Abstract
Rudolph Schoenheimer’s concept of the “dynamic state of body constituents” has existed since the 1940s, but the idea that heart muscle cells renew themselves from within is relatively new. Many studies have elucidated the interaction of metabolic pathways for energy provision and contraction of the heart, and work in the field has uncovered novel metabolic regulators of enzyme action. However, the impact of myocardial energy metabolism on myocardial protein turnover has received little attention. Here, we review recent findings which identify metabolic signals as regulators of myocardial protein turnover and seek to broaden the role of energy substrate metabolism from a provider of ATP to a regulator of self-renewal of the cardiomyocyte.
Introduction
Heart failure is the leading cause of death in the United States, claiming the lives of more than 500,000 Americans every year (Roger, et al., 2011). Accepted features in the development of heart failure are cardiac hypertrophy and impaired ATP production, which develop in response to both endogenous (genetic) and exogenous (environmental) changes. While it is well established that structural remodeling, at both the intracellular and extracellular level, occurs in response to these changes (Hill and Olson, 2008), we have early evidence suggesting that metabolic remodeling (which is potentially reversible) precedes, triggers and sustains structural and functional remodeling of the heart (Goodwin, et al., 1998, Taegtmeyer, et al., 2004, Young, et al., 2007). Excessive intracellular remodeling results in the enlargement of cardiomyocytes, which contributes to an overall increase in heart size. The transition from hypertrophy to heart failure or the transition from adaptation to maladaptation of the heart remains elusive. Consequently it is of interest to know more about the mechanisms that control rebuilding of the cardiomyocyte.
In general terms, we propose that metabolic signals are putative regulators of myocardial remodeling, and more specifically regulators of protein turnover. A vast number of molecular targets have been investigated as regulators of myocardial hypertrophy (Frey and Olson, 2003) and atrophy (Baskin and Taegtmeyer, 2011). However, there is good evidence that metabolism is the first responder to any form of stress (Goodwin, et al., 1998, Neely, et al., 1967). We have extended these observations to investigate the hypothesis that modulation of metabolism provides a means to trigger structural remodeling of cardiomyocytes, at least in part, by regulating protein degradation. The identification of metabolic signals which govern cardiac remodeling will set us on the path to develop novel strategies aiming at specific metabolic intermediaries as modulators of cardiomyocyte size. In a broad sense we seek to establish mechanisms underlying the self-renewal of the intact cardiomyocyte.
Intracellular Protein Turnover in Perspective
The intrinsic mechanism of self-renewal of the cardiomyocyte requires the regulated degradation of damaged, misfolded, or useless proteins, and their replacement by new and functional proteins. Protein turnover therefore constitutes a major line of defense for protein quality control of the cardiomyocyte (Wang and Robbins, 2006). The rate of myocardial protein turnover is probably faster than has generally been assumed, with the half-life of individual proteins ranging from several hours to several days (Morgan, et al., 1979). Although the importance of protein degradation in the heart has been appreciated for some time (Morgan, et al., 1974), the specific proteolytic systems been investigated in detail only recently.
Intracellular protein degradation in cardiomyocytes is controlled by independent but interrelated processes: ubiquitin proteasome system (UPS)-mediated proteolysis and autophagy. While autophagy can degrade whole organelles (Zheng, et al., 2009), individual proteins are degraded through the UPS (Glickman and Ciechanover, 2002, Razeghi, et al., 2006). The UPS is comprised of three enzymatic steps responsible for the charging and transferring of ubiquitin (Hershko and Ciechanover, 1998). Ubiquitin ligases confer specificity to the system by the selective ubiquitination of target proteins which are then degraded by the proteasome (Willis, et al., 2010).
Over 600 ubiquitin ligases have been identified in mammalian cells, but several key ligases have emerged as important tissue-specific regulators of cell size (Nagy and Dikic, 2010). Two muscle-specific ubiquitin ligases, Muscle Atrophy F-box (MAFbx/atrogin-1) and Muscle Ring Finger-1 (MuRF1), are essential regulators of UPS-mediated protein degradation in muscle and in the heart. The initial discovery of Atrogin-1 and MuRF1 and their requirement for skeletal muscle atrophy (Bodine, et al., 2001, Gomes, et al., 2001), were the first of many studies on the mechanisms of UPS-mediated protein degradation in muscle. Subsequent investigations have characterized the expression of these ubiquitin ligases under many disease conditions and have identified some of their protein targets, as reviewed in (Foletta, et al., 2011, Portbury, et al., 2011). Much of what we know about Atrogin-1 and MuRF1 emanates from studies performed in skeletal muscle, while comparatively less is known about their function in the heart.
