Abstract
A limited set of phosphoinositide membrane lipids regulate diverse cellular functions including proliferation, differentiation, and migration. We developed two techniques based on rapamycin-induced protein dimerization to rapidly change the concentration of plasma membrane phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2]. First, we increased PI(4,5)P2 synthesis from phosphatidylinositol 4-phosphate [PI(4)P] using a membrane recruitable form of PI(4)P 5-kinase, and found that COS-7, HeLa, and HEK293 cells formed bundles of motile actin filaments known as actin comets. In contrast, a second technique that increased the concentration of PI(4,5)P2 without consuming PI(4)P induced membrane ruffles. These distinct phenotypes were mediated by dynamin-mediated vesicular trafficking and mutually inhibitory crosstalk between the small guanosine triphosphatases Rac and RhoA. Our results indicate that the effect of PI(4,5)P2 on actin reorganization depends on the abundance of other phosphoinositides, such as PI(4)P. Thus, combinatorial regulation of phosphoinositide concentrations may contribute to the diversity of phosphoinositide functions.
INTRODUCTION
Complexity in signaling networks is often derived from a limited set of molecules that are co-opted for multiple tasks. Understanding how cells achieve such sophisticated processing using a finite set of molecules within a confined space is critical to understanding their biology. A paradigmatic example of such multitasking molecule is phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2], a member of the phosphoinositide family of membrane lipids that is produced by means of the phosphorylation of phosphatidylinositol 4-phosphate [PI(4)P] by PI(4)P 5-kinase [PI(4)P5K]. Long known as a component of the cellular plasma membrane (PM), where it constitutes 1~2% of total lipids, PI(4,5)P2 has recently been identified as a second messenger that can trigger such diverse cellular functions as membrane trafficking, ion channel activation, cytokinesis, phagocytosis, and cell migration (1-8). Consistent with this pleiotropic role, a series of lipid kinases and phosphatases dynamically control the concentration of PI(4,5)P2 with high spatio-temporal precision in response to various stimuli (10). Mutations in genes encoding these enzymes result in various human diseases, including renal, neuromuscular, and developmental disorders, many types of cancers, and diabetes (11, 12). However, the question of how this single lipid can drive multiple cellular functions remains unresolved.
The technical difficulty of rapidly and specifically manipulating PI(4,5)P2 in living cells has contributed to this lack of understanding. Rapid and inducible manipulation of membrane lipids can help identify the direct consequences of the perturbation by avoiding the possible compensations often observed among these membrane lipids (13, 14).
To begin to understand the multiple roles of PI(4,5)P2 at the molecular level, we developed a technique to rapidly manipulate PI(4,5)P2 concentration based on the rapamycin-triggered dimerization of FK506 binding protein (FKBP) and FKBP-rapamycin binding protein (FRB) and used it to study dynamic actin phenotypes such as ruffles and comets. Ruffles provide the driving force for the motility of various cell types, including leukocytes, lymphocytes, fibroblasts, epithelial cells, and neurons (15). They are often observed at the leading edge of migrating cells where they develop into pseudopodia. In contrast, endocytic vesicles can be rapidly propelled through the cell by actin comet tails (16), suggesting that they facilitate vesicular trafficking. Using the FKBP-FRB dimerization technique, we can rapidly target a protein of interest to a predetermined intracellular location, thereby inducing changes in the activity of various signaling molecules on a timescale of seconds (17). Here, we describe using this approach to manipulate phosphoinositides and to distinguish between the effects of increasing the concentration of PI(4,5)P2 and decreasing that of PI(4)P, which is a signaling molecule in its own right (18).
RESULTS
PI(4,5)P2 concentration can be increased in cells independently of its synthesis from PI(4)P
We used two approaches to increase the concentration of PI(4,5)P2. In the first approach, we anchored FRB at the PM by means of an N-terminal signal sequence from a Lyn kinase (Lyn11-FRB), and used rapamycin to rapidly recruit a YFP-FKBP-PI(4)P5K construct to the PM to phosphorylate PI(4)P (Fig. 1). When the Lyn11-FRB labelled with CFP (Lyn11-CFP-FRB) and YFP-FKBP-PI(4)P5K were transected in COS-7 cells, YFP-FKBP-PI(4)P5K translocated from the cytosol to the PM on a timescale of seconds upon addition of rapamycin (Fig. 2A, Supplementary Movie 1).
Figure 1. Schematic illustration of two techniques to rapidly manipulate PI(4,5)P2 using chemically inducible dimerization.
PI(4,5)P2 synthesis from PI(4)P: Rapamycin recruits cytosolic FKBP-PI(4)P5K to the plasma membrane where FRB is anchored through a myristoylation and palmitoylation modification sequence, resulting in formation of the tripartite FRB-Rapamycin-FKBP complex. This places PI(4)P5K near its substrate PI(4)P, enabling PI(4)P phosphorylation at the D5 position to produce PI(4,5)P2. PI(4,5)P2 liberation: Prior to rapamycin addition, FKBP-PH(PLCδ) preferentially localizes at the plasma membrane through its interaction with PI(4,5)P2. Rapamycin recruits FKBP-PH(PLCδ) from the PM to mitochondria, where FRB is anchored via the transmembrane motif of monoamine oxidase A (MoA).
Figure 2. PI(4,5)P2 concentration is increased in cells independently of its synthesis from PI(4)P using PI(4,5)P2 liberation.
