Abstract
Interleukin-34 (IL-34) and colony stimulating factor-1 (CSF-1) both signal through the CSF-1R receptor tyrosine kinase, but they have no sequence homology, and their functions and signaling activities are not identical. We report the crystal structures of mouse IL-34 alone and in complex with the N-terminal three immunoglobulin-like domains (D1-D3) of mouse CSF-1R. IL-34 is structurally related to other helical hematopoietic cytokines, but contains two additional helices integrally associated with the four shared helices. The non-covalently linked IL-34 homodimer recruits two copies of CSF-1R on the sides of the helical bundles, with an overall shape similar to the CSF-1:CSF-1R complex, but the flexible linker between CSF-1R D2 and D3 allows these domains to clamp IL-34 and CSF-1 at different angles. Functional dissection of the IL-34:CSF-1R interface indicates that the hydrophobic interactions, rather than the salt bridge network, dominate the biological activity of IL-34. To degenerately recognize two ligands with completely different surfaces, CSF-1R apparently takes advantage of different subsets of a chemically inert surface that can be tuned to fit different ligand shapes. Differentiated signaling between IL-34 and CSF-1 is likely achieved by the relative thermodynamic independence of IL-34 vs. negative cooperativity of CSF-1 at the receptor-recognition sites, in combination with the difference in hydrophobicity which dictates a more stable IL-34:CSF-1R complex compared to the CSF-1:CSF-1R complex.
Keywords: interleukin-34, colony stimulating factor-1 receptor, growth factor, receptor tyrosine kinase, X-ray crystallography, ligand/receptor binding
1. Introduction
The colony-stimulating factor-1 receptor (CSF-1R) is a cell-surface glycoprotein encoded by the c-fms proto-oncogene in humans [1, 2]. Its ligand CSF-1, also named macrophage-colony stimulating factor (M-CSF), is the primary regulator of the survival, proliferation and differentiation of mononuclear phagocytes, such as macrophages, osteoclasts, microglia, and cells of the female reproductive tract [3–6]. Recently, interleukin-34 (IL-34) was identified as an alternative ligand of CSF-1R, although it shares no apparent sequence homology with CSF-1 [7]. IL-34 acts as a regulator of myeloid lineage differentiation, proliferation and survival, and triggers phosphorylation of CSF-1R and ERK1/2 that may also be induced by CSF-1, indicating that IL-34 and CSF-1 generate signals that at least partially overlap [7–9]. However, since different anti-CSF-1R monoclonal antibodies demonstrated different blocking effects on CSF-1R signaling, it was suggested that IL-34 and CSF-1 are not identical in biological activity and signal activation kinetics, and that macrophage phenotype and function are differentially regulated by the two ligands [10]. The different spatiotemporal expression patterns of IL-34 and CSF-1 also suggests that these ligands have complementary rather than identical functions [8].
CSF-1R is a member of the class III receptor tyrosine kinases (RTKs), which also include KIT (the receptor for Stem Cell Factor, or SCF), Flt3 (the receptor for Flt3 Ligand, or Flt3L), PDGFRα and PDGFRβ (the receptors for Platelet-Derived Growth Factors, or PDGFs)[11]. The class III RTKs are composed of a glycosylated extracellular region comprising five immunoglobulin (Ig)-like domains, a single transmembrane segment, and a split intracellular kinase domain. The structures of SCF:KIT, PDGF-B: PDGFRβ, CSF-1:CSF-1R, and Flt3L:Flt3 complexes have indicated that the N-terminal three Ig-like domains of these receptors are responsible for ligand recognition, and the membrane-proximal domains are responsible for complex stabilization and accurate positioning which are required for activation [12–16]. Both KIT and Flt3 have only one ligand, but akin to CSF-1R, each PDGFR can recognize multiple PDGF ligands [17].
Given the lack of homology between IL-34 and CSF-1, it has been puzzling how receptor sharing is achieved for these two CSF-1R ligands. Comparative co-evolution analysis with all vertebrate genes suggests that the two ligands interact with distinct regions of the CSF-1R [18], but it is unclear how this paradigm is compatible with overlapping but distinct functions of IL-34 and CSF-1. Here, we report the crystal structures of IL-34 alone and in complex with the ligand recognition domains of CSF-1R. Combined with thermodynamic and functional data, our studies reveal a basis for CSF-1R cross-reactivity as aided by flexibility in both conformation and recognition chemistry, which may explain the differentiated functional outcomes of the two ligands, IL-34 and CSF-1.
2. Materials and Methods
2.1. Cloning, cell culture and BacMam virus preparation
The following proteins were prepared: human IL-34 (hIL-34), the full-length ectodomain of human CSF-1R (hCSF-1R D1-D5), the N-terminal three Ig-like domains of human CSF-1R (hCSF-1R D1-D3), the extracellular membrane-proximal two Ig-like domains of human CSF-1R (hCSF-1R D3-D5), mouse IL-34 (mIL-34), the N-terminal three Ig-like domains of mouse CSF-1R (mCSF-1R D1-D3), the full-length ectodomain of mouse CSF-1R (mCSF-1R D1-D5). These cDNA sequences were attached to a C-terminal 7-His tag, PCR-amplified and subcloned into the baculovirus transfer vector for mammalian cell expression as described previously [25]. The expression constructs and the BacVector-3000 baculovirus DNA (EMD) were used to co-transfect adherent sf9 cells in the presence of Insect GeneJuice (EMD). 5 days later, the resulting low-titer virus stock was harvested, and then used to infect sf9 cells at a density of 2×106 cells per ml for making high-titer viruses.
