SUMMARY
Dysregulated metabolism is a hallmark of cancer cell lines, but little is known about the fate of glucose and other nutrients in tumors growing in their native microenvironment. To study tumor metabolism in vivo, we used an orthotopic mouse model of primary human glioblastoma (GBM). We infused 13C-labeled nutrients into mice bearing three independent GBM lines, each with a distinct set of mutations. All three lines displayed glycolysis, as expected for aggressive tumors. They also displayed unexpected metabolic complexity, oxidizing glucose via pyruvate dehydrogenase and the citric acid cycle, and using glucose to supply anaplerosis and other biosynthetic activities. Comparing the tumors to surrounding brain revealed obvious metabolic differences, notably the accumulation of a large glutamine pool within the tumors. Many of these same activities were conserved in cells cultured ex vivo from the tumors. Thus GBM cells utilize mitochondrial glucose oxidation during aggressive tumor growth in vivo.
INTRODUCTION
Malignant transformation is driven by genetic mutations that promote unchecked proliferation, abrogate cell death programs, and reprogram metabolism (DeBerardinis et al., 2008a; Hanahan and Weinberg, 2011). Tumor metabolism is now an area of intense research to identify novel therapeutic targets and biomarkers for next-generation imaging (Kurhanewicz et al., 2011; Tennant et al., 2010). Exploring the clinical applications of cancer metabolism requires an accurate picture of metabolic pathways of human tumors in situ. Cell culture studies have produced insights into how tumorigenic mutations can stimulate a metabolic phenotype characterized by high rates of glycolysis relative to oxidative metabolism (Ramanathan et al., 2005), and detailed studies using isotope-tracer methods have described the intricacies of metabolic flux in cancer cell lines grown in culture (Forbes et al., 2006). However, caution is necessary when extending such findings to the in vivo setting, since primary tumor cells acquire extensive genomic and gene expression changes when adapted to growth in culture (Lee et al., 2006; Mehrian Shai et al., 2005) and may lose the influence of the native microenvironment on metabolism. The importance of this latter issue is emphasized in recent work demonstrating roles for adjacent, normal cells in supporting tumor cell metabolism (Nieman et al., 2011; Zhang et al., 2012).
Glioblastoma (GBM), the most common primary malignant brain tumor in humans, is highly metabolically active and evades multimodal therapy, leading to short survival times. The prevailing view of GBM metabolism is that these tumors have high rates of glycolysis and glutaminolysis with minimal oxidation of glucose in the mitochondrial citric acid cycle (CAC). This perception is largely based on studies of glioma cell lines grown under conditions of maximal proliferation (DeBerardinis et al., 2007; Portais et al., 1996). Clinical targeting of metabolism in GBM is under investigation based on the assumption that glucose oxidation is suppressed in vivo (Michelakis et al., 2010). However, recent work on intact human brain tumors demonstrated that their metabolism involves extensive glucose oxidation in vivo (Maher et al., 2012). In that study, patients were infused with 13C-glucose during resection of solitary brain masses, and metabolites extracted from the tumors were analyzed by 13C NMR spectroscopy to detect the metabolic fates of glucose. All tumors analyzed, including high-grade gliomas and brain metastases, demonstrated avid oxidation of glucose by pyruvate dehydrogenase (PDH) and the CAC.
Here we studied GBM metabolism by infusing 13C-labeled nutrients into mice bearing human GBM cells that were isolated from freshly-resected tumors and implanted directly into the NOD-SCID mouse brain, without prior adaptation to cell culture (referred to as human GBM orthotopic tumors, HOTs). This model is of particular interest because of growing evidence that GBMs are organized by a cellular hierarchy in which only a subpopulation of cells drive tumor growth. Glioma stem cells, or tumor initiating cells, are uniquely capable of forming tumors when transplanted into the brains of immunocompromised mice (Singh et al., 2004). Thus the HOT model system provides a unique opportunity to study metabolism of the most relevant cell types in human GBM under conditions similar to their growth in the human brain. Our goals were two-fold: first, to identify the metabolic fates of glucose and glutamine, the two nutrients most rapidly consumed by GBM cell lines; and second, to identify definitive metabolic differences between the tumor and surrounding brain, reasoning that metabolic activities differentiating these two compartments could provide targets for cancer therapy and/or tumor-specific imaging. The data show that HOTs derived from multiple independent human GBMs exhibit glucose oxidation via PDH and the CAC, anaplerosis, accumulation of a large glutamine pool consisting in part of glutamine synthesized de novo from glucose, and minimal glutaminolysis.
RESULTS
Genetically diverse, orthotopically-transplanted human GBMs avidly consume glucose
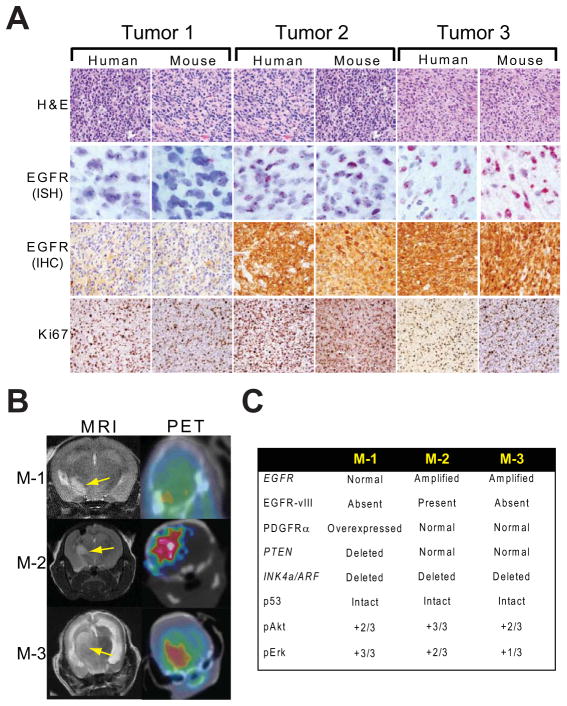
Tumor cells were isolated during surgical resection of human GBMs, and implanted into the putamen-caudate region of the brains of NOD-SCID mice. After 2–3 months, the mice became symptomatic as a result of an expanding intracranial mass. Dissection and re-implantation of tumor cells from pre-morbid mice, without adaptation to culture, enabled the tumors to be propagated indefinitely in the mouse brain. Comparing the genomic profile (array CGH, Nimblegen) of each of the HOTs to the patient tumors from which they were derived revealed that they were indistinguishable (data not shown). In addition, HOTs retained phenotypic features of the parental tumors, including diffuse infiltration, high mitotic (Ki67) index (Figure 1A) and 18fluoro-2-deoxyglucose (18FDG) uptake (Figure 1B).
Figure 1. Human orthotopic tumors (HOTs) established from three primary human GBMs.
(A) Hematoxylin and eosin (H&E) stains, EGFR in situ hybridization (ISH) and immunohistochemistry (IHC), and Ki67 staining of parental human GBMs and representative mouse HOTs.
(B) MRI and 18FDG-PET of representative mouse HOTs (M-1, M-2, M-3) derived from each parental line. Arrows indicate GBM masses in the right hemisphere.
(C) Summary of genomic and IHC data from HOTs M-1, M-2 and M-3.