Like in skeletal muscle, Atrogin-1 and MuRF1 are important determinants of cardiac protein degradation and myocardial mass. Studies in vivo demonstrate that overexpressing atrogin-1 in the heart attenuates the development of hypertrophy (Li, et al., 2004), while the deletion of MuRF1 results in increased hypertrophy (Arya, et al., 2004, Willis, et al., 2007). These experiments highlight the importance of atrogin-1 and MuRF1 in regulating heart size. However, the mechanisms by which the ligases themselves are regulated in the heart are not completely understood.
Metabolic Homeostasis in the Cardiomyocyte
Cardiomyocytes liberate the energy contained in organic molecules to support their function. This highly regulated process generates ATP from fatty acids, glucose, lactate, ketone bodies, and (under extreme circumstances) amino acids (Taegtmeyer, et al., 1980). For a given environment the source of ATP production can be changed from one fuel to another, and the heart has therefore been termed a “metabolic omnivore” (Taegtmeyer, 1985). In the fasted state and under resting conditions the heart prefers fatty acids while carbohydrates become a major fuel for the stressed heart (Goodwin, et al., 1998).
A key enzyme involved in the metabolic response to stress in the heart is AMP-activated protein kinase (AMPK). AMPK, also known as the “fuel gauge” of the cell, is activated under a variety of conditions that decrease the ratio of [ATP]/[AMP]. AMPK regulates processes involved in ATP production, such as glucose uptake, glycolysis, fatty acid oxidation, and mitochondrial biogenesis. At the same time, AMPK inhibits processes involved in ATP consumption, such as protein synthesis, glycogen synthesis, gluconeogenesis, and fatty acid synthesis (Hardie and Carling, 1997). While the amount of ATP turnover of the heart is higher than any other organ (Taegtmeyer, 1994 #37), AMPK is an essential regulator of metabolic homeostasis in the cardiomyocyte (Dyck, 2006 #410).
We have started to elucidate the mechanisms whereby metabolic remodeling precedes, triggers and sustains structural and functional remodeling of the heart. We propose that under conditions of metabolic stress in the heart, the ensuing activation of AMPK contributes to structural remodeling of the cardiomyocyte. Here, we review our recent findings that AMPK regulates protein degradation in the heart, in part by regulating ubiquitin ligases (Baskin and Taegtmeyer, 2011).
The Ubiquitin Proteasome System and AMPK
Early studies in the heart in vivo demonstrated that nutrient deprivation decreases protein synthesis and increases rates of protein degradation (Samarel, et al., 1987). Starvation decreases the intracellular concentration of ATP and, consequently, AMP-activated protein kinase (AMPK) is activated in order to provide energy to maintain normal cellular function (Kim, et al., 2009). It is well established that AMPK regulates energy substrate metabolism (Hardie and Carling, 1997), inhibits protein synthesis (Bolster, et al., 2002), and regulates transcription of metabolic genes (Bungard, et al., 2010, McGee, et al., 2008, Stoppani, et al., 2002). Although it has been reported that starvation induces autophagy in cardiomyocytes through AMPK (Matsui, et al., 2007), until recently a role of AMPK in the cardiac UPS has never been considered.
To investigate the metabolic regulation of protein degradation in cardiomyocytes we first verified that substrate deprivation in vitro enhances protein degradation, as has been shown already in vivo (Samarel, et al., 1987). Protein degradation was enhanced in cardiomyocytes during starvation, but decreased with bortezomib, a proteasome inhibitor, or with 3-methyladenine (3-MA), an inhibitor of autophagy. These results suggest that, like autophagy (Matsui, et al., 2007), proteasome-mediated protein degradation is important during nutrient starvation in cardiomyocytes. Atrogin-1 and MuRF1, important mediators of cardiac protein degradation and cardiac size, were significantly increased with starvation, which also correlated with enhanced AMPK activity and increased AMP levels. We therefore asked whether AMPK regulates the ubiquitin ligases directly, independent of nutrient starvation. Pharmacologic and genetic activation of AMPK increased both atrogin-1 and MuRF1 expression, which was significantly impaired with AMPK inhibition. Thus, AMPK is a regulator of the ubiquitin ligases atrogin-1 and MuRF1 in the heart (Baskin and Taegtmeyer, 2011).
