Abstract
Fibroblast growth factor receptors (FGFRs) play critical roles in craniofacial and skeletal development via multiple signaling pathways including MAPK, PI3K/AKT, and PLC-γ. FGFR-mediated signaling is modulated by several regulators. Proteins with leucine-rich repeat (LRR) and/or immunoglobulin (IG) superfamily domains have been suggested to interact with FGFRs. In addition, fibronectin leucine-rich repeat transmembrane protein 3 (FLRT3) has been shown to modulate the FGFR-mediated signaling via the fibronectin type III (FNIII) domain. Therefore proteins with LRR, IG, and FNIII are candidate regulators of the FGFRs. Here we identify leucine-rich repeat, immunoglobulin-like and transmembrane domain 3 (LRIT3) as a regulator of the FGFRs.
Keywords: Craniofacial development, ER export, FGFR regulation, FGF signaling, non-syndromic craniosynostosis
INTRODUCTION
Fibroblast growth factor (FGF)-signaling plays crucial roles in cell proliferation and differentiation. FGFs activate the different isoforms of the FGF receptors (FGFR1, 2, 3 and 4) [1]. All FGFRs are type I membrane proteins that are synthesized in the endoplasmic reticulum (ER). Heparin or heparan sulfate forms a bridge between FGF and FGFR and is necessary for efficient FGF signaling [2]. Upon exposure to FGFs, FGFRs dimerize, resulting in activation of the tyrosine kinase (TK) activity and transautophosphorylation of tyrosine residues on the intracellular portion of the receptor [3]. Phosphorylated FGFR activates a variety of cellular signaling pathways including the mitogen-activated protein kinase (MAPK), PLC-γ, and PI3K/AKT [1,4,5].
Missense mutations or amplification of FGFR have been implicated in cancer as well as in other developmental diseases including craniosynostosis (CS), or premature fusion of cranial sutures [1,6]. Especially, gain-of-function mutations of FGFR cause syndromic craniosynostosis (SC) where CS is associated with other developmental anomalies and inherited in a Mendelian fashion [7–9]. However, SC only accounts for a small fraction of CS cases and the majority of CS cases occur as sporadic findings without other associated anomalies [10]. Considering the major impact of aberrant FGF-signaling in SC, it is possible that some NSC cases are caused by alterations in proteins that regulate or mediate FGF-signaling.
Multiple cellular factors such as Sprouty (SPRY), MAP kinase phosphatase 3 (MKP3), Similar Expression to FGFs (SEF), and fibronectin-leucine-rich transmembrane protein 3 (FLRT3) have been shown to regulate the FGF-signaling pathway [11–13]. In addition, a recent study has shown that proteins with leucine-rich repeat (LRR) and/or immunoglobulin (IG) superfamily domains may interact with FGFRs [14]. In the case of FLRT3, interaction with FGFR1 is via its fibronectin type III (FNIII) domain and activation of the MAPK signaling pathway [13]. Thus, families of proteins with FNIII, LRR, and IG domains may serve as regulators of FGFRs and other growth factor receptors.
We sought to identify new regulators of FGFR1. We focused on fibronectin leucine-rich repeat transmembrane protein 3 (LRIT3) as it contains the suggested domains for FGFR interaction. Our results suggest that LRIT3 regulates maturation and signaling of FGFR1.
MATERIALS AND METHODS
Antibodies
The following antibodies were used for immunoblotting: Mouse anti-Myc (Millipore, USA; 1/1,000), rabbit anti-beta-tubulin (Cell signaling Tech, USA, 1/1,000), rabbit anti-phospho-ERK (Cell signaling Tech, USA, 1/1,000), rabbit anti-ERK (Cell signaling Tech, USA, 1/1,000), mouse anti-LRIT-3 (Novus Bioscience, USA, 1/1,000), rabbit anti-phospho-AKT (Cell signaling Tech, USA, 1/1,000), rabbit anti-AKT (Cell signaling Tech, USA, 1/1,000), rabbit anti-PLC-γ (Cell signaling Tech, USA, 1/1,000), rabbit anti-phospho-PLC-γ (Cell signaling Tech, USA, 1/1,000), anti-rabbit IgG conjugated with horse radish peroxidase (Amersham Bioscience, USA, 1/5,000), rabbit anti- FGFR1 (Cell signaling Tech, USA, 1/1,000).
Subjects and Clinical Data
Informed consent was obtained from all patients and/or their parents. This study was approved by the Institutional Review Boards of the University of California, Davis, and was conducted in accordance with institutional guidelines.
