2-SF3b and U1 snRNP is ~45% of the volume of the native spliceosome. The accuracy of this estimate is limited by the significant differences in the resolutions of the structures. Nevertheless, it appears that substantial percentage of the volume of the native spliceosome is free. Some of this free volume accommodates the U2-SF3a and additional spliceosomal components. Yet, the existence of unassigned density regions near the snRNPs suggests that structural modulations of the snRNPs can be tolerated while keeping the integrity of the spliceosome assembly.
U5 loop I, which aligns the exons to be spliced and thus is part of the catalytic core of the spliceosome, is exposed at the surface of the deepest and most sheltered pocket within the large cavity of the native spliceosome. It is located near the centroid of the spliceosome (indicated in Figure 2) as was found common for the active sites of enzymes (Ben-Shimon and Eisenstein, 2005). Moreover, the small U1 snRNP is positioned near loop I of U5 snRNA, further supporting the notion that the large cavity is the pre-mRNA binding and processing site. In the deep pocket within the cavity the catalytic process is sheltered from the surrounds. In the supraspliceosome the openings of the large cavities of the native spliceosomes are exposed and the inter-particle contacts are maintained by the small subunits (Cohen-Krausz et al., 2007), thus facilitating access of the pre-mRNA into the cavity and allowing structural remodeling within the large subunits.
Although we used NMA to represent the structural flexibility for U5 snRNP, U4/U6.U5 tri-snRNP and SF3b, our final model is static, depicting a single arrangement of the spliceosomal snRNPs, as would an X-ray structure do. We find that the 5’ end of U1 snRNA, which binds to the intronic 5’ splice site in the early stages of the splicing reaction, is at the surface of the spliceosomal cavity, as is U5 snRNA loop I. The predicted arrangement shares some features with the in vitro assembled B-complex (Will and Luhrmann, 2010). Thus, the 5’ end of U2 snRNA, which is located near protein SF3b49, is positioned in our model near the region attributed to the 5’ end of U4 snRNA and therefore also near the segment of U6 snRNA that shifts contacts to U2 snRNA. Another segment of the U2 snRNA, between nucleotides 30 and 40, was shown to bind to the intron branch site together with protein p14 (Golas et al., 2003), requiring an open conformation of SF3b that exposes p14. Our model includes the closed form of SF3b with one valve adjusted to U4/U6.U5 tri-snRNP. The other valve can move to occupy an unassigned nearby density region and expose the density protrusion assigned to p14 to the spliceosomal cavity.
For splicing complexes assembled in-vitro, the modulation of RNA-RNA interactions in the activated B-complex, as compared with the B-complex, is accompanied with changes in composition. Thus, the U6 snRNA segment that binds to U4 snRNA in the B-complex, binds near the 5’ end of U2 snRNA in the activated B-complex, leading to release of U4 snRNP (Will and Luhrmann, 2010). However, U4 snRNP was found associated with catalytically active native spliceosomes and supraspliceosomes (Azubel et al., 2006; Sperling et al., 2008), indicating that the significant conformation changes in the U2 and U6 snRNAs, and possibly U4 snRNA, which destabilize the base-pairing interactions of U4/U6 snRNAs, do not require release of U4 snRNP from the spliceosome. The above-mentioned structural changes in the U6 snRNA also facilitate its interaction with the 5’ end of the intron; U1 snRNA at this stage would be only weakly associated with the rest of the spliceosome, and in vitro it is released (Will and Luhrmann, 2010). We find that U1 snRNP is close to the outer surface of the spliceosome and it may be released; yet, like U4 snRNP, U1 snRNP was found to be part of the active native spliceosome and supraspliceosome (Azubel et al., 2006; Sperling et al., 2008). The resolution of our model is limited; for example, we cannot distinguish between the in vitro B-complex and activated B-complex formed by rearrangement of the RNA-RNA contacts. The accuracy of the model is affected mostly by the resolution of the EM maps and of the experimental contact data. For example, SF3b has an approximately circular center with many small protrusions; its position within the spliceosome is similar in all our models hence its orientation is determined by the position of SF3b49. In contrast, the translational grid interval and the clustering limit (13Å) are very fine relative to the size of the spliceosome and are not likely to affect the accuracy. Experiments for direct localization of U snRNP components within the native spliceosome are planned for validation of the model.
