SUMMARY
The timing and mechanisms of mitochondrial DNA (mtDNA) segregation and transmission in mammals are poorly understood. Genetic bottleneck in female germ cells has been proposed as the main phenomenon responsible for rapid intergenerational segregation of heteroplasmic mtDNA. We demonstrate here that mtDNA segregation occurs during primate preimplantation embryogenesis resulting in partitioning of mtDNA variants between daughter blastomeres. A substantial shift toward homoplasmy occurred in fetuses and embryonic stem cells (ESCs) derived from these heteroplasmic embryos. We also observed a wide range of heteroplasmic mtDNA variants distributed in individual oocytes recovered from these fetuses. Thus, we present here evidence for a previously unknown mtDNA segregation and bottleneck during preimplantation embryo development, suggesting that return to the homoplasmic condition can occur during development of an individual organism from the zygote to birth, without a passage through the germline.
INTRODUCTION
Mitochondria are cellular organelles responsible for the production of most cellular energy. Importantly, mitochondria contain their own relatively autonomous genome (mtDNA) encoding critical proteins, rRNAs and tRNAs for ATP generation by oxidative phosphorylation (OXPHOS) (Hudson and Vinograd, 1967; Wallace, 1999). The genetics of mtDNA is markedly different from the nuclear genome including uniparental (exclusively maternal) inheritance, the presence of hundreds to thousands of mtDNA copies per cell and uneven or random distribution of mtDNA to daughter cells (Giles et al., 1980; Taylor and Turnbull, 2005). Mitochondrial genes are also more prone to mutations than nuclear DNA, possibly due to the proximity to mitochondrial reactive oxygen species (ROS) sources, lack of protective histones and limited mtDNA repair capacity (Linnane et al., 1989; Mason et al., 2003). Mutations in mtDNA often occur in the somatic cell lineage during development and aging, and have been linked to age-related diseases such as Parkinson’s disease, cancer and diabetes (Wallace, 2010). However, mtDNA mutations may also arise in the female germline resulting in maternally-inherited human diseases (Taylor and Turnbull, 2005; Tuppen et al., 2010).
The multicopy nature and high mutation rate of mtDNA contributes to another important feature of mitochondrial genetics – heteroplasmy - the presence of wild-type and mutated mtDNA copies within a cell. In such cells, there is a critical threshold level of mutation load above which ability to produce energy is compromised and clinical symptoms of the disease manifest. However, the threshold level varies for specific mutations, typically ranging from 60%–90% of mutant mtDNA. In addition, the likelihood that children will inherit mtDNA diseases is difficult to forecast due to unpredictable segregation pattern of heteroplasmic mtDNA variants in various cells, tissues and organs during development.
Studies of intergenerational mtDNA inheritance in heteroplasmic cattle and mice revealed a rapid shift towards homoplasmy within just a few or even one generation (Ashley et al., 1989; Jenuth et al., 1996; Wai et al., 2008). While, the precise mechanism by which this rapid genetic shift occurs remains largely unclear, these observations suggested the existence of so-called mtDNA bottleneck. Mitochondrial bottleneck is believed to take place in the germline during early oogenesis, resulting in rapid segregation of mtDNA variants into homoplasmic conditions in individual oocytes (Hauswirth and Clayton, 1985; Hauswirth and Laipis, 1982; Hauswirth and Laipis, 1985; Hauswirth et al., 1984; Jenuth et al., 1996; Michaels et al., 1982; Olivo et al., 1983). However, it remains to be established whether mtDNA segregation and bottleneck also occurs during early embryo development, i.e. in precursors of somatic and germ cells (Laipis et al., 1988).
