Abstract
The ubiquitin-dependent proteasome system plays a critical role in many cellular processes and pathogenesis of various human diseases, including cancer. Although there are a large number of E3 ubiquitin ligases, the majority are RING-finger type E3s. Pirh2, a target of p53 transcription factor, contains a highly conserved C3H2C3 type RING domain. Importantly, Pirh2 was found to regulate a group of key factors dedicated to the DNA damage response, such as p53, p73, PolH, and c-Myc. Interestingly, Pirh2 was upregulated or downregulated in different types of cancers. These suggest that Pirh2 is implicated in either promoting or suppressing tumor progression in a tissue-dependent manner. This review will focus on the major findings in these studies and discuss the potential to explore Pirh2 as a cancer therapeutic target.
Keywords: Pirh2, E3 ligase, tumorigenesis, cancer therapy
Introduction
The ubiquitin-proteasome system is one of the critical mechanisms controlling protein turnover and thus maintains cellular protein homeostasis. Although protein ubiquitination is catalyzed by a highly ordered enzymatic cascade, including ubiquitin-activating enzyme E1s, ubiquitin-conjugating enzyme E2s, and ubiquitin ligase E3s, the last of which primarily determine the substrate specificity [1]. E3 ubiquitin ligases contain three major groups, RING (the real interesting new gene) finger domain containing E3s, HECT (the homologous to E6-AP carboxyl terminus) domain E3s, and U-box proteins. Intriguingly, the majority of identified E3 ubiquitin ligases contain a RING-finger domain. Evidence showed that RING-finger E3 ubiquitin ligases target substrate proteins involved in many cellular processes, such as cell proliferation, differentiation, DNA repair, apoptosis, and metabolism [2]. As a result, aberrant activities of RING-finger E3 ubiquitin ligases are correlated with the pathogenesis of various human diseases, including cancer. For example, in multiple cancers, decreased expression of the p53 tumor suppressor is correlated with amplified expression of its ubiquitin ligases, such as Mdm2 and COP1 [3,4]. Therefore, understanding how E3s are implicated in tumorigenesis will provide clues for developing anti-E3s based cancer therapeutics. Indeed, small molecular inhibitors of Mdm2 have shown to be a promising treatment for tumors with elevated Mdm2 but decreased p53 [5–7]. However, the importance of Pirh2, a target and a ubiquitin E3 ligase of p53, in tumorigenesis is emerging. It appears that in addition to p53, Pirh2 regulates multiple other factors, including p73, p27, PolH (DNA polymerase eta), and c-Myc [8–12]. In this review, we will provide an overview of current findings on Pirh2 and discuss the potential to develop Pirh2 as a new target for cancer therapy.
1. Pirh2 and its isoforms
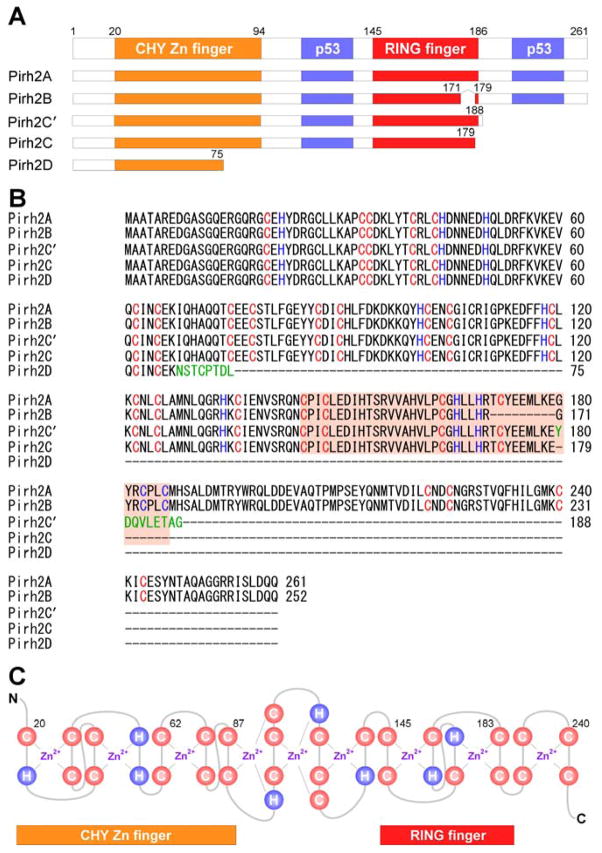
Human Pirh2 (p53-induced RING-H2 protein), also known as Rchy1 (RING-finger and CHY-zinc-finger domain-containing protein 1), is encoded by the RCHY1 gene which contains nine exons and located on the chromosome 4p21.1. To date, p53 is the only transcription factor known to directly activate the promoter of the RCHY1 gene [13]. Two p53 homologous, p63 and p73, share high sequence and structure identity with p53 and regulate some p53 target genes [14–16]. Therefore, p63 and p73 are likely to regulate Pirh2 expression. Evidence also showed that at least five isoforms of Pirh2 protein, named as Pirh2A, B, C, C′ (also called Pirh2b), and D, are generated by alternative splicing (Fig. 1). Pirh2A (full-length Pirh2) is composed of 261 amino acids and contains the N-terminal CHY-Zn-finger domain, the central RING-finger domain, and the C-terminal domain (CTD) (Fig. 