(A and B) Time-series confocal fluorescence microscopy images of COS-7 cells co-expressing either Lyn11-CFP-FRB and YFP-FKBP-PI(4)P5K to achieve PI(4,5)P2 synthesis (A), or CFP-FRB-MoA and YFP-FKBP-PH(PLCδ) to achieve PI(4,5)P2 liberation (B). Cell images were collected every 15 s and are shown before (−60 s) and after (15 s and 90 s) rapamycin addition (100 nM). Scale bars, 20 μm. (C and D) Currents through KCNQ4 potassium channels transfected in tSA201 cells were elicited by depolarization from −60 mV to −20 mV for 300 ms and quantified as tail current amplitude. Note that density of expressed channels varies considerably between cells (22). (C) Expression of KCNQ4 with the components for PI(4,5)P2 synthesis. (D) Expression of KCNQ4 with the components for PI(4,5)P2 liberation. Translocation of YFP-PH(PLCδ) to CFP-FRB-MoA is simultaneously reported by FRET (YFP emission with CFP excitation / CFP emission with CFP excitation) from the same cells. Bar graphs in C and D represent summary data (mean +/− SEM) of the percent increase in KCNQ4 tail current in response to rapamycin treatment for n = 7 cells for PI(4,5)P2 synthesis and n = 9 cells for PI(4,5)P2 liberation. (E and F) TIRF measurements of plasma membrane PI(4)P with PI(4,5)P2 manipulation. Normalized fluorescence intensity of GFP-PH(OSBP) was collected every 15 s from COS-7 cells expressing Lyn11-FRB with mCherry-FKBP-PI(4)P5K (blue) or mCherry-FKBP-PI(4)P5K-KD (gray) (F), where rapamycin was added on the 60th second. Normalized fluorescence intensity of GFP-PH(OSBP) for COS-7 cells expressing FRB-MoA and mCherry-FKBP-PH(PLCδ) with addition of rapamycin (red) or DMSO (0.1%, gray) (G). ΔF/F0 indicates fluorescence change divided by initial fluorescence (n=3). Error bars indicate SEM.
In the second approach, we developed a technique that we call PI(4,5)P2 “liberation,” which increases PI(4,5)P2 without decreasing PI(4)P (Fig. 1). Much of PI(4,5)P2 at the PM is bound to proteins (19). We fused the pleckstrin homology (PH) domain of phospholipase Cδ (PLCδ) to FKBP tagged with fluorescent protein (FP) to create a PI(4,5)P2-binding protein (FKBP-PH(PLCδ)) and used rapamycin to rapidly remove FKBP-PH(PLCδ) from the PM, thus increasing the amount of free PI(4,5)P2. We recruited FKBP-PH(PLCδ) to FRB anchored at the cytosolic surface of mitochondria through the transmembrane motif of monoamine oxidase A (MoA). We chose mitochondria because they are the second largest intracellular organelle, and thereby provide sufficient capacity to sequester FKBP-PH(PLCδ), and because they have little role in phosphoinositide signaling (20).
To determine whether the FKBP modification affected the PI(4,5)P2-binding property of PH(PLCδ), we coexpressed YFP-FKBP-PH(PLCδ) with a non-FKBP-modified form of PH(PLCδ), CFP-PH(PLCδ). Like CFP-PH(PLCδ), YFP-FKBP-PH(PLCδ) localized to the PM (fig. S1A), supporting the previous report that PI(4,5)P2 preferentially exists at the PM (21). When we stimulated cells with platelet-derived growth factor (PDGF) to activate endogenous PLC, which cleaves PI(4,5)P2, FKBP-PH(PLCδ) transiently dissociated from the PM, with kinetics and extent very similar to those of CFP-PH(PLCδ) (fig. S1B). Thus the FKBP modification did not appear to alter the efficacy of PI(4,5)P2 binding by PH(PLCδ).
When FKBP-PH(PLCδ) was transfected together with mitochondria-targeted FRB (FRB-MoA), rapamycin induced the rapid translocation of FKBP-PH(PLCδ) from the PM to mitochondria (Fig. 1B, Supplementary Movie 2).
To evaluate PI(4,5)P2 concentration at the PM in response to PI(4,5)P2 synthesis or PI(4,5)P2 liberation, we measured current through KCNQ2 and 3 (KCNQ2/3) potassium channels, which increases with increasing PI(4,5)P2 concentration but is insensitive to PI(4)P (13, 22-26). We first confirmed that KCNQ2/3 current increases with PI(4,5)P2 synthesis – as previously shown (13), and also with PI(4,5)P2 liberation (fig. S2A-D, left bar in fig. S2E). However, sequestration of a phosphatidylserine binding protein such as the C2 domain of lactadherin [FKBP-C2(Lact)] from the PM to mitochondria did not lead to a striking increase in KCNQ2/3 current (right bar in fig. S2E, p < 0.05 between PH+MoA and C2+MoA). Subsequently, we used KCNQ4 channels which have lower affinity for PI(4,5)P2 (27) and are thus less likely to be saturated with increasing PI(4,5)P2. KCNQ4 current increased with PI(4,5)P2 synthesis and PI(4,5)P2 liberation by 74 ± 13 % and 46 ± 7 %, respectively (Fig. 1, C and D), confirming that PI(4,5)P2 increases with both manipulations. The time course of FKBP-PH(PLCδ) translocation to the mitochondria, as determined by Förster resonance energy transfer (FRET) between YFP on FKBP-PH(PLCδ) and CFP on FRB-MoA, coincided with that of the increase in KCNQ current (Fig. 1D, fig. S2). The similar time course and extent of the increase in PI(4,5)P2 with the two manipulations was produced by a computational simulation (fig. S2F and G, Supplemental Table S1 and 2) using previously established parameters of phosphoinositide metabolism (23).
We used total internal reflection fluorescence (TIRF) microscopy, using the PH domain of oxysterol binding protein (OSBP)-labeled with FP as a PI(4)P biosensor [FP-PH(OSBP)] (28), to evaluate the effects of both manipulations on PI(4)P. TIRF microscopy measures fluorescence signals originating from the PM and its immediate vicinity but not from deeper in the cytoplasm. We found GFP-PH(OSBP) rapidly dissociated from the PM upon membrane recruitment of PI(4)P5K, but not of a catalytically-inactive D253A mutant form (PI(4)P5K-KD) (Fig. 1E), consistent with depletion of its substrate PI(4)P. In contrast, the GFP-PH(OSBP) signal was unaffected by PI(4,5)P2 liberation (Fig. 1F). Based on our computational simulation of PI(4,5)P2 liberation (fig. S2G) – which is informed by experimental measurements of PH(PLCδ) translocation and PI(4,5)P2 metabolism (23), we expected that a fraction of the liberated PI(4,5)P2 might be dephosphorylated into PI(4)P by ambient 5-phosphatases. However, if this small PI(4)P increase occurred, it did not result in a visible translocation of FP-PH(OSBP) to the PM.