2.2. Protein expression and purification
The amplified viruses were used to infect 1–6 liters of HEK-293H cells (for expressing proteins used for ITC study and cell-based assays) or GnTI− HEK-293S cells (for expressing proteins used for crystallization) [26] at a density of 1.0–1.5 × 106 cells/ml. After 72–96 hours, the cells were pelleted by centrifugation. The supernatants were concentrated and buffer-exchanged to HBS (Hepes buffered saline, 10mM Hepes pH 7.2, 150mM NaCl). The recombinant proteins in the supernatant were captured by Ni-NTA metal affinity resin (QIAGEN) and eluted with HBS in the presence of 300 mM imidazole (pH 7.5). The eluted proteins were then applied to size-exclusion chromatography (Superdex-200 HR column, GE healthcare) for further purification.
2.3. Crystallization, data collection and processing
For the crystallization of mIL-34 in complex with mCSF-1R D1-D3, the HEK293S-derived proteins were incubated with carboxypeptidase-A (Sigma) and Endo-H (New England Biolabs) overnight for trimming His-tag and glycan, respectively. For crystallizing mIL-34 alone, the protein was only His-tag-trimmed prior to crystallization. The Wizard Crystallization Screen Series I and II (Emerald BioSystems) were used for initial screening. Crystallization was performed at 20°C using the sitting-drop vapor diffusion method. X-ray quality crystals were obtained from drops composed of 0.5 μl reservoir solution and 0.5 μl protein solution (7–10mg/ml) equilibrated against 1 ml reservoir solution. The mIL-34 crystals grew in the condition: 1.4 M ammonium sulfate and 0.1 M HEPES, pH 7.5. The mIL-34: mCSF-1R D1-D3 complex was crystallized in the condition: 20% (w/v) PEG3000, 0.1 M sodium acetate, pH 5.0 and 0.2 M lithium sulfate.
Prior to being flash frozen in liquid nitrogen, all the crystals were cryo-protected using the respective crystallization conditions supplemented with 20–25% ethylene glycol. To make a heavy atom derivative, mIL-34 crystals were soaked in the cryo-protectant containing 0.2 M NaI for 30 seconds. X-ray diffraction datasets were collected at 100 K at the Life Sciences Collaborative Access Team (LS-CAT) beamline 21-ID-D, the Advanced Photon Source, Argonne, Illinois, USA. The data were processed with HKL2000 [27]. The statistics are summarized in Table S1.
2.4. Structure determination and refinement
The structure of mIL-34 was determined using the single isomorphous replacement with anomalous scattering (SIRAS) method. Eight iodide ions per asymmetric unit were found using the program SOLVE [28]. The resulting SIRAS phases were improved using the RESOLVE program, and a model was automatically rebuilt by RESOLVE with about 50% IL-34 residues assigned [28]. Solvent flattening and phase extension was carried out using CNS [29], and the model was rebuilt manually using COOT [30]. The structure of mIL-34:mCSF-1R complex was solved by the molecular replacement method using PHASER [31], with the refined mIL-34 model and the structure of mCSF-1R D1-D3 (PDB ID: 3EJJ) as the search models. The structure models of both crystals were subject to simulated annealing, minimization, and group B factor refinements with CNS [29]. As guided by the SIGMAA weighted Fo-Fc difference density map, the carbohydrate moieties (Man)5(GlaNAc)2 were built to two glycosylation sites Asn76 and Asn100 on IL-34 in the mIL-34 crystal structure. For the IL-34-CSF-1R complex crystal structure, only a single GlcNAc residue was modeled to each glycosylation site on IL-34 and CSF-1R. Water molecules were automatically introduced using CNS and then manually edited. A summary of the refinement statistics is given in Table S1.
2.5. Preparation of mIL-34 mutants
Based on the IL-34-CSF-1R complex structure, we designed seven mutants of full-length mouse IL-34: D36R, Y40E, E111R, K117E, Q123R, L125E and E127R. Mutations were generated using overlap extension PCR and confirmed by DNA sequencing. Subcloning, baculovirus generation, protein expression and purification of these mutants were performed using the same protocols as used for the wide-type full-length mIL-34.
2.6. Isothermal titration calorimetry
The proteins used for ITC were expressed from HEK293H cells, and purified on gel filtration column equilibrated with the same batch of buffers to reduce dilution heat. All samples were thoroughly degassed before titration. ITC was carried out on a VP-ITC calorimeter (MicroCal, Northhampton, MA) at 30°C. The titration data were processed with MicroCal Origin software, Version 5.0.
2.7. Cell proliferation assay
Activities of mIL-34 mutants on cell proliferation were evaluated using the M-NFS-60 cell line, which was derived from a myelogenous leukemia induced with the Cas-Br-MuLV wild mouse ecotropic retrovirus. M-NFS-60 cells were cultured in the presence of mIL-34 or its mutants at different concentrations for 4 days. Then the amount of ATP released from lysed cells was determined by CellTiter-Glo Luminescent Cell Viability Assay Kit (Promega, No. G7571), following the manufacturer’s protocol.