At present there are no cures for inherited mtDNA disorders; thus, families increasingly seek assisted reproductive technology options to prevent transmission of mtDNA mutations to their children. One of the most promising recent advancements is the development of a technique for efficient replacement of mutant mtDNA with normal mtDNA in mature oocytes or zygotes (Craven et al., 2010; Tachibana et al., 2009), which may become a clinical therapy for patients in the future. Among other options currently available, preimplantation genetic diagnosis (PGD) has been introduced, where cleaving 8-cell embryos are created in vitro and one or two cells (blastomeres) are biopsied and analyzed for mtDNA mutation load (Poulton and Bredenoord, 2010; Steffann et al., 2006). Embryos with low mutation content are then selected to start a pregnancy. Although promising, PGD for mtDNA disorders cannot be applied to women carrying homoplasmic mtDNA mutations. Another potential problem is that the proportional levels of mutant to wildtype mtDNA quantified in the biopsied cells must represent the levels in the remaining blastomeres. However, there are conflicting reports in the mouse model regarding the distribution of mtDNA between the blastomeres in heteroplasmic embryos (Dean et al., 2003; Meirelles and Smith, 1998).
Available data on human preimplantation embryos are limited and suffer from small sample size and technical limitations (Monnot et al., 2011; Steffann et al., 2006; Tajima et al., 2007) emphasizing the need to conduct thorough studies in nonhuman primate models.
We generated heteroplasmic rhesus monkey oocytes carrying two wild-type mtDNA haplotypes, henceforth termed "resident" and "alien" originating from nuclear and cytoplasm donors, respectively (Figure 1). Oocytes were then fertilized, and transmission of mtDNA haplotypes was analyzed in individual blastomeres of cleaving embryos. We also studied mtDNA transmission into somatic and germ cells of offspring generated from these oocytes. We observed a rapid mtDNA segregation occurring during preimplantation embryo development. This segregation preceded the drastic shift of mtDNA variants towards homoplasmy for either alien or resident mtDNA in somatic cells of the embryo proper.
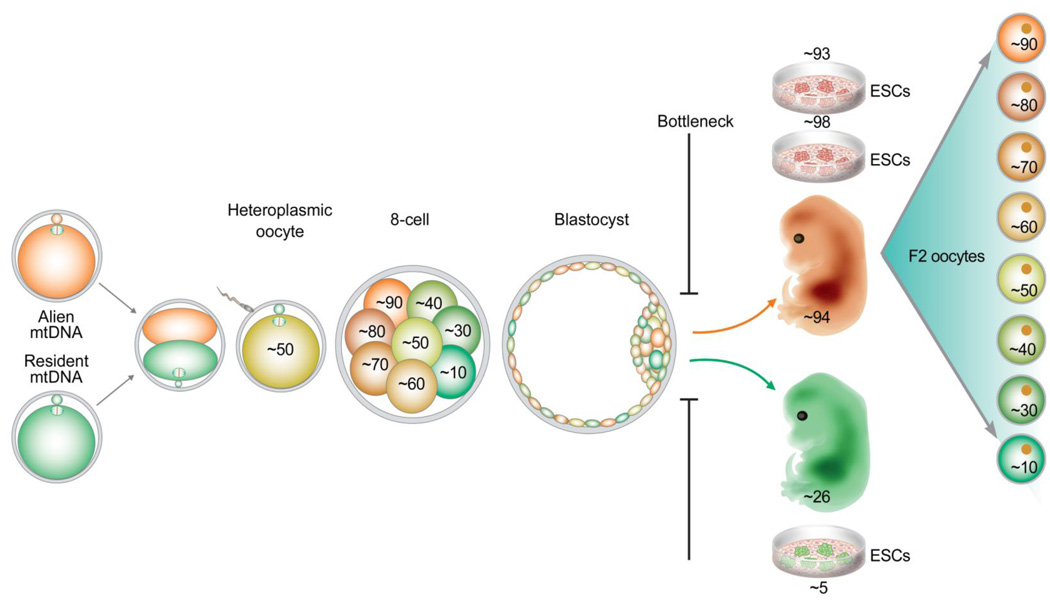
Figure 1. Schematic model demonstrating mtDNA segregation and bottleneck in primates.
Heteroplasmic rhesus monkey oocytes with equal mixture of two wild-type mtDNA haplotypes were constructed and mtDNA transmission to preimplantation embryos, fetuses and germ cells was followed. We demonstrate rapid segregation of mtDNA variants between daughter blastomeres in preimplantation embryos. However, fetuses and ESCs derived from these embryos shifted towards homoplasmic conditions. This bottleneck suggests that possibly a few cells within an ICM contribute to the somatic cell lineage of embryo proper. In contrast, individual fetal oocytes (F2) showed a wide range of heteroplasmy. This model also implies that the majority of ICM cells may contribute to the germline.