PCR, DNA sequencing, and Sequence Analyses
A total of 431 individuals with non-syndromic craniosynostosis were selected for sequencing of LRIT3. Peripheral blood or saliva samples were collected from individuals, and genomic DNA was isolated as per conventional protocols with PureGene (5 Prime Inc.) or Oragene (Nalgene). LRIT3 exons were amplified by polymerase chain reactions. PCR products were purified with Shrimp Alkaline Phosphatase and Exonuclease I (USB Corporation, Cleveland, OH). PCR primers are available in Table S1. Purified DNA fragments were sent to UC Davis Sequencing Facility and electropherograms were analyzed with VectorNTI™ Version 11 computer program. The 5’- and 3’-untranslated regions of LRIT3, as well as at least 100 base pairs of flanking intronic sequence for each exon were included in the sequencing analysis. The observed variants were confirmed by independent PCRs and sequencing of the reverse DNA strands. Parental samples (when available) were sequenced. Single nucleotide polymorphisms (SNPs) were considered novel if not described in the NCBI SNP database.
Taqman Assays
5 Custom TaqMan® SNP Genotyping Assays manufactured by Applied Biosystems were designed to detect the novel polymorphic variants on Human Random Control DNA Panels 1 to 5 (European Collection of Cell Cultures, kind gifts from Michael L. Cunningham, University of Washington) using the ABI 7900HT QPCR machine. QPCR primers, probes, and conditions are available upon request. Allelic Discrimination was performed to classify the zygosities of the targeted templates by analyzing the fluorescence signals in each reaction well.
Construction of a human calvarial osteoblast cDNA library
Total RNA from human calvarial osteoblasts was isolated using Trizol reagent (Invitrogen, USA) RNA extraction reagent. cDNA was synthesized from 1 μg of total RNA using SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen, USA). The DNA fragment containing the new exon 1 and a part of the previously known exon 1 of human LRIT3 was amplified using osteoblast cDNA library with following synthetic oligonucleotide pairs (forward, 5’-ATGCATCTCTTTGCATGTCTGTGC-3’; reverse, 5’-CACGGGGAGGTTCGTAGGCAGCTCGTTCATATC-3’). The PCR product was confirmed by DNA sequencing.
Cell Culture and Transient Transfection
The HEK 293T cells were cultured in DMEM media containing 10% fetal bovine serum and maintained in a water-jacketed incubator at 37°C with 5% CO2 enrichment (Boyd et al, 2006). Sub-cultured cells were maintained in DMEM media with 10% fetal bovine serum and split 1:5 weekly or when confluent. The plasmid DNAs were transiently transfected into HEK 293T cells using Lipofectamine and Plus according to the manufacturer’s protocol (Invitrogen, USA).
Immunoblotting
Cells were washed in cold PBS and lysed in radioimmunoprecipitation assay buffer (25 mM Tris-HCl, pH7.6, 150 mM NaCl, 1% NP-40, 0.1% Sodium dodecyl sulfate, 1% Sodium deoxycholate and 5 mM EDTA) containing protease inhibitors (Roche, USA). The proteins concentration of cell lysates was determined with a bicinchroninic acid assay according to the manufacturer’s protocol (Pierce, USA). Protein lysates were resolved in SDS-PAGE, transferred to PVDF membrane, probed with primary antibodies, incubated with secondary antibodies conjugated with horse radish peroxidase (HRP), and visualized with ECL plus.
Site-directed Mutagenesis and Plasmid Construction
The human LRIT3 coding region was amplified from pCR-Blunt II-LRIT-3 (Open Biosystems, USA) using synthetic oligonucleotides pairs (5’-GGCTAACTAGAGAACCCACTG-3’ and 5’-GATTCTAGATTACAGGTCCTCCTCTGAGAT-3’). The amplified fragments were digested with Nae I and Xba I and inserted into mammalian expression vector pCMV-SPORT6 (Invitrogen, USA). The resulting plasmid has a Myc-tag at the C terminus. The mutagenic primers for LRIT3 (T53M, S494T, and C592Y) were as follows: sense LRIT3 T53M, 5’-CCCGCTAGCATGGATATGAACGAGCTGCCTATGAACCTC-3’; antisense LRIT3 T53M, 5’-GAGGTTCATAGGCAGCTCGTTCATATCCATGCTAGCGGG-3’; sense LRIT3 S494T, 5’-GCAATAGAAAACCTCAGGGTGGTCACTGAGACTAAAG-3’; antisense LRIT3 S494T 5’-CGTCAATGTCACACTCTCTTTAGTCTCAGTGACCAC-3’; sense LRIT3 C592Y 5’-GACCAG ACTGCCTATGTTGTTATC-3’ ; antisense LRIT3 C592Y 5’-GATAACAACATA GGCAGTACTGGTC. To incorporate a signal sequence for LRIT3, an oligonucleotide (5’-ATGCATCTCTTTGCATGTCTGTGCATTGTCCTTAGCTTTTTGGAAGGAGTGGGCTGTT TGTGTCCTTCACAGTGCACCTGTGATTATCACGGCAGAAATGACGGCTCAGGATCAA GGTTGGTGCTATGTAATGAC-3’) was used. The sequence was confirmed by DNA sequencing.
