Abstract
Haemagglutinin is a determinant of many viral properties, and successful adaptation to a human-like form is thought to be an important step toward pandemic influenza emergence. The availability of structurally distinct sialic acid linked receptors in the sites of human and avian influenza infection are generally held to account for the differences observed, but the relevance of other selection pressures has not been elucidated. There is evidence for genetic and structural constraints of haemagglutinin playing a role in restricting haemagglutinin adaptation, and also for differences in the selection pressure to alter binding, specifically when considering virus replication within host compared to transmission between hosts. Understanding which characteristics underlie such adaptations in humans is now possible in greater detail by using glycan arrays. However, results from these assays must also interpreted in context of an as yet still to be determined detailed knowledge of the structural diversity of sialic acids in the human respiratory tract. A clearer understanding of the evolutionary benefits conveyed by different haemagglutinin properties would have substantial impact and would affect the risk we allocate to viral propagation in different species, such as swine and poultry. Relevant to the H5N1 threat, current evidence also suggests that mortality associated with any emergent pandemic from current strains may be reduced if haemagglutinin specificity changes, further emphasising the importance of understanding how and if selection pressures in the human will cause such an alteration.
Introduction
The ability to predict the emergence of new pandemic strains is a topical issue. When the World Health Organisation (WHO) declared the H1N1 swine-flu outbreak a pandemic, it became the fourth recorded influenza pandemic in a century to emerge from a strain previously sustained outside the human population. Previous pandemics have consisted of the zoonosis of another subtype of influenza which, due to genetic diversity, is antigenically different from the currently circulating strains. Without prior exposure or anticipatory vaccine cover, the antigenic novelty allows the new strain to transmit rapidly through an immunogenically naive global population, inevitably resulting in increased mortality. This change in influenza subtype or 'antigenic shift' had been the major paradigm for future pandemics. With the exception of the reintroduction of H1N1 in humans in the early 1970s, the new subtype results in the replacement of the previous circulating subtype. The H2 subtype replaced H1 in 1957 and was subsequently replaced by H3 in 1968 [1]. Given the recent circulation of H1N1 in humans, the emergence of swine origin H1N1 in 2009 was unexpected. Although the 2009 pH1N1 strains were antigenically different from the human H1N1 strains, which warranted a novel pH1N1 vaccine, there is evidence of prior immunity in the population [2]. This demonstrates the difficulty in accurately predicting how close any particular strain may actually be to achieving sustained human spread and forces us to reconsider the barriers to avian or swine to human zoonosis.
Currently, the H5 subtype is still of great concern, with the current total number of confirmed human cases at 562 [3], but subtypes H7, H9 & H10 have also been reported in humans [4] and any new subtype could theoretically be the foundation of the next pandemic. Although there have already been suspected cases of human-human transmission of H5N1 [5–7], circumstances are generally exceptional and the transmission has not been sustained [8].
In response to pandemic concerns, structural and genetic properties characteristic of human viral strains are beginning to be defined. A hope is that through the identification of the key features that avian viruses require to achieve pandemic spread, we will be able to better evaluate the true risk posed by current avian strains. Haemagglutinin (HA) has been the focus of a large area of investigation and many people have reviewed the features of HA found in viruses that have successfully invaded the human population [9–12]. These properties are being defined in increasing detail and potential explanations for the evolutionary forces that shape these characteristics have been put forward. Rarely, however, has the relevance of each of these evolutionary factors to viral success been discussed in relation to each other. Here we describe the functional significance of HA in pandemic emergence before considering evidence for the existence and relative contribution of a variety of selection pressures imposed upon it. Ways in which more detailed descriptions of these factors are relevant to prediction and prevention concerning pandemic strains are then explored, highlighting avenues for further research.
The function and significance of the Influenza Haemagglutinin protein
Influenza haemagglutinin (HA) is a glycoprotein coded in the HA gene segment of the influenza virus and along with neuraminidase, is expressed as a trimer on the surface of the viral capsid. HA allows the recognition of cells in the upper respiratory tract or erythrocytes by binding to glycans containing the monosaccharide sialic acid. The virus is subsequently engulfed by the cell into an endosome and the lower pH of the endosome facilitates a structural rearrangement of HA. HA then fuses with the membrane of the endosome and allows release of the ribonucleoproteins (RNPs) into the cytoplasm. The RNPs are transported into the nucleus followed by transcription, and replication of the viral genome. The HA, along with the other newly generated viral proteins and a replicated genome, is then incorporated into the envelope of the influenza virion as it buds from an infected host cell. The neuraminidase gene of the virus is then required for efficient release of the newly formed virion.