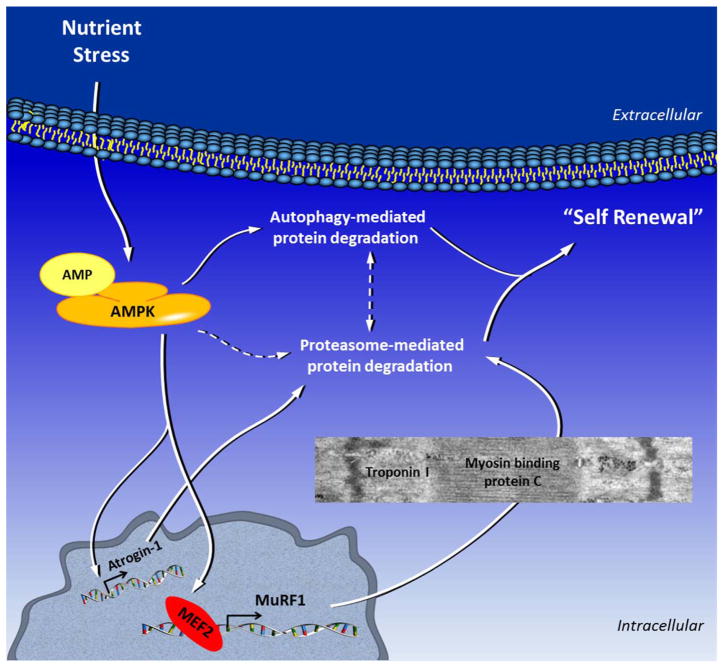
We further defined a mechanism by which AMPK regulates MuRF1 expression in cardiomyocytes. Using several approaches we demonstrated that AMPK regulates MuRF1 transcription through the transcription factor myocyte enhancer factor 2 (MEF2) in vitro and in vivo. Long term starvation-induced protein degradation in the heart is detrimental to cardiac function, while the absence of MuRF1 preserves cardiac function during starvation. Furthermore, proteasome-mediated protein degradation during starvation, downstream of AMPK, requires MuRF1. We propose that the activation of AMPK during starvation, leads to MuRF1-mediated degradation of several contractile proteins in cardiomyocytes. Consequently this would reduce energy consumption at the expense of a reduction in cardiac contractile function (Baskin and Taegtmeyer, 2011). These findings are summarized in Figure 1.
Figure 1.
Nutrient deprivation or other forms of environmental stress result in increased intracellular AMP levels and activation of AMPK. AMPK increases protein degradation via several different mechanisms, including autophagy and proteasome-mediated protein degradation by upregulating ubiquitin ligases. Specifically, AMPK regulates MuRF1 expression through the transcription factor MEF2, and regulates Atrogin-1 expression through an as yet unknown mechanism. During starvation, MuRF1 degrades myosin binding protein C and troponin I, which may contribute to decreased cardiac function. MuRF1 also degrades metabolic enzymes like aldolase a and pyruvate dehydrogenase (not shown in the figure). Hence, protein degradation is a mediator of cardiomyocyte remodeling, particularly during metabolic stress in the cardiomyocyte. See text for details. Dashed lines refer to possible crosstalk or as yet unknown mechanisms.