RESULTS
Segregation of mtDNA haplotypes between individual blastomeres of cleaving embryos
We initially screened a colony of Indian- and Chinese-origin rhesus macaque females and identified animals carrying different wild-type mtDNA haplotypes based on the sequence polymorphism within the mitochondrial D-loop hypervariable region 1 as described previously (Tachibana et al., 2009). Next, we selected and paired animals carrying distinct mtDNA haplotypes for ovarian stimulation and collected mature metaphase II (MII) stage oocytes (Smith, 2005). In order to generate heteroplasmic oocytes, we removed approximately half of the cytoplasm (cytoplast, without the spindle-chromosomal complex) from an oocyte derived from one female and fused it with another halved oocyte containing spindle-chromosomal complex (karyoplast) recovered from a second female (Figure 1). We measured the diameters of cytoplasts (carrying alien mtDNA) and karyoplasts (resident mtDNA) and calculated that the volume ratio was on average 50%:50%. This suggested that resulting composite oocytes contained approximately 50% of mtDNA from each haplotype. To corroborate this estimate, actual heteroplasmy in individual reconstructed oocytes was analyzed after fertilization using an amplification refractory mutation system quantitative PCR (ARMS-qPCR) assay as we previously reported (Steinborn et al., 2000).
The mean heteroplasmy level for the alien mtDNA haplotype in 15 analyzed zygotes was 54.9 ± 10.1%, close to the estimated ratio by the cytoplasmic volume. We also carried out an independent ARMS-qPCR using a different set of primers to measure the resident mtDNA in these zygotes and confirmed our results. Overall, we concluded that this approach is capable of producing oocytes with significant heteroplasmy levels suitable to study mtDNA segregation and bottleneck.
Therefore, we next generated heteroplasmic oocytes from 18 unrelated females and subsequently fertilized them by intracytoplasmic sperm injection (ICSI). Resulting zygotes were cultured in vitro under our standard conditions and mtDNA segregation was investigated in individual blastomeres of the 2-, 4-, and 8-cell stage cleaving embryos. Embryos at each stage were selected, freed of zona pellucida and blastomeres separated by brief exposure to trypsin-EDTA. Initially, we determined the proportion of the alien mtDNA in each blastomere in fourteen individual 2-cell embryos using ARMS-qPCR. Approximately half of the 2-cell embryos contained blastomeres with equal or comparable mtDNA heteroplasmy levels within each embryo (Table S1). However, heteroplasmy levels between blastomeres in the remaining embryos varied significantly. For example, two blastomeres of the embryo #14 contained alien mtDNA heteroplasmy levels of 36% and 70%. To corroborate these observations we measured the amount of resident mtDNA a confirmed the heteroplasmy levels for each of these blastomeres.