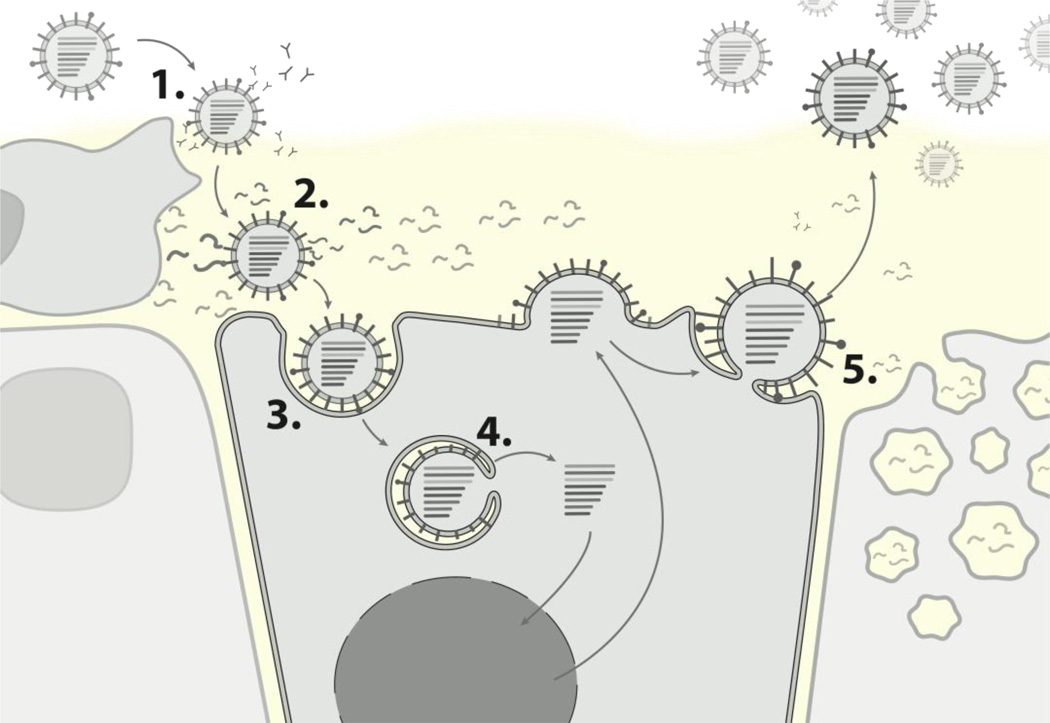
Despite the focus of this review on HA, the relevance of adaptation in other viral proteins as part of the generation of pandemic strains should certainly not be forgotten. Even in the wider context of the whole viral lifecycle however, HA is a uniquely significant component of the virus; structural variations greatly affect viral antigenicity, while its function in cellular attachment through membrane fusion makes it an important determinant of other viral properties including host specificity, replication potential, transmissibility and pathogenicity [13]. Ultimately, HA is involved in a number of steps that may represent many of the most substantial barriers to productive human infections by otherwise avian strains (summarised in Figure 1).
Figure 1. The role of HA in steps to productive human infection.
1. Evasion of acquired immunity, through novel antigenic features. 2. Avoidance of innate immunity, including possible loss of 'decoy' receptor binding to mucin. 3. Productive SA binding, specifically to a2–6 linkages prevalent in the human upper respiratory tract. 4. Effective membrane fusion. 5. Efficient viral budding, for example through an appropriate "HA/NA balance".
In seasonal influenza, the most significant currently understood predictor of viral strain success is the extent to which HA differs antigenically from the currently circulating strains. This knowledge has meant that direct measurements of antigenic similarity can be employed to make effective predictions as to which viral strains are likely to propagate most effectively through the global population [14, 15]. Even ignoring the exceptionally deadly 1918 pandemic, the last two pandemics of the 20th century caused over one million deaths each [16]. The value of pre-empting the emergence of pandemic strains in a similar way to the efforts with seasonal viral strains is self-evident. To date, however, the properties that contribute to the success of novel avian strains in human hosts are far less clearly defined, but are equally important in our understanding of how new viral strains invade the global population. In this respect, the direct effect HA has upon a number of viral characteristics has made it an important area for investigation.
Avian and human receptor specificities
HA has been implicated as a key determinant of host specificity and consequently, adaptation of avian HA to humans was an important stage in the evolution of previous pandemic strains. In the life cycle of the virus, HA functions to bind sialic acids (SA) on host cell surfaces, the first stage in a process that leads to internalisation of the virus and invasion into the cell body. SA are a diverse family of sugars with a nine carbon backbone, which exist in a variety of arrangements, branched and unbranched, and with a range of associated chemical groups. They are typically found at the distal end of glycan chains, which are a feature of all cell types. Consequently, a capability to bind to the kinds of SA present in the human respiratory tract is an important feature of human influenza strains [17].
In 1983, Rogers and Paulson first demonstrated that H3 avian viruses showed a significant preference for binding erythrocytes derivatised to contain SA with an α2–3 linkage, compared to those with an α2–6 linkage [18]. In contrast, human isolates were shown to have the reverse preference. Later, this pattern was also identified in larger studies, including studies investigating H1 & H2 subtypes [10, 19]. Further results from Paulson and colleagues, which demonstrated an apparent abundance of SA with α2–6 linkages in the human respiratory tract, have often been taken as evidence that avian viruses cannot infect humans because of a paucity of suitable SA on human respiratory cells. Perhaps for this reason it has been commonly assumed that, in terms of HA, adaptation to bind the more abundant α2–6 linkages has been the overriding evolutionary barrier to human infection for avian strains. In support of this hypothesis, experiments have correlated changes in HA receptor preference strongly with changes in transmissibility [20–23]. However, it is always important to consider that it does not necessarily follow that binding to α2–6 linkages on mammalian cells is the most significant HA property that has been affected under such modification.