3. Results
3.1. IL-34 is a structurally unique dimeric cytokine with 6 helices
We cloned and expressed the N-terminal globular domain of mouse IL-34 for structural studies. IL-34 has a ~40 amino acid (aa) C-terminal tail which is Pro-Ser-Thr-rich, a typical feature of flexible mucin-like O-linked glycosylation-rich sequences. In additional to the potential O-linked glycosylation, IL-34 carries N-linked glycans at Asn76 and Asn100 positions. To facilitate crystallization, we tested trimming these N-linked glycans using endoglycosidase-F1, but the protein immediately formed aggregates, indicating that the N-linked glycans are vital for IL-34 stability. We therefore used only fully N-glycosylated IL-34 in subsequent studies. We first crystallized free IL-34, and solved its structure using the Single Isomorphous Replacement with Anomalous Scattering (SIRAS) method (Table S1, Figure 1). IL-34 exists as a dimer in both crystal and solution (Figure 1A), consistent with previous observations [7].
Figure 1.
Structure of IL-34. (A) Ribbon representation of an IL-34 dimer. The glycans N-linked to N76 and N100 are depicted as green sticks. (B) Cartoon representation of an IL-34 monomer, with a-helices demonstrated as cylinders. The classical 4-helix bundle is colored in violet, whereas the “extra” two helices, α2 and α5, are colored in blue. Disulfide bonds are shown as yellow sticks. (C) Topological diagram of the “up-down-up-down-up-down” fold of IL-34.
The IL-34 protomer folds into a six-helix bundle with short loops and two short β-strands connecting the helices (Figure 1B). The six helices α–helices (α1 for residues 38–57, α2 for 74–82, α3 for 87–108, α4 for 115–134, α5 for 140–150 and α6 for 161–180) in the bundle are oriented in an “up-down-up-down-up-down “fashion (Figure 1C), with a kink disrupting the first helix into two stretches. The longer α1, α3, α4 and α6 helices roughly correspond to the four-helix core structure characteristic of helical cytokines, including CSF-1[19]. The other two helices (α2 and α5), packed against α3 and α6, are associated with the 4 longer helices through continuous hydrophobic interactions. Together, the six-helix bundle in IL-34 is an integral module. The conserved Asn76 glycan serves to fill the cavity between the α2 and α5 helices, explaining why the dependence of IL-34 on glycosylation for stability in solution. In addition to the inter-helical hydrophobic interactions, the six-helix bundle of IL-34 is further locked by the Cys35-Cys180 disulfide bridge between the first and the last helices (α1 and α6) at the fatter end of the bundle. Another disulfide bridge, Cys177-Cys191, locks the C-terminal extension (183–194) to the end of the α6 helix (Figure 1B, Figure S1). The protomer structure of IL-34 modestly resembles CSF-1. However, IL-34’s four core helices are longer and more parallel to each other than CSF-1, and their remaining parts are vastly different from each other (Figure S2).
To form a dimer, two IL-34 protomers pack against each other with the thinner ends of their helical bundles, in a head-to-head fashion akin to SCF [20, 21], Flt3L [16] and CSF-1 [22]. Formed by four intertwined loops, the IL-34 dimer interface buries ~1300 Å2 solvent accessible surface, and is primarily mediated by hydrophobic interactions which include Tyr57, Pro59, ILe60 and Tyr62 from the loop connecting the α1 helix and the β1 strand, and Val108, Leu110 and Pro114 from the loop connecting helices α3 and α4 (Figure S1B). Unlike CSF-1, no inter-molecular disulfide bridge is used to covalently cross-link two monomers in the IL-34 dimer.
3.2. Mapping the CSF-1R domains for IL-34-binding
We used isothermal titration calorimetry (ITC) to map which domains of CSF-1R are responsible for binding IL-34. Previously, based on antibody-blocking data, it was postulated that IL-34 binds to different domains of CSF-1R from CSF-1 [10], as was also suggested by comparative sequence and co-evolution analysis [6]. We measured the binding of recombinant human IL-34 (hIL-34) to either human CSF-1R D1-D5 (Ig-like domains 1–5), D1-D3, or D3-D5. IL-34 binds to CSF-1R D1-D5 with ~400-fold higher affinity than to CSF-1R D1-D3 (Figure 2A), and no detectable binding was observed between IL-34 and CSF-1R D3-D5. This indicates that the primary IL-34-bindings site in CSF-1R is located in D1-D3. In particular, without D2, CSF-1R D3-D5 could not bind to IL-34, suggesting that D2 is required for IL-34:CSF-1R recognition. D4-D5 also contributes to the high affinity between IL-34 and the full extracellular domain of CSF-1R, but by itself cannot engage IL-34. Importantly, with or without D4-D5, CSF-1R binds IL-34 with 1:1 (or 2:2) stoichiometry, suggesting that IL-34 is capable of dimerizing CSF-1R-D1-D3, unlike CSF-1 which cannot dimerize CSF-1R-D1-D3 without the help of D4-D5 [12].
Figure 2.