PI(4,5)P2 synthesis and PI(4,5)P2 liberation induce distinct actin phenotypes
To evaluate the effects of the rapid PI(4,5)P2 manipulations on the actin cytoskeleton, we used a regulator of actin assembly, Ena/VASP, labeled with FP (FP-Ena/VASP-like, or FP-Evl) that can visualize various types of actin structures(29). Using confocal fluorescence microscopy imaging of FP-Evl in living cells, we confirmed that recruitment of PI(4)P5K to the PM robustly induced actin comets (Fig. 3A, Supplementary Movie 3). We failed to observe actin comet formation when PI(4)P5K-KD was recruited to the membrane (fig. S3), indicating that a catalytic reaction [i.e., PI(4,5)P2 synthesis] is responsible for comet formation. In contrast, PI(4,5)P2 liberation through sequestration of FKBP-PH(PLCδ) from the PM to the mitochondria did not elicit the formation of actin comets, but instead induced membrane ruffling (Fig. 3B, Supplementary Movie 4). When PI(4,5)P2 liberation was achieved through sequestration of a different (4,5)P2-binding protein, Tubby (2, 30), cells exhibited the same ruffling phenotype (fig. S4A and B). However, sequestration of a phosphatidylserine-binding protein [FKBP-C2(Lact)] from the PM to mitochondria, did not elicit membrane ruffling (fig. S4C and D).
Figure 3. Distinct actin phenotypes induced by PI(4,5)P2 synthesis and PI(4,5)P2 liberation.
(A and C) Confocal fluorescence microscopy images of actin cytoskeleton before and after PI(4,5)P2 synthesis in the absence (A) or presence (C) of overexpressed mCherry-PH(PLCδ). (B and D) Confocal fluorescence microscopy images of actin cytoskeleton before and after PI(4,5)P2 liberation in the absence (B) or presence (D) of overexpressed mCherry-PI(4)P5K. Actin phenotypes were visualized by YFP-Evl. Images were collected at indicated time points. Right panel images highlight sites that show morphological changes (actin comets in A and C, membrane ruffle in B and D, highlighted by arrowheads and arrows, respectively). Scale bars in A are 10 μm and represent for A-D. (E) Quantification of actin phenotypes (% of cells showing comets and ruffles) induced by PI(4,5)P2 manipulation. Values represent mean ± SEM (n ≥ 30, from three independent experiments). Asterisks indicate p<0.01. All p-values are summarized in Table S3.
Actin comets are preferentially generated from lipid rafts (9). To determine whether the distinct actin phenotypes we observed with PI(4,5)P2 synthesis and its liberation were due to the PI(4,5)P2 production at different membrane domains, we created a construct in which FRB was fused to a K-Ras CAAX tail [as a PM-targeting sequence that is not associated with any particular domain (31)] instead of Lyn11, which is thought to be associated with lipid rafts (31). We observed that, PI(4,5)P2 synthesis achieved with the FRB-CAAX construct resulted in the same actin comet phenotype as that associated with Lyn11-FRB (fig. S3). The actin comets and ruffles induced by these PI(4,5)P2 manipulations were also apparent with phalloidin staining of fixed cells in which FP-Evl was not overexpressed (fig. S5), and were consistently observed in different cell types, including HEK293T and HeLa cells (fig. S6).
Overexpression of PH(PLCδ) sequesters substantial amounts of PI(4,5)P2 and alters some aspects of PI(4,5)P2 signaling and metabolism (32). Therefore, the cells used to investigate the effects of PI(4,5)P2 liberation might show aberrant PI(4,5)P2 signaling before addition of rapamycin [and independent of PI(4,5)P2 liberation]. Similarly, overexpression of may increase PI(4,5)P2 signaling despite the deliberate mislocalization of the FKBP-PI(4)P5K construct to the cytoplasm (13). To exclude these possibilities, we conducted PI(4,5)P2 synthesis or PI(4,5)P2 liberation in the presence of overexpressed PH(PLCδ) or PI4P5K, respectively. In both cases, we observed the predicted actin phenotypes (i.e., actin comets with synthesis or membrane ruffles with liberation, respectively) (Fig. 3, C and D). Occurrence of the actin phenotypes induced by the two different PI(4,5)P2 manipulations appeared to be mutually exclusive (Fig. 3E).
Crosstalk between Rho GTPases regulates PI(4,5)P2-induced actin remodeling
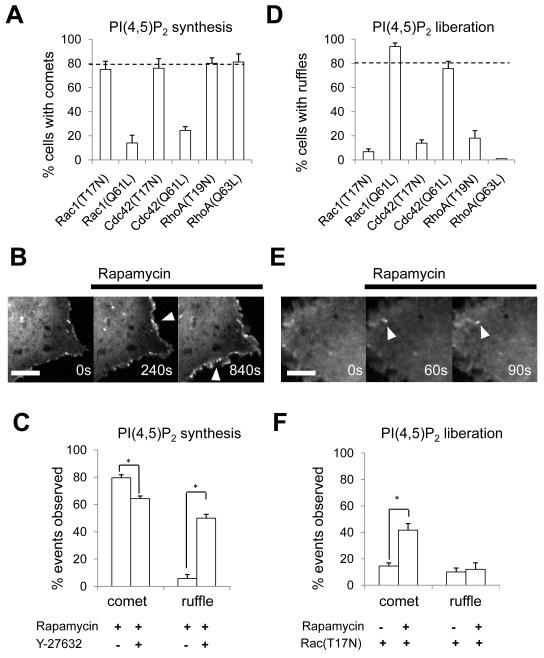
Next, w e addressed the molecular mechanisms that give rise to these distinct actin phenotypes. The Rho-family guanosine triphosphatases (GTPases), which include Rac, Cdc42, and RhoA, act as molecular switches capable of orchestrating actin dynamics (15, 33). The activity of the Rho GTPases is regulated by phosphoinositides (33) suggesting that they could provide a link between the effects of our manipulations on phosphoinositide concentration and the actin phenotypes we observed. Therefore, we employed constitutively active or dominant negative mutants of Rho GTPases to dissect the mechanisms underlying the PI(4,5)P2-induced actin phenotypes. Constitutively active Rac or Cdc42 significantly attenuated PI(4,5)P2 synthesis-induced formation of actin comets (Fig. 4A, p < 0.01, see Table S3), whereas dominant negative mutants of Rac or Cdc42 did not affect actin comet formation. This combination suggests that Rac and Cdc42 are not required for comet formation but that the activity of Rac and Cdc42 must be suppressed for comet formation. Neither constitutively active nor dominant negative forms of RhoA affected actin comet formation (Fig. 4A), suggesting that either RhoA does not play a role in comet formation. As these dominant mutant experiments do not directly test involvement of Rho GTPase-activating (GAP) proteins, it is also possible that RhoA plays a role by being activated through suppression of Rho GAPs. To test the latter possibility, we inhibited the RhoA downstream effector Rho-associated kinase (ROCK). In the presence of the ROCK inhibitor Y-27632, we observed phenotype conversion; cells exhibited membrane ruffling and diminished actin comet formation with PI(4,5)P2 synthesis (Fig. 4, B and C). This suggests that ROCK activity (and, therefore, RhoA signaling) is required for formation of actin comets and may need to be suppressed for membrane ruffling. Y-27632 administration over long periods of time can decrease PI(4,5)P2 synthesis, suggesting that PI(4)P5K could be a downstream target of Rho-ROCK signaling (34). However, we observed no effect of Y-27632 on the recovery of KCNQ2/3 currents after rapid PI(4,5)P2 dephosphorylation at the D5 position, one measure of PI4(P)5K activity (23) (fig. S7), supporting the hypothesis that ROCK acts downstream of PI(4,5)P2.