Although widespread, the correlation first observed by Paulson et al. is not absolute and there are examples of avian isolate strains that have been shown to possess a preference for binding SA with α2–6 linkages [24], while equally, some human isolates have retained an α2–3 linkage preference, an example being recent H5N1 viruses isolated from humans [25]. With some qualification, however, α2–3 and α2–6 binding preference has remained an important phenomenon since although α2–3 binders can infect humans in exceptional circumstances, all strains successfully propagating as an epidemic or pandemic in the human population appear to have an α2–6 preference, or at least joint α2–6, α2–3 affinity. Some early H2 epidemic strains were described with a preference for α2–3 linked SA [26, 27] but such reports often identify serial egg passaging as part of the viral preparation, a technique that can create an artificial switch in receptor preference of human viral strains to one for α2–3 linked SA [28]. Significantly, no contemporary epidemic strains have displayed such characteristics.
Selection pressures during infection and transmission
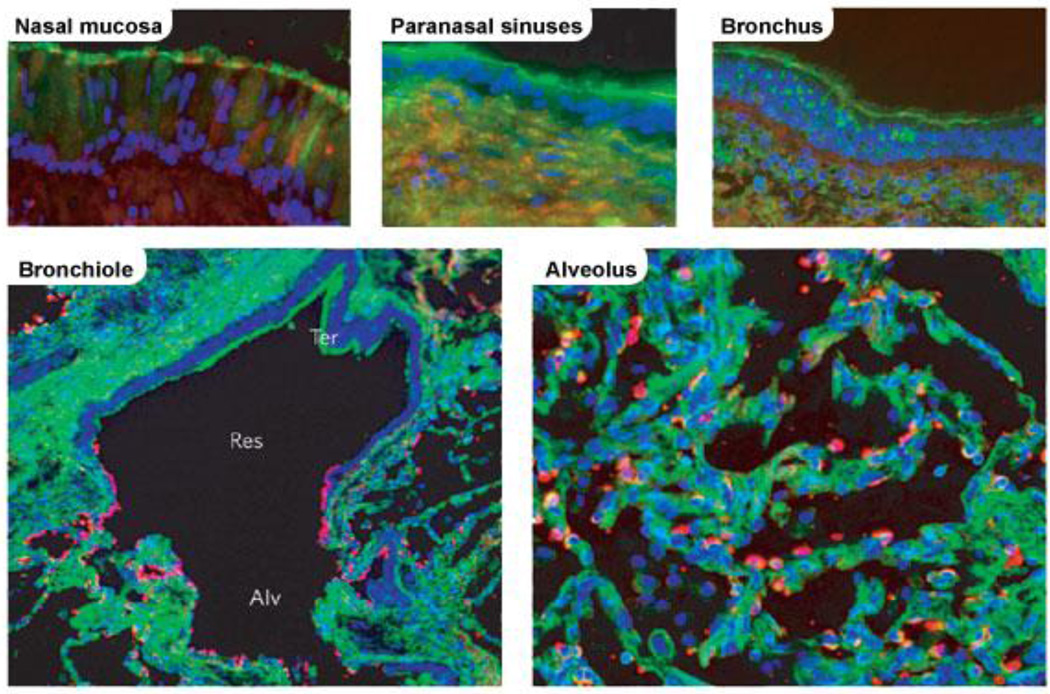
Importantly, α2–3 and α2–6 linked SA are heterogeneously distributed in the human respiratory tract. The prevalence of α2–6 linkages in the upper respiratory tract (URT) declines with depth, until it reaches an approximately equal ratio to that of SA with α2–3 linkages in the alveoli (Figure 2) [29]. It has been shown that initially, human viruses preferentially infected nonciliated cells, which predominantly contain α2–6 SA, whereas avian viruses mainly infected ciliated cells in the LRT, which predominantly contain α2–3 SA [30, 31]. Later in an infection, only the human viruses were able to infect all cell types. Although avian viruses can infect human epithelium, their replication and subsequent spread was limited [30, 31]. The fact that receptor specificity of avian strains does not alter during the course of a human infection can be explained by at least two hypotheses: a lack of time or sufficient pressure to select for mutants with a different binding profile; and/or genetic or structural constraints against the adoption of mutations necessary to achieve this.
Figure 2. Reactivity of human respiratory tissues with lectins specific for different sialic acid linkages.
Res, respiratory bronchiole (adjacent to alveoli); Ter, terminal bronchiole (distal to alveoli); Alv, alveolus. The green shows reaction with Sambucus nigra lectin, which indicates the presence of sialic acid linked to galactose by an α2–6-linkage. The red shows reaction with Maackia amurensis lectin, indicating the presence of sialic acid linked to galactose by an α2–6-linkage. (Reprinted by permission from Macmillan Publishers Ltd: images reproduced from Shinya et al. 2006 [29])
Strains with an α2–3 binding preference may not experience a pressure to alter binding once they begin successfully replicating if they have already located to the lower respiratory tract (LRT) where the advantage of α2–6 binding is lower. Unusually, high viral titres of H5N1 infection have been isolated from the nose and throat of infected individuals [32] and there is no indication that these isolates had a human α2–6 preference. Whether such reports represent actual viral replication or simply contamination of this area from lower areas is unclear and is worthy of further investigation. However, ferrets display a similar virus attachment pattern in the respiratory tract compared to humans and although H5N1 viruses bind to the lower respiratory tract tissue of ferrets, H5N1 infection does result in robust replication of H5N1 in the UTR [21, 33, 34]. Another possibility is that selection pressures against virions produced without an α2–6 preference are not as relevant once infection has established, even where the environment is prevalent in α2–6 linkages.