Deglycosylation experiments
Cleared cell lysates (30 μg) obtained from transiently transfected cells with either LRIT3 or FGFR1 or both were heat treated in 1× Glycoprotein Denaturing Buffer at 100 °C for 10 minutes according to the manufacturer's instructions (New England Biolabs Inc, Beverly, MA, USA). The denatured proteins were treated with peptide N glycosidase F (PNGase F) or endoglycosidase H (Endo H) at 37°C 1h. The resulting proteins were analyzed by immunoblotting after separation with 10% SDS–PAGE.
RESULTS
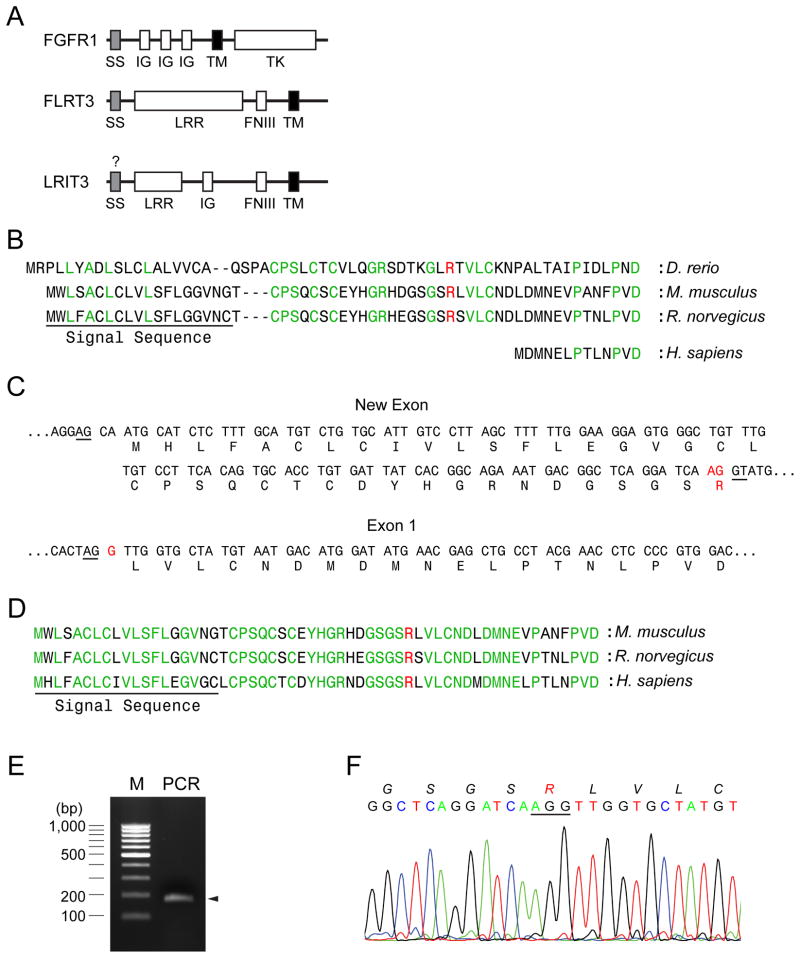
When we compared the amino terminal sequence of human LRIT3 with those of the piscine and rodent LRIT3, it was noted that human LRIT3 lacked a signal sequence and its immediate flanking region (Fig. 1A and B). Simian and bovine LRIT3 also lacked a signal sequence and its flanking region (Fig. S1A). We reasoned that an exon(s) encoding these regions must exist in the genomic sequence of LRIT3 and searched for the exon in the 5’upstream sequence (about 6 kb) of human LRIT3. We anticipated that the new exon would encodes a signal sequence and its flanking region that would be highly homologous to those of rodent LRIT3s. Indeed, we were able to identify a sequence stretch with such features approximately 3.3 kb upstream of the previous exon 1 (Fig. 1C). This newly identified putative exon is bordered by splice acceptor and donor sequences (AG and GT, respectively) and is in-frame with the subsequent exon (the previous exon 1). The newly assembled N-terminus of human LRIT3 is highly homologous to those of rodent LRIT3s (Fig. 1D). Although there may be additional exons for the 5’untranslated region at the 5’ upstream of this sequence, we believe that the complete N-terminal polypeptide sequence of LRIT3 is encoded in this new exon because the current sequence shows typical characteristics of a signal sequence [15,16].