Figure 4. Crosstalk between Rho GTPases regulates PI(4,5)P2-induced actin remodeling.
(A and D) Effect of dominant mutants of Rho GTPases on comet formation in response to PI(4,5)P2 synthesis (A) or ruffle formation in response to PI(4,5)P2 liberation (D). The bar graphs represent scoring of actin phenotype in cells expressing CFP-FKBP-PI(4)P5K, Lyn11-FRB, YFP-Evl, and mCherry-tagged dominant negative or constitutively active Rho GTPases (A) and CFP-FKBP-PH(PLCδ), mCherry-FRB-MoA and YFP tagged dominant negative or constitutively active Rho GTPase (D). The following dominant mutants were used: constitutively active: RacQ61L, Cdc42Q61L, RhoAQ63L, dominant negative: RacT17N, Cdc42T17N, RhoAT19N. Values represent mean ± SEM (n ≥ 30, from three to four independent experiments). (B-F) Conversion of actin phenotypes induced by perturbation of Rho GTPase signaling. Confocal fluorescence microscopy images of actin cytoskeleton in COS-7 cells before and after PI(4,5)P2 synthesis in the presence of Y-27632 (B), or PI(4,5)P2 liberation in the presence of Rac(T17N) (E). Arrowheads indicate actin phenotypes. Scale bars, 10 μm. The observed actin phenotypes were quantified by counting the number of cells showing comets or ruffles under these conditions (C and F). Values represent mean ± SEM (n ≥ 30 cells, from three independent experiments). Asterisks indicate p<0.01. Single-letter abbreviations for the amino acid residues are as follows: L, Leu; N, Asn; Q, Gln; T, Thr. All p-values are summarized in Table S3.
In contrast to their lack of effect on comet formation with PI(4,5)P2 synthesis, dominant negative Rac or Cdc42 suppressed membrane ruffling in response to PI(4,5)P2 liberation (Fig. 4D), suggesting that Rac and Cdc42 are necessary for this process. Both constitutively active and dominant negative forms of RhoA suppressed membrane ruffling in response to PI(4,5)P2 liberation, suggesting that either a cycle of GDP-GTP exchange is required for ruffle formation, or that there is a narrow concentration window at which active RhoA promotes ruffle formation (35). In the presence of dominant negative Rac, cells developed actin comets in response to PI(4,5)P2 liberation (Fig. 4, E and F). Together with the ruffles observed in response to PI(4,5)P2 synthesis in the presence of the ROCK inhibitor (Fig. 4, B and C), these phenotypic conversions underline the mutually exclusive actin phenotypes shown in Fig. 3E.
To further characterize the pathways, we visualized the dynamics of actin binding proteins such as N-WASP and cofilin, major participants in PI(4,5)P2-dependent actin reorganization (8). N-WASP accumulated at the tip of comets in response to PI(4,5)P2 synthesis, whereas cofilin localized at the shaft region of comets (fig. S8A). In contrast, both N-WASP and cofilin localized to ruffles induced by PI(4,5)P2 liberation (fig. S8B).
Endocytosis is required for PI(4,5)P2-induced actin remodeling
PI(4,5)P2 also interacts with dynamin and promotes endocytosis (36), and actin comets are closely associated with regions of endocytic and exocytic activity (9). To investigate the possible role of membrane trafficking in the actin phenotypes we observed in response to PI(4,5)P2 manipulation, we used a dominant-negative mutant form of dynamin 2 [Dyn2(K44A)] to inhibit endocytosis. Dyn2(K44A) significantly inhibited the formation of PI(4,5)P2 synthesis-induced actin comets (Fig. 5A, p. <0.01, see Table S3), but not that of PI(4,5)P2 liberation-induced ruffles (Fig. 5B), suggesting that synthesis of PI(4,5)P2 triggers endocytosis through dynamin. If PI(4,5)P2 liberation induces endocytosis, this process is not required for the subsequent ruffling activity.
Figure 5. Endocytosis regulates PI(4,5)P2-induced actin remodeling.