Structure of the IL-34:CSF-1R complex. (A) Thermodynamic profiles of IL-34 binding to CSF-1R D1-D5 (left panel) and CSF-1R D1-D3 (right panel) measured by ITC. The stoichiometry (N), affinity (Kd), enthalpy change (ΔH) and entropy change (ΔS) are listed by the fitted curves. (B) Overall structure of a 2:2 IL-34:CSF-1R D1-D3 complex in ribbons. IL-34, violet and simon; CSF-1R, blue and slate. (C) Overlapping of CSF-1-bound CSF-1R (blue) and IL-34-bound CSF-1R (cyan). The V296 Cα atoms of two CSF-1R molecules are depicted as balls to indicate the shift.
3.3. Structure of the IL-34:CSF-1R complex
We crystallized the complex between mouse IL-34 and CSF-1R D1-D3, and determined its structure using molecular replacement (Figure 2, Table S1). The complex contains an IL-34 dimer recruiting 2 copies of CSF-1R D1-D3 on the opposite sides. The overall shape of the complex is reminiscent of the symmetrical 2:2 SCF:KIT and CSF-1:CSF-1R complexes. The receptor-bound IL-34 is similar to free IL-34, except a few differences at the terminal extensions which are used in receptor-binding (Figure S3). The CSF-1R D1 domain is rotated ~100° towards the D2 domain at the D1-D2 hinge, forming an integral D1-D2 module similar to KIT [13, 14] and PDGFRβ [15], as well as the CSF-1-bound CSF-1R [12]. The D3 domain is loosely associated with the D2 domain through a long linker peptide (residues 197–210). The flexible D2-D3 linker enables easy hinge movement, which allows CSF-1R to adopt different D2-D3 orientations upon binding to different ligands (discussed below). To form the IL-34:CSF-1R complex, the convex surface formed by the α1/α3/α4 helices of IL-34 is clamped by the CSF-1R D2 and D3 domains near the D2-D3 junction. There is no direct contact between IL-34 and CSF-1R D1. Assuming CSF-1R D3-D5 domains are orientated similarly to KIT D3-D5, it is unlikely for CSF-1R D4-D5 to contact IL-34 directly. Therefore, the ~400 fold increase of IL-34:CSF-1R affinity by D4-D5 is likely a contribution from receptor-to-receptor interactions.
The IL-34:CSF-1R complex is significantly different from the CSF-1:CSF-1R complex. First, the relative positions of the two ligand-binding domains in CSF-1R, D2 and D3, are different. Overlaying CSF-1-bound CSF-1R and IL-34-bound CSF-1R revealed a ~20° rotation difference of their D3 domains when their D2 domains were superimposed (Figure 2C). Secondly, CSF-1 is clamped deeper by the CSF-1R D2-D3 junction than IL-34. Consequently, overlapping but different sets of CSF-1R segments are used in binding CSF-1 and IL-34 (discussed below). Thirdly, to compensate for the loss of intimacy toward CSF-1R-D2, IL-34 adopts N-terminal and C-terminal extensions to contact CSF-1R D3.
3.4. The interface between IL-34 and CSF-1R
Formation of the interface within each pair of IL-34 and CSF-1R D1-D3 buries ~2200 Å2 of solvent-accessible surface area, which is ~460 Å2 larger than each CSF-1: CSF-1R interface. On the IL-34 side it involves the α1, α3, and α4 helices, the N-terminal segment, and the C-terminal loop; on the CSF-1R side, it involved the CD loop and the F strand of the D2 domain, and the C, D, E strand and BC, DE loops of the D3 domain. The interface can be divided into two spatially discontinuous sub-interfaces: a 900 Å2 sub-interface I involving CSF-1R D2, and a 1300 Å2 sub-interface II involving CSF-1R D3. At sub-interface I, the primary contact is the CD loop of CSF-1R D2 forming a network of charge-charge interactions with the α3 and α4 helices of IL-34 (Figure 3A). Salt bridges are formed between IL-34 Glu111 and CSF-1R Arg150, IL-34 Glu127 and CSF-1R Arg146, and IL-34 Glu127 and CSF-1R Arg142. Notably, in IL-34, Glu103 and Asp107 from the α3 helix, Glu121 and Glu127 from α4, Glu143 from α5, as well as Glu111, Asp135 and Glu137 from the helix-connecting loops, are distributed around the surface formed by the α3/α4/α5 helices, collaboratively form a large negatively-charged surface to face the D2 domain of CSF-1R (Figure S4), suggesting longer-range charge-charge attractions in addition to the above described close salt bridges. Collectively, this sub-interface is highly hydrophilic and electrostatically complemented.
Figure 3.
The IL-34:CSF-1R interface. (A) Sub-interface I which involves the α3 and α4 helices of IL-34 and the D2 domain of CSF-1R. Salt bridges are shown as dashes. (B) Sub-interface II which involves the α1 and α4 helices, the N- and C-terminal segments of IL-34, and the D3 domain of CSF-1R. (C) Biological activities of wild-type and mutant IL-34 in the M-NFS-60 cell proliferation assay. The luminescence signal (relative light unit), against the dose of proteins, is generated from reaction with luciferin in the presence of luciferase. Each data point represents the mean of triplicate and the standard deviation values are shown. The EC50 value for each protein was derived from the dose response curves.