For metabolic studies, we chose three lines (M-1, M-2 and M-3) that contained mutations commonly observed in human GBM (Figure 1C). These included genomic amplification of the epidermal growth factor receptor (EGFR) gene with high levels of EGFR protein (M-2 and M-3, Figure 1A); expression of the EGFR variant EGFRvIII (M-2, Figure S1A); and high expression of platelet-derived growth factor receptor-α (M-1, Figure S1A). Homozygous deletion of PTEN was observed in M-1. All 3 lines had loss of the INK4A/ARF locus and intact p53 (data not shown). As expected for tumors with enhanced growth factor receptors and/or loss of PTEN, all three lines displayed constitutive activation of Akt/mTOR signaling, and M-3 also had activation of ERK (Figure S1A). We examined the tumors for hypoxia by determining the abundance of the α-subunits of hypoxia-inducible factors-1 and -2 (HIF-1α and HIF-2α). A few HOT lines contained detectable levels of HIF-1α, but this was not a consistent feature in M-1, M-2 or M-3, and no tumors displayed significant amounts of HIF-2α (Figure S1B). Furthermore, histopathological analysis demonstrated infiltrating GBM cells in close proximity to host brain microvasculature, which maintained the normal pattern of tight-junction associated proteins (ZO-1, Occludin-1) and associated pericytes (Figure S1C). These observations suggest that the HOTs may co-opt preexisting host cerebrovasculature for nutrient support, perhaps avoiding significant hypoxia.
A 13C-glucose infusion method to establish isotopic steady-state within orthotopic GBM tumors
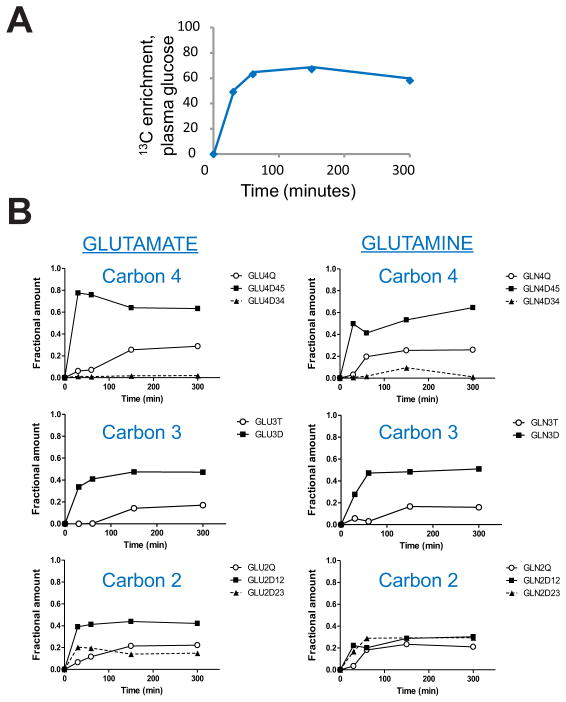
Infusion with 13C-glucose and other isotopically-labeled nutrients has been used to analyze metabolism in vivo, including in human tumors (Fan et al., 2009; Landau et al., 1996; Maher et al., 2012). In order to establish the time course of maximal 13C-enrichment and isotopic steady state in the HOTs, we infused 13C-glucose into several mice bearing tumors derived from the same human GBM. All mice received a bolus of [U-13C]glucose followed by a continuous infusion of up to 300 minutes. At the end of each infusion, 13C enrichment was determined in plasma glucose, and the tumor was rapidly dissected from the surrounding brain. Metabolites extracted from the tumor were analyzed by NMR to determine labeling patterns at each time point. More than 50% of the plasma glucose was 13C labeled within 60 minutes, and this level was maintained for the duration of the infusion (Figure 2A). In the tumor, 13C labeling in carbons 2, 3 and 4 of glutamate and glutamine were monitored as markers of isotopic steady state (Malloy et al., 1987). All of these carbons were labeled within 30 minutes. A smooth evolution of labeling emerged from these samples, despite the fact that they were obtained from several different mice (Figure 2B). This demonstrates that metabolic activity was highly conserved among these individual tumors. There was no appreciable change in the labeling pattern after 150 minutes, demonstrating that isotopic steady state had been reached (Figure 2B). All subsequent experiments used a bolus followed by a continuous infusion of 150 minutes.
Figure 2. Time course for 13C-glucose infusions in HOT-bearing mice.
(A) Four mice bearing HOTs derived from the same parental tumor were infused with [U-13C]glucose for the indicated times. The time course shows 13C enrichment (in %) of plasma glucose in the individual mice used in this time course experiment. Enrichment at time 0 was assumed to be 0%. All mice received a bolus of [U-13C]glucose over 1 minute followed by a continuous [U-13C]glucose infusion as described in Experimental Procedures.
(B) NMR isotopomer analysis for carbons 2, 3 and 4 of glutamate and glutamine. None of these carbons demonstrated any appreciable change in 13C labeling after 150 minutes of [U-13C]glucose infusion.
Orthotopic GBMs simultaneously use glycolysis and glucose oxidation
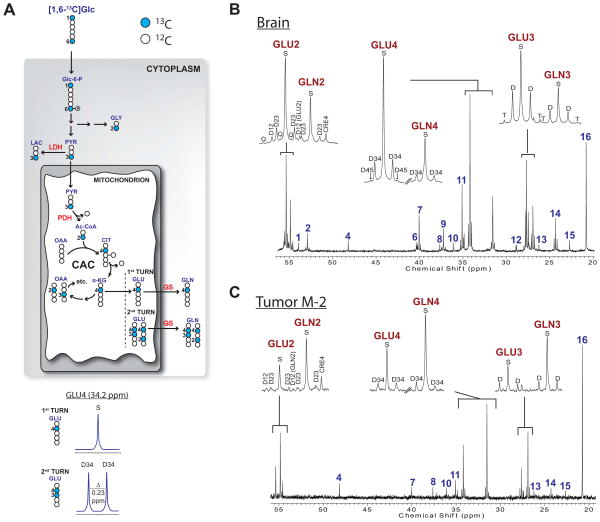
To identify the metabolic fates of glucose within these tumors and to compare metabolic activity of the tumor to that of the surrounding brain tissue, mice from each line were infused with [1,6-13C]glucose. This tracer probes many pathways supplied by glucose, including glycolysis, the CAC and biosynthesis of amino acids (Figure 3A; see Supplemental Methods for explanation of 13C NMR spectra). 13C NMR spectroscopy of tissue extracts from brain and tumor revealed 13C signal in many metabolites (Figure 3, S2). The brain spectra were well-resolved and contained the expected 13C-labeling of metabolites exchanging with the CAC, including glutamate, glutamine, aspartate and γ-aminobutryic acid (GABA) (Figure 3B, S2A,C). Because pyruvate derived from [1,6-13C]glucose via glycolysis is labeled at carbon 3 (C3), 13C-13C multiplets in CAC-related metabolites only occur when both oxaloacetate and acetyl-CoA are 13C enriched. This occurs when the CAC turns over (cycles) normally, as expected in the brain. 13C-lactate was apparent in all brain samples as a result of glycolysis (Figure 3B, S2A,C).
Figure 3. Metabolism of [1,6-13C]glucose in HOTs and surrounding brain.