(A and B) Confocal fluorescence microscopy images of COS-7 cells expressing either wild type dynamin2 [Dyn2(WT), top panels] or dominant negative [Dyn2(K44A), bottom panels] before and after PI(4,5)P2 manipulation: PI(4,5)P2 synthesis (A) and PI(4,5)P2 liberation (B). Images are from YFP-Evl (A) or YFP-labeled Dyn2(WT) and Dyn2(K44A) (B). The observed actin phenotypes were quantified by counting the number of cells showing comets in response to PI(4,5)P2 synthesis (A) or ruffles in response to PI(4,5)P2 liberation (B). (C) Proposed model for the distinct actin phenotypes elicited by PI(4)P and PI(4,5)P2. (D) Reconstituting actin comet formation. Confocal fluorescence microscopy images of actin cytoskeleton of cells expressing inactive “Mock” protein (mCherry, top rows), RhoA(Q63L) (middle rows), or Rac1(T17N) (bottom rows) before and after rapid activation of Arf6. Arrowheads indicate actin phenotypes. Scale bars, 20 μm. The observed actin phenotypes were quantified on the right by counting the number of cells showing comets or ruffles under these conditions. Values represent mean ± SEM (n ≥ 30, from three independent experiments). Asterisks indicate p<0.01. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; K, Lys. All p-values are summarized in Table S3.
Collectively, or results suggested a working model for distinct roles for PI(4)P and PI(4,5)P2 in actin remodeling by (Fig. 5C). In this model, a PI(4,5)P2 increase alone is sufficient to trigger membrane ruffling through activation of the Rac and Cdc42 pathway. In contrast, PI(4,5)P2 increase combined with a PI(4)P reduction drives actin comet formation in a manner that requires two signaling processes: PI(4,5)P2-induced endocytosis and PI(4)P-mediated activation of the RhoA- ROCK pathway. Our model predicts that a decrease in PI(4)P alone will not elicit comets or ruffles. In order to test this, we treated cells with wortmannin, which decreases PI(4)P at the PM but not PI(4,5)P2 (21, 37), though wortmannin is also a potent inhibitor of PI3K. Consistent with the model, we did not observe comets or ruffles in response to wortmannin treatment (fig. S9).
Dynamin associates with the head region of actin comets (38, 39), raising the possibility that dynamin plays a role in comet formation through direct actin remodeling independent of its role in endocytosis. In order to further define the role of dynamin in comet formation, we rapidly induced endocytosis in the presence of active RhoA (fig. S10A) through a procedure that did not involve manipulation of phosphoinositides but relied on activation of the small GTPase Arf6 at the PM (fig. S10B). We recruited an Arf6 GEF (guanine nucleotide exchange factor, i.e. an activator) fused to FKBP (FKBP-Sec7) to PM-targeted FRB, a manipulation that induced endocytosis (fig. S8C and D). When we rapidly induced endocytosis with FKBP-Sec7in the presence of constitutively active RhoA, we observed actin comet formation (Fig. 5D). We also observed actin comets in the presence of dominant negative Rac1 (Fig. 5D), further supporting the reciprocal antagonism of RhoA and Rac GTPases (Fig. 5C).
DISCUSSION
Mutually inhibitory Rac-RhoA signaling has been observed in biological processes including cell polarization, where such crosstalk helps to spatially segregate of Rac and RhoA activity (40). In this study, we found a signaling network dominated by strong mutual inhibition between components mediating actin comets (RhoA, ROCK, and endocytosis) and components mediating membrane ruffles (Rac and Cdc42). This crosstalk seems to function as a driving force to sharply define actin phenotypes based on differential input signals originating from membrane lipids in a substrate-product relationship.
Under physiological conditions, there are four main ways whereby extracellular stimuli regulate PI(4,5)P2 concentration. These are: i) PLC-mediated conversion of PI(4,5)P2 into inositol phosphates and diacylglycerol, ii) PI(4)P5K-mediated phosphorylation of PI(4)P, iii) PI3K-mediated phosphorylation of PI(4,5)P2, and iv) masking and unmasking of PI(4,5)P2 by binding proteins such as myristoylated alanine-rich C-kinase substrate (5). Each mode is associated with a unique combination of changes in the concentration of lipid products. Previous work has shown that decreased PI(4,5)P2 concentration in response to acetylcholine receptor stimulation (mode i) deactivate ion channel activity (13). Here, we manipulated PI(4,5)P2 synthetically to mimic modes ii and iv and found that PI(4)P and PI(4,5)P2 play distinct roles in remodeling of the actin cytoskeleton. Our findings indicate that the consequence of an increase in PI(4,5)P2 will depend on accompanying changes in different but closely related signaling molecules such as PI(4)P. Such a combinatorial regulation of downstream signals may at least partly explain the ability of a limited set of membrane lipids to exhibit such functional diversity.
Our model proposes that RhoA and ROCK signaling are activated downstream of PI(4)P. There seems to be no report of direct regulation of RhoA by PI(4)P. However, there are dozens of GEFs and GAPs for RhoA that contain lipid binding domains (41), making them ideal candidates for PI4P-mediated RhoA regulation. Indeed, phospholipids have been reported to modulate activity of p190 RhoGAP, one of the Rho GAPs (42). PI4P has long been viewed as a precursor of PI(4,5)P2, and has only recently begun to be appreciated as a signaling molecule (18). Our data provide support for such a role.
A Lowe syndrome is a genetic disease with defective 5-phosphatase activity for PI(4,5)P2 that causes various developmental defects including eye, kidney and brain problems. Of particular note, actin comets have been observed in fibroblasts from Lowe syndrome patients (43). These fibroblasts also exhibited cell migration defects (44). As membrane ruffles offer the driving force for the cell motility, it is possible that the cell migration defects of these fibroblasts are due to a lack of ruffles through the mutual inhibition between actin comets and ruffles. Our study provides not only a powerful technology for probing the complexity of phosphoinositide signaling, but also insights into the molecular mechanism of this currently incurable disease.
Materials and Methods
Cell culture, transfection, and time-lapse fluorescence imaging
Culture of HeLa, tsA201, and NIH3T3 cells was performed as described previously (13, 45). HEK293 and COS-7 cells were maintained in DMEM (Dulbecco’s modified Eagle’s medium; GIBCO 11995) supplemented with 10% (v/v) fetal calf serum, 50 units/ml penicillin and 50 μg/ml streptomycin. Cells were passaged every other day. Plasmid transfection of Cos7, HeLa, HEK293, and NIH3T3 cells was performed with Lipofectamine2000. After plating on poly-L-lysine-coated coverslips, all cells were maintained in DMEM supplemented with 10% FBS for 12-24 h before fluorescence imaging started. Typically, we took images every 15 s at room temperature and processed them using analysis software (Metamorph, Molecular Devices). In all experiments concerning PI(4,5)P2, cells were incubated for ≥3 hours in serum-free conditions before imaging. Phalloidin staining was conducted as follows: Cells were incubated with 100 nM rapamycin for 15 min at room temperature prior to fixation with 1% glutaraldehyde. The cells were then stained with Alexa488-Phalloidin with 0.1% Triton-X100.