At sub-interface II, the dominant feature appears to be a cluster of hydrophobic interactions. On the CSF-1R D3 side, this cluster involves Val231 and Phe233 from the BC loop, and Phe252 and Tyr257 from the DE loop; on the IL-34 side, it involves, Leu37 and Tyr40 from the α1 helix, and Leu125 and the Thr124 Cγ2 atom from the α4 helix (Figure 3B). In addition to this hydrophobic cluster, at the lower (membrane-proximal) edge of this sub-interface, the C-terminal segment of IL-34, with its trajectory restrained by two disulfide bonds (C35-C180 and C177-C191), contacts the D strand of CSF-1R D3 with hydrophobic interactions (Ile185 and Leu186 from IL-34, and Pro247 and Leu248 from CSF-1R) (Figure 3B, Table S2). Notably, a buried salt bridge is formed between IL-34 Asp36 and CSF-1R Lys259, flanked by the above groups of hydrophobic interactions (Figure 3B).
3.5. Functional dissection of the IL-34:CSF-1R interface
We verified the functional significance of the potential “hotspot” IL-34 residues at the IL-34:CSF-1R interface, as assessed by site-directed mutagenesis and M-NFS-60 cell-based assay (Figure 3C). The effects of mutating these residues on cell proliferation activities of IL-34 are generally consistent with the apparent structural roles of these residues at the interface. For instance, the mutant K117E demonstrated identical activity to the wild–type (Figure 3C), consistent with the fact that the salt bridge between IL-34 Lys117 and CSF-1R Asp254 is located at the periphery of the IL-34:CSF-1R interface. Interestingly, none of the individual hydrophilic residues of IL-34 interacting with CSF-1R D2 is essential, as evidenced by the fact that the individual point mutations in sub-interface I, including E111R, Q123R, E127R, designed to antagonize the charge-charge interactions, essentially had no effect on the biological activity of IL-34 (Figure 3C). In contrast, disturbing the sub-interface II interactions between CSF-1R-D3 and IL-34 had much larger biological effects. For instance, the Y40E mutation completely abolished the IL-34 bio-activity, and the L125E mutation reduced IL-34 activity by 80-fold. CSF-1RIn addition, the buried salt bridge between IL-34 Asp36 and CSF-1R Lys259 is also critical, as the D36R mutant of IL-34 has 74-fold reduced activity (Figure 3C).
Hence, it appears that sub-interface II plays a more important role than sub-interface I in the biological responses induced by IL-34:CSF-1R interaction. Given that the D2 domain of CSF-1R was indispensable for binding according to our calorimetric data, but each single mutation of IL-34 residues interacting with this domain had only minor effect on cell proliferation, it is likely that the strong charge-charge attraction provided by CSF-1R D2 is used to first capture IL-34, and the more geometrically specific interactions (hydrophobic interactions and buried salt bridge) provided by CSF-1R D3 is formed subsequently to stabilize the IL-34:CSF-1R complex, and to define accurate positions of the CSF-1R D3 domains which are important for signaling.
4. Discussion
4.1. Mechanism of degenerate CSF-1R recognition of non-homologous ligands
IL-34 and CSF-1 bear no sequence similarity, despite that our structures show that these two cytokines are actually structurally related. Both IL-34 and CSF-1 have the 4-helix core, but IL-34 has two additional helices integrally bundled to the core, as well as unique terminal extensions that are used in receptor binding. The overall modes of receptor engagements are similar between IL-34 and CSF-1, which use overlapping epitopes to associate with the same D2 and D3 domains of CSF-1R. Nevertheless, with each residue on the ligand surfaces being different from each other, no single interaction with CSF-1R is shared between CSF-1 and IL-34. How does CSF-1R recognize two ligands with completely different surfaces, yet both achieve efficient signaling [7, 9]? The comparison of the IL-34:CSF-1R and CSF-1:CSF-1R binding revealed two unique strategies to achieve the extraordinary cross-reactivity.
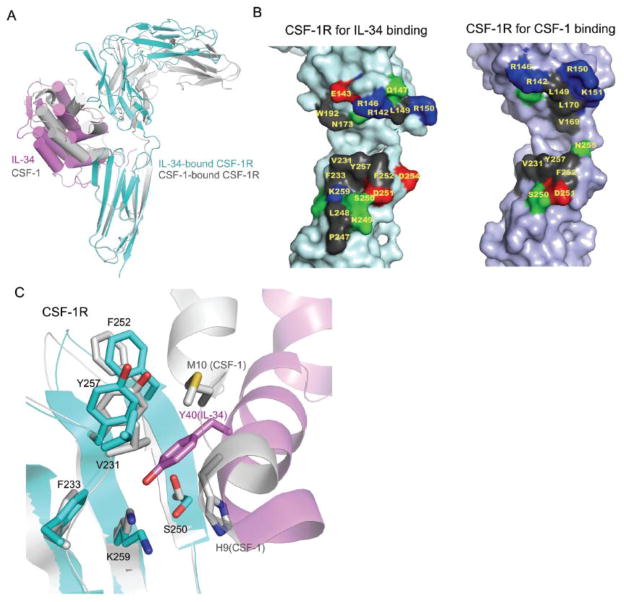
The first strategy is to utilize overlapping but different subsets of receptor surfaces to recognize different ligands. This is apparently aided by the capability of CSF-1R D2 and D3 domains to rotate around the flexible D2-D3 linker. As the linker is located at the far end of the D2-D3 junction, it offers the opportunity of D2 and D3 domains to clamp ligands at different levels of openness, and correspondingly, the ligands to be clamped in different depth. CSF-1 sits deeper in the clamp than IL-34 (Figure 4A). Consequently, CSF-1R-D2 uses both the CD (outer) and EF (inner) loops to contact CSF-1, but only needs the CD loop to contact IL-34 (Figure 4A and 4B). In addition, CSF-1R D3 has a large flat face above its CD loop, which is configured to embed most of its hydrophilic residues, including Asn234, Ser250, Lys259 and Arg261. The embedding of usually flexible hydrophilic side chains greatly reduces conformational entropy, which is favorable for protein-protein interactions. This large area can be selectively used in recognizing ligands. For the smaller CSF-1, only the upper part of this surface is used; for the larger IL-34 which has extra extensions at the N- and C-termini, an expanded area of this surface, notably towards the membrane direction, is used (Figure 4B).