(A) Illustration of [1,6-13C]glucose metabolism. Filled symbols are 13C and open symbols are 12C. The diagram shows the positions of 13C after glucose is metabolized through glycolysis, glycine synthesis, and multiple turns of the CAC. Numbers refer to carbon positions. At the bottom, the spectra demonstrate the appearance of the 13C NMR spectra for glutamate labeled in position 4 alone (S, singlet), or in positions 3 and 4 (D34, 3–4 doublet), as detailed in Supplemental Experimental Procedures. Abbreviations: Glc, glucose; Glc-6-P, glucose-6-phosphate; GLY, glycine; PYR, pyruvate; LAC, lactate; Ac-CoA, acetyl-CoA; CIT, citrate; α-KG, α-ketoglutarate; OAA, oxaloacetic acid; GLU, glutamate; GLN, glutamine; LDH, lactate dehydrogenase; PDH, pyruvate dehydrogenase; GS, glutamine synthetase.
(B) Brain and (C) tumor spectra from a mouse with an M-2 HOT infused with [1,6-13C]glucose. Insets are GLU and GLN C2, C3 and C4. Chemical shift assignments are the same for all spectra in the paper: 1, NAA C2; 2, Aspartate C2; 3, Alanine C2; 4, Taurine C1; 5, Glycine C2; 6, NAA C3; 7, GABA C4; 8, Creatine C2; 9, Aspartate C3; 10, Taurine C2; 11, GABA C2; 12–13, unassigned; 14, GABA C3; 15, NAA C6; 16, Lactate C3. Abbreviations: S, singlet; D, doublet; T, triplet; Q, quartet; ppm, parts per million.
Despite the small size of the tumor samples (avg. 230 mg), the tumor spectra were well resolved with excellent signal-to-noise ratios and extensive labeling in lactate, glutamate and glutamine (Figure 3C, S2B,D). These spectra contained minimal labeling in GABA, which is synthesized in a sub-population of GABAergic neurons, suggesting that the tumor fragment was highly enriched with GBM cells. This was verified in each HOT by histopathology and neuron-specific immunohistochemistry (IHC), which demonstrated >90% tumor cellularity and loss of normal neurons (data not shown). Label in C4 of glutamate and glutamine, observed in all three spectra, results from the activity of pyruvate dehydrogenase (PDH) in the tumor (Figure 3A). 13C-13C multiplets were also present in glutamate and glutamine in all three spectra (Figure 3C, S2B,D), demonstrating normal turnover of the CAC in the tumors. Furthermore, labeling in glutamine demonstrated that the tumors synthesized glutamine de novo from glucose-derived glutamate. Compared to the brain, all three tumors had much higher labeling in glutamine relative to glutamate, with the ratio of glutamine:glutamate areas for C2, C3 and C4 measuring 1.11±0.23, 1.11±0.21 and 1.01±0.26 in the tumors and 0.43±0.02, 0.49±0.04 and 0.42±0.02 (mean ± S.E.M.) in the brains. Therefore, all three HOTs have intact pathways of glucose oxidation, and the differences in GABA, glutamine and glutamate labeling establish that the tumors are metabolically distinct from the brain in vivo with respect to their handling of glucose-derived metabolites.
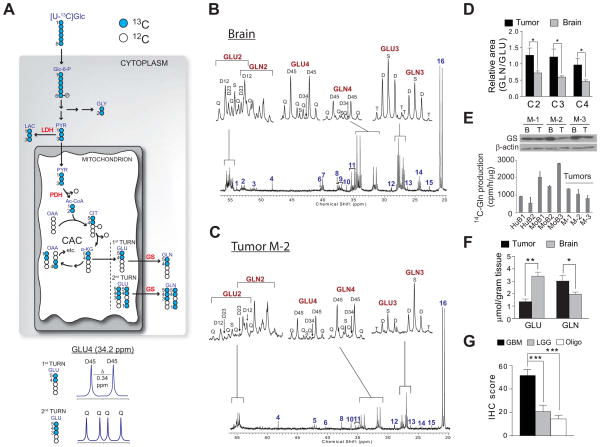
Two mice each bearing HOTs M1-M3 were then infused with [U-13C]glucose (glucose labeled in all six positions with 13C). In this scheme (Figure 4A), glucose-derived pyruvate and acetyl-CoA are uniformly 13C labeled, so that multiplets occur during the first CAC turn. In all mice, spectra from both the brain and the tumor contained prominent 4–5 doublets in glutamate and glutamine, confirming the activity of PDH in both compartments (Figure 4B,C, S3A–D). Evidence for complete turnover of the CAC included the quartet (Q) in glutamate C4 and glutamine C4. Consistent with data from the [1,6-13C]glucose infusion, glutamine was produced de novo from glucose-derived glutamate in both compartments. Again, the overall signal in glutamine relative to glutamate was higher in the tumor than in the brain (Figure 4D). All three tumors expressed glutamine synthetase (GS), the enzyme that converts glutamate to glutamine, and tumor tissue extracts possessed GS enzyme activity (Figure 4E). Measuring total glutamine and glutamate content in all mice infused with either [1,6-13C]glucose or [U-13C]glucose (n=9) revealed that the tumors contained significantly more glutamine per gram of tissue, and significantly less glutamate, than the brain (Figure 4F). We also evaluated GS expression in a human tissue microarray of 150 gliomas. GS was highly expressed in GBM relative to low-grade gliomas and oligodendrogliomas (Figure 4G), suggesting that expression of this enzyme and the ability to synthesize glutamine from glucose-derived glutamate are prominent metabolic features of GBMs.
Figure 4. Metabolism of [U-13C]glucose in HOTs and surrounding brain.
(A) Illustration of [U-13C]glucose metabolism. See legend to Fig. 3A and Supplemental Experimental Procedures for details and abbreviations.
(B) Brain and (C) tumor spectra from a mouse with an M-2 HOT infused with [U-13C]glucose. Insets are glutamate (GLU) and glutamine (GLN) C2, C3 and C4.
(D) Ratio of glutamine area to glutamate area for carbons 2, 3 and 4 in tumor and surrounding brain. Data are the average and S.E.M. for six individual mice, two for each of the three HOT lines. Statistical analysis: Wilcoxon signed rank test. *, p<0.05.
(E) Top, Glutamine synthetase (GS) western blot in tumor (T) and surrounding brain (B) of HOT lines. Bottom, GS enzyme activity in two human brain samples (HuB1, HuB2), three mouse brain samples (MoB1 – MoB3), and three HOTs (M-1 to M-3). MoB3 is the brain tissue surrounding tumor M-1. Data are the average and S.D. for three replicates from each sample.
(F) Total GLN and GLU abundance in tumor and brain extracts (n=9), measured by high-performance liquid chromatography. Data are the average and S.D. *, p<0.05; **, p<0.005, Student’s t-test.
(G) Immunohistochemistry (IHC) scores for GS expression in a tissue microarray consisting of GBM (n=81), low-grade (Grade II–III) gliomas (LGG, n=37) and oligodendrogliomas (Oligo, n=33). Data are the average and S.E.M. ***, p<0.001, one-way ANOVA with Dunnett’s post-hoc test.
Since the spectra included fully-resolved multiplets, we used glutamate isotopomer analysis to estimate some of the major metabolic activities related to glucose oxidation. Assuming that the tissue under investigation is metabolically homogeneous, isotopomer analysis calculates specific metabolic parameters from relative peak areas within glutamate multiplets (Malloy et al., 1987; Sherry and Malloy, 2002). This analysis revealed that plasma glucose was the major source of acetyl-CoA in the tumor, consistent with the robust glucose oxidation and PDH activity observed in this model (Figure S3E). The contribution of glucose to acetyl-CoA in the tumors was similar to that of the brain (Figure S3E). Anaplerosis, the process by which the CAC generates excess intermediates for neurotransmitter synthesis in the brain and for macromolecular synthesis during cell growth (Owen et al., 2002), also contributed significantly to overall tumor CAC activity in both compartments (Figure S3F).