1A). Due to alternative splicing, Pirh2B lacks exon 7, which encodes amino acids 171 to 179 of the RING domain [17] (Fig. 1B). Due to the usage of a second donor site in intron 7 and generation of a premature stop codon, Pirh2C lacks the last seven amino acids (180 to 186) of the RING domain and the entire downstream C-terminal sequence [17] (Fig. 1B). Due to the usage of an alternative 5′ splice site in exon 8 and a 38-nucleotide deletion in the 5′ end of exon 8 along with a premature stop codon, Pirh2C′ contains identical amino acids to Pirh2C plus additional 9 unique amino acids at the C-terminus [18] (Fig. 1B). Due to insertion of an “A” that shifts the reading frame and generation of a premature stop codon, Pirh2D only contains 75 amino acids from the N-terminal of Pirh2A plus 8 unique amino acids [19] (Fig. 1B). Pirh2 contains nine zinc binding sites, with six in the CHY-Zn-finger domain, two in the RING domain, and one in the CTD [20] (Fig. 1C). Since the consensus RING-H2 (C3H2C3) finger domain is required for Pirh2 to act as an E3 ubiquitin ligase in vitro and in vivo [21], Pirh2B, Pirh2C, Pirh2C′, and Pirh2D lack the intrinsic ubiquitin ligase function because of truncation or deletion of the RING domain [17–19]. However, Pirh2B, Pirh2C, and Pirh2C′ are still capable of binding to p53. Interestingly, it has been shown that Pirh2B and Pirh2C not only promote p53 ubiquitination and degradation through Mdm2, but also inhibit p53 transcriptional activity [17]. However, it is not clear how Pirh2C′ and Pirh2D regulate p53 activity. Importantly, these alternative spliced isoforms of Pirh2 are naturally expressed in multiple tissues. Therefore, it is likely that all Pirh2 isoforms play a role in tumorigenesis, but the mechanism needs to be further explored.
Fig. 1.
Schematic representation of Pirh2 and isoforms. (A) The domains of the human Pirh2 protein and the binding sites of p53 are indicated. (B) Sequence alignment of full-length Pirh2 and its isoforms using ClustalW2 multiple sequence alignment program. Pirh2C′ and Pirh2D have an additional unique amino acid (shown in green). (C) Secondary sequence organization of the CHY-zinc-finger/RING-finger domain. The cysteine and histidine are labeled as C and H, respectively. There are nine potential interleaved zinc binding sites.
2. The binding partners and potential substrates/targets of Pirh2
Pirh2 was originally identified as an androgen receptor (AR) N-terminal interacting protein (ARNIP) [21], suggesting that Pirh2 is involved in the AR signaling pathway. Indeed, Pirh2 was found to enhance the recruitment of AR to the promoter of the PSA (prostate specific antigen) gene or promote the degradation of AR corepressor HDAC1 (histone deacetylase 1) [22]. In addition, Pirh2 is also found to inhibit androgen-dependent secretion of PSA via ubiquitination and degradation of -COP (-subunit of coatmer complex) [23]. These indicate that Pirh2 might promote prostate cancer formation. The oncogenic activity of Pirh2 is consistent with the findings that Pirh2 ubiquitinates and degrades multiple tumor suppressors, including p53, p73, and p27 [9,12,13,20]. By contrast, Hakem et al., showed that the c-Myc oncoprotein is a new Pirh2 binding partner and degraded by Pirh2-mediated polyubiquitination [8]. Meanwhile, it was found that Pirh2−/− mice with elevated expression of c-Myc are predisposed to plasma cell hyperplasia and tumorigenesis, which is further enhanced by p53-deficiency [8]. Together, Pirh2 might play a role in cell cycle progression via regulating checkpoint proteins and disrupting the balance between pro- and anti-cell proliferation activities of Pirh2 contributes to tumor formation in a tissue-specific manner.