In keeping with this hypothesis, several studies have shown that MuRF1 degrades key enzymes involved in ATP production (including aldolase a and pyruvate dehydrogenase) (Hirner, et al., 2008). During fasting, intracellular glucose 6-phosphate and fructose 6-phosphate in the heart are increased, and glucose phosphorylation is decreased, most likely by allosteric inhibition of hexokinase (Newsholme and Randle, 1961). Additionally, glycogen deposition and citrate levels are increased in the heart (Adrouny, 1969). This is most likely a protective mechanism of the heart: it stores enough endogenous substrate for temporarily use during extreme conditions in order to maintain contractile function. Citrate inhibits phosphofructokinase, a regulatory enzyme in the glycolytic pathway which phosphorylates fructose-6-phosphate to fructose-1,6-bisphosphate, putting a brake on glycolysis. The possible degradation of aldolase a, a glycolytic enzyme downstream of phosphofructokinase, by MuRF1 may also be an adaptive mechanism to remove enzymes that are no longer needed during starvation. This may be the case with pyruvate dehydrogenase as well.
During atrophy, MuRF1 is upregulated and phosphofructokinase is downregulated (Bodine, et al., 2001). Because MuRF1 and phosphofructokinase localize to the M-line of sarcomeres, it is tempting to speculate that MuRF1 ubiquitinates phosphofructokinase thereby leading to its degradation. As suggested above, upregulation of MuRF1 during starvation could lead to the downregulation of metabolic enzymes, decreased ATP production from glycolysis and/or inhibition of the conversion of pyruvate to acetyl-CoA, and ultimately decreased cardiac function.
Although we have demonstrated that AMPK-mediated protein degradation by MuRF1 is detrimental to cardiac function during starvation, we have not yet investigated AMPK-mediated protein degradation through MuRF1 in other settings of metabolic stress in the heart. AMPK is known to be protective during myocardial ischemia (Dyck and Lopaschuk, 2006). MuRF1 has also recently been shown to be protective during ischemia reperfusion (I/R). MuRF1 inhibits JNK signaling through ubiquitin-mediated degradation of c-Jun, thus decreasing cell death (Li, et al., 2011). While the overexpression of MuRF1 in cardiomyocytes decreases apoptosis in response to I/R, it is still unclear whether endogenous protein levels of MuRF1 are upregulated in response to I/R, and whether this is cardioprotective.
Conclusions and Perspective
The work reviewed here extends the long established concept of the “dynamic state of body constituents” (Schoenheimer, 1942) to a specific situation when the heart adapts to changes in its metabolic environment. The heart is a dynamic organ that adapts metabolically and structurally to various types of stress. This remodeling process is critical for survival because cardiomyocytes are terminally differentiated cells. Although there is evidence for the existence of sub-populations of cardiac progenitor cells, they have been suggested to generate and replace cardiomyocytes at a very low rate compared to other cell types. Thus, intracellular homeostasis, in a broad sense, is vital for cardiomyocyte survival and, ultimately, cardiac function.
Intracellular protein degradation is a complex and highly controlled process that is integrated with the environment of the cell. Although important, protein turnover is an underappreciated mechanism of adaptation in the heart. Therefore, it is of interest to understand how protein degradation is regulated in the cardiomyocyte under various circumstances. There is evidence that markers of the UPS are upregrulated in the heart in several settings of cardiac remodeling (Razeghi, et al., 2006), but the extent of how the markers themselves are regulated is not completely understood. AMPK regulates cellular homeostasis in part by decreasing protein synthesis (Bolster, et al., 2002) and activating autophagy (Matsui, et al., 2007). While AMPK itself has recently been found to be regulated by the UPS (Zungu, et al., 2011), we now have evidence demonstrating that AMPK regulates protein degradation through the UPS in the heart, in part by regulating ubiquitin ligases (Baskin and Taegtmeyer, 2011).
We do not know at present to what extent metabolic signals regulate protein degradation and protein synthesis. We have preliminary evidence, based on earlier work which suggests that metabolic signals, i.e. changes in intracellular metabolite levels in response to stress, may activate pathways of protein degradation and protein synthesis (Taegtmeyer, et al., 2004, Young, et al., 2007). While we have only uncovered one group of metabolic signals which regulate protein degradation, there certainly remain many more that have yet to be defined. Our ideas advance a new role for cardiac metabolism. In addition to energy provision (Taegtmeyer, et al., 1980), intermediary metabolism of energy providing substrates is an integral part of the self-renewing cardiomyocyte. We consider altered fuel metabolism (leading to either a decrease or an increase of certain metabolic signals) as a root cause for altered rates of intracellular protein turnover and, hence, self-renewal of the cardiomyocyte. Taken together, we propose that the “metabolic” approach to myocardial protein synthesis and degradation provides a new framework that will expose new regulators driving self-renewal of cardiomyocytes from within.