In summary, although we tested thousands of conformers and millions of possible configurations, and despite the low resolution of the structures, we obtained a single biologically relevant model of the arrangement of the five U snRNPs within the native spliceosome. Based on the model presented here, we can conclude that the native spliceosome can accommodate all five snRNPs, leaving free volume for the additional spliceosomal components, and allowing for local conformation changes. Furthermore, the functional domains of the snRNPs form a continuous region that lines the surface of the deepest part of the large spliceosomal cavity, thereby providing a sheltered environment for the pre-mRNA binding and for the splicing reaction.
Methods
A stepwise model building approach was used in this study, starting with the largest substructures, which would fill up most of the volume of the native spliceosome and limit the search volume for the smaller substructures. In each step the position of the assembled complex within the native spliceosome was refined and, where possible, the growing complex was validated by docking of its components. Furthermore, in each step the inter-component interactions were tested against the available experimental findings.
Converting EM maps into 3D objects
EM maps were downloaded from the EM database (Henrick et al., 2003) or kindly provided by the research groups. The maps were converted into lists of virtual atoms as previously described (Frankenstein et al., 2008). The virtual atoms lists could be split into domains to be used in the FitEM2EM matching and docking tools or converted into an elastic network model to be used by the NMA tool. The electron density thresholds were determined manually to match the published structural features.
Rigid body matching and docking
Matching and docking were performed with our in-house tools FitEM2EMIN and FitEM2EMOUT, which match and dock low resolution structures using their shape attributes (Frankenstein et al., 2008). These tools evaluate the surface similarity between two EM objects by calculating correlation scores. Thus, higher positive scores indicate more extensive overlap of the surfaces and little or no protrusions. The scores have arbitrary units and depend on the grid interval used in the scan; hence, only scans with the same grid interval can be significantly compared (Kowalsman and Eisenstein, 2007). The surface thickening and smoothing feature in FitEM2EM is controlled by the surface extension parameter w, which is measured in units of grid interval, and whose value can be predicted from the resolutions of the EM maps (Frankenstein et al., 2008). Yet, w settings within 0.3 units of the predicted value confer distinction between correct and false models, and therefore several values of w, within the 0.3 units limit, were used in this study. The predicted w + 0.3 were consistently used in the matching of normal modes conformers (see below). The translational grid interval was based on the grid interval of the experimental EM maps and the rotational grid interval was set to 12° (Frankenstein et al., 2008).
Weighted matching
Weighted matching scans were conducted only for the small U1 snRNP. Weighted scans reward interface contacts that were detected by other means, thereby increasing the fraction of solutions that are in accord with the external data, and elevating their ranks (Ben-Zeev and Eisenstein, 2003). The extra weight given to the relevant surface grid points was set to 0.5, thus contributing 1.5 to the matching score per overlapping grid point instead of 1. The weight parameter was determined empirically; it was made large enough to significantly increase the score of likely models without interpenetration of the docked objects.
Refinement and clustering
In each modeling step the acceptable matches were refined (Eisenstein et al., 1997). Starting from the predicted pose, small rotations about the x, y and z axes were performed in steps of ±3° up to a limit of ±12°. Refined poses were clustered based on the root mean square deviation (RMSD) between the models. A cluster contained models with pairwise RMSDs below 13Å, a limit determined by comparing sets of NMA models, as described below. Each cluster was represented by the highest scoring model within it.