To date, Pirh2 has been shown to associate with well over 20 proteins: some are Pirh2 targets and others are Pirh2 regulators (Table 1). For examples, histone acetyltransferase Tip60 (Tat-interactive protein of 60 kDa) and PLAGL2 (Pleomorphic adenoma gene like 2) [24,25] inhibit auto-ubiquitination and degradation of Pirh2 [17]. In addition, Pirh2 can enhance Mdm2 expression by interacting with SCYL1-BP1 (SCY1-like 1 binding protein 1), a regulator of Mdm2 auto-ubiquitination [26,27]. Moreover, Pirh2 can be increased by Mdm2 and COP1 coexpression [28], although the mechanism is not clear. Interestingly, we have showed that Pirh2 associates with and promotes ubiquitin-independent 20S proteasomal degradation of PolH, a Y-family translesion DNA polymerase required for bypassing UV-induced DNA damage [11]. In addition, we showed that Pirh2 monoubiquitinates PolH, which blocks PolH association with PCNA and prevents PolH from bypassing UV-induced DNA lesions [10].
Table 1.
Reported Pirh2-binding partners.
| Protein | Function | Substrate |
|---|---|---|
| AIG1 | Activates NFAT signaling pathway [68] | n.d. |
| AR | Mediates androgen signaling in prostate development and differentiation [21,22] | no |
| ARF4 | Stimulates the ADP-ribosyltransferase activity [69] | n.d. |
| Axin | Acts as a tumor suppressor and a negative regulator of Wnt-signaling [45] | no |
| CaMKII | Serine/threonine-specific protein kinase that is regulated by calmodulin complex [70] | no |
| c-Myc | An oncogenic transcription factor that is activated by mitogenic signals [8] | yes |
| COP1 | A negative regulator of p53 [28] | n.d. |
| PolH | A DNA Polymerase involved in DNA repair by translesion synthesis [10,11] | yes |
| -COP | A subunit of COPI coatomer complex that is involved in Golgi-trafficking [23] | yes |
| HDAC1 | A histone deacetyltransferase that is responsible for the deacetylation of histone [22] | yes |
| K8/18 | A major components of intermediate filaments in single layer epithelial cells [71] | no |
| Mdm2 | A major negative regulator of p53 [28] | n.d. |
| Mdmx | A enzymatically inactive component of Mdm2 complex [28] | n.d. |
| MV P | Contributes to innate immune evasion [72] | no |
| PCV2 ORF3 | Induces apoptosis and regulates p53 ubiquitination [73,74] | n.d. |
| p27 | Inhibits G1-S transition [9] | yes |
| p53 | A tumor suppressor [13,20] | yes |
| p73 | A member of p53-superfamily proteins [12,63] | yes |
| PLAGL2 | Acts as a transcription factor and suppresses cellular differentiation [25] | n.d. |
| SCYL1-BP1 | A member of the Golgin family which is involved in the secretary pathway [26,27] | yes |
| SR | A docking protein that is involved in targeting secretary protein to ER [75] | yes |
| Tip60 | A histone acetyltransferase that has a role in the DNA damage response [24] | n.d. |
AIG1, androgen induced gene 1; AR, androgen receptor; ARF4, ADP-ribosylation factor 4; CaMKII, calmodulin kinase II; COP1, constitutive photomorphogenesis protein1; PolH, DNA polymerase eta; HDAC1, histone deacetylase 1; Mdm2, murine double minute 2; PCV2 ORF3, porcine circovirus type 2 open reading frame 3; PLAGL2, pleomorphic adenoma gene like 2; SCYL1-BP1, SCY1-like 1 binding protein 1; SR, signal recognition particle receptor ; Tip60, tat-interactive protein of 60 kDa.
Additionally, there exist a few potential target proteins for Pirh2. First, p21 might be regulated by Pirh2. Previously, we showed that p21 is increased by Pirh2 knockdown, whereas decreased by Pirh2 overexpression in H1299 cells [11]. Since p21 degradation is regulated by several E3 ubiquitin ligases [29], the relationship among which needs to be further investigated. Second, p63 is a potential target protein for Pirh2-mediated degradation. p53, p63, and p73 share high structural similarity and can form mixed tetramers [30–32]. Since Pirh2 promotes degradation of p53 and p73, it is likely that Pirh2 regulates p63 stability.