Lack of glutamine catabolism in orthotopic GBMs
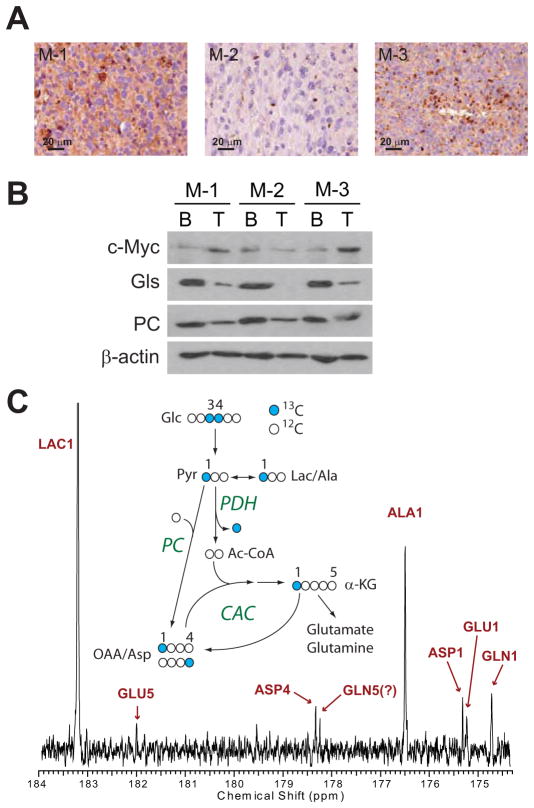
We next examined potential mechanisms of anaplerosis in the tumors. Glutamine is the major anaplerotic nutrient for many cancer cells (DeBerardinis et al., 2007; Portais et al., 1996), and cells with high levels of c-Myc have high rates of glutamine catabolism, and require glutamine for survival and growth in culture (Wise et al., 2008; Yuneva et al., 2007). c-Myc regulates levels of glutaminase (GLS), one of the enzymes that converts glutamine to glutamate in cells that use glutamine catabolism for anaplerosis (Gao et al., 2009). However, other tumor cells use a glucose-dependent anaplerotic pathway that requires pyruvate carboxylase (PC), and these cells do not need to express GLS or catabolize glutamine (Cheng et al., 2011). Relative to the surrounding brain, HOTs M-1 and M-3 had abundant c-Myc expression by IHC and Western blot (Figure 5A,B). As expected, M-1 and M-3 also expressed GLS, while M-2 did not. However, in all cases GLS was much more abundant in the surrounding brain than in the tumor. By contrast, PC was expressed in all tumors at levels more comparable to the surrounding brain (Figure 5B). Examination of the 13C spectra from all tumors infused with [U-13C]glucose revealed the presence of a 2–3 doublet in glutamate C2, and this feature is consistent with PC activity (Cheng et al., 2011). However, other pathways, including multiple turns of the CAC, may also produce this doublet. To test definitively for PC activity, a mouse with a HOT M-1 tumor was infused with [3,4-13C]glucose. Because glucose labeled in this manner produces [1-13C]pyruvate, 13C only enters the CAC if PC is active (Figure 5C). The resulting tumor 13C spectrum contained several features consistent with PC activity, including labeling of carbons 1 and 4 in aspartate, and an excess of label in carbon 1 relative to carbon 5 of glutamate and glutamine (Figure 5C). Together the data suggest that PC contributes to anaplerosis in these tumors.
Figure 5. Anaplerosis in HOTs.
(A) c-Myc IHC in HOT tumors.
(B) Protein abundance of c-Myc and two anaplerotic enzymes, glutaminase (GLS) and pyruvate carboxylase (PC) in HOT tumors (T) and surrounding brain (B).
(C) 13C NMR spectrum from an M-1 tumor infused with [3,4-13C]glucose. The diagram illustrates that in this infusion, PC is active if C1 signal exceeds C5 signal in glutamate and glutamine. GLN5 was not definitively assigned; it is either the indicated peak, or co-resonant with ASP4, both of which are smaller than the GLN1 peak.
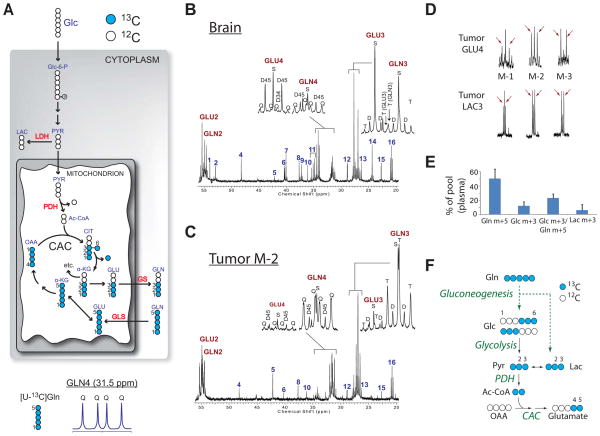
The accumulation of glucose-derived glutamine in all three HOTs, coupled with PC activity, raised the question of whether they engaged in glutamine catabolism. To address this, one mouse from each line was infused with [U-13C]glutamine (Figure 6, S4). Presence of [U-13C]glutamine in tumor and brain is indicated by quartets in glutamine C2 and C4 and the triplet in C3 (Figure 6B,C and S4A–D). In the tumor, glutamine multiplets far exceeded glutamate labeling, whereas the reverse occurred in surrounding brain. The overall pattern revealed that blood-borne glutamine is taken up by the tumors, but not metabolized significantly. Interestingly, all three tumors infused with [U-13C]glutamine also contained prominent 4–5 doublets at glutamate C4 (Figure 6D, S4B,D). These occur when C4 and C5 but not C3 are labeled, and thus the pattern cannot be produced from the direct conversion of [U-13C]glutamine to [U-13C]glutamate. The data suggested that 13C-labeled glucose or lactate entered the tumor and was oxidized. Because glutamine is an excellent gluconeogenic precursor (Nurjhan et al., 1995), we used mass spectrometry to measure 13C enrichment in plasma glucose during [U-13C]glutamine infusions (Figure 6E). Each mouse had a significant fraction of glucose m+3, the expected product if pyruvate derived from [U-13C]glutamine was converted to glucose in the liver or elsewhere (Figure 6F). The enrichment of glucose normalized for the enrichment of glutamine, calculated by the ratio of glucose m+3/glutamine m+5, was 23 ± 6% (Figure 6E). Two of the plasma samples also contained lactate m+3, which could have been produced either through glutaminolysis or glycolysis of glutamine-derived glucose. The 2-3 doublet at lactate C3, observed in all three tumors (Figure 6D), also reflects metabolism of [U-13C]glutamine in other tissues followed by delivery of 13C-glucose and/or 13C-lactate to the tumor. These observations reinforce the notion that glucose is preferred over glutamine for oxidative metabolism in these tumors.
Figure 6. HOTs use glucose, not glutamine, to supply the citric acid cycle.
(A) Illustration of [U-13C]glutamine metabolism. See legend to Fig. 3A and Supplemental Experimental Procedures for details and abbreviations.
(B) Brain and (C) tumor spectra from a mouse with an M-2 HOT infused with [U-13C]glutamine. Insets are glutamate (GLU) and glutamine (GLN) C3 and C4.