Stepwise construction of a multi-component complex
The DifferenceGrid tool calculates the difference between the grid representations of the matched objects and provides an estimate of the remaining free volume (Frankenstein et al., 2008). Matching another substructure into the difference map produces models of a complex that consists of the previously matched (and subtracted) substructure and the one matched to the difference map. For example, matching SF3b into the difference map between the native spliceosome and the U4/U6.U5 tri-snRNP produced models of the U2-SF3b.[U4/U6.U5] complex. The surface complementarity between the components in the new complex was further evaluated by docking with the FitEM2EMOUT tool, testing small angular deviations from the predicted matching pose, in steps of 3°, up to a limit of ±12°. Translational deviations up to 13Å from the starting pose were accepted.
Representing gross structural modifications
We used the ElNemo web server (Suhre and Sanejouand, 2004) to obtain conformational snapshots. The elastic network model depends on a preset cutoff distance between atoms, or virtual atoms as in our study, and suitable cutoffs were determined for different EM map resolutions (Tama et al., 2002). We tested cutoff distances within these ranges, just above the range or just below it, and found that similar ensembles of conformers were produced with RMSD below 7Å between corresponding conformers.
The "conformer selection" scheme: Combining gross structural modifications with rigid body matching
Matching of all the NMA conformers to the native spliceosome using exhaustive 3D FitEM2EMIN scans is computationally impractical and therefore a new procedure was devised that consisted of rigid body matching of the largest rigid domain to the target entity, superposition of the ensemble of NMA conformers on each of the high-scoring rigid-body matching poses and evaluation of the shape complementarity to the target entity. This fast procedure consists of the following main steps:
Creating ensembles of conformers using NMA. The NMA calculations provide the directions of motions but not their amplitudes. We therefore selected several snapshots along the trajectory of each mode or bimode, applying incremental perturbations until sterical clashes between the mobile domains were observed or their structures became significantly distorted. Distortions were detected by computing the RMSD between the individual domains in each snapshot and in the unperturbed structure. Snapshots with RMSD ≤ 13Å were retained as they preserved the domains shapes. This RMSD limit was also used in model clustering. Sterical clashes between the mobile domains were detected using the DifferenceGrid tool. Snapshots with domain overlap below 1% of the volume of the smaller domain were accepted.
Dividing subcomplexes into rigid domains. U5 snRNP, U4/U6.U5 tri-snRNP, and BΔU1 consist each of two large domains, whose observed relative rigid body motions (Boehringer et al., 2004; Sander et al., 2006) are supported by our NMA results. These subcomplexes were divided into "head" and "body" domains, which were assumed to be rigid in the subsequent computations. In every case the body domain was considerably larger than the head domain.
Matching of the body domain. Several rigid body matching scans of the body domain into the larger entity were performed using different w settings. The distributions of scores followed an extreme value distribution (Levitt and Gerstein, 1998) allowing estimate of the mean score (μ) and the standard deviation (σ). Only matches with score exceeding μ+0.5σ were considered in the next step. In the SF3b case the whole particle was used for rigid body matching. Only a small number of poses with positive score were obtained and all were considered in the next step.
Selecting the best matching conformers. Each conformer (NMA snapshot from step 2) was placed within the larger entity by superposing its body domain onto each of the matching poses produced for the unperturbed body domain in step 3. In the SF3b case, the whole particle was used for superposition. The complementarity between each superposed conformer and the larger entity was evaluated with FitEM2EMIN, which also provided the best relative translation.
Supplementary Material
Highlights.
We present a computational procedure for fast matching of thousands of conformers.
We obtained a unique model localizing the snRNPs within the native spliceosome.
The 5 snRNPs are accommodated mostly within the large subunit of the spliceosome.
The large spliceosomal cavity emerges as the site of mRNA binding and splicing.