Figure 4.
Degenerate recognition of non-homologous ligands by CSF-1R. (A) Overlay of the IL-34:CSF-1R complex (magenta and cyan) and the CSF-1:CSF-1R complex (gray) by superimposing the D3 domains of CSF-1R, showing that CSF-1 and IL-34 are clamped at the CSF-1R D2-D3 junction in different depth. (B) Different subsets of surfaces of CSF-1R are used to recognize IL-34 and CSF-1. Hydrophobic and aromatic residues at the interface are colored in grey; basic residues in blue; acidic residues in red, and other polar residues in green. (C) Slight re-arrangement of a CSF-1R surface patch, results in the recognition of vastly different residues of IL-34 and CSF-1.
The second strategy is chemical promiscuity, which uses the same set of residues in different ways to meet different needs in ligand binding. For CSF-1R D2, it takes advantage of the intrinsic flexibility of the long-chain positive-charged residues, such as Arg142, Arg146 and Arg150, using different rotamer conformations of their side chains to form salt bridges with different patterns of negative charges. Importantly, these positively charged residues are positioned to protrude in the outset loop at the edge of the sub-interface I, generating the possibilities to accept negative-charges from a wide range of directions. Such degenerate charge-charge interactions are enthapically favorable, but can be entropically detrimental.
To corroborate the enthapically favorable but entropically unfavorable interactions provided by CSF-1R D2, at sub-interface II, CSF-1R D3 forms a flat and rigid ligand-binding surface with a mixed set of hydrophobic and hydrophilic residues. This creates chemical inertness that when multiple ligand surfaces are presented, for each type of ligand surface, the fitting pattern between ligand and receptor can be stably dictated by the distributed pattern of the hydrophobic/hydrophilic interactions, but for different type of ligand surfaces, different interaction patterns can easily be achieved through minor side chain shifts. For instance, the Val231, Phe257, Ser250 and Lys259 patch can make a hydrophobic pocket for the aromatic ring of mouse IL-34 Tyr40 (Phe40 in human); by squeezing each side chain of this pocket slightly closer, this patch can form flat interactions with Met10 and the Cβ atom of His9 of CSF-1 (Figure 4C). This type of degenerate recognition by chemical inertness is akin to that of gp130, a shared receptor for over 10 types of low-homology cytokines [23].
The cross-reactivity strategies of CSF-1R are clearly different from PDGFRα, another class III RTK that is capable of recognizing several ligands, including homodimeric PDGF-AA, PDGF-BB, and PDGF-CC, and heterodimeric PDGF-AB. For PDGFR, there is no domain conformational flexibility of the ligand-binding domains, and the PDGF-PDGFR interface is almost exclusively hydrophobic [15]. The cross-reactivity of PDGF-PDGFR mostly takes advantage of the intrinsic lack of directionability of hydrophobic interactions, as well as the moderate-to-high sequence identities among both receptors and ligands. The PDGFR-type, rather than the CSF-1R-type, of cross-reactivity is more commonly observed for multi-member ligand/receptor families [24].
4.2. Non-identical activation of the same CSF-1R receptor by CSF-1 and IL-34
Despite the degenerate yet convergent recognition of their receptor, IL-34 and CSF-1 are not identical in functions. Aside from their differences in spatiotemporal expression patterns [8], they have different abilities to induce chemokine production in primary macrophages, and to induce morphological changes of TF1-fms cells, suggesting differences in the CSF-1R receptor activation and signaling [10]. Our calorimetric data showed that in the presence of D4-D5, CSF-1R binds IL-34 7-fold stronger than CSF-1, although both at high affinities [12] (Figure 2); in the absence of D4-D5, CSF-1R binds CSF-1 and IL-34 with comparable affinities, but IL-34 is capable of recruiting two CSF-1R-D1-D3, and CSF-1 is capable of recruiting only one. The second site of CSF-1:CSF-1R-D1-D3 binding is compromised by negative cooperativity [12], which is not obvious in IL-34:CSF-1R binding. The negative co-operativity of the CSF-1 dimer is most likely a result of the inter-molecular disulfide bond, which can pass the inter-helical minor shifts of one monomer, upon receptor-binding, to the other monomer through the covalent pulling. In comparison, the lack of inter-molecular disulfide bond in IL-34 makes the conformational changes of the monomers easily buffered around the dimerization interface. Hence, the two monomers in the IL-34 dimer can behave more independently. Indeed, the free IL-34 dimer and the CSF-1R-bound IL-34 dimer are very similar (Figure S3B), but the CSF-1 dimers are very different in free and receptor-bound states [12].