(D) Expansion of GLU4 and LAC3 multiplets in three tumors infused with [U-13C]-glutamine. Arrows highlight the 2–3 doublet in lactate and the 4–5 doublet in glutamate.
(E) 13C enrichment in plasma glutamine, glucose and lactate of mice infused with [U-13C]glutamine. Data are the average ± S.D. of three HOT-bearing mice.
(F) Schematic of metabolic activities occurring outside of the tumor by which [U-13C]-glutamine is converted to glucose and lactate (dashed green arrows), which are detected in the plasma. Subsequent metabolism in the tumor (black arrows) uses 13C glucose and/or lactate to supply the CAC.
Glutamine independence of cells derived from orthotopic GBMs
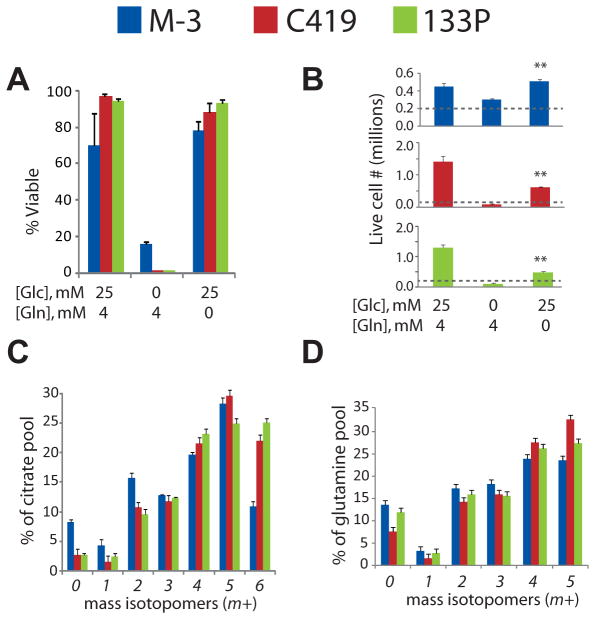
Finally, to determine whether these activities were cell-autonomous, we isolated GBM cells from HOT M-3 and briefly cultured them as neurospheres. Successful neurosphere isolation supports the view that the HOTs contain cancer stem cells and represent the truly malignant GBM cell type. These primary GBM cells were highly dependent on glucose, but consistent with the lack of glutamine catabolism in vivo, they did not require glutamine for survival and were able to maintain at least a low rate of growth over one week of culture without any access to glutamine (Figure 7A,B). Two additional neurosphere cell lines derived from other HOTs displayed similar properties (Figure 7A,B). All three neurosphere lines expressed similar levels of c-Myc, PC and GS, but none expressed high levels of GLS (Figure S5A). The three lines were then cultured with [U-13C]glucose, and metabolites were analyzed by mass spectrometry to determine isotopic enrichment. In all three lines, the CAC was highly enriched by glucose, with well over half of the citrate molecules in these cells containing 13C in at least four of the six carbons (Figure 7C). Glutamate, malate and aspartate were also highly enriched (Figure S5B). Glutamine was avidly produced from glucose (Figure 7D), as it was in vivo. Thus, these neurospheres are metabolically programmed to produce glutamine from glucose, and to survive and grow without the need for extracellular glutamine.
Figure 7. Cells from HOTs survive and grow without exogenous glutamine.
(A) Viability and (B) live cell number in neurosphere cultures derived from three HOTs (M-3, C419 and 133P) in medium containing both glucose and glutamine, glutamine alone, or glucose alone. The dashed line indicates the number of cells plated at the start of the experiment. Data are the average ± S.D. of three independent cultures (**, p<0.005, Student’s t-test).
(C) Mass isotopomer distribution of citrate in neurospheres cultured in medium with [U-13C]glucose. Data are the average ± S.D. of three independent cultures.
(D) Mass isotopomer distribution of glutamine in the same cells. Data are the average ± S.D. of three independent cultures.
DISCUSSION
We analyzed nutrient metabolism in a biologically-accurate mouse model of GBM. Contrary to the prevailing view of diminished mitochondrial function in tumor cells, the data demonstrate that oxidation in the CAC is a significant fate of glucose carbon in these 18FDG-PET-positive tumors. The overall metabolic phenotype of GBM involved allocation of glucose carbon into oxidative and non-oxidative pathways to produce both energy and macromolecular precursors. Thus, our finding rules out the possibility that tumor metabolism is confined to aerobic glycolysis during aggressive growth in vivo, in particular in GBMs with diffuse infiltration, which ensures access to the nutrient- and oxygen-rich environment of the brain. Flux through PDH was intact in each tumor analyzed and was the major source of carbon entering the CAC, as it was in the surrounding brain. This observation is at odds with the prevailing hypothesis that PDH is suppressed in high-grade tumors (Michelakis et al., 2010). The differences between metabolism of HOTs in vivo and in cancer cell lines in vitro may reflect adaptation to long-term culture, loss of microenvironmental factors that influence metabolism, or the fact that some metabolic hallmarks of cancer cell lines, particularly the Warburg effect, reflect maximal rates of cell proliferation in culture rather than malignancy per se (Deberardinis et al., 2008b). Importantly, the HOTs studied here displayed numerous metabolic similarities to primary human GBMs, which also use PDH, the CAC and GS in vivo (Maher et al., 2012).
An intriguing finding is that the three HOTs studied closely in this work had similar metabolic features despite being derived from independent human tumors with different driver mutations. Although both EGFR and PTEN regulate PI3K signaling, which in turn controls glucose metabolism in tumor cells (Elstrom et al., 2004; Liu et al., 1998; Myers et al., 1998), the convergence of these three HOTs onto a similar metabolic phenotype suggests that neither EGFR gain-of-function nor PTEN loss-of-function is necessary for robust glucose uptake and oxidation. Rather, the relationship between genomic information and tumor metabolism may reflect the tumor grade, or may depend on intricate cross-talk among growth regulatory pathways operating in a cell-context specific fashion under the influence of the local microenvironment. Examination of additional HOTs with different patterns of signature GBM mutations, application of other methods to investigate a broader swath of the metabolome, and loss-of-function studies to examine the specific role of particular signaling pathways, will be necessary to determine the full spectrum of GBM metabolism in vivo, and the precise role of oncogenes in orchestrating metabolism in these tumors.
The comparison of 13C metabolism in the tumors and surrounding brain demonstrated distinct metabolic profiles, the most striking of which was a large glutamine pool in the tumors. The tumors and tumor-derived neurospheres mimicked the metabolism of a subset of cancer cell lines that rely on glucose to provide carbon and anaplerotic flux to the CAC, and do not require exogenous glutamine for growth (Cheng et al., 2011). In the normal brain, glutamine exchanges rapidly with glutamate between neurons and astrocytes, but this cycle was markedly perturbed within the HOTs. Based on the labeling data, it is clear that all three HOTs synthesize glutamine like normal astrocytes, perhaps reflecting their cellular origins. However, de novo glutamine synthesis did not appear to be sufficient to explain the large glutamine pools, because the HOTs did not possess more GS enzyme activity than the surrounding brain. Glutamine accumulation likely resulted from a combination of activities including de novo synthesis, uptake of glutamine from the blood supply, and perhaps a reduced rate of glutamine export relative to normal astrocytes. The dense tumor cellularity and consequential paucity of neurons in the tumor mass may have also led to the appearance of glutamine accumulation. We speculate that the high glutamine content within GBMs could serve as an imaging biomarker using 18F-labeled glutamine analogs for PET, or with high-field magnetic resonance spectroscopy, as has been demonstrated for glycine and 2-hydroxyglutarate (Choi et al., 2011; Choi et al., 2012; Qu et al., 2011).