An alternative hypothesis for a lack of HA adaptation is that genetic barriers or structural constraints prevent a switch from readily occurring. HAmutations permitting adaptation to egg environments have been described but these frequently require a number of passages to appear [28]. The capacity for adaptation of H9N2 HA following repeated passages has also been shown in ferrets, although similarly, this took time, with anything up to 9 inoculations of the same viral strain through a sequence of hosts [35]. Both of these results suggest that HA adaptation may be slowed by the need for simultaneous mutations to retain fitness. In support of these results, there is evidence that part of the reason H5N1 has so far failed to achieve sustained human-human transmission is that adaptation to URT binding in the human requires such a set of mutations, each shown to reduce viral fitness if occurring separately [36]. In a recent exception, Chutinimitkul et al. demonstrated that in one avian viral strain tested, only one amino acid substitution was required to acquire both a human receptor specificity and binding profile [37]. Significantly, however, although replicative capacity was maintained, this mutation was still associated with a reduction in viral fitness. This result suggests that complementary mutations in other gene segments may also be required before such a mutant could dominate in a host. To date, although potentially achieved in isolation, it still cannot be ruled out that the appearance of HA adaptation could be substantially slowed by the requirement for complementary mutations.
When answering the question of why human tracheal binding characteristics are not always readily adopted during human infection, there is reason to believe that both hypotheses outlined above play a role, but neither has been fully elucidated. An emphasis is often placed in the literature on the role of genetic factors in preventing the appearance of human tracheal binding, but currently there are no experiments that can quantify their relative importance accurately enough. The extent to which selection pressures differ throughout each stage of human infection and across different areas of the respiratory tract should also not be ignored.
Understanding each of these considerations may have implications for the risk we allocate to human infections. For example, if it were only genetic or structural barriers that constrained successful adaptation of viruses, each individual infected with an avian virus would represent a pressure selecting for binding to the URT, characteristic of more efficient transmission. Each case therefore increases the chance of a strain accumulating the correct set of mutations to make the step toward pandemic emergence.
What underlies the avian-like α2–3 and human-like α2–6 preference?
Efforts have also been made to describe more precisely how SA features of the human respiratory tract give rise to host specificities. Recently, Chandrasekaran et al. [38] suggested that it is not the α2–6 linkage itself, but a characteristic structural topography, which in part determined by the linkage, that enables binding of HA to human SA in the URT. The authors distinguish two structural conformations: cone-like and umbrella- like. At the heart of their analysis is the notion that the angle constrained by the α2–6 linkage causes it to adopt the umbrella-like form, but only if the SA has sufficient length. Shorter SA with this linkage will adopt the less spread out cone-like topology. Due to a straighter profile, all SA with an α2–3 linkage will also adopt a cone-like structure (Figure 3). Their conclusion is that only viruses capable of binding the umbrella-like conformation gain potential for human transmission. It is therefore mutations that convey binding to the umbrella conformation that are positively selected for in human strains, and not binding to the α2–6 linkage itself. They propose that strains which display an ability to bind α2–6 linkages but do not transmit efficiently can be explained in that they bind only the shorter SA variants. Others have also found a tendency for human adapted viruses to bind longer oligosaccharide chains, but the distinction is not so absolute, with exceptions such as disaccharide binding [39]. However, as shown by Stevens et al. [40] following their own investigation into viral glycan binding characteristics, it is becoming increasingly evident that beyond a general preference for α2–6 linkages, human viruses bind to a structurally diverse set of α2–6 SA. Although Chandrasekaran implies that longer α2–6 linked SA are characteristic of the URT, the fact is that the detailed structures of the vast majority of human or avian SA remain obscure. Additionally, the cone-like topography shared by both α2–3 and short α2–6 linked SA fails to account for those viruses, at least as seen in glycan arrays, which are capable of binding α2–3 but not α2–6 in any form.
Figure 3. The cone-like (left) and umbrella-like (right) topologies presented by Chandrasekaran et al.
The umbrella conformation is unique to sialic acids with an α2–6 linkage and chains of sufficient length. As illustrated above, the topology is alleged to result from both the angle constrained by the linkage and the open spread allowed by the longer chain. (Reprinted by permission from Macmillan Publishers Ltd: Chandrasekaran et al. Nat Biotechnol 2008 Jan;26(1):107–13).
Glycan arrays are a good tool to investigate how HA binding preferences vary for the increasingly specific types of SA. However, Kumari et al. [39] found that H3N2 viruses with identical binding patterns on glycan arrays did not always have similar agglutination results. The authors suggest that variable HA density, rather than SA affinity, could explain these results. Studies like this demonstrate how successful viral replication in cells cannot always be inferred directly from binding preference alone. If we are to understand which HA features are positively selected for (or negatively against), it must be acknowledged that factors other than receptor binding and SA specificity can influence the probability of successful viral-cell attachment in the human respiratory environment.