The negative co-operativity of CSF-1 vs. relative thermodynamic independence of IL-34 at the CSF-1R-binding sites supports a more stable IL-34:CSF-1R association than the CSF-1:CSF-1R association. In addition, the CSF-1:CSF-1R interface is mostly hydrophilic, but the IL-34:CSF-1R interface entails large amount of hydrophobic area. Even at similar affinities, the disassociation of hydrophobic contacts is kinetically slower than the disassociation of hydrophilic contacts. Therefore once the ligands are recognized, the IL-34:CSF-1R complex on the cell-surface is likely to have a longer timespan to produce transmembrane signals than the CSF-1:CSF-1R complex.
Supplementary Material
Figure S1. The disulfide bond pattern and the dimerization pattern of IL-34. (A) The disulfide bonds in an IL-34 monomer. Note that Cys35-Cys180 and Cys177-Cys191 help to lock the helices and to stabilize the C-terminal segment respectively. Cys168 is located in the core and is paired with a buried serine (Ser98). Cys179 is exposed and is possibly linked to the cysteine (Cys199) in the O-linked glycosylation-rich tail. (B) The dimerization of IL-34 entails a large number of hydrophobic interactions but no inter-molecular disulfide bond.
Figure S2. Comparison of the protomer structures of IL-34 (left) and CSF-1(right).
Figure S3. Comparison of free IL-34 and the CSF-1R-bound-IL34. (A) Conformational differences in the N-terminal segment and C-terminal loop. The D2-D3 region of CSF-1R is shown as molecular surface (gray) for reference. (B) The orientation between the protomers in the IL-34 dimer is similar in free and bound states.
Figure S4. Charge-charge attractions at the IL-34:CSF-1R interface, especially between CSF-1R D2 and IL-34. The blue color indicates positively charges and red indicates negatively charges.
Table S1. Crystallographic data collection and refinement statistics
Table S2. Contacts between CSF-1R and IL-34
Highlights.
Structures of mouse interleukin-34 and its complex with receptor
The basis of CSF-1R cross-reactivity towards non-homologous ligands is revealed
Functional dissection of the interleukin-34: CSF-1R interface reveals hotspots
Conformational flexibility and chemical inertness underline CSF-1R degeneracy
Acknowledgments
We thank P.J. Focia and Z. Wawrzak for support in data collection and S. Gomes for support in generating IL-34 mutant clones. X.H. is supported by the NIH grant 1R01GM078055. The Structural Biology Facility is supported by the R.H. Lurie Comprehensive Cancer Center of Northwestern University. Data were measured at the LS-CAT beamline 21-ID-D at the Advanced Photon Source (APS), Argonne, IL.
Abbreviations
- IL-34
interleukin-34
- CSF1
colony stimulating factor-1
- CSF-1R
colony stimulating factor-1 receptor
- ITC
isothermal titration calorimetry
Footnotes
The authors declare competing financial interests. C.L., B.W., L.T.W., and H.L. are employees of Five Prime Therapeutics, Inc.
Data deposition: The structure factor amplitudes and coordinates of mIL34 and the mIL-34:mCSF-1R complex are being deposited in the Protein Data Bank, www.pdb.org, and will be immediately released upon publication.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Donner L, Fedele LA, Garon CF, Anderson SJ, Sherr CJ. McDonough feline sarcoma virus: characterization of the molecularly cloned provirus and its feline oncogene (v-fms) J Virol. 1982;41:489–500. doi: 10.1128/jvi.41.2.489-500.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sherr CJ, Rettenmier CW, Sacca R, Roussel MF, Look AT, Stanley ER. The c-fms proto-oncogene product is related to the receptor for the mononuclear phagocyte growth factor, CSF-1. Cell. 1985;41:665–676. doi: 10.1016/s0092-8674(85)80047-7. [DOI] [PubMed] [Google Scholar]
- 3.Chitu V, Stanley ER. Colony-stimulating factor-1 in immunity and inflammation. Curr Opin Immunol. 2006;18:39–48. doi: 10.1016/j.coi.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Dai XM, Ryan GR, Hapel AJ, Dominguez MG, Russell RG, Kapp S, Sylvestre V, Stanley ER. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002;99:111–120. doi: 10.1182/blood.v99.1.111. [DOI] [PubMed] [Google Scholar]
- 5.Teitelbaum SL, Ross FP. Genetic regulation of osteoclast development and function. Nat Rev Genet. 2003;4:638–649. doi: 10.1038/nrg1122. [DOI] [PubMed] [Google Scholar]
- 6.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, Merad M. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin H, Lee E, Hestir K, Leo C, Huang M, Bosch E, Halenbeck R, Wu G, Zhou A, Behrens D, Hollenbaugh D, Linnemann T, Qin M, Wong J, Chu K, Doberstein SK, Williams LT. Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science. 2008;320:807–811. doi: 10.1126/science.1154370. [DOI] [PubMed] [Google Scholar]
- 8.Wei S, Nandi S, Chitu V, Yeung YG, Yu W, Huang M, Williams LT, Lin H, Stanley ER. Functional overlap but differential expression of CSF-1 and IL-34 in their CSF-1 receptor-mediated regulation of myeloid cells. Journal of leukocyte biology. 2010;88:495–505. doi: 10.1189/jlb.1209822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Droin N, Solary E. Editorial: CSF1R, CSF-1, and IL-34, a “menage a trois” conserved across vertebrates. Journal of leukocyte biology. 2010;87:745–747. doi: 10.1189/jlb.1209780. [DOI] [PubMed] [Google Scholar]