Fig. 1.
Identification of signal sequence of LRIT3. (A) Comparison of domain structures of FGFR1, FLRT3 and LRIT3. SS, IG, LRR, FNIII, TM, and TK denote signal sequence, immunoglobulin-like, leucin-rich repeat, fibronectin type III, transmembrane, and tyrosine kinase domains, respectively. (B) Human LIRT3 (NP_940908.2) from the NCBI database lacks a signal sequence and its flanking sequences compared to rodent and piscine LRIT3s (XP_143529.7, XP_578083.3, and NP_001139102.1). The putative signal sequence is underlined and the signal peptide cleavage site was predicted by using a program, SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/). Arginine shown in red represents the putative splicing juncture. (C) A putative exon encoding the signal sequence and the early mature region was found at the 5’-upstream of LRIT3 from human genomic sequence. Putative splicing donor and acceptor sequences are underlined. Arginine shown in red represents the putative splicing juncture. (D) The proposed N-terminus of LRIT3 is shown. Residues in green are conserved in rodent and human LRIT3s. Arginine shown in red represents the putative splicing juncture. The putative signal sequence is underlined. (E) A PCR reaction was performed using a forward primer that anneals at the new exon and a reverse primer that anneals at the exon 1 using a cDNA library generated from human osteoblasts (see Materials and Methods for detail). An 171 bp-fragment is expected to be amplified from this reaction. (F) Chromatogram near the splicing juncture of the new exon and the exon 1 from the 171 bp-fragment amplified from the cDNA library. Amino acid sequences are italicized. Arginine shown in red represents the putative splicing juncture.
To verify correct transcription and splicing of this new sequence, we designed a primer pair for the new exon (a forward primer) and the previously known exon1 (a reverse primer) that can amplify the portion of the spliced form (171 bp). Using this primer pair and a cDNA library generated from human calvarial osteoblasts, we were able to amplify the expected 171 bp fragment and to verify its sequence (Fig. 1E and F). This result shows that the human LRIT3 also contains a signal sequence and its flanking sequence that are highly homologous to those of the rodent LRIT3s. In addition, we were able to identify the equivalent sequences for simian and bovine LRIT3s from their genomic sequences (Fig. S1B and C). The nucleotide sequence of this new human LRIT3 has been deposited into the GenBank database (accession number: JQ354981).
When LRIT3 and FGFR1 were expressed in HEK293 cells, two interesting observations were made. Firstly, when they were co-expressed, the levels of the two proteins were increased compared to those of individual expression (Fig. 2A, compare LRIT3 in lane 3 with that in lane 6; compare FGFR1 in lane 4 with that in lane 5 or 6). The increase in the levels of FGFR1 was more pronounced than the increase in the levels of LRIT3. Secondly, the slowly migrating FGFR1 species was enhanced when LRIT3 was present. FGFR1 is N-glycosylated and exists as Golgi-modified and ER-modified forms [17]. The enhancement of the slowly migrating FGFR1 species likely represents an increase in Golgi-modified forms. To corroborate this possibility, HEK293 cells were transfected with LRIT3 and/or FGFR1 and the resulting cell lysates were treated with endoglycosidase H (Endo H) and peptide N-glycosidase F (PNGase F) (Fig. 2B). Endo H removes N-glycans from ER-modified forms and PNGase F removes N-glycans from ER-modified forms as well as from Golgi-modified forms. As expected, the slowly migrating FGFR1 species was Endo H-resistant and PNGase F-sensitive, indicating that LRIT3 enhances Golgi-modified forms of FGFR1. Surprisingly, LRIT3 existed predominantly as ER-modified forms. These results suggest that LRIT3 facilitates exit of FGFR1 from the ER.
Fig. 2.