Confocal fluorescence microscopy
Time-lapse live cell imaging using the confocal fluorescence microscope was conducted as previously described (17). Briefly, CFP and YFP excitations were conducted with helium-cadmium laser and argon laser (CVI-Melles Griot), respectively. The two lasers were fiber-coupled (OZ optics) to the spinning disk confocal unit (CSU10; Yokogawa) mounted with dual dichroic mirrors for CFP and YFP (Semrock). The lasers were processed with appropriate filter sets for CFP and YFP (Chroma Technology) to capture fluorescence images with a CCD camera (Orca ER, Hamamatsu Photonics), driven by Metamorph 7.5 imaging software (Molecular Devices). Images were taken using a 40× objective (Zeiss, NA 1.30) mounted on an inverted Axiovert 200 microscope (Zeiss).
Total Internal Reflection Fluorescence Microscopy
TIRF imaging was done with a Zeiss AxioObserver microscope (Zeiss) (46). The excitation laser was a Coherent Sapphire (488 nm, 50 mW, Coherent). The laser was coupled to a Zeiss TIRF slider via fiber optics (KineFLEX, Point Source). A Z488RDC dichroic mirror (Chroma Technology) was used to reflect the incoming laser onto a Zeiss a plan 100× objective (N.A. = 1.45, Zeiss). An ET525/50 emission filter was used for GFP-PH(OSBP) fluorescence detection (Chroma Technology). An EMCCD camera (ImagEM C9100-13; Hamamatsu Photonics) was used as detector. To detect dim signals, the EMCCD gain was set to maximal. The camera was maintained at −85 °C during the experiment using a JULABO HF25-ED heating and refrigerated circulator (JD Instruments). A Uniblitz LS6ZM2 shutter controlled by VCM-D1 (Vincent Associates) was integrated between the laser head and the fiber launcher to control the laser. Data were acquired using SlideBook (Intelligent Imaging Innovations). F/F0 traces were calculated from the TIRF signal intensity (F) of GFP-PH(OSBP), averaged over the region of interest, and the initial fluorescence intensity (F0).
Electrophysiology
KCNQ currents were recorded as previously (13, 23) in a whole-cell, gigaseal voltage clamp of tsA201 cells transiently transfected with KCNQ4 K channels. Internal solution contained (in mM) 175 KCl, 5 MgCl2, 5 HEPES, 0.1 K4BAPTA, 3 Na2ATP, 0.1 Na3GTP, pH 7.4 (KOH). External solution contained (in mM) 160 NaCl, 2.5 KCl, 2 CaCl2, 1 MgCl2, 10 HEPES, 8 glucose, pH 7.4 (NaOH). Recordings used an EPC9 amplifier with Patchmaster 2.35 software (HEKA). Currents were filtered at 2.9 kHz. Sample intervals were 200 μs. Holding potential was −60 mV. KCNQ current was quantified by measuring tail currents: Every 5 s, the membrane potential was depolarized to −20 mV for 300 ms and then repolarized to −60 mV. KCNQ2/3 current activates slowly upon depolarization and deactivates slowly upon repolarization. KCNQ2/3 tail currents were measured by comparing current at 20 ms and 400 ms after repolarization to −60 mV. Simultaneously we recorded FRET (Förster resonance energy transfer) time courses as previously (23) using a two-channel photometry system with Polychrome IV (TILL Photonics) for excitation, a three-color dichroic mirror (89006bs “CFP, YFP, mCherry”, Chroma) and two photodiodes (TILL Photonics) with D480/40 and ET535/30 emission filters (Chroma). We recorded CFPC (440 nm excitation, 480 nm emission), and raw YFPC (440 nm excitation, 535 nm emission) every 5 s and calculated FRETr = YFPC/CFPC (in arbitrary units, AU) after subtracting background and correcting raw YFPC for CFP emission at 535 nm (YFPC = raw YFPC - 0.7*CFPC).
Modeling
A kinetic model of rapamycin-induced translocation and phosphoinositide metabolism was formulated as previously (23) as a compartmental model in the Virtual Cell framework (University of Connecticut). The Virtual Cell Model, “Ueno2011”, is publicly available at http://www.vcell.org/ under shared models/bfalken. Model equations and initial conditions are listed in supplemental Table S2.
Design of constructs
Sequences encoding the human FRB (T2098L) domain of mTOR and human FKBP12 were used for generating the following dimerization constructs. For CFP-FKBP-PH(PLCδ) and YFP-FKBP-PH(PLCδ) vector construction, YFP of YFP-PH(PLCδ) vector was replaced with sequence encoding NheI and EcoRI digested fragment of CFP-FKBP vector or YFP-FKBP vector. For Tubby-CFP-FKBP or Tubby-YFP-FKBP vector construction, a PCR product encoding Tubby domain of Tubby protein (a kind gift from Dr. Andrew Tinker, University College London, UK) (47) was digested using EcoRI and BamHI, and then inserted into the artificially introduced digestion site of the pEGFP-C1 vector in which EGFP was replaced with sequence encoding CFP-FKBP or YFP-FKBP. For CFP-FKBP-C2(Lact) and YFP-FKBP-C2(Lact) vector construction, C2 domain of lactadherin from YFP-C2(Lact) (a kind gift from Dr. Won Do Heo, KAIST, Korea) was subjected to PCR to provide EcoRI and BamHI cleavage sites at its 5′ and 3′ ends. The PCR product was inserted using EcoRI and BamHI restriction enzymes into CFP-FKBP and YFP-FKBP plasmids. For CFP-FKBP-C2(Lact) or YFP-FKBP-C2(Lact) vector construction, a PCR product encoding C2 domain of lactadherin was digested using EcoRI and BamHI, and then inserted into the multiple cloning site of the pEGFP-C1 vector in which EGFP was replaced with sequence encoding CFP-FKBP or YFP-FKBP. For mCherry-PH(PLCd) and mCherry-PI(4)P5K vector construction, YFP of YFP-PH(PLCδ) and CFP-FKBP of CFP-FKBP-PI(4)P5K(13) was replaced with sequence encoding mCherry. For the construction of CFP-FKBP-Sec7, the ArfGEF domain (Sec7: cytohesin-3 77-258aa) from human cytohesin-3 (a kind gift from Tobias Meyer, Stanford University, USA, NM_004227) was subcloned into Cerulean2-FKBP plasmid using XhoI and KpnI after performing PCR reaction for Sec7 to have these restriction enzyme sites, and then inserted into the multiple cloning site of pEGFP C1 vector in which EGFP was replaced with sequence encoding Cerulean2-FKBP. (Cerulean2 is a variant of CFP, a kind gift from Mark A Rizzo, University of Maryland Baltimore, USA).