Acknowledgements
We thank the US NIH, grant GM079549 to R.S. and J.S., and the Helen and Milton Kimmelman Center for Biomolecular Structure and Assembly at the Weizmann Institute of Science (J.S. and M.E.) for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ast G, Weiner AM. A novel U1/U5 interaction indicates proximity between U1 and U5 snRNAs during an early step of mRNA splicing. Rna. 1997;3:371–381. [PMC free article] [PubMed] [Google Scholar]
- Azubel M, Habib N, Sperling J, Sperling R. Native spliceosomes assemble with pre-mRNA to form supraspliceosomes. J Mol Biol. 2006;356:955–966. doi: 10.1016/j.jmb.2005.11.078. [DOI] [PubMed] [Google Scholar]
- Azubel M, Wolf SG, Sperling J, Sperling R. Three-dimensional structure of the native spliceosome by cryo-electron microscopy. Mol Cell. 2004;15:833–839. doi: 10.1016/j.molcel.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Bahar I, Rader AJ. Coarse-grained normal mode analysis in structural biology. Curr Opin Struct Biol. 2005;15:586–592. doi: 10.1016/j.sbi.2005.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J, Dube P, Will CL, Urlaub H, Stark H, et al. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. Embo J. 2007;26:1737–1748. doi: 10.1038/sj.emboj.7601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shimon A, Eisenstein M. Looking at enzymes from the inside out: the proximity of catalytic residues to the molecular centroid can be used for detection of active sites and enzyme-ligand interfaces. J Mol Biol. 2005;351:309–326. doi: 10.1016/j.jmb.2005.06.047. [DOI] [PubMed] [Google Scholar]
- Ben-Zeev E, Eisenstein M. Weighted geometric docking: incorporating external information in the rotation-translation scan. Proteins. 2003;52:24–27. doi: 10.1002/prot.10391. [DOI] [PubMed] [Google Scholar]
- Boehringer D, Makarov EM, Sander B, Makarova OV, Kastner B, Luhrmann R, Stark H. Three-dimensional structure of a pre-catalytic human spliceosomal complex B. Nat Struct Mol Biol. 2004;11:463–468. doi: 10.1038/nsmb761. [DOI] [PubMed] [Google Scholar]
- Brow DA. Allosteric cascade of spliceosome activation. Annu Rev Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- Burge CB, Tuschl TH, Sharp PA. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. second edition. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- Chacon P, Tama F, Wriggers W. Mega-Dalton biomolecular motion captured from electron microscopy reconstructions. J Mol Biol. 2003;326:485–492. doi: 10.1016/s0022-2836(02)01426-2. [DOI] [PubMed] [Google Scholar]
- Cohen-Krausz S, Sperling R, Sperling J. Exploring the architecture of the intact supraspliceosome using electron microscopy. J Mol Biol. 2007;368:319–327. doi: 10.1016/j.jmb.2007.01.090. [DOI] [PubMed] [Google Scholar]
- Donmez G, Hartmuth K, Kastner B, Will CL, Luhrmann R. The 5' end of U2 snRNA is in close proximity to U1 and functional sites of the pre-mRNA in early spliceosomal complexes. Mol Cell. 2007;25:399–411. doi: 10.1016/j.molcel.2006.12.019. [DOI] [PubMed] [Google Scholar]
- Dybkov O, Will CL, Deckert J, Behzadnia N, Hartmuth K, Luhrmann R. U2 snRNA-protein contacts in purified human 17S U2 snRNPs and in spliceosomal A and B complexes. Mol Cell Biol. 2006;26:2803–2816. doi: 10.1128/MCB.26.7.2803-2816.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein M, Shariv I, Koren G, Friesem AA, Katchalski-Katzir E. Modeling supra-molecular helices: extension of the molecular surface recognition algorithm and application to the protein coat of the tobacco mosaic virus. J Mol Biol. 1997;266:135–143. doi: 10.1006/jmbi.1996.0773. [DOI] [PubMed] [Google Scholar]