3. The role of Pirh2 in the p53-Mdm2 axis
Mdm2, a RING-finger domain E3 ligase, is the first identified non-viral E3 ligase degrading p53 via the ubiquitin-proteasome system [33–37]. In addition, Mdm2 is also capable of regulating its own levels through auto-polyubiquitination [38]. As a transcriptional target of p53, Mdm2 acts as a negative feedback regulator of p53 to maintain low levels of p53 expression [39,40]. In addition to polyubiquitinating p53, Mdm2 mediates p53 monoubiquitination leading to nuclear export of p53 [41]. However, Mdm2 loses its biological activity to regulate p53 upon DNA damage. On the one hand, DNA damage-induced Mdm2 phosphorylation at Ser-395 disrupts Mdm2 interaction with p53 due to its conformational change [42]. On the other hand, DNA damage-induced phosphorylation of p53 at Ser-15 and Ser-20 dissociates p53 from Mdm2 [43,44]. Therefore, the p53-Mdm2 regulatory loop has been proposed to be a critical mechanism for degrading p53 in an unstressed condition (Fig. 2).
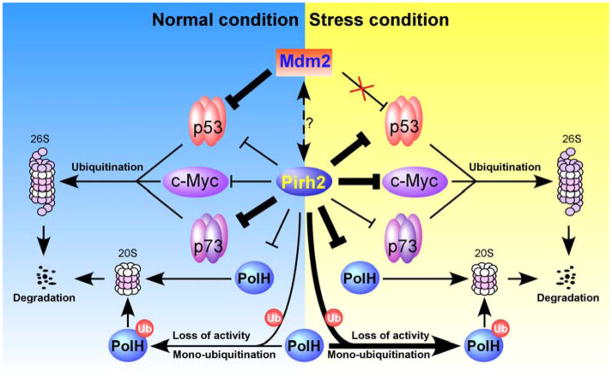
Fig. 2.
A model for the biological role of Pirh2 in the p53-Mdm2 axis. Under normal conditions, p53 is associated with Pirh2 and Mdm2 and thereby degraded by the 26S proteasome. Upon stresses, Mdm2 is released from p53, whereas Pirh2 become a primary negative regulator of p53.
Pirh2 is identified as the second E3 ligase promoting p53 degradation (Fig. 2). Like Mdm2, Pirh2 is a transcriptional target of p53 and can degrade p53 protein through the proteasomal degradation pathway [13,20]. Thus, p53 and Pirh2 constitute a negative regulatory feedback loop. Unlike Mdm2, Pirh2 is able to downregulate p53 in response to DNA damage. It has been shown that upon moderate DNA damage, Pirh2 suppresses Axin-HIPK2-mediated p53 phosphorylation at Ser-46 by competing with HIPK2 for binding to Axin, and thus inhibits p53-mediated apoptosis [45]. However, upon lethal DNA damage, Tip60 competes with and dissociates Pirh2 from Axin, and consequently, the assembly of Axin-Tip60-HIPK2-p53 protein complex induces p53-mediated apoptosis [45]. In addition, Pirh2−/− mice display enhanced p53 expression and DNA damage response [8]. Together, these suggest that Pirh2 acts as a negative regulator of p53 in both unstressed and stressed conditions. As mentioned above, Pirh2 isoforms lacking the RING domain are still able to degrade p53. Therefore, whether Pirh2-mediated ubiquitination is essential for p53 degradation and whether other factors are implicated in the p53-Pirh2 feedback loop need to be further investigated.
Similar to Mdm2 and Pirh2, COP1 (constitutively photomorphogenic 1), a p53 target, is a RING-finger E3 ligase that targets p53 for degradation [46]. It has been shown that Mdm2, Pirh2, and COP1 are capable of forming a dimer and mutually enhance their protein stability [28]. In addition, COP1 coordinates with Mdm2 and Pirh2 to enhance p53 degradation [46]. Thus, a proper level of p53 is ensured by a set of E3 ligases. Interestingly, p73 turnover is regulated by Pirh2, but not by Mdm2 and COP1 [47–49]. This suggests that Pirh2 also regulates its own substrates different from Mdm2 and COP1. However, whether Mdm2 and COP1 modulate the ability of Pirh2 to target Pirh2-specific substrates needs to be further investigated.
4. Pirh2 as a novel anticancer target
Pirh2 expression was found to be upregulated in multiple cancers, including lung, prostate, head and neck cancers, and hepatocellular carcinoma [22,50–52], indicating that Pirh2 might promote tumor formation. However, the view that Pirh2 acts as an oncogene has been challenged by the finding that Pirh2 exhibits tumor suppression activity in some types of cancer via degrading the c-Myc oncoprotein [8]. In addition, reduced expression of Pirh2 in several human cancers, such as lung, ovarian, breast, and bladder cancers, is correlated with poor survival [8,53–56]. Therefore, Pirh2 might serve as a novel prognostic biomarker for specific types of cancers.