Evidence for selection against α2–3 binding in human strains
It is still not apparent whether the selective pressure on HA binding in humans is chiefly for gaining affinity to α2–6 linked SA, or if there are also additional pressures against the binding of SA with an α2–3 linkage. It has been shown that fully adapted human viruses bind to the α2–6 rich URT more readily than avian viruses [41] with the reverse being true [42]. An analysis of the alterations in HA receptor binding properties from influenza strains isolated soon after introduction into mammals found that, besides increased affinity for α2–6 linked SA, there was a substantially decreased affinity for α2–3 linked SA [11].
Evaluating the role of intermediate hosts and genetics
Pigs are often thought to play a role in pandemic emergence because of their permissiveness for infection with both avian and human influenza strains and subsequent potential for reassortment of gene segments from both viruses [43, 44]. Genetic analysis of the 2009 pandemic showed evidence of multiple such reassortant events in pigs occurring over a number of years [45]. The respiratory tract of pigs contains a mixture of both α2–3 and α2–6 forms (Figure 4) [46] and avian origin H3N2 strains with an α2–6 receptor affinity have been identified in swine populations [47]. Significantly, although pigs were once thought to support a distribution of strains that included both viruses with avian-like and viruses with human-like binding preferences, this is now less certain and such receptor switching may be commonplace. An analysis of the binding preference of H1 and H3 of swine strains cultured in MDCK cells revealed that none of the strains showed any α2–3 binding, in contrast to other swine strains which had been cultured in eggs [8]. An analysis of the amino acid sequence of the egg cultured swine strains revealed that all had a Asp225Gly substitution in HA, an adaptation known to alter SA specificity [48]. However, since none of the swine strains were cultured in both MDCK cells and eggs, a direct interpretation of the role of such egg adaptation is difficult.
Figure 4. Lectin staining in the pig trachea.
M. amurensis lectin was used to detect SA with α2–3 linkages, while S. nigra was used to detect SA with α2–6 linkages. The green and orange in the images indicates both are present in the pig trachea. Blue staining in the connective tissue represents autofluorescence. (Image reproduced from Ito et al. 1998 with permission from American Society for Microbiology [46])
Whether results such as this could be indicative of an even more important role of pigs in facilitating human adaptation will depend upon how far the shared preference for α2–6 linkages in swine and human propagated strains translates to similar viral attributes beyond this. Importantly, other than this apparent benefit they create for an α2–6 binding preference, the extent to which the selection pressures in pigs mirror those within humans has not been described. Patterns of viral binding in the respiratory epithelium of the pig do closely mirror those found in the human but they are not identical (Table 1) [42]
Table 1.
Virus Attachment in Respiratory Tract of Humans and Five Animal Species
Results summarising virus attachment of a variety of strains, in the respiratory tract of humans and five animal species. (reproduced from van Riel et al. 2007 [42] with permission from Elsevier)
| Trachea |
Bronchus |
Bronchiole |
Alveolus |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Predominant | Predominant | Predominant | Predominant | ||||||
| Virus | Species | Score | cell type | Score | cell type | Score | cell type | Score | cell type |
| H3N2 | Human | ++† | cil | ++† | cil | + | cil | + | Type I |
| Mouse | − | − | − | ± | |||||
| Ferret | ++* | cil | ±* | ± | + | Type I | |||
| Macaque | − | − | − | − | |||||
| Pig | ++† | cil | ++† | cil | ++ | cil | + | Type I | |
| Cat | − | − | − | − | |||||
| H1N1 | Human | ++† | cil | ++† | cil | + | cil | + | Type I |
| Mouse | − | − | − | ± | |||||
| Ferret | +* | cil | +* | cil | ± | + | Type I | ||
| Macaque | − | − | − | − | |||||
| Pig | ++† | cil | ++† | cil | ++ | cil | + | Type I | |
| Cat | − | − | − | − | |||||
| H5N1 | Human | −* | +* | cil | + | non-cil | +‡ | Type II | |
| Mouse | ++ | Both | + | non-cil | + | non-cil | + | Type II | |
| Ferret | − | − | ± | + | Type II | ||||
| Macaque | − | ± | ± | + | Type I | ||||
| Pig | − | − | − | + | Type II | ||||
| Cat | − | − | + | non-cil | +‡ | Type II | |||
| H5N9 | Human | −* | +* | cil | + | non-cil | +‡ | Type II | |
| Mouse | ++ | Both | ++ | non-cil | ++ | non-cil | + | Type II | |
| Ferret | − | − | − | ± | |||||
| Macaque | − | − | ± | ± | |||||
| Pig | − | − | ± | ± | |||||
| Cat | − | − | ± | +‡ | Type II | ||||
| H6N1 | Human | ±* | ±* | + | non-cil | +‡ | Type II | ||
| Mouse | ++ | Both | + | non-cil | + | non-cil | + | ||
| Ferret | − | − | − | ± | Type II | ||||
| Macaque | − | ± | ± | + | Type I | ||||
| Pig | − | − | ± | + | Type II | ||||
| Cat | − | − | ± | non-cil | +‡ | Type II | |||
The mean abundance of cells to which virus attached was scored as follows: −, no attachment; ±, attachment to rare or few cells; +, attachment to a moderate number of cells; ++, attachment to many cells. Where possible, the predominant cell type to which virus attached is indicated: ciliated cells (cil), nonciliated cuboidal cell (non-cil), type I pneumocytes (type I), or type II pneumocytes (type II).