- Frankenstein Z, Sperling J, Sperling R, Eisenstein M. FitEM2EM--tools for low resolution study of macromolecular assembly and dynamics. PLoS One. 2008;3:e3594. doi: 10.1371/journal.pone.0003594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golas MM, Sander B, Will CL, Luhrmann R, Stark H. Molecular architecture of the multiprotein splicing factor SF3b. Science. 2003;300:980–984. doi: 10.1126/science.1084155. [DOI] [PubMed] [Google Scholar]
- Golas MM, Sander B, Will CL, Luhrmann R, Stark H. Major conformational change in the complex SF3b upon integration into the spliceosomal U11/U12 di-snRNP as revealed by electron cryomicroscopy. Mol Cell. 2005;17:869–883. doi: 10.1016/j.molcel.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Hacker I, Sander B, Golas MM, Wolf E, Karagoz E, Kastner B, Stark H, Fabrizio P, Luhrmann R. Localization of Prp8, Brr2, Snu114 and U4/U6 proteins in the yeast tri-snRNP by electron microscopy. Nat Struct Mol Biol. 2008;15:1206–1212. doi: 10.1038/nsmb.1506. [DOI] [PubMed] [Google Scholar]
- Heinrich B, Zhang Z, Raitskin O, Hiller M, Benderska N, Hartmann AM, Bracco L, Elliott D, Ben-Ari S, Soreq H, et al. Heterogeneous nuclear ribonucleoprotein G regulates splice site selection by binding to CC(A/C)-rich regions in pre-mRNA. J Biol Chem. 2009;284:14303–14315. doi: 10.1074/jbc.M901026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrick K, Newman R, Tagari M, Chagoyen M. EMDep: a web-based system for the deposition and validation of high-resolution electron microscopy macromolecular structural information. J Struct Biol. 2003;144:228–237. doi: 10.1016/j.jsb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Igel H, Wells S, Perriman R, Ares M., Jr Conservation of structure and subunit interactions in yeast homologues of splicing factor 3b (SF3b) subunits. Rna. 1998;4:1–10. [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Jurica MS, Sousa D, Moore MJ, Grigorieff N. Three-dimensional structure of C complex spliceosomes by electron microscopy. Nat Struct Mol Biol. 2004;11:265–269. doi: 10.1038/nsmb728. [DOI] [PubMed] [Google Scholar]
- Kowalsman N, Eisenstein M. Inherent limitations in protein-protein docking procedures. Bioinformatics. 2007;23:421–426. doi: 10.1093/bioinformatics/btl524. [DOI] [PubMed] [Google Scholar]
- Kramer A, Gruter P, Groning K, Kastner B. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J Cell Biol. 1999;145:1355–1368. doi: 10.1083/jcb.145.7.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Nagai K, Li J. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature. 2011;473:536–539. doi: 10.1038/nature09956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M, Gerstein M. A unified statistical framework for sequence comparison and structure comparison. Proc Natl Acad Sci U S A. 1998;95:5913–5920. doi: 10.1073/pnas.95.11.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- Malca H, Shomron N, Ast G. The U1 snRNP base pairs with the 5' splice site within a penta-snRNP complex. Mol Cell Biol. 2003;23:3442–3455. doi: 10.1128/MCB.23.10.3442-3455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell TS, Steitz JA. Proximity of the invariant loop of U5 snRNA to the second intron residue during pre-mRNA splicing. Embo J. 2001;20:3577–3586. doi: 10.1093/emboj/20.13.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrail JC, O'Keefe RT. The U1, U2 and U5 snRNAs crosslink to the 5' exon during yeast pre-mRNA splicing. Nucleic Acids Res. 2008;36:814–825. doi: 10.1093/nar/gkm1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medalia O, Typke D, Hegerl R, Angenitzki M, Sperling J, Sperling R. Cryoelectron microscopy and cryoelectron tomography of the nuclear pre-mRNA processing machine. J Struct Biol. 2002;138:74–84. doi: 10.1016/s1047-8477(02)00027-8. [DOI] [PubMed] [Google Scholar]
- Müller S, Wolpensinger B, Angenitzki M, Engel A, Sperling J, Sperling R. A supraspliceosome model for large nuclear ribonucleoprotein particles based on mass determinations by scanning transmission electron microscopy. J Mol Biol. 1998;283:383–394. doi: 10.1006/jmbi.1998.2078. [DOI] [PubMed] [Google Scholar]