Small molecule compounds, such as Nutlin-3a, RITA, and MI-219, have been identified as potent Mdm2 inhibitors [5–7]. These small molecule compounds disrupt Mdm2-mediated p53 degradation and thus lead to tumor regression by inducing p53-mediated cell cycle arrest and cell death [5,7,57]. However, these small molecules are not effective in some cases [58–60]. Similar to Mdm2, Pirh2 is a negative regulator for p53. Thus, small molecule compounds targeting Pirh2 might provide a solution for cancers resistant to Mdm2 inhibitors. In addition to Mdm2-like activities, Pirh2 regulates its own targets. For instance, high Pirh2 and low p27 have been observed in several cancers [9,52]. Since p27 inhibits the G1/S progression in a p53-independent manner, activation of p27 by inhibiting Pirh2 would be an alternative approach to treat cancers deficient in p53. Similarly, p73, a p53 family member, exerts tumor suppressor activity in tumors lacking p53 [61,62]. We and others showed that Pirh2 promotes p73 polyubiquitination and degradation [12,63]. Thus, it is likely that activation of p73 is another therapeutic strategy in the absence of p53. In addition, we showed that Pirh2 is a major factor that inhibits PolH activity in TLS (translesion DNA synthesis) [10,11]. Since downregulation of PolH is known to predispose XPV patients to UV-induced skin cancers [64,65], selective small molecule inhibitors targeting the Pirh2-PolH loop may be explored for malignancies associated with PolH deficiency. However, low Pirh2 was correlated with c-Myc activation in certain types of cancers. Since induction of cell death and/or cellular senescence upon c-Myc inactivation leads to regression of some tumors lacking functional p53 [8,66], small molecules activating Pirh2 may be feasible for c-Myc-based cancer therapy. Nevertheless, Pirh2-targeting molecules may provide a useful tool to uncover how Pirh2 promotes and inhibits tumorigenesis and eventually help to develop clinically applicable approaches target Pirh2-overexpressing/deficient tumors.
5. Concluding remarks
The ubiquitin proteasome pathway plays a critical role in regulating many signaling pathways important for cancer cell growth and survival. Inhibition of proteasome function has been postulated to be a new direction for developing anti-cancer drugs. The feasibility of this hypothesis was approved by preclinical studies on a proteasome inhibitor Bortezomib [67]. However, much work is needed to improve the specificity and efficiency of proteasome inhibitors. Since the specificity of the ubiquitin proteasome pathway is primarily determined by E3 ligases, developing E3 ligase inhibitors would be one solution to improve pharmacological properties of proteasome inhibitors. For example, Nutlin-3a, a small molecule that interrupts Mdm2-mediated p53 ubiquitination and degradation, is one potential drug to treat cancers with increased Mdm2 and decreased p53. However, it is not applicable to all cancers especially tumors with mutant p53. Thus, it is necessary to identify new targets. Evidence showed that expression levels of Pirh2 are altered in various human cancers. Along with this, evidence showed that Pirh2 regulates a growing number of key factors for cell survival and cell death, including p53, p73, p27, PolH, and c-Myc. Therefore, Pirh2 may be explored as a target for cancer therapeutics. However, there are several issues remained to be answered. For example, how does Pirh2 exert two opposing cellular functions as an oncoprotein or a tumor suppressor in different tissues? Are there other regulators or targets of Pirh2 in tumorigenesis? How is p53 activity affected by the elevated Pirh2 in breast and lung cancers? Does Pirh2:p53 dysregulation represent a new mechanism for cellular transformation in certain types of cancers? Do Pirh2 isoforms directly or indirectly regulate Pirh2 substrates by interfering with Pirh2 function? Does Pirh2 exert E3 ubiquitin ligase-independent activities? Addressing these questions will facilitate our understanding of Pirh2 functions and provide a foundation for developing Pirh2-based cancer therapies.