Statistical analysis
Bars and markers in the figures represent mean ± SEM. Statistical analysis was performed using an unpaired two-tailed Student’s t-test assuming the two populations have the same variances. All analyses involved at least 30 cells in at least 3 separate experiments; statistical analyses and p-values are summarized in Supplemental Table S3.
Supplementary Material
Acknowledgements
We thank T. Balla for the PH(OSBP) plasmid, T. Andrew for the Tubby plasmid, W.D. Heo for the dominant mutants of Rho GTPases and C2(Lact) plasmids, J. Donaldson for the Dynamin2 (WT and K44A) plasmids, M. Rizzo for the mCerulean2 plasmid, and ARIAD Pharmaceuticals, Inc. for FKBP and FRB plasmids. We extend our appreciation to R. Huganir and Y. Araki for help with TIRF measurements, also to D. Montell, A. Ewald, B. Hille, M. Fivaz, and Inoue lab members for valuable discussion and comments on the manuscript. Funding: Our research is supported by NIH grants DK090868, MH084691 and GM092930 (to T.I.). T.U. is a recipient of a fellowship from the Japan Society for the Promotion of Science. B.F. is a long-term fellow of the Human Frontier Science Program and funded by NIH grant NS08174 (to B. Hille). The Virtual Cell is supported by NIH grant P41RR013186 from the National Center for Research Resources.
Footnotes
Author contributions: T.U. and T.I. conceived an original idea of PI(4,5)P2 manipulation techniques. T.U., B.H.F., C.P. and T.I. designed and conducted experiments with following data analysis. T.U., B.H.F., and T.I. wrote the manuscript.
Competing interest: The authors declare no competing interest.
REFERENCES AND NOTES
- 1.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 2.Field SJ, Madson N, Kerr ML, Galbraith KA, Kennedy CE, Tahiliani M, Wilkins A, Cantley LC. PtdIns(4,5)P2 functions at the cleavage furrow during cytokinesis. Curr Biol. 2005;15:1407–1412. doi: 10.1016/j.cub.2005.06.059. [DOI] [PubMed] [Google Scholar]
- 3.Ling K, Schill NJ, Wagoner MP, Sun Y, Anderson RA. Movin’ on up: the role of PtdIns(4,5)P(2) in cell migration. Trends Cell Biol. 2006;16:276–284. doi: 10.1016/j.tcb.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Martin TF. PI(4,5)P(2) regulation of surface membrane traffic. Curr Opin Cell Biol. 2001;13:493–499. doi: 10.1016/s0955-0674(00)00241-6. [DOI] [PubMed] [Google Scholar]
- 5.McLaughlin S, Wang J, Gambhir A, Murray D. PIP(2) and proteins: interactions, organization, and information flow. Annu Rev Biophys Biomol Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- 6.Suh BC, Hille B. Regulation of ion channels by phosphatidylinositol 4,5-bisphosphate. Curr Opin Neurobiol. 2005;15:370–378. doi: 10.1016/j.conb.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Yeung T, Ozdamar B, Paroutis P, Grinstein S. Lipid metabolism and dynamics during phagocytosis. Curr Opin Cell Biol. 2006;18:429–437. doi: 10.1016/j.ceb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Yin HL, Janmey PA. Phosphoinositide regulation of the actin cytoskeleton. Annu Rev Physiol. 2003;65:761–789. doi: 10.1146/annurev.physiol.65.092101.142517. [DOI] [PubMed] [Google Scholar]
- 9.Rozelle AL, Machesky LM, Yamamoto M, Driessens MH, Insall RH, Roth MG, Luby-Phelps K, Marriott G, Hall A, Yin HL. Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr Biol. 2000;10:311–320. doi: 10.1016/s0960-9822(00)00384-5. [DOI] [PubMed] [Google Scholar]
- 10.Santarius M, Lee CH, Anderson RA. Supervised membrane swimming: small G-protein lifeguards regulate PIPK signalling and monitor intracellular PtdIns(4,5)P2 pools. Biochem J. 2006;398:1–13. doi: 10.1042/BJ20060565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCrea HJ, De Camilli P. Mutations in phosphoinositide metabolizing enzymes and human disease. Physiology (Bethesda) 2009;24:8–16. doi: 10.1152/physiol.00035.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ooms LM, Horan KA, Rahman P, Seaton G, Gurung R, Kethesparan DS, Mitchell CA. The role of the inositol polyphosphate 5-phosphatases in cellular function and human disease. Biochem J. 2009;419:29–49. doi: 10.1042/BJ20081673. [DOI] [PubMed] [Google Scholar]
- 13.Suh BC, Inoue T, Meyer T, Hille B. Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 2006;314:1454–1457. doi: 10.1126/science.1131163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varnai P, Thyagarajan B, Rohacs T, Balla T. Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J Cell Biol. 2006;175:377–382. doi: 10.1083/jcb.200607116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 16.Merrifield CJ, Moss SE, Ballestrem C, Imhof BA, Giese G, Wunderlich I, Almers W. Endocytic vesicles move at the tips of actin tails in cultured mast cells. Nat Cell Biol. 1999;1:72–74. doi: 10.1038/9048. [DOI] [PubMed] [Google Scholar]
- 17.Komatsu T, Kukelyansky I, McCaffery JM, Ueno T, Varela LC, Inoue T. Organelle-specific, rapid induction of molecular activities and membrane tethering. Nat Methods. 7:206–208. doi: 10.1038/nmeth.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Angelo G, Vicinanza M, Di Campli A, De Matteis MA. The multiple roles of PtdIns(4)P -- not just the precursor of PtdIns(4,5)P2. J Cell Sci. 2008;121:1955–1963. doi: 10.1242/jcs.023630. [DOI] [PubMed] [Google Scholar]