- 10.Chihara T, Suzu S, Hassan R, Chutiwitoonchai N, Hiyoshi M, Motoyoshi K, Kimura F, Okada S. IL-34 and M-CSF share the receptor Fms but are not identical in biological activity and signal activation. Cell death and differentiation. 2010;17:1917–1927. doi: 10.1038/cdd.2010.60. [DOI] [PubMed] [Google Scholar]
- 11.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Liu H, Focia PJ, Shim AH, He X. Structure of macrophage colony stimulating factor bound to FMS: diverse signaling assemblies of class III receptor tyrosine kinases. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18267–18272. doi: 10.1073/pnas.0807762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Chen X, Focia PJ, He X. Structural basis for stem cell factor-KIT signaling and activation of class III receptor tyrosine kinases. EMBO J. 2007;26:891–901. doi: 10.1038/sj.emboj.7601545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuzawa S, Opatowsky Y, Zhang Z, Mandiyan V, Lax I, Schlessinger J. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130:323–334. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]
- 15.Shim AH, Liu H, Focia PJ, Chen X, Lin PC, He X. Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11307–11312. doi: 10.1073/pnas.1000806107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verstraete K, Vandriessche G, Januar M, Elegheert J, Shkumatov AV, Desfosses A, Van Craenenbroeck K, Svergun DI, Gutsche I, Vergauwen B, Savvides SN. Structural insights into the extracellular assembly of the hematopoietic Flt3 signaling complex. Blood. 2011;118:60–68. doi: 10.1182/blood-2011-01-329532. [DOI] [PubMed] [Google Scholar]
- 17.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes & development. 2008;22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garceau V, Smith J, Paton IR, Davey M, Fares MA, Sester DP, Burt DW, Hume DA. Pivotal Advance: Avian colony-stimulating factor 1 (CSF-1), interleukin-34 (IL-34), and CSF-1 receptor genes and gene products. Journal of leukocyte biology. 2010;87:753–764. doi: 10.1189/jlb.0909624. [DOI] [PubMed] [Google Scholar]
- 19.Bazan JF. Emerging families of cytokines and receptors. Current biology : CB. 1993;3:603–606. doi: 10.1016/0960-9822(93)90009-d. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Z, Zhang R, Joachimiak A, Schlessinger J, Kong XP. Crystal structure of human stem cell factor: implication for stem cell factor receptor dimerization and activation. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7732–7737. doi: 10.1073/pnas.97.14.7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang X, Gurel O, Mendiaz EA, Stearns GW, Clogston CL, Lu HS, Osslund TD, Syed RS, Langley KE, Hendrickson WA. Structure of the active core of human stem cell factor and analysis of binding to its receptor kit. EMBO J. 2000;19:3192–3203. doi: 10.1093/emboj/19.13.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandit J, Bohm A, Jancarik J, Halenbeck R, Koths K, Kim SH. Three-dimensional structure of dimeric human recombinant macrophage colony-stimulating factor. Science. 1992;258:1358–1362. doi: 10.1126/science.1455231. [DOI] [PubMed] [Google Scholar]
- 23.Boulanger MJ, Bankovich AJ, Kortemme T, Baker D, Garcia KC. Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130. Molecular cell. 2003;12:577–589. doi: 10.1016/s1097-2765(03)00365-4. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Lupardus P, Laporte SL, Garcia KC. Structural biology of shared cytokine receptors. Annual review of immunology. 2009;27:29–60. doi: 10.1146/annurev.immunol.24.021605.090616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dukkipati A, Park HH, Waghray D, Fischer S, Garcia KC. BacMam system for high-level expression of recombinant soluble and membrane glycoproteins for structural studies. Protein Expr Purif. 2008;62:160–170. doi: 10.1016/j.pep.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger TC, Berendzen J. Automated structure solution for MIR and MAD. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The disulfide bond pattern and the dimerization pattern of IL-34. (A) The disulfide bonds in an IL-34 monomer. Note that Cys35-Cys180 and Cys177-Cys191 help to lock the helices and to stabilize the C-terminal segment respectively. Cys168 is located in the core and is paired with a buried serine (Ser98). Cys179 is exposed and is possibly linked to the cysteine (Cys199) in the O-linked glycosylation-rich tail. (B) The dimerization of IL-34 entails a large number of hydrophobic interactions but no inter-molecular disulfide bond.
Figure S2. Comparison of the protomer structures of IL-34 (left) and CSF-1(right).
Figure S3. Comparison of free IL-34 and the CSF-1R-bound-IL34. (A) Conformational differences in the N-terminal segment and C-terminal loop. The D2-D3 region of CSF-1R is shown as molecular surface (gray) for reference. (B) The orientation between the protomers in the IL-34 dimer is similar in free and bound states.
Figure S4. Charge-charge attractions at the IL-34:CSF-1R interface, especially between CSF-1R D2 and IL-34. The blue color indicates positively charges and red indicates negatively charges.
Table S1. Crystallographic data collection and refinement statistics
Table S2. Contacts between CSF-1R and IL-34