Subrnucosal glands positive.
Goblet cells occasionally positive.
Alveolar macrophages positive.
Understanding more clearly the importance of potential HA selection pressures such as decoy receptor evasion would help assess the risk posed by various species. For example, H5N1 viruses isolated from domestic poultry have been shown to have a lower affinity for α2–3 linked SA [25] with no concomitant increase in α2–6 affinity. These strains may be less affected by factors such as mucin binding.
Aside from the inter-species differences in influenza infection, incidences of H5N1 often include more than one family member and there has been speculation that this is suggestive of shared genetic characteristics, despite the fact that similar exposure can also account for this phenomenon [49]. The extent of the role of innate immunity in protecting against avian strains is poorly understood. Variation of SA found in an individual’s respiratory tract would be a clear way in which susceptibility for avian strains could be increased. Congenital variations in SA expression are a well-documented cause of pathology and the notion that SA can be used to predict the risk of certain diseases is not a new concept [50].
Predicting the pathogenicity of emergent strains
A final important implication of HA adaptation, and an understanding of its drivers, is the potential to predict not only properties necessary for human transmission, but also how properties such as the pathogenicity are likely to alter upon the acquisition of pandemic capability. Pathogenicity itself has been a notably variable feature in previous influenza pandemics and is a property that proved very difficult to predict in the most recent H1N1 pandemic. Currently, H5N1 is approximated to have a mortality of 59% [3] and this has obviously been of great concern [50]. Despite this, and although such pathogenicity is undoubtedly a multigenic trait, there is reason to link it at least in part to HA binding specificity. In particular, one hypothesis states that in making the switch from preference from α2–3 to α2–6 binding, the severity of the disease will decrease since it is likely to infect the URT rather than, as currently assumed, in the LRT (Figure 5) [34, 52]. In support of the hypothesis that an avian binding profile causes an increased mortality, clinical reviews of H5N1 infections identify the most common cause of death as respiratory failure due to diffuse alveolar damage [52, 53]. Additionally, experiments investigating cell tropism of H5N1 have regularly identified a strong binding preference in the alveoli for type II pneumocytes [34, 42]. The high metabolic activity (and therefore potentially increased viral production) associated with these cells and their role in surfactant production has led to speculation that, beyond simply LRT localisation, even tropism to this particular cell may be an important factor in the severity of lower respiratory complications associated with H5N1.
Figure 5. Attachment of H5N1 virus to regions of the human respiratory tract.
Red is representative of viral binding, which is shown to increase with depth in the respiratory tract. Image reproduced from van Riel et al. 2007 [42] with permission from Elsevier)
Concluding remarks
There is evidence that the human respiratory environment exerts several types of selection pressures upon influenza HA, beyond simply receptor type and availability. Each appears to influence the fate of viral strains during human infection, yet it is still unclear to what extent each dictates viral success in the human. In particular, the identification of further complexities behind HA features such as α2–3 and α2–6 binding preferences thought classically human and avian, illustrates how important it is that analysis of these features is paralleled by direct exploration, rather than inference, of the nature of the selective pressures that have maintained them. This knowledge will not only help in the explanation of the characteristics observed in current human strains, but will also help focus the expectations we can reasonably have of the alterations that will accompany avian strains should they successfully mutate to achieve pandemic potential. In the future, more detailed characterisation of the structure of SA in the human respiratory tract will aid in the interpretation of the increasingly sensitive binding comparisons resulting from glycan microarray technology. A twinned approach, investigating properties of the virus as well as the viral environment should help with more reliable identification of the genetic and structural attributes that form the markers of efficient human-human transmission.
Highlights.
Haemagglutinin is a determinant of many viral properties.
Specificity derived from different human and avian sialic acid linked receptors.
There is evidence for genetic and structural constraints on haemagglutinin.
Mortality of emergent pandemic may be reduced if haemagglutinin specificity changes.
Acknowledgements
M.G. is a Marie Curie fellow and funded under contract PIEF-GA-2009-237505. D.J.S. and D.F.B. were supported in part by an NIH Director’s Pioneer Award, part of the NIH roadmap for medical research, through grant DP1-OD000490-01; 223498 EMPERIE, an FP7 grant from the European Union; and program grant P0050/2008 from the Human Frontier Science Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kilbourne ED. Influenza pandemics of the 20th century. Emerg Infect Dis. 2006 Jan;12(1):9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Itoh Y, Shinya K, Kiso M, Watanabe T, Sakoda Y, Hatta M, et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009 Aug 20;460(7258):1021–1025. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. Cumulative Number of Confirmed Human Cases of Avian Influenza A/(H5N1) Reported to WHO. 2011 22/06/2011. [cited 2011 29/06/2011];Cumulative Number of Confirmed Human Cases of Avian Influenza A/(H5N1) Reported to WHO up before 22nd June 2011. Available from: . [cited; Available from: http://www.who.int/csr/disease/avian_influenza/country/cases_table_2011_06_22/en/index.html.