Acknowledgments
This work was supported by NIH grant CA076069, CA081237, CA102188, and CA123227.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–78. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 2.Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer. 2011;11:629–43. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dornan D, et al. COP1, the negative regulator of p53, is overexpressed in breast and ovarian adenocarcinomas. Cancer Res. 2004;64:7226–30. doi: 10.1158/0008-5472.CAN-04-2601. [DOI] [PubMed] [Google Scholar]
- 4.Manfredi JJ. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 2010;24:1580–9. doi: 10.1101/gad.1941710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shangary S, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci U S A. 2008;105:3933–8. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, Selivanova G. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–8. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 7.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 8.Hakem A, et al. Role of Pirh2 in mediating the regulation of p53 and c-Myc. PLoS Genet. 2011;7:e1002360. doi: 10.1371/journal.pgen.1002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hattori T, Isobe T, Abe K, Kikuchi H, Kitagawa K, Oda T, Uchida C, Kitagawa M. Pirh2 promotes ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor p27Kip1. Cancer Res. 2007;67:10789–95. doi: 10.1158/0008-5472.CAN-07-2033. [DOI] [PubMed] [Google Scholar]
- 10.Jung YS, Hakem A, Hakem R, Chen X. Pirh2 E3 ubiquitin ligase monoubiquitinates DNA polymerase eta to suppress translesion DNA synthesis. Mol Cell Biol. 2011;31:3997–4006. doi: 10.1128/MCB.05808-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung YS, Liu G, Chen X. Pirh2 E3 ubiquitin ligase targets DNA polymerase eta for 20S proteasomal degradation. Mol Cell Biol. 2010;30:1041–8. doi: 10.1128/MCB.01198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung YS, Qian Y, Chen X. The p73 tumor suppressor is targeted by Pirh2 RING finger E3 ubiquitin ligase for the proteasome-dependent degradation. J Biol Chem. 2011;286:35388–95. doi: 10.1074/jbc.M111.261537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leng RP, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–91. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 14.Zhu J, Jiang J, Zhou W, Chen X. The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res. 1998;58:5061–5. [PubMed] [Google Scholar]
- 15.Harms K, Nozell S, Chen X. The common and distinct target genes of the p53 family transcription factors. Cell Mol Life Sci. 2004;61:822–42. doi: 10.1007/s00018-003-3304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dohn M, Zhang S, Chen X. p63alpha and DeltaNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene. 2001;20:3193–205. doi: 10.1038/sj.onc.1204427. [DOI] [PubMed] [Google Scholar]
- 17.Corcoran CA, Montalbano J, Sun H, He Q, Huang Y, Sheikh MS. Identification and characterization of two novel isoforms of Pirh2 ubiquitin ligase that negatively regulate p53 independent of RING finger domains. J Biol Chem. 2009;284:21955–70. doi: 10.1074/jbc.M109.024232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu G, Sun M, Zhang L, Zhou J, Wang Y, Huo K. A novel hPirh2 splicing variant without ubiquitin protein ligase activity interacts with p53 and is down-regulated in hepatocellular carcinoma. FEBS Lett. 2010;584:2772–8. doi: 10.1016/j.febslet.2010.04.075. [DOI] [PubMed] [Google Scholar]
- 19.Shi J, Huang Y, Sheikh MS. Identification of Pirh2D, an Additional Novel Isoform of Pirh2 Ubiquitin Ligase. Mol Cell Pharmacol. 2010;2:21–23. doi: 10.4255/mcpharmacol.10.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheng Y, et al. Molecular basis of Pirh2-mediated p53 ubiquitylation. Nat Struct Mol Biol. 2008;15:1334–42. doi: 10.1038/nsmb.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beitel LK, Elhaji YA, Lumbroso R, Wing SS, Panet-Raymond V, Gottlieb B, Pinsky L, Trifiro MA. Cloning and characterization of an androgen receptor N-terminal-interacting protein with ubiquitin-protein ligase activity. J Mol Endocrinol. 2002;29:41–60. doi: 10.1677/jme.0.0290041. [DOI] [PubMed] [Google Scholar]
- 22.Logan IR, Gaughan L, McCracken SR, Sapountzi V, Leung HY, Robson CN. Human PIRH2 enhances androgen receptor signaling through inhibition of histone deacetylase 1 and is overexpressed in prostate cancer. Mol Cell Biol. 2006;26:6502–10. doi: 10.1128/MCB.00147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maruyama S, Miyajima N, Bohgaki M, Tsukiyama T, Shigemura M, Nonomura K, Hatakeyama S. Ubiquitylation of epsilon-COP by PIRH2 and regulation of the secretion of PSA. Mol Cell Biochem. 2008;307:73–82. doi: 10.1007/s11010-007-9586-3. [DOI] [PubMed] [Google Scholar]