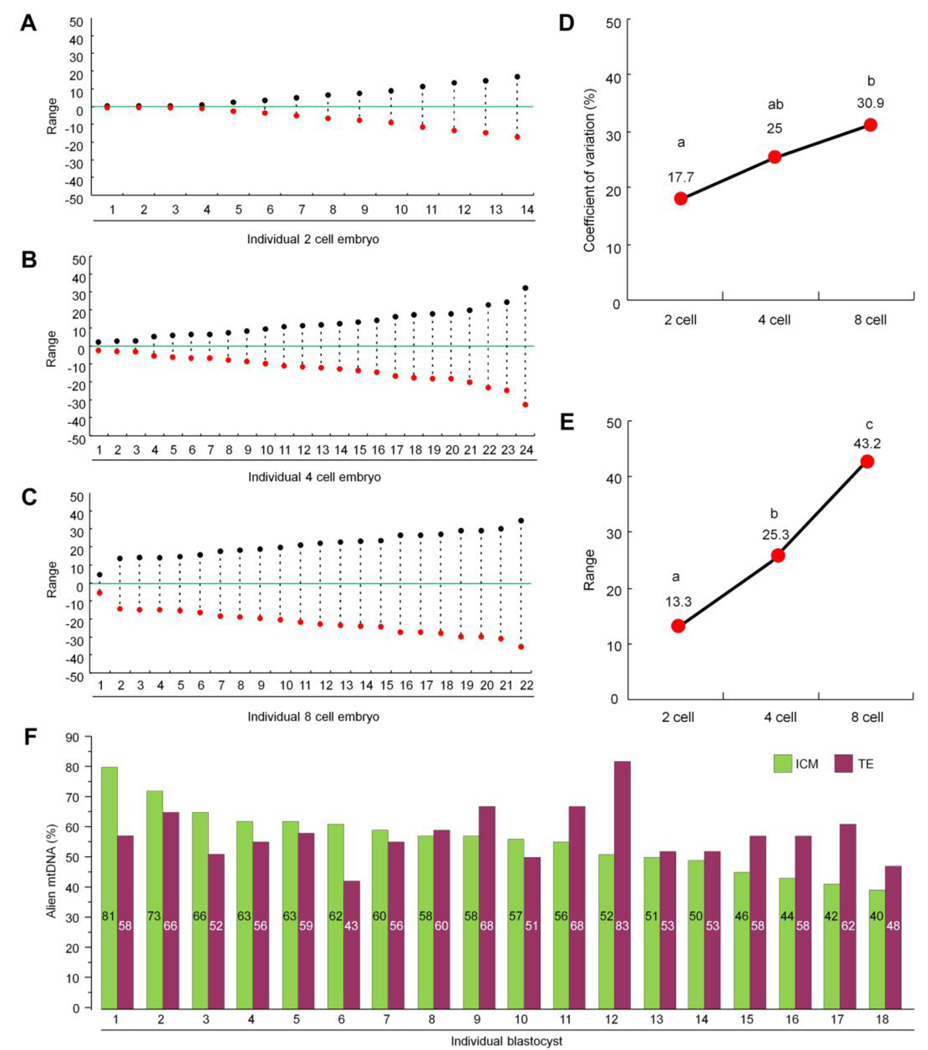
Next, we collected and sampled individual blastomeres in twenty-four 4-cell and twenty-two 8-cell stage embryos. Analysis of heteroplasmy in individual blastomeres revealed that majority of embryos in these groups contained highly segregated mtDNA haplotypes (Figures 2A–C). Moreover, the dispersion of mtDNA variants between daughter blastomeres within each embryo, calculated as the coefficient of variation (Figure 2D) and the range (Figure 2E), increased considerably compared to the 2-cell group (P < 0.05). For instance, heteroplasmy levels in four blastomeres of the embryo #24 varied between 12, 31, 34 and 77% (Table S2). Segregation was even more dramatic in the 8-cell embryos ranging from 10% to 80% in some embryos (embryo #22, Table 1). We also found that the mean value of alien mtDNA heteroplasmy calculated for each embryo stage were similar and comparable to the value in parental heteroplasmic zygotes (zygote, 54.9 ± 10.1%; 2-cell, 54.6 ± 9.7%; 4-cell, 51.2 ± 12.3%; and 8-cell, 49.2 ± 7.9%), suggesting that the original mtDNA heteroplasmy levels in oocytes are conserved in pooled embryos during early preimplantation development. However, our results suggest a significant increase in mtDNA heteroplasmy dispersion between daughter blastomeres during progressive cleavage from the 2-, to 4- and 8-cell stages.
Figure 2. Segregation of mtDNA in individual blastomeres of the 2-, 4- and 8-cell embryos.
Segregation of alien mtDNA between daughter blastomeres of individual (A) 2-cell, (B) 4-cell and (C) 8-cell embryos expressed by range (maximum or minimum alien mtDNA values minus the mean median values). Comparison of mtDNA dispersal between 2-, 4- and 8-cell embryos based on the (D) coefficient of variation and (E) range. Different letters indicates P values <0.05. (F) MtDNA segregation between the ICM and TE in individual blastocysts. The mean alien mtDNA in ICM and TE were 56.8 ± 10.7% and 58.4 ± 9.0%, respectively. Data are represented as mean±s.d. See also Tables S1–S3.