The potential to develop metabolically-directed cancer therapies assumes that tumors have metabolic preferences and vulnerabilities that distinguish them from normal tissue in vivo. However, direct evidence for this hypothesis is scant. Warburg’s classical experiments were performed ex vivo on tumor slices (Koppenol et al., 2011), and the vast majority of subsequent data was derived from cancer cell lines. These are informative approaches, but they lack the crucial component of observing metabolism in live tumors before separating them from their natural microenvironment and supply of nutrients and oxygen. 13C isotopic studies are straightforward, and our data demonstrate that important and potentially translatable information can be produced from small tissue samples. Furthermore, there are no barriers to generalizing this approach to other tumor types, because a similar strategy was used to characterize metabolism qualitatively in lung tumors (Fan et al., 2009; Fan et al., 2011). Therefore we anticipate that metabolic flux studies using stable isotopes in vivo will greatly facilitate the investigation of bona fide tumor metabolism in mouse models of cancer and in human patients, perhaps enabling the identification of targets for therapy or diagnostic imaging.
EXPERIMENTAL PROCEDURES
Human orthotopic tumor (HOT) model
Tumor samples were obtained from patients at the University of Texas - Southwestern Medical Center after written informed consent under a protocol approved by the Institutional Review Board. The animal portion of this study was performed under a protocol approved by the Institutional Animal Care and Use Committee. Tumor tissue was dissociated with papain and a suspension of 5 × 104 cells was stereotactically injected into the right caudate of 6-week-old NOD/SCID mice as previously described (Marian et al., 2010). Tumor growth was monitored using MRI (see below) and by following the neurological status of the animals.
MRI of mouse brain and tumors
MR imaging was conducted with a 7-T small animal MR system (Varian, Inc, Palo Alto, CA) with a 40 mm (I.D.) Horizontal Millipede™ Coil as previously described (Marin-Valencia et al., 2012).
18FDG-PET imaging
18FDG-PET imaging was performed in a small animal PET scanner with a full-width at half-maximum resolution of 2 mm as previously described (Marin-Valencia et al., 2012)
Infusions of 13C-labeled nutrients and dissection of brains and tumors
Tracer studies were initiated 8-10 weeks after injection of tumor cells, when the mice began to develop neurological symptoms (lack of grooming, cachexia, seizures, focal deficits) according to previously published methods (Marin-Valencia et al., 2012). Three mice (one from each tumor line) were infused with [1,6-13C]glucose (99% enrichment; Cambridge Isotope Laboratories, Andover, MA) and six (two from each tumor line) were infused with [U-13C]glucose (99% enrichment; Cambridge Isotope Laboratories, Andover, MA) as follows: a bolus of 0.4mg/g body weight (0.3 mL) was infused in ~1 min, followed by a continuous infusion of 0.012 mg/g/min at 150 μL/hr for 150 min. This protocol yielded a final plasma glucose enrichment of 65 ± 15% (mean ± S.D.). Three mice (one from each tumor line) were infused with [U-13C]glutamine (97–99% enrichment; Cambridge Isotope Laboratories, Andover, MA) as a bolus of 0.28 mg/g body weight (0.3 mL) administered over ~1 min, followed by a continuous infusion of 0.005 mg/g for 180 min. This protocol yielded a final plasma glutamine enrichment of 51 ± 13% (mean ± S.D.). At the end of the infusions, 100–150 μL of blood was collected from the orbit prior to decapitation, and this plasma was used to determine the enrichment of the relevant nutrient pool. After decapitation, the tumors and surrounding brain tissue were rapidly dissected, weighed, frozen in liquid nitrogen, and stored at −85°C. The tumors weighed 235 ± 130 mg and the surrounding brain tissue weighed 406 ± 103 mg (mean ± SE).
Metabolite extraction
Tumors and surrounding brains were finely ground in a mortar under liquid nitrogen and prepared for NMR analysis as previously described (Marin-Valencia et al., 2012; Maher et al., 2012).
13C NMR spectroscopy
Proton decoupled 13C spectra were acquired either on a 400 MHz (Varian) or 600 MHz (Oxford) system. For the 400 MHz system, a VNMRS Direct Drive console with a 5-mm auto-switchable broadband probe (Varian Instruments, Palo Alto, CA) was used. Proton decoupling was performed at 2.3 kHz using a Waltz-16 sequence. 13C NMR parameters included a 45 degree flip angle per transient, a relaxation delay of 1.5 sec, an acquisition time of 1.5 sec, and spectral width of 24.5 kHz. Samples were spun at 20 Hz with the temperature regulated at an ambient 25° C. A 2H field-frequency lock was used. For the 600 MHz system, a Varian VNMRS Direct Drive console using a 3-mm broadband probe (Varian Instruments, Palo Alto, CA) was used. Proton decoupling was performed at 2.3 kHz using a Waltz-16 sequence. 13C NMR spectroscopy parameters included a 45° flip angle per transient, a relaxation delay of 1.5 s, an acquisition time of 1.5 s, and a spectral width of 36.7 kHz. The samples were spun at 20 Hz and 25°C. A heteronuclear 2H lock was used to compensate for magnet drift during data acquisition. To achieve adequate signal-to-noise in 13C NMR spectra, the number of scans acquired was typically 15,000. NMR spectral analysis was performed with ACD/Spec Manager 11.0 software (Advanced Chemistry Development, Inc., Toronto ON, Canada). Free induction decays were zero-filled and windowed with an exponential weighting function prior to Fourier transformation. Resonances were assigned based on chemical shift position referenced to the glutamate C4 singlet at 34.2 ppm. Each signal was fitted with a Gauss-Lorentz function and the area measurements for each fitted resonance peak and the multiplet areas were estimated as previously described (Malloy et al., 1987; Malloy et al., 1990).
Measurement of 13C fractional enrichments in blood
Blood samples were processed to measure 13C enrichment in glucose, glutamine and lactate by gas chromatography-mass spectrometry (GC-MS), as previously described (Marin-Valencia et al., 2012). A three-point standard curve was prepared for each metabolite by mixing un-enriched glucose with [1,6-13C2]glucose or [U-13C6]glucose; un-enriched glutamine with [U-13C5]glutamine; and unenriched lactate with [13C3]lactate, such that 0, 50 or 100% of each metabolite was 13C-labeled. GC-MS was performed using an Agilent 6890N Gas Chromatograph coupled to an Agilent 5973 Mass Selective Detector (Agilent Technologies, Santa Clara, CA). One microliter of each standard or sample was injected and analyzed in scan mode. Fragment ions of m/z 204 (unenriched) and 205 (enriched) for [1,6-13C2]-glucose; 435 (unenriched) and 441 (enriched) for [U-13C6]-glucose; and 258 (unenriched) and 263 (enriched) for [U-13C5]glutamine were quantified for each standard and experimental sample. Linear regression was used to calculate the enrichment of each plasma sample.
Western blots
50 μg of whole tumor tissue extracts was used for Western blot analysis. Primary antibodies were used according to manufacturer’s protocols. Horseradish peroxidase conjugated sheep anti-mouse and donkey anti-rabbit or antigoat secondary antibodies (Cell Signaling, Danvers, MA) were used at 1:2000 and 1:5000 dilutions, respectively. The signal was developed with Super-Signal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL).