- 24.Logan IR, Sapountzi V, Gaughan L, Neal DE, Robson CN. Control of human PIRH2 protein stability: involvement of TIP60 and the proteosome. J Biol Chem. 2004;279:11696–704. doi: 10.1074/jbc.M312712200. [DOI] [PubMed] [Google Scholar]
- 25.Zheng G, Ning J, Yang YC. PLAGL2 controls the stability of Pirh2, an E3 ubiquitin ligase for p53. Biochem Biophys Res Commun. 2007;364:344–50. doi: 10.1016/j.bbrc.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Li J, Wang C, Ma Y, Huo K. A new human gene hNTKL-BP1 interacts with hPirh2. Biochem Biophys Res Commun. 2005;330:293–7. doi: 10.1016/j.bbrc.2005.02.156. [DOI] [PubMed] [Google Scholar]
- 27.Yan J, Zhang D, Di Y, Shi H, Rao H, Huo K. A newly identified Pirh2 substrate SCYL1-BP1 can bind to MDM2 and accelerate MDM2 self-ubiquitination. FEBS Lett. 2010;584:3275–8. doi: 10.1016/j.febslet.2010.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, He G, Zhang P, Wang X, Jiang M, Yu L. Interplay between MDM2, MDMX, Pirh2 and COP1: the negative regulators of p53. Mol Biol Rep. 2010 doi: 10.1007/s11033-010-0099-x. [DOI] [PubMed] [Google Scholar]
- 29.Jung YS, Qian Y, Chen X. Examination of the expanding pathways for the regulation of p21 expression and activity. Cell Signal. 2010;22:1003–12. doi: 10.1016/j.cellsig.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joerger AC, Rajagopalan S, Natan E, Veprintsev DB, Robinson CV, Fersht AR. Structural evolution of p53, p63, and p73: implication for heterotetramer formation. Proc Natl Acad Sci U S A. 2009;106:17705–10. doi: 10.1073/pnas.0905867106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coutandin D, et al. Conformational stability and activity of p73 require a second helix in the tetramerization domain. Cell Death Differ. 2009;16:1582–9. doi: 10.1038/cdd.2009.139. [DOI] [PubMed] [Google Scholar]
- 32.Natan E, Joerger AC. Structure and kinetic stability of the p63 tetramerization domain. J Mol Biol. 2012;415:503–13. doi: 10.1016/j.jmb.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–45. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 34.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–60. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 35.Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010;17:86–92. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 37.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 38.Michael D, Oren M. The p53 and Mdm2 families in cancer. Curr Opin Genet Dev. 2002;12:53–9. doi: 10.1016/s0959-437x(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 39.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. Embo J. 1993;12:461–8. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–32. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 41.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–5. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 42.Maya R, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15:1067–77. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187:112–26. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 44.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–34. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 45.Li Q, et al. Axin determines cell fate by controlling the p53 activation threshold after DNA damage. Nat Cell Biol. 2009;11:1128–34. doi: 10.1038/ncb1927. [DOI] [PubMed] [Google Scholar]
- 46.Dornan D, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 47.Ongkeko WM, Wang XQ, Siu WY, Lau AW, Yamashita K, Harris AL, Cox LS, Poon RY. MDM2 and MDMX bind and stabilize the p53-related protein p73. Curr Biol. 1999;9:829–32. doi: 10.1016/s0960-9822(99)80367-4. [DOI] [PubMed] [Google Scholar]
- 48.Zeng X, et al. MDM2 suppresses p73 function without promoting p73 degradation. Mol Cell Biol. 1999;19:3257–66. doi: 10.1128/mcb.19.5.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watson IR, Irwin MS. Ubiquitin and ubiquitin-like modifications of the p53 family. Neoplasia. 2006;8:655–66. doi: 10.1593/neo.06439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duan W, Gao L, Druhan LJ, Zhu WG, Morrison C, Otterson GA, Villalona-Calero MA. Expression of Pirh2, a newly identified ubiquitin protein ligase, in lung cancer. J Natl Cancer Inst. 2004;96:1718–21. doi: 10.1093/jnci/djh292. [DOI] [PubMed] [Google Scholar]
- 51.Wang XM, Yang LY, Guo L, Fan C, Wu F. p53-induced RING-H2 protein, a novel marker for poor survival in hepatocellular carcinoma after hepatic resection. Cancer. 2009;115:4554–63. doi: 10.1002/cncr.24494. [DOI] [PubMed] [Google Scholar]