LRIT3 influences the stability and glycosylation of FGFR1. (A) HEK293 cells were transfected with FGFR1 and LRIT3 as indicated. The transfected cells were analyzed by immunoblotting. β-tubulin was also probed as a loading control. A mouse monoclonal anti-LRIT3 antibody was used to visualize LRIT3. Note that endogenous FGFR1 and LRIT3 are not detectable in HEK cells under our conditions. (B) The LRIT3/FGFR1-transfected HEK293 cells were lyzed in RIPA buffer and treated with endoglycosidase H (Endo H) or peptide N glycosidase F (PNGase F). Endo H removes ER forms of N-glycans and PNGase F removes ER/Golgi forms of N-glycans. A mouse monoclonal anti-LRIT3 antibody was used to visualize LRIT3. LRIT3 became unstable during deglycoyslation procedure and difficult to detect. LRIT3 was detectable only in lanes 7–8. (C) HEK293 cells transfected with the indicated plasmid constructs were incubated with or without bFGF1 for 30 min. An anti-Myc antibody was used to visualize LRIT3. The anti-Myc antibody also labels a nonspecific band (asterisk) above the LRIT3 band position.
We then expected that the increase in Golgi-modified forms of FGFR1 results in enhancement in FGF-signaling (Fig. 2C). Expression of FGFR1 alone induced robust phosphorylation of ERK1/2 when the cells were treated with bFGF1 (Fig. 2C, compare lane 3 with lane 4). However, we did not detect enhanced phosphorylation of AKT nor phosphorylated PLC-γ in the presence of bFGF1. Coexpression of FGFR1 and LRIT3 did not cause an additional increase in phosphorylation of ERK1/2 compared to expression of FGFR1 alone (Fig. 2C, compare lane 4 with lane 8). It is likely that even when FGFR1 is expressed alone this amount of FGFR1 (ER forms or Golgi forms) can efficiently saturate the capacity of the cells to phosphorylate ERK1/2. Thus, more Golgi-modified forms of FGFR1 may not cause additional increase in the ERK1/2 phosphorylation. In addition, we did not observe any change in phosphorylation of AKT by LRIT3 expression. Interestingly, however, phosphorylation of PLC-γ was detectable when FGFR1 and LRIT3 were coexpressed. However, phosphorylation of PLC-γ was not dependent on bFGF1. These results suggest that LRIT3 can aberrantly activate the PLC-γ branch of the FGFR1-signaling pathway.
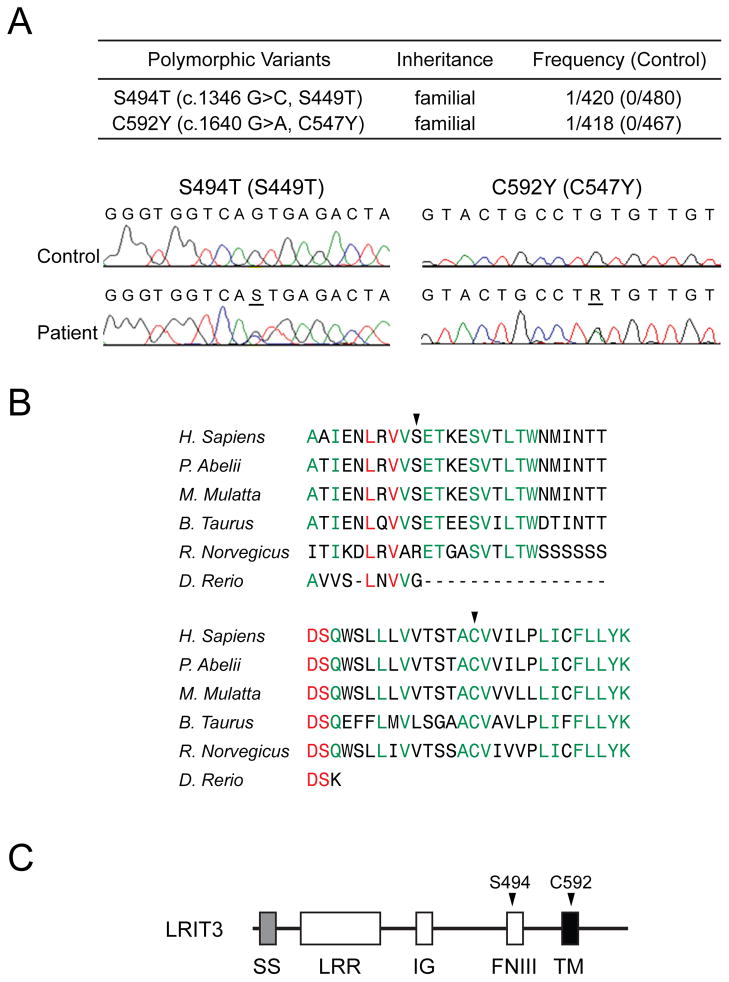
Over-activation of FGFR has been implicated in syndromic CS [18,19]. Furthermore, constitutive RTK- PLC-γ signaling has been also implicated in murine CS [20]. Thus, it seemed reasonable to propose that LRIT3 mutations that can aberrantly modulate FGFR1-signaling might also be found in patients with NSC. We therefore sequenced all coding exons of LRIT3 in more than 400 NSC patients and identified two probands with sagittal NSC with previously unreported LRIT3 mutations: S494T (S449T, based on the previous numbering) and C592Y (C547Y, based on the previous numbering) (Fig. 3A). These residues are highly conserved (Fig. 3B) and not present in either in 400 control chromosomes nor reported among the more than 10,700 alleles of LRIT3 reported in the University of Washington Exome Variant Server (http://evs.gs.washington.edu/EVS/). The S494 residue is located in the FNIII domain, and the C592 residue in the TM domain (Fig. 3C). It is noteworthy that the FNIII domain of FLRT3 is implicated in FGFR1 interaction [21].