- 19.Golebiewska U, Nyako M, Woturski W, Zaitseva I, McLaughlin S. Diffusion coefficient of fluorescent phosphatidylinositol 4,5-bisphosphate in the plasma membrane of cells. Mol Biol Cell. 2008;19:1663–1669. doi: 10.1091/mbc.E07-12-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammond GR, Schiavo G, Irvine RF. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2) Biochem J. 2009;422:23–35. doi: 10.1042/BJ20090428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falkenburger BH, Jensen JB, Hille B. Kinetics of M1 muscarinic receptor and G protein signaling to phospholipase C in living cells. J Gen Physiol. 2010;135:81–97. doi: 10.1085/jgp.200910344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falkenburger BH, Jensen JB, Hille B. Kinetics of PIP2 metabolism and KCNQ2/3 channel regulation studied with a voltage-sensitive phosphatase in living cells. J Gen Physiol. 2010;135:99–114. doi: 10.1085/jgp.200910345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez CC, Falkenburger B, Shapiro MS. Affinity for phosphatidylinositol 4,5-bisphosphate determines muscarinic agonist sensitivity of Kv7 K+ channels. J Gen Physiol. 2009;134:437–448. doi: 10.1085/jgp.200910313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suh BC, Hille B. Recovery from muscarinic modulation of M current channels requires phosphatidylinositol 4,5-bisphosphate synthesis. Neuron. 2002;35:507–520. doi: 10.1016/s0896-6273(02)00790-0. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Craciun LC, Mirshahi T, Rohacs T, Lopes CM, Jin T, Logothetis DE. PIP(2) activates KCNQ channels, and its hydrolysis underlies receptor-mediated inhibition of M currents. Neuron. 2003;37:963–975. doi: 10.1016/s0896-6273(03)00125-9. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Gamper N, Hilgemann DW, Shapiro MS. Regulation of Kv7 (KCNQ) K+ channel open probability by phosphatidylinositol 4,5-bisphosphate. J Neurosci. 2005;25:9825–9835. doi: 10.1523/JNEUROSCI.2597-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balla A, Tuymetova G, Tsiomenko A, Varnai P, Balla T. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol Biol Cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- 30.Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L. G-protein signaling through tubby proteins. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- 31.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 32.Jensen JB, Lyssand JS, Hague C, Hille B. Fluorescence changes reveal kinetic steps of muscarinic receptor-mediated modulation of phosphoinositides and Kv7.2/7.3 K+ channels. J Gen Physiol. 2009;133:347–359. doi: 10.1085/jgp.200810075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto M, Hilgemann DH, Feng S, Bito H, Ishihara H, Shibasaki Y, Yin HL. Phosphatidylinositol 4,5-bisphosphate induces actin stress-fiber formation and inhibits membrane ruffling in CV1 cells. J Cell Biol. 2001;152:867–876. doi: 10.1083/jcb.152.5.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Symons M, Settleman J. Rho family GTPases: more than simple switches. Trends Cell Biol. 2000;10:415–419. doi: 10.1016/s0962-8924(00)01832-8. [DOI] [PubMed] [Google Scholar]
- 36.Schmid SL, McNiven MA, De Camilli P. Dynamin and its partners: a progress report. Curr Opin Cell Biol. 1998;10:504–512. doi: 10.1016/s0955-0674(98)80066-5. [DOI] [PubMed] [Google Scholar]
- 37.Nakanishi S, Catt KJ, Balla T. A wortmannin-sensitive phosphatidylinositol 4-kinase that regulates hormone-sensitive pools of inositolphospholipids. Proc Natl Acad Sci U S A. 1995;92:5317–5321. doi: 10.1073/pnas.92.12.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee E, De Camilli P. Dynamin at actin tails. Proc Natl Acad Sci U S A. 2002;99:161–166. doi: 10.1073/pnas.012607799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orth JD, Krueger EW, Cao H, McNiven MA. The large GTPase dynamin regulates actin comet formation and movement in living cells. Proc Natl Acad Sci U S A. 2002;99:167–172. doi: 10.1073/pnas.012607899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iden S, Collard JG. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat Rev Mol Cell Biol. 2008;9:846–859. doi: 10.1038/nrm2521. [DOI] [PubMed] [Google Scholar]
- 41.Moon SY, Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003;13:13–22. doi: 10.1016/s0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
- 42.Ligeti E, Dagher MC, Hernandez SE, Koleske AJ, Settleman J. Phospholipids can switch the GTPase substrate preference of a GTPase-activating protein. J Biol Chem. 2004;279:5055–5058. doi: 10.1074/jbc.C300547200. [DOI] [PubMed] [Google Scholar]
- 43.Allen PG. Actin filament uncapping localizes to ruffling lamellae and rocketing vesicles. Nat Cell Biol. 2003;5:972–979. doi: 10.1038/ncb1059. [DOI] [PubMed] [Google Scholar]
- 44.Coon BG, Mukherjee D, Hanna CB, Riese DJ, 2nd, Lowe M, Aguilar RC. Lowe syndrome patient fibroblasts display Ocrl1-specific cell migration defects that cannot be rescued by the homologous Inpp5b phosphatase. Hum Mol Genet. 2009;18:4478–4491. doi: 10.1093/hmg/ddp407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Araki Y, Lin DT, Huganir RL. Plasma membrane insertion of the AMPA receptor GluA2 subunit is regulated by NSF binding and Q/R editing of the ion pore. Proc Natl Acad Sci U S A. 2010;107:11080–11085. doi: 10.1073/pnas.1006584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quinn KV, Behe P, Tinker A. Monitoring changes in membrane phosphatidylinositol 4,5-bisphosphate in living cells using a domain from the transcription factor tubby. J Physiol. 2008;586:2855–2871. doi: 10.1113/jphysiol.2008.153791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.