- 52.Shimada M, et al. High expression of Pirh2, an E3 ligase for p27, is associated with low expression of p27 and poor prognosis in head and neck cancers. Cancer Sci. 2009;100:866–72. doi: 10.1111/j.1349-7006.2009.01122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chin K, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–41. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 54.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 55.Berchuck A, et al. Patterns of gene expression that characterize long-term survival in advanced stage serous ovarian cancers. Clin Cancer Res. 2005;11:3686–96. doi: 10.1158/1078-0432.CCR-04-2398. [DOI] [PubMed] [Google Scholar]
- 56.Raponi M, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66:7466–72. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 57.Cao C, Shinohara ET, Subhawong TK, Geng L, Woon Kim K, Albert JM, Hallahan DE, Lu B. Radiosensitization of lung cancer by nutlin, an inhibitor of murine double minute 2. Mol Cancer Ther. 2006;5:411–7. doi: 10.1158/1535-7163.MCT-05-0356. [DOI] [PubMed] [Google Scholar]
- 58.Wade M, Wong ET, Tang M, Stommel JM, Wahl GM. Hdmx modulates the outcome of p53 activation in human tumor cells. J Biol Chem. 2006;281:33036–44. doi: 10.1074/jbc.M605405200. [DOI] [PubMed] [Google Scholar]
- 59.Hu B, Gilkes DM, Farooqi B, Sebti SM, Chen J. MDMX overexpression prevents p53 activation by the MDM2 inhibitor Nutlin. J Biol Chem. 2006;281:33030–5. doi: 10.1074/jbc.C600147200. [DOI] [PubMed] [Google Scholar]
- 60.Patton JT, Mayo LD, Singhi AD, Gudkov AV, Stark GR, Jackson MW. Levels of HdmX expression dictate the sensitivity of normal and transformed cells to Nutlin-3. Cancer Res. 2006;66:3169–76. doi: 10.1158/0008-5472.CAN-05-3832. [DOI] [PubMed] [Google Scholar]
- 61.Flores ER, et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–73. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 62.Tomasini R, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22:2677–91. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu H, Zeinab RA, Flores ER, Leng RP. Pirh2, a ubiquitin E3 ligase, inhibits p73 transcriptional activity by promoting its ubiquitination. Mol Cancer Res. 2011;9:1780–90. doi: 10.1158/1541-7786.MCR-11-0157. [DOI] [PubMed] [Google Scholar]
- 64.Alt A, Lammens K, Chiocchini C, Lammens A, Pieck JC, Kuch D, Hopfner KP, Carell T. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science. 2007;318:967–70. doi: 10.1126/science.1148242. [DOI] [PubMed] [Google Scholar]
- 65.Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat Rev Cancer. 2005;5:564–73. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]
- 66.Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci U S A. 2007;104:13028–33. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crawford LJ, Walker B, Irvine AE. Proteasome inhibitors in cancer therapy. J Cell Commun Signal. 2011;5:101–10. doi: 10.1007/s12079-011-0121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu G, Sun M, Zhang W, Huo K. AIG1 is a novel Pirh2-interacting protein that activates the NFAT signaling pathway. Front Biosci (Elite Ed) 2011;3:834–42. doi: 10.2741/E291. [DOI] [PubMed] [Google Scholar]
- 69.Zhang W, Wu G, Yan X, Shi H, Huo K. Preliminary Study on the Interaction between Human PIRH2b and ARF4. J Med Mol Biol. 2007;4:469–474. [Google Scholar]
- 70.Duan S, Yao Z, Hou D, Wu Z, Zhu WG, Wu M. Phosphorylation of Pirh2 by calmodulin-dependent kinase II impairs its ability to ubiquitinate p53. EMBO J. 2007;26:3062–74. doi: 10.1038/sj.emboj.7601749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Duan S, Yao Z, Zhu Y, Wang G, Hou D, Wen L, Wu M. The Pirh2-keratin 8/18 interaction modulates the cellular distribution of mitochondria and UV-induced apoptosis. Cell Death Differ. 2009;16:826–37. doi: 10.1038/cdd.2009.12. [DOI] [PubMed] [Google Scholar]
- 72.Chen M, Cortay JC, Logan IR, Sapountzi V, Robson CN, Gerlier D. Inhibition of ubiquitination and stabilization of human ubiquitin E3 ligase PIRH2 by measles virus phosphoprotein. J Virol. 2005;79:11824–36. doi: 10.1128/JVI.79.18.11824-11836.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karuppannan AK, Liu S, Jia Q, Selvaraj M, Kwang J. Porcine circovirus type 2 ORF3 protein competes with p53 in binding to Pirh2 and mediates the deregulation of p53 homeostasis. Virology. 2010;398:1–11. doi: 10.1016/j.virol.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 74.Liu J, et al. The ORF3 protein of porcine circovirus type 2 interacts with porcine ubiquitin E3 ligase Pirh2 and facilitates p53 expression in viral infection. J Virol. 2007;81:9560–7. doi: 10.1128/JVI.00681-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abe K, Hattori T, Isobe T, Kitagawa K, Oda T, Uchida C, Kitagawa M. Pirh2 interacts with and ubiquitylates signal recognition particle receptor beta subunit. Biomed Res. 2008;29:53–60. doi: 10.2220/biomedres.29.53. [DOI] [PubMed] [Google Scholar]