Immunohistochemistry
Paraffin-embedded GBM specimens were obtained from the Division of Neuropathology at UT-Southwestern Medical Center. The GBM tissue and HOT-containing mouse brains were fixed in 10% formalin and embedded in paraffin. Sections were cut at 4-μm thickness and deparaffinized. The sections were incubated in 3% H2O2 for 10 minutes to block endogenous peroxidase. The primary antibodies and their dilutions were as follows: Neurofilament (1:2,000; 2F11; M0762; Dako, Carpinteria, CA), GAD67 (Millipore, Billerica, MA), pAKT, pERK, EGFR, (Cell Signaling, Danvers, MA). All immunostains were performed in a Benchmark XT stainer using the CC1 pretreatment solution (95°C for 32 min) and XT-UltraView Universal DAB detection system (Ventana Medical Systems, Tucson, AZ). The sections were counterstained with hematoxylin.
GS enzyme assay
50 mg of tissue (tumor or brain) was homogenized by electronic polytron in 0.5 mL of homogenization solution (0.25 M sucrose, 0.2 mM EDTA, 2 mM 2-mercaptoethanol). The resulting homogenate was centrifuged at 20,000g, 4°C for 15 min. Protein concentration of the supernatant was determined by BCA Protein Assay (Thermo Scientific, Rockford, IL). GS activity was measured using a modification of a published method (McDermott and Butler, 1993). Briefly, an extract containing 25–100 μg protein was incubated with 2.5 mM L-[U-14C]glutamic acid (0.68 Ci/mol), 7.5 mM ATP, 30 mM MgCl2, 25 mM NH4Cl and 50 mM imidazole/HCl buffer (pH 7.5) in a volume of 250 μl. Background was determined by including a GS inhibitor (methionine sulfoxime) in reactions with each extract. After 1 hour at 37°C, the reaction was terminated by adding 1 mL of ice-cold 20 mM imidazole/HCl buffer (pH 7.5). One mL of this sample was applied to a 1 mL bed volume column of AG 1-X8 anion-exchange resin (200–400 mesh, 8% crosslinked, chloride form; Bio-Rad, Hercules, CA) which had been previously equilibrated with 4 mL 20 mM imidazole/HCl (pH 7.5). 14C-glutamine was eluted with 3 mL of the same buffer, and quantified by scintillation counting.
Cell culture metabolism and mass isotopomer analysis
Cells from an M-3 tumor were enzymatically disengaged and cultured in DMEM/F12 medium supplemented with 2% (v/v) B27 serum-free supplement, 1% (v/v) insulin-transferrin supplement, 20ng/mL epidermal growth factor, 10ng/mL basic fibroblast growth factor, 20ng/mL progesterone, 4ug/mL doxycycline hyclate, 0.1% Fungizone and 1% penicillin-streptomycin. Cells were initially plated at ~1×106 cells/dish in 10 mL of medium, and grew as neurospheres. When the culture reached ~5×106 cells/dish, the spheres were collected by centrifugation at 1000 g for 5 minutes at 4°C. A single cell suspension was produced by incubating the pellet in 0.5 mL of 0.25% trypsin-EDTA and 0.5 mL of 0.2% (w/v) collagenase for 10 minutes. This reaction was terminated by adding 0.5 mL trypsin inhibitor (4mg/mL). The cells were then washed once with 10 mL PBS and re-plated. For 13C metabolism experiments, 1×106 cells were seeded in a 6-cm dish with 4 mL of growth medium. After 2 days, the spheres were collected by centrifugation, rinsed once in PBS and re-suspended in 2 mL of medium containing 10mM [U-13C]glucose for 24 hours. Metabolites were extracted by lysing the cells in 0.5 mL of cold (−20°C) 50% methanol and subjecting to three or more freeze-thaw cycles and centrifuged to remove debris. GC/MS was used to analyze enrichment in citrate, aspartate, malate and glutamate. Samples were evaporated and derivatized in 100 μL of a trimethylsilyl donor (Thermo Scientific, Rockford, IL), then 3 μL were injected onto an Agilent 6890N GC coupled to an Agilent 5973 Mass Selective Detector. The retention of each metabolite was validated using pure standards. Fragment ions of interest were monitored and quantified, and mass isotopomer distribution analysis was performed as described previously (Cheng et al., 2011; Fernandez et al., 1996). To analyze intracellular glutamine, we used a different approach because the GC/MS method converted glutamine and other intracellular metabolites into L-pyrrolidone-5-carboxylate, making it difficult to determine 13C enrichment in glutamine conclusively. Extracts were evaporated and resuspended in 10 mM tributylamine/15 mM acetic acid pH 5.0, then filtered through a 0.2 μ spin column and injected onto a Shimadzu Prominence LC20/SIL-20AC high-performance liquid chromatograph coupled to an AB SCIEX 3200 QTRAP triple quadrupole mass spectrometer. Metabolites were separated chromatographically on a C18-based column with polar embedded groups (Synergi Fusion, 150 x 2.0 mm 4 μ, Phenomenex, Torrance, CA) using a tributylammonium acetate/methanol gradient and subjected to MS/MS analysis based on published methods (Luo et al., 2007; Tu et al., 2007). The flow rate was 0.5 mL/min using the following method: Buffer A: 10 mM tributylamine adjusted with 15 mM acetic acid to pH 5.0, Buffer B: 100% methanol. T = 0 min, 0% B; T = 2 min, 0% B; T = 13 min, 100% B; T = 15 min, 100% B; T = 16 min, 0% B, T = 20 min, 0% B; T = 20.1 min, stop. The two best MRM transitions in negative mode for glutamine were 145/127 and 145/109 as identified through quantitative optimization. Glutamine standards were injected to confirm MRM transitions and elution time. From analysis of yeast grown in 13C glucose, the 127 and 109 MS/MS daughter fragments of glutamine each contained all 5 carbons. Thus, the MRM transitions used to quantitate 13C-labeled forms of glutamine were (M+1): 146/128, 146/110; (M+2): 147/129, 147/111; (M+3): 148/130, 148/112; (M+4): 149/131, 149/113; (M+5): 150/132, 150/114. Peaks were quantitated and normalized against the total ion count using Analyst 1.4 software.
Statistical Analysis
For NMR and IHC experiments (Figures 4D, 4G, S3E and S3F), error bars correspond to the S.E.M. In all other experiments, error bars correspond to the S.D. For data sets involving S.D., statistical significance was determined using Student’s t-test (two-tailed, equal variance). Other data sets used either the Wilcoxon signed rank test (Figure 4D) or the one-way ANOVA with Dunnett’s post hoc test (Figure 4G).
Supplementary Material
RESEARCH HIGHLIGHTS.
Glioma-bearing mice were infused with 13C to study tumor metabolism in vivo.
The tumors were metabolically complex, and distinct from surrounding brain.
Tumor metabolism was very different from that of established glioma cell lines.
This versatile method will be simple to apply to many mouse models of cancer.
Acknowledgments
Changho Choi, Mark Jeffrey, and members of the DeBerardinis and Bachoo laboratories critically evaluated the data. Carolina Cardona purified the glutaminase antibody. Osamu Togao, Koji Sagiyama, and Masaya Takahashi performed the MRI scans and Xiankai Sun performed the PET scans. The authors were supported by NIH grants RR02584 and RC1NS0760675. In addition, IMV was supported by the Fundación Caja Madrid and Billingsley Fund. RJD was supported by grants from the NIH (DK072565, CA157996), CPRIT (HIRP100437 and RP101243), Robert A. Welch Foundation (I-1733) and Damon-Runyon Cancer Research Foundation. JMM is supported by grants from Ministerio de Ciencia y Tecnología of Spain (SAF2010-17573) and Junta de Andalucia (CVI-6656).