- 4.CDC. Avian Influenza A Virus Infections of Humans. 2008 23/05/2008. [cited 2011 29/06/2011];Information on confirmed cases of avian influenza A virus infections in humans. Available from: http://www.cdc.gov/flu/avian/gen-info/avian-flu-humans.html.
- 5.Wang H, Feng Z, Shu Y, Yu H, Zhou L, Zu R, et al. Probable limited person-to-person transmission of highly pathogenic avian influenza A (H5N1) virus in China. Lancet. 2008 Apr 26;371(9622):1427–1434. doi: 10.1016/S0140-6736(08)60493-6. [DOI] [PubMed] [Google Scholar]
- 6.Ungchusak K, Auewarakul P, Dowell SF, Kitphati R, Auwanit W, Puthavathana P, et al. Probable person-to-person transmission of avian influenza A (H5N1) N Engl J Med. 2005 Jan 27;352(4):333–340. doi: 10.1056/NEJMoa044021. [DOI] [PubMed] [Google Scholar]
- 7.Kandun IN, Wibisono H, Sedyaningsih ER, Yusharmen Hadisoedarsuno W, Purba W, et al. Three Indonesian clusters of H5N1 virus infection in 2005. N Engl J Med. 2006 Nov 23;355(21):2186–2194. doi: 10.1056/NEJMoa060930. [DOI] [PubMed] [Google Scholar]
- 8.Abdel-Ghafar AN, Chotpitayasunondh T, Gao Z, Hayden FG, Nguyen DH, de Jong MD, et al. Update on avian influenza A (H5N1) virus infection in humans. N Engl J Med. 2008 Jan 17;358(3):261–273. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- 9.Bean WJ, Schell M, Katz J, Kawaoka Y, Naeve C, Gorman O, et al. Evolution of the H3 influenza virus hemagglutinin from human and nonhuman hosts. J Virol. 1992 Feb;66(2):1129–1138. doi: 10.1128/jvi.66.2.1129-1138.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994 Nov 15;205(1):17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 11.Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, et al. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000 Sep;74(18):8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matrosovich MGA, Klenk H. Receptor specificity of influenza viruses and its alteration during interspecies transmission. Avian Influenza: Karger Publishers; 2008. pp. 134–155. [Google Scholar]
- 13.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992 Mar;56(1):152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004 Jul 16;305(5682):371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 15.Fouchier RA, Smith DJ. Use of antigenic cartography in vaccine seed strain selection. Avian Dis. 2010 Mar;54(1 Suppl):220–223. doi: 10.1637/8740-032509-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 16.Potter CW. A history of influenza. J Appl Microbiol. 2001 Oct;91(4):572–579. doi: 10.1046/j.1365-2672.2001.01492.x. [DOI] [PubMed] [Google Scholar]
- 17.Bouvier NM, Palese P. The biology of influenza viruses. Vaccine. 2008 Sep 12;26(Suppl 4):D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogers GN, Paulson JC. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983 Jun;127(2):361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 19.Rogers GN, D'Souza BL. Receptor binding properties of human and animal H1 influenza virus isolates. Virology. 1989 Nov;173(1):317–322. doi: 10.1016/0042-6822(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 20.Baum LG, Paulson JC. Sialyloligosaccharides of the respiratory epithelium in the selection of human influenza virus receptor specificity. Acta Histochem Suppl. 1990;40:35–38. [PubMed] [Google Scholar]
- 21.Bodewes R, Kreijtz JH, van Amerongen G, Fouchier RA, Osterhaus AD, Rimmelzwaan GF, et al. Pathogenesis of Influenza A/H5N1 virus infection in ferrets differs between intranasal and intratracheal routes of inoculation. Am J Pathol. 2011 Jul;179(1):30–36. doi: 10.1016/j.ajpath.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solorzano A, Pappas C, et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007 Feb 2;315(5812):655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- 23.Van Hoeven N, Pappas C, Belser JA, Maines TR, Zeng H, Garcia-Sastre A, et al. Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc Natl Acad Sci U S A. 2009 Mar 3;106(9):3366–3371. doi: 10.1073/pnas.0813172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matrosovich MN, Krauss S, Webster RG. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology. 2001 Mar 15;281(2):156–162. doi: 10.1006/viro.2000.0799. [DOI] [PubMed] [Google Scholar]
- 25.Gambaryan A, Tuzikov A, Pazynina G, Bovin N, Balish A, Klimov A. Evolution of the receptor binding phenotype of influenza A (H5) viruses. Virology. 2006 Jan 20;344(2):432–438. doi: 10.1016/j.virol.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 26.Carroll SM, Higa HH, Paulson JC. Different cell-surface receptor determinants of antigenically similar influenza virus hemagglutinins. J Biol Chem. 1981 Aug 25;256(16):8357–8363. [PubMed] [Google Scholar]
- 27.Choppin PW, Tamm I. Two kinds of particles with contrasting properties in influenza A virus strains from the 1957 pandemic. Virology. 1959 Aug;8:539–542. doi: 10.1016/0042-6822(59)90059-5. [DOI] [PubMed] [Google Scholar]