Acknowledgments
We thank Roxy A. Tate for assistance with the manuscript preparation and Romain Harmancey for helpful discussions. The studies reviewed here were supported in part, by a grant from the National Heart, Lung, and Blood Institute (5R01HL061483-9) of the US Public Health Service. K.K.B. received a predoctoral fellowship from the American Heart Association, National Center (11PRE5200006).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adrouny GA. Differential patterns of glycogen metabolism in cardiac and skeletal muscles. Am J Physiol. 1969;217:686–93. doi: 10.1152/ajplegacy.1969.217.3.686. [DOI] [PubMed] [Google Scholar]
- Arya R, Kedar V, Hwang JR, et al. Muscle ring finger protein-1 inhibits PKC{epsilon} activation and prevents cardiomyocyte hypertrophy. J Cell Biol. 2004;167:1147–59. doi: 10.1083/jcb.200402033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin KK, Taegtmeyer H. AMP-Activated Protein Kinase Regulates E3 Ligases in Rodent Heart. Circ Res. 2011;109:1153–61. doi: 10.1161/CIRCRESAHA.111.252742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin KK, Taegtmeyer H. Taking pressure off the heart: the ins and outs of atrophic remodelling. Cardiovasc Res. 2011;90:243–50. doi: 10.1093/cvr/cvr060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–8. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Bolster DR, Crozier SJ, Kimball SR, Jefferson LS. AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. J Biol Chem. 2002;277:23977–80. doi: 10.1074/jbc.C200171200. [DOI] [PubMed] [Google Scholar]
- Bungard D, Fuerth BJ, Zeng PY, et al. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–5. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyck JR, Lopaschuk GD. AMPK alterations in cardiac physiology and pathology: enemy or ally? J Physiol. 2006;574:95–112. doi: 10.1113/jphysiol.2006.109389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foletta VC, White LJ, Larsen AE, Leger B, Russell AP. The role and regulation of MAFbx/atrogin-1 and MuRF1 in skeletal muscle atrophy. Pflugers Arch. 2011;461:325–35. doi: 10.1007/s00424-010-0919-9. [DOI] [PubMed] [Google Scholar]
- Frey N, Olson EN. Cardiac hypertrophy: the good, the bad, and the ugly. Annu Rev Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci U S A. 2001;98:14440–5. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin GW, Taylor CS, Taegtmeyer H. Regulation of energy metabolism of the heart during acute increase in heart work. J Biol Chem. 1998;273:29530–9. doi: 10.1074/jbc.273.45.29530. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Carling D. The AMP-activated protein kinase--fuel gauge of the mammalian cell? Eur J Biochem. 1997;246:259–73. doi: 10.1111/j.1432-1033.1997.00259.x. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–80. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- Hirner S, Krohne C, Schuster A, et al. MuRF1-dependent regulation of systemic carbohydrate metabolism as revealed from transgenic mouse studies. J Mol Biol. 2008;379:666–77. doi: 10.1016/j.jmb.2008.03.049. [DOI] [PubMed] [Google Scholar]
- Kim AS, Miller EJ, Young LH. AMP-activated protein kinase: a core signalling pathway in the heart. Acta Physiol (Oxf) 2009;196:37–53. doi: 10.1111/j.1748-1716.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- Li HH, Du J, Fan YN, et al. The ubiquitin ligase MuRF1 protects against cardiac ischemia/reperfusion injury by its proteasome-dependent degradation of phospho-c-Jun. Am J Pathol. 2011;178:1043–58. doi: 10.1016/j.ajpath.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Kedar V, Zhang C, et al. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J Clin Invest. 2004;114:1058–71. doi: 10.1172/JCI22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y, Takagi H, Qu X, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–22. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- McGee SL, van Denderen BJ, Howlett KF, et al. AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5. Diabetes. 2008;57:860–7. doi: 10.2337/db07-0843. [DOI] [PubMed] [Google Scholar]