Fig. 3.
Identification of rare LRIT3 mutations from a cohort of non-syndromic CS patients. (A) Two rare mutations of LRIT3 were found from DNA samples of more than 200 non-syndromic sagittal CS patients. Chromatograms show the altered bases (underlined). Note that numbering of the LRIT3 residues is according to the new sequence in Fig. 1. Previous numbering is shown in parenthesis. (B) The affected positions are highly conserved in vertebrates (arrowheads). Absolutely conserved sequences were shown in red and the residues conserved in five species in green. The Clustal W analysis was performed using LRIT3 sequences from NP_940908.2 (Homo sapiens), XP_002815104.1 (Pongo abelii), XP_001088511.1 (Macaca mulatta), DAA28914.1 (Bos taurus), XP_143529.7 (Mus musculus), XP_578083.3 (Rattus norvegicus), and NP_001139102.1 (Danio rerio). (C) The relative positions of the two mutations identified from this study were shown in the domain structure of LRIT3 with arrowheads.
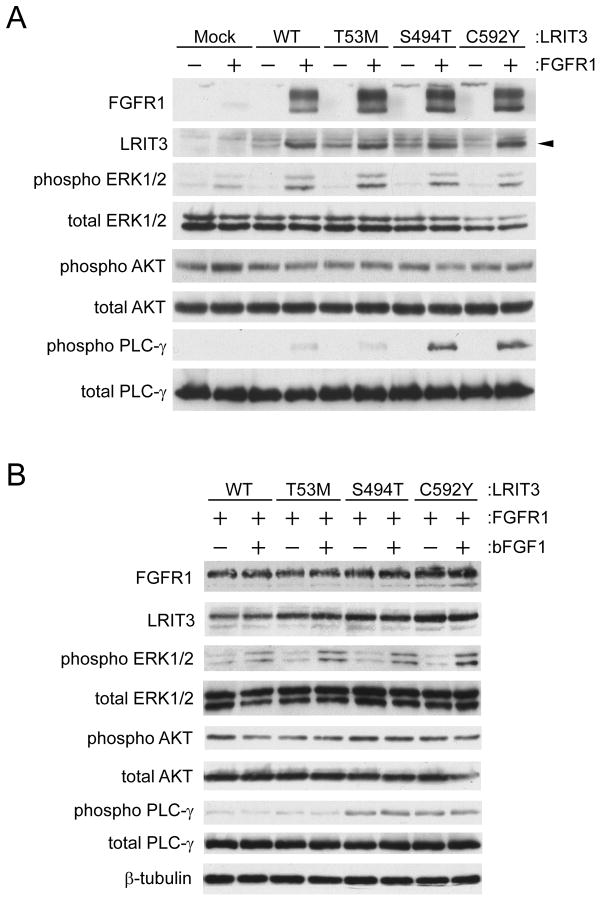
We then tested whether these mutations affect FGF-signaling (Fig. 4). We also generated a T53M (previously known as T8M) LRIT3 construct as a control. This is a known variant that may or may not alter the function of LRIT3. All LRIT3 variants stabilized FGFR1 and increased Golgi-modifed forms of FGFR1 just like wild-type LRIT3 (Fig. 4A). This result suggests that these mutations do not perturb the roles of LRIT3 for stabilization and glycosylation of FGFR1. Mutant LRIT3 did not affect phosphorylation of ERK1/2 or AKT, compared to wild-type LRIT3. Remarkably, S494T and C592Y LRIT3 further enhanced phosphorylation of PLC-γ than WT or T53M LRIT3. This increase in PLC-γ phosphorylation required FGFR1. However, phosphorylation of PLC-γ was not additionally stimulated by bFGF1 (Fig. 4B). Thus, we conclude that LRIT3 can influence maturation and signaling of FGFR1.
Fig. 4.