Table 1.
Heteroplasmy in blastomeres of 8-cell embryos
| Embryo | Alien mtDNA (%) in blastomere | Mean | S.D. | CV (%) | Range | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | ||||||
| 1 | 45 | 48 | 51 | 52 | 52 | 53 | 53 | 55 | 51.1 | 3.2 | 6.2 | 10 | |
| 2 | 30 | 31 | 39 | 49 | 51 | 57 | 58 | 58 | 46.6 | 11.8 | 25.2 | 28 | |
| 3 | 27 | 29 | 32 | 33 | 34 | 34 | 43 | 56 | 36.0 | 9.4 | 26.0 | 29 | |
| 4 | 17 | 24 | 25 | 35 | 42 | 43 | 43 | 46 | 34.4 | 11.0 | 31.9 | 29 | |
| 5 | 35 | 39 | 43 | 45 | 50 | 54 | 62 | 65 | 49.1 | 10.7 | 21.7 | 30 | |
| 6 | 32 | 40 | 44 | 45 | 45 | 46 | 49 | 64 | 45.6 | 9.0 | 19.8 | 32 | |
| 7 | 26 | 28 | 30 | 39 | 39 | 43 | 57 | 62 | 40.5 | 13.2 | 32.6 | 36 | |
| 8 | 44 | 45 | 48 | 50 | 59 | 64 | 73 | 81 | 58.0 | 13.7 | 23.7 | 37 | |
| 9 | 17 | 33 | 37 | 39 | 41 | 45 | 48 | 55 | 39.3 | 11.5 | 29.2 | 39 | |
| 10 | 40 | 43 | 44 | 47 | 50 | 72 | 72 | 80 | 56.0 | 15.9 | 28.4 | 40 | |
| 11 | 36 | 36 | 42 | 42 | 57 | 71 | 73 | 79 | 54.5 | 17.8 | 32.7 | 43 | |
| 12 | 36 | 43 | 46 | 47 | 47 | 49 | 50 | 81 | 49.9 | 13.3 | 26.7 | 45 | |
| 13 | 39 | 44 | 48 | 49 | 78 | 80 | 84 | 85 | 63.4 | 20.0 | 31.5 | 46 | |
| 14 | 17 | 25 | 30 | 37 | 40 | 42 | 46 | 64 | 37.6 | 14.3 | 38.0 | 47 | |
| 15 | 36 | 47 | 48 | 48 | 48 | 51 | 52 | 84 | 51.8 | 13.9 | 26.9 | 48 | |
| 16 | 34 | 35 | 39 | 45 | 49 | 53 | 57 | 88 | 50.0 | 17.4 | 34.9 | 54 | |
| 17 | 30 | 42 | 42 | 46 | 51 | 71 | 74 | 84 | 55.0 | 19.0 | 34.5 | 54 | |
| 18 | 27 | 34 | 38 | 47 | 57 | 64 | 65 | 82 | 51.8 | 18.5 | 35.8 | 55 | |
| 19 | 16 | 38 | 42 | 43 | 48 | 61 | 70 | 75 | 49.1 | 19.1 | 38.9 | 59 | |
| 20 | 24 | 35 | 45 | 47 | 52 | 63 | 83 | 83 | 54.0 | 21.2 | 39.3 | 59 | |
| 21 | 29 | 35 | 38 | 71 | 74 | 77 | 80 | 90 | 61.8 | 23.8 | 38.5 | 61 | |
| 22 | 10 | 20 | 29 | 32 | 65 | 69 | 77 | 80 | 47.8 | 27.9 | 58.4 | 70 | |
| Mean | 49.2 | 15.3 | 30.9 | 43.2 | |||||||||
| S.D. | 7.9 | 14.0 | |||||||||||
| CV (%) | 16.0 | ||||||||||||
S.D, standard deviation; CV, coefficient of variation; Range, differences between maximum and minimum values in daughter blastomeres.