- Morgan HE, Rannels DE, Kao RL. Factors controlling protein turnover in heart muscle. Circ Res. 1974;35(Suppl 3):22–31. [PubMed] [Google Scholar]
- Nagy V, Dikic I. Ubiquitin ligase complexes: from substrate selectivity to conjugational specificity. Biol Chem. 2010;391:163–9. doi: 10.1515/bc.2010.021. [DOI] [PubMed] [Google Scholar]
- Neely JR, Liebermeister H, Morgan HE. Effect of pressure development on membrane transport of glucose in isolated rat heart. Am J Physiol. 1967;212:815–22. doi: 10.1152/ajplegacy.1967.212.4.815. [DOI] [PubMed] [Google Scholar]
- Newsholme EA, Randle PJ. Regulation of glucose uptake by muscle. 5. Effects of anoxia, insulin, adrenaline and prolonged starving on concentrations of hexose phosphates in isolated rat diaphragm and perfused isolated rat heart. Biochem J. 1961;80:655–62. doi: 10.1042/bj0800655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portbury AL, Willis MS, Patterson C. Tearin' up my heart: proteolysis in the cardiac sarcomere. J Biol Chem. 2011;286:9929–34. doi: 10.1074/jbc.R110.170571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razeghi P, Baskin KK, Sharma S, et al. Atrophy, hypertrophy, and hypoxemia induce transcriptional regulators of the ubiquitin proteasome system in the rat heart. Biochem Biophys Res Commun. 2006;342:361–4. doi: 10.1016/j.bbrc.2006.01.163. [DOI] [PubMed] [Google Scholar]
- Roger VL, Go AS, Lloyd-Jones DM, et al. Heart disease and stroke statistics--2011 update: a report from the American Heart Association. Circulation. 2011;123:e18–e209. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarel AM, Parmacek MS, Magid NM, Decker RS, Lesch M. Protein synthesis and degradation during starvation-induced cardiac atrophy in rabbits. Circ Res. 1987;60:933–41. doi: 10.1161/01.res.60.6.933. [DOI] [PubMed] [Google Scholar]
- Stoppani J, Hildebrandt AL, Sakamoto K, et al. AMP-activated protein kinase activates transcription of the UCP3 and HKII genes in rat skeletal muscle. Am J Physiol Endocrinol Metab. 2002;283:E1239–48. doi: 10.1152/ajpendo.00278.2002. [DOI] [PubMed] [Google Scholar]
- Taegtmeyer H. Carbohydrate interconversions and energy production. Circulation. 1985;72:IV1–8. [PubMed] [Google Scholar]
- Taegtmeyer H, Golfman L, Sharma S, Razeghi P, van Arsdall M. Linking gene expression to function: metabolic flexibility in the normal and diseased heart. Ann N Y Acad Sci. 2004;1015:202–13. doi: 10.1196/annals.1302.017. [DOI] [PubMed] [Google Scholar]
- Taegtmeyer H, Hems R, Krebs HA. Utilization of energy-providing substrates in the isolated working rat heart. Biochem J. 1980;186:701–11. doi: 10.1042/bj1860701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99:1315–28. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- Willis MS, Ike C, Li L, et al. Muscle ring finger 1, but not muscle ring finger 2, regulates cardiac hypertrophy in vivo. Circ Res. 2007;100:456–9. doi: 10.1161/01.RES.0000259559.48597.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis MS, Townley-Tilson WH, Kang EY, Homeister JW, Patterson C. Sent to destroy: the ubiquitin proteasome system regulates cell signaling and protein quality control in cardiovascular development and disease. Circ Res. 2010;106:463–78. doi: 10.1161/CIRCRESAHA.109.208801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ME, Yan Z, Razeghi P, et al. Proposed regulation of gene expression by glucose in rodent heart. Gene Reg Systems Biol. 2007;1:251–262. doi: 10.4137/grsb.s222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Li J, Wang X. Interplay between the ubiquitin-proteasome system and autophagy in proteinopathies. Int J Physiol Pathophysiol Pharmacol. 2009;1:127–142. [PMC free article] [PubMed] [Google Scholar]
- Zungu M, Schisler JC, Essop MF, et al. Regulation of AMPK by the ubiquitin proteasome system. Am J Pathol. 2011;178:4–11. doi: 10.1016/j.ajpath.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]