- Newman AJ, Nagai K. Structural studies of the spliceosome: blind men and an elephant. Curr Opin Struct Biol. 2010;20:82–89. doi: 10.1016/j.sbi.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Ohi MD, Ren L, Wall JS, Gould KL, Walz T. Structural characterization of the fission yeast U5.U2/U6 spliceosome complex. Proc Natl Acad Sci U S A. 2007;104:3195–3200. doi: 10.1073/pnas.0611591104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raitskin O, Angenitzki M, Sperling J, Sperling R. Large nuclear RNP particles--the nuclear pre-mRNA processing machine. J Struct Biol. 2002;140:123–130. doi: 10.1016/s1047-8477(02)00541-5. [DOI] [PubMed] [Google Scholar]
- Raitskin O, Cho DS, Sperling J, Nishikura K, Sperling R. RNA editing activity is associated with splicing factors in lnRNP particles: The nuclear pre-mRNA processing machinery. Proc Natl Acad Sci USA. 2001;98:6571–6576. doi: 10.1073/pnas.111153798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander B, Golas MM, Makarov EM, Brahms H, Kastner B, Luhrmann R, Stark H. Organization of core spliceosomal components U5 snRNA loop I and U4/U6 Di-snRNP within U4/U6.U5 Tri-snRNP as revealed by electron cryomicroscopy. Mol Cell. 2006;24:267–278. doi: 10.1016/j.molcel.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Sperling J, Azubel M, Sperling R. Structure and function of the Pre-mRNA splicing machine. Structure. 2008;16:1605–1615. doi: 10.1016/j.str.2008.08.011. [DOI] [PubMed] [Google Scholar]
- Sperling R, Koster AJ, Melamed-Bessudo C, Rubinstein A, Angenitzki M, Berkovitch-Yellin Z, Sperling J. Three-dimensional image reconstruction of large nuclear RNP (lnRNP) particles by automated electron tomography. J Mol Biol. 1997;267:570–583. doi: 10.1006/jmbi.1997.0898. [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Stark H, Dube P, Luhrmann R, Kastner B. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature. 2001;409:539–542. doi: 10.1038/35054102. [DOI] [PubMed] [Google Scholar]
- Stark H, Luhrmann R. Cryo-electron microscopy of spliceosomal components. Annu Rev Biophys Biomol Struct. 2006;35:435–457. doi: 10.1146/annurev.biophys.35.040405.101953. [DOI] [PubMed] [Google Scholar]
- Suhre K, Sanejouand YH. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004;32:W610–W614. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JS, Manley JL. A novel U2-U6 snRNA structure is necessary for mammalian mRNA splicing. Genes Dev. 1995;9:843–854. doi: 10.1101/gad.9.7.843. [DOI] [PubMed] [Google Scholar]
- Tama F, Wriggers W, Brooks CL., 3rd Exploring global distortions of biological macromolecules and assemblies from low-resolution structural information and elastic network theory. J Mol Biol. 2002;321:297–305. doi: 10.1016/s0022-2836(02)00627-7. [DOI] [PubMed] [Google Scholar]
- Tirion MM. Large Amplitude Elastic Motions in Proteins from a Single-Parameter, Atomic Analysis. Phys Rev Lett. 1996;77:1905–1908. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Kolev NG, Conard NK, Fok V, Steitz JA. The ever-growing world of small nuclear ribonucleoproteins. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. third edition. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2006. pp. 327–368. [Google Scholar]
- Will CL, Luhrmann R. Spliceosome Structure and Function. Cold Spring Harb Perspect Biol. 2010 doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yitzhaki S, Miriami E, Sperling J, Sperling R. Phosphorylated Ser/Arg-rich proteins: Limiting factors in the assembly of 200S large nuclear ribonucleoprotein particles. Proc Natl Acad Sci USA. 1996;93:8830–8835. doi: 10.1073/pnas.93.17.8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.