- 28.Ito T, Suzuki Y, Takada A, Kawamoto A, Otsuki K, Masuda H, et al. Differences in sialic acid-galactose linkages in the chicken egg amnion and allantois influence human influenza virus receptor specificity and variant selection. J Virol. 1997 Apr;71(4):3357–3362. doi: 10.1128/jvi.71.4.3357-3362.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006 Mar 23;440(7083):435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 30.Matrosovich M, Matrosovich T, Uhlendorff J, Garten W, Klenk HD. Avian-virus-like receptor specificity of the hemagglutinin impedes influenza virus replication in cultures of human airway epithelium. Virology. 2007 May 10;361(2):384–390. doi: 10.1016/j.virol.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 31.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci U S A. 2004 Mar 30;101(13):4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006 Oct;12(10):1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maines TR, Chen LM, Van Hoeven N, Tumpey TM, Blixt O, Belser JA, et al. Effect of receptor binding domain mutations on receptor binding and transmissibility of avian influenza H5N1 viruses. Virology. 2011 Apr 25;413(1):139–147. doi: 10.1016/j.virol.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, et al. H5N1 Virus Attachment to Lower Respiratory Tract. Science. 2006 Apr 21;312(5772):399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- 35.Sorrell EM, Wan H, Araya Y, Song H, Perez DR. Minimal molecular constraints for respiratory droplet transmission of an avian-human H9N2 influenza A virus. Proc Natl Acad Sci U S A. 2009 May 5;106(18):7565–7570. doi: 10.1073/pnas.0900877106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ayora-Talavera G, Shelton H, Scull MA, Ren J, Jones IM, Pickles RJ, et al. Mutations in H5N1 influenza virus hemagglutinin that confer binding to human tracheal airway epithelium. PLoS One. 2009;4(11):e7836. doi: 10.1371/journal.pone.0007836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chutinimitkul S, van Riel D, Munster VJ, van den Brand JM, Rimmelzwaan GF, Kuiken T, et al. In vitro assessment of attachment pattern and replication efficiency of H5N1 influenza A viruses with altered receptor specificity. J Virol. 2010 Jul;84(13):6825–6833. doi: 10.1128/JVI.02737-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chandrasekaran A, Srinivasan A, Raman R, Viswanathan K, Raguram S, Tumpey TM, et al. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat Biotechnol. 2008 Jan;26(1):107–113. doi: 10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]
- 39.Kumari K, Gulati S, Smith DF, Gulati U, Cummings RD, Air GM. Receptor binding specificity of recent human H3N2 influenza viruses. Virol J. 2007;4:42. doi: 10.1186/1743-422X-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, et al. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol. 2006 Feb 3;355(5):1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 41.van Riel D, den Bakker MA, Leijten LM, Chutinimitkul S, Munster VJ, de Wit E, et al. Seasonal and pandemic human influenza viruses attach better to human upper respiratory tract epithelium than avian influenza viruses. Am J Pathol. 2010 Apr;176(4):1614–1618. doi: 10.2353/ajpath.2010.090949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, et al. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am J Pathol. 2007 Oct;171(4):1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castrucci MR, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster RG. Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology. 1993 Mar;193(1):503–506. doi: 10.1006/viro.1993.1155. [DOI] [PubMed] [Google Scholar]
- 44.Ma W, Kahn RE, Richt JA. The pig as a mixing vessel for influenza viruses: Human and veterinary implications. J Mol Genet Med. 2008;3(1):158–166. [PMC free article] [PubMed] [Google Scholar]
- 45.Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009 Jun 25;459(7250):1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 46.Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998 Sep;72(9):7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma W, Vincent AL, Gramer MR, Brockwell CB, Lager KM, Janke BH, et al. Identification of H2N3 influenza A viruses from swine in the United States. Proc Natl Acad Sci U S A. 2007 Dec 26;104(52):20949–20954. doi: 10.1073/pnas.0710286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gambaryan AS, Karasin AI, Tuzikov AB, Chinarev AA, Pazynina GV, Bovin NV, et al. Receptor-binding properties of swine influenza viruses isolated and propagated in MDCK cells. Virus Res. 2005 Dec;114(1–2):15–22. doi: 10.1016/j.virusres.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 49.Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, de Jong MD, et al. Avian influenza A (H5N1) infection in humans. N Engl J Med. 2005 Sep 29;353(13):1374–1385. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 50.Varki A. Sialic acids in human health and disease. Trends Mol Med. 2008 Aug;14(8):351–360. doi: 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webster R WE. The world is teetering on the edge of a pandemic that could kill a large fraction of the human population. American Scientist. 2003;2003:122–123. [Google Scholar]
- 52.Kuiken T, Taubenberger JK. Pathology of human influenza revisited. Vaccine. 2008 Sep 12;26(Suppl 4):D59–D66. doi: 10.1016/j.vaccine.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uyeki TM. Human infection with highly pathogenic avian influenza A (H5N1) virus: review of clinical issues. Clin Infect Dis. 2009 Jul 15;49(2):279–290. doi: 10.1086/600035. [DOI] [PubMed] [Google Scholar]