Footnotes
Supplemental Information contains an overview of the 13C NMR approach, and five figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cheng T, Sudderth J, Yang C, Mullen AR, Jin ES, Mates JM, DeBerardinis RJ. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc Natl Acad Sci U S A. 2011;108:8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C, Ganji SK, DeBerardinis RJ, Dimitrov IE, Pascual JM, Bachoo R, Mickey BE, Malloy CR, Maher EA. Measurement of glycine in the human brain in vivo by 1H-MRS at 3 T: application in brain tumors. Magn Reson Med. 2011;66:609–618. doi: 10.1002/mrm.22857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C, Ganji SK, Deberardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, Yang XL, Mashimo T, Raisanen JM, Marin-Valencia I, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18:624–629. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008a;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008b;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- Fan TW, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, Miller DM. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM) Mol Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TW, Lane AN, Higashi RM, Yan J. Stable isotope resolved metabolomics of lung cancer in a SCID mouse model. Metabolomics. 2011;7:257–269. doi: 10.1007/s11306-010-0249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom. 1996;31:255–262. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Forbes NS, Meadows AL, Clark DS, Blanch HW. Estradiol stimulates the biosynthetic pathways of breast cancer cells: detection by metabolic flux analysis. Metab Eng. 2006;8:639–652. doi: 10.1016/j.ymben.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- Kurhanewicz J, Vigneron DB, Brindle K, Chekmenev EY, Comment A, Cunningham CH, Deberardinis RJ, Green GG, Leach MO, Rajan SS, et al. Analysis of cancer metabolism by imaging hyperpolarized nuclei: prospects for translation to clinical research. Neoplasia. 2011;13:81–97. doi: 10.1593/neo.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau BR, Wahren J, Chandramouli V, Schumann WC, Ekberg K, Kalhan SC. Contributions of gluconeogenesis to glucose production in the fasted state. J Clin Invest. 1996;98:378–385. doi: 10.1172/JCI118803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Liu AX, Testa JR, Hamilton TC, Jove R, Nicosia SV, Cheng JQ. AKT2, a member of the protein kinase B family, is activated by growth factors, v-Ha-ras, and v-src through phosphatidylinositol 3-kinase in human ovarian epithelial cancer cells. Cancer Res. 1998;58:2973–2977. [PubMed] [Google Scholar]
- Luo B, Groenke K, Takors R, Wandrey C, Oldiges M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J Chromatogr A. 2007;1147:153–164. doi: 10.1016/j.chroma.2007.02.034. [DOI] [PubMed] [Google Scholar]
- Maher EA, Marin-Valencia I, Bachoo RM, Mashimo T, Raisanen J, Hatanpaa K, Jindal A, Jeffrey FM, Choi C, Madden C, et al. Metabolism of [U-13C]glucose in human brain tumors in vivo. NMR Biomed. 2012 doi: 10.1002/nbm.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malloy CR, Sherry AD, Jeffrey FM. Carbon flux through citric acid cycle pathways in perfused heart by 13C NMR spectroscopy. FEBS Lett. 1987;212:58–62. doi: 10.1016/0014-5793(87)81556-9. [DOI] [PubMed] [Google Scholar]
- Malloy CR, Thompson JR, Jeffrey FM, Sherry AD. Contribution of exogenous substrates to acetyl coenzyme A: measurement by 13C NMR under non-steady-state conditions. Biochemistry. 1990;29:6756–6761. doi: 10.1021/bi00481a002. [DOI] [PubMed] [Google Scholar]
- Marian CO, Cho SK, McEllin BM, Maher EA, Hatanpaa KJ, Madden CJ, Mickey BE, Wright WE, Shay JW, Bachoo RM. The telomerase antagonist, imetelstat, efficiently targets glioblastoma tumor-initiating cells leading to decreased proliferation and tumor growth. Clin Cancer Res. 2010;16:154–163. doi: 10.1158/1078-0432.CCR-09-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Valencia I, Cho SK, Rakheja D, Hatanpaa KJ, Kapur P, Mashimo T, Jindal A, Vemireddy V, Good LB, Raisanen J, et al. Glucose metabolism via the pentose phosphate pathway, glycolysis and Krebs cycle in an orthotopic mouse model of human brain tumors. NMR Biomed. 2012 doi: 10.1002/nbm.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott RH, Butler M. Uptake of glutamate, not glutamine synthetase, regulates adaptation of mammalian cells to glutamine-free medium. J Cell Sci. 1993;104 (Pt 1):51–58. doi: 10.1242/jcs.104.1.51. [DOI] [PubMed] [Google Scholar]
- Mehrian Shai R, Reichardt JK, Ya-Hsuan H, Kremen TJ, Liau LM, Cloughesy TF, Mischel PS, Nelson SF. Robustness of gene expression profiling in glioma specimen samplings and derived cell lines. Brain Res Mol Brain Res. 2005;136:99–103. doi: 10.1016/j.molbrainres.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Michelakis ED, Sutendra G, Dromparis P, Webster L, Haromy A, Niven E, Maguire C, Gammer TL, Mackey JR, Fulton D, et al. Metabolic modulation of glioblastoma with dichloroacetate. Sci Transl Med. 2010;2:31ra34. doi: 10.1126/scitranslmed.3000677. [DOI] [PubMed] [Google Scholar]
- Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, Wigler MH, Downes CP, Tonks NK. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc Natl Acad Sci U S A. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB, Hotamisligil GS, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17:1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurjhan N, Bucci A, Perriello G, Stumvoll M, Dailey G, Bier DM, Toft I, Jenssen TG, Gerich JE. Glutamine: a major gluconeogenic precursor and vehicle for interorgan carbon transport in man. J Clin Invest. 1995;95:272–277. doi: 10.1172/JCI117651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- Portais JC, Voisin P, Merle M, Canioni P. Glucose and glutamine metabolism in C6 glioma cells studied by carbon 13 NMR. Biochimie. 1996;78:155–164. doi: 10.1016/0300-9084(96)89500-9. [DOI] [PubMed] [Google Scholar]
- Qu W, Zha Z, Ploessl K, Lieberman BP, Zhu L, Wise DR, Thompson CB, Kung HF. Synthesis of optically pure 4-fluoro-glutamines as potential metabolic imaging agents for tumors. J Am Chem Soc. 2011;133:1122–1133. doi: 10.1021/ja109203d. [DOI] [PubMed] [Google Scholar]
- Ramanathan A, Wang C, Schreiber SL. Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proc Natl Acad Sci U S A. 2005;102:5992–5997. doi: 10.1073/pnas.0502267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry AD, Malloy CR. 13C isotopomer analysis of glutamate: a NMR method to probe metabolic pathways intersecting in the TCA cycle. In: Berliner LJ, Robitaille P-ML, editors. In Vivo Carbon-13 NMR. New York, U.S.A: Kluwer Academic/Plenum Publishers; 2002. pp. 59–97. [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci U S A. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Trachootham D, Liu J, Chen G, Pelicano H, Garcia-Prieto C, Lu W, Burger JA, Croce CM, Plunkett W, et al. Stromal control of cystine metabolism promotes cancer cell survival in chronic lymphocytic leukaemia. Nat Cell Biol. 2012;14:276–286. doi: 10.1038/ncb2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.