Functional consequences of the LRIT3 mutations. The T53M (T8M) mutation of LRIT3 is a known single nucleotide polymorphism (rs181200721). T53M LRIT3 was used as a control. (A and B) LRIT3 and/or FGFR1 were transfected into HEK293 cells. Transfected cells were analyzed by immunoblotting. Expression and phosphorylation status of indicated molecules were examined in the absence (A) or presence (B) of basic FGF1. A mouse monoclonal anti-LRIT3 antibody was used to visualize LRIT3. Note that all LRIT3 variants increased Golgi-modifed forms of FGFR1.
DISCUSSION
We have identified full-length LRIT3, have shown that this LRIT3 may influence maturation and signaling of FGFR1 when over-expressed in cultured cells, and have pinpointed FNIII and TM of LRIT3 as critical domains for influencing FGFR1-signaling. LRIT3 predominantly exists in ER-modified forms and can increase the ratio of Golgi-modified forms to ER-modified forms of FGFR1. In particular, we have observed bFGF1-independent activation of the PLC-γ branch of the FGFR-signaling pathway. Perhaps highly over-expressed FGFR1s can interact and phosphorylate each other at the cell surface without a ligand [22]. Alternatively, stabilization and over-expression of FGFR1 simply allow cells to respond to even small amounts of growth factors in the culture medium (serum) rather than these cells become truly FGF-independent. Although the exact mechanism of action is not clear, our results strongly suggest that LRIT3 facilitates ER export of FGFR1, that FGFR1 exits the ER not in a constitutive fashion, but in a regulated fashion, and that LRIT3 modulates the FGFR-signaling pathway.
The LRIT3 mutations we have identified are inherited from the clinically unaffected parents of the probands. Therefore, these mutations alone are not sufficient to cause the disease. Interestingly, expression of auto-activated PDGFRα causes CS in mice [20]. In these mice, PLC-γ is hyper-phosphorylated. Furthermore, inhibitors of PLC-γ prevent the mineralization of synthetic bone matrix. Consistent with this finding, osteoblasts isolated from Apert CS patients (FGFR2 S252W) display up-regulation of PLC-γ signaling [23]. These results suggest that over-activation of PLC-γ is involved in CS. We report here that mutant LRIT3s stimulate the PLC-γ branch of the FGFR1 pathway. Perhaps, aberrant PLC-γ signaling may contribute to NSC in concert with yet undefined phenotypic contributors.
To avoid modifications by Golgi enzymes, LRIT3 should constantly stay in the ER or cycles between the ER and the cis-Golgi rapidly. There are multiple possibilities regarding how a mutation in LRIT3 affects PLC-γ. One possibility is that mutant LRIT3s themselves are slightly defective in ER exit, resulting in an ER export defect of FGFR1. ER retained FGFR1 may aberrantly activate PLC-γ. Alternatively, mutant LRIT3s fail to assist FGFR1 folding, resulting in misfolded FGFR1 that can constitutively activate PLC-γ. It is also possible that FGFR1-LRIT3 complex constitutively activates PLC-γ in cis-Golgi.
Our results, in combination with others, suggest that FGFRs are regulated at various steps (i.e., the biosynthetic route and the endocytic pathway). Multiple defects in these regulatory steps may synergistically influence the FGFR-signaling pathway, leading to skeletal diseases or cancer.
Supplementary Material
Highlights.
We identified the signal sequence and its flanking region for human LRIT3.
LRIT3 facilitates maturation of FGFR1.
LRIT3 modulates the PLC-γ branch of the FGFR-signaling pathway
FNIII and TM domains of LRIT3 can influence FGFR1-signaling.
Acknowledgments
This work was supported in part by the Children’s Miracle Network Endowed Chair and NIH R01 D016886 to SAB, an Australian NHMRC overseas postdoctoral Fellowship to TR, a Marie Curie Fellowship from the European Union Directorate of Science to MFB, and an Academic Senate Research Award to JK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–49. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Spivak-Kroizman T, et al. Heparin-induced oligomerization of FGF molecules is responsible for FGF receptor dimerization, activation, and cell proliferation. Cell. 1994;79:1015–24. doi: 10.1016/0092-8674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 3.Lemmon MA, Schlessinger J. Regulation of signal transduction and signal diversity by receptor oligomerization. Trends Biochem Sci. 1994;19:459–63. doi: 10.1016/0968-0004(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 4.Kouhara H, Hadari YR, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 5.Rhee SG, Bae YS. Regulation of phosphoinositide-specific phospholipase C isozymes. J Biol Chem. 1997;272:15045–8. doi: 10.1074/jbc.272.24.15045. [DOI] [PubMed] [Google Scholar]
- 6.Simon R, et al. High-throughput tissue microarray analysis of 3p25 (RAF1) and 8p12 (FGFR1) copy number alterations in urinary bladder cancer. Cancer Res. 2001;61:4514–9. [PubMed] [Google Scholar]
- 7.Jabs E. Toward understanding the pathogenesis of craniosynostosis through clinical and molecular correlates. Clin Genet. 1998;53:79–86. doi: 10.1111/j.1399-0004.1998.tb02648.x. [DOI] [PubMed] [Google Scholar]
- 8.Passos-Bueno MR, Serti Eacute AE, Jehee FS, Fanganiello R, Yeh E. Genetics of craniosynostosis: genes, syndromes, mutations and genotype-phenotype correlations. Front Oral Biol. 2008;12:107–43. doi: 10.1159/000115035. [DOI] [PubMed] [Google Scholar]
- 9.Wilkie A. Epidemiology and genetics of craniosynostosis. Am J Med Genet. 2000;90:82–84. doi: 10.1002/(sici)1096-8628(20000103)90:1<82::aid-ajmg15>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 10.Boyadjiev SA. Genetic analysis of non-syndromic craniosynostosis. Orthod Craniofac Res. 2007;10:129–37. doi: 10.1111/j.1601-6343.2007.00393.x. [DOI] [PubMed] [Google Scholar]
- 11.Tsang M, Dawid IB. Promotion and attenuation of FGF signaling through the Ras-MAPK pathway. Sci STKE. 2004;2004:e17. doi: 10.1126/stke.2282004pe17. [DOI] [PubMed] [Google Scholar]
- 12.Thisse B, Thisse C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev Biol. 2005;287:390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Bottcher RT, Pollet N, Delius H, Niehrs C. The transmembrane protein XFLRT3 forms a complex with FGF receptors and promotes FGF signalling. Nat Cell Biol. 2004;6:38–44. doi: 10.1038/ncb1082. [DOI] [PubMed] [Google Scholar]
- 14.Sollner C, Wright GJ. A cell surface interaction network of neural leucine-rich repeat receptors. Genome Biol. 2009;10:R99. doi: 10.1186/gb-2009-10-9-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izard JW, Kendall DA. Signal peptides: exquisitely designed transport promoters. Mol Microbiol. 1994;13:765–73. doi: 10.1111/j.1365-2958.1994.tb00469.x. [DOI] [PubMed] [Google Scholar]
- 16.von Heijne G. The signal peptide. J Membr Biol. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- 17.Raivio T, et al. Impaired fibroblast growth factor receptor 1 signaling as a cause of normosmic idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2009;94:4380–90. doi: 10.1210/jc.2009-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neilson KM, Friesel RE. Constitutive activation of fibroblast growth factor receptor-2 by a point mutation associated with Crouzon syndrome. J Biol Chem. 1995;270:26037–40. doi: 10.1074/jbc.270.44.26037. [DOI] [PubMed] [Google Scholar]
- 19.Robertson SC, Meyer AN, Hart KC, Galvin BD, Webster MK, Donoghue DJ. Activating mutations in the extracellular domain of the fibroblast growth factor receptor 2 function by disruption of the disulfide bond in the third immunoglobulin-like domain. Proc Natl Acad Sci U S A. 1998;95:4567–72. doi: 10.1073/pnas.95.8.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moenning A, Jager R, Egert A, Kress W, Wardelmann E, Schorle H. Sustained platelet-derived growth factor receptor alpha signaling in osteoblasts results in craniosynostosis by overactivating the phospholipase C-gamma pathway. Mol Cell Biol. 2009;29:881–91. doi: 10.1128/MCB.00885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karaulanov EE, Bottcher RT, Niehrs C. A role for fibronectin-leucine-rich transmembrane cell-surface proteins in homotypic cell adhesion. EMBO Rep. 2006;7:283–90. doi: 10.1038/sj.embor.7400614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wesche J, Haglund K, Haugsten EM. Fibroblast growth factors and their receptors in cancer. Biochem J. 2011;437:199–213. doi: 10.1042/BJ20101603. [DOI] [PubMed] [Google Scholar]
- 23.Lemonnier J, Hay E, Delannoy P, Lomri A, Modrowski D, Caverzasio J, Marie PJ. Role of N-cadherin and protein kinase C in osteoblast gene activation induced by the S252W fibroblast growth factor receptor 2 mutation in Apert craniosynostosis. J Bone Miner Res. 2001;16:832–45. doi: 10.1359/jbmr.2001.16.5.832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.