Due to smaller cell sizes and difficulties in cell dispersal, single cell mtDNA heteroplasmy analysis in more advanced cleaving or blastocyst stages was not feasible. However, we were able to isolate the inner cell mass (ICM) and the trophectoderm (TE) from each expanded blastocyst by immunosurgery as we previously described (Mitalipov et al., 2006). We analyzed eighteen blastocysts generated from heteroplasmic oocytes and results demonstrated that heteroplasmy levels in some blastocysts varied between the ICM and TE. For example, alien mtDNA heteroplasmy for blastocyst #12 was measured at 52% in the ICM and 83% in the TE (Figure 2F and Table S3). However, less segregation between the ICM and TE was observed in other embryos. The mean heteroplasmy levels calculated for ICMs and TE (56.8± 10.7% and 58.4 ± 9.0%, respectively) were comparable to cleaving embryos.
MtDNA segregation in the somatic lineage of the embryo proper and in ESCs
To relate the dramatic mtDNA segregation seen in blastomeres of cleaving embryos with postimplantation development, we transplanted blastocysts derived from heteroplasmic oocytes into recipients. Two singleton pregnancies were generated, carrying one male and one female fetuses that were recovered by caesarean section at day 105 and 109 post embryo transfer. We analyzed heteroplasmy levels in all major fetal organs and tissues as well as in the placenta. Intriguingly, all samples in the female fetus carried predominantly alien mtDNA (93.8 ± 3.8%, Table 2) whereas the major component in male fetus was the resident mtDNA (alien mtDNA heteroplasmy 26.3 ± 5.2%, Table 2). In the female fetus, most organs and tissues exhibited comparable heteroplasmy levels ranging from 91.1% in the blood to 98.4% in kidneys. In contrast, blood, right lung, esophagus and pituitary tissues from the male fetus showed relatively higher heteroplasmy levels compared to other organs. Heteroplasmy in placental samples representing an extraembryonic lineage in both pregnancies was similar to the fetal tissues. To validate our ARMS-qPCR results we also performed an independent restriction fragment length polymorphism (RFLP) assay as we described earlier (Tachibana et al., 2009) and confirmed our observations (Figure S1).
Table 2.
mtDNA segregation in fetuses derived from heteroplasmic oocytes, related to Figure S1
| Organs and tissues | Alien mtDNA (%) in female fetus |
Alien mtDNA (%) in male fetus |
|---|---|---|
| Cord blood | 91.1 | 34.2 |
| Placenta | 97.2 | 20.8 |
| Cerebrum | 95.4 | 28.9 |
| Cerebellum | 94.1 | 22.4 |
| Spinal cord | 89.2 | 21.8 |
| Eye (lens) | 96.4 | 28.4 |
| Retina | 95.5 | 27.6 |
| Right-lung | 96.2 | 33.9 |
| Left-lung | 96.7 | 24.7 |
| Heart | 81.7 | 21.2 |
| Tongue | 94.9 | 25.2 |
| Esophagus | 94.0 | 37.8 |
| Stomach | 92.3 | 28.5 |
| Small intestine | 92.6 | 33.2 |
| Colon | 95.2 | 32.1 |
| Rectum | 95.9 | 27.6 |
| Liver | 82.9 | 31.9 |
| Pancreas | 91.7 | 21.8 |
| Pituitary gland | 94.3 | 36.6 |
| Adrenal gland | 91.6 | 19.2 |
| Thyroid | 91.2 | 24.9 |
| Kidney-R | 98.4 | 20.9 |
| Kidney-L | 98.4 | 24.3 |
| Bladder | 94.9 | 24.1 |
| Uterus | 94.5 | NA |
| Testis | NA | 28.8 |
| Prostate | NA | 19.4 |
| Spleen | 94.4 | 24.3 |
| Thymus | 95.5 | 21.7 |
| Tonsil | 92.3 | 23.4 |
| Skin | 97.6 | 21.5 |
| Skeletal muscle | 97.4 | 23.3 |
NA, not applicable.
To gain further insights into when this dramatic shift in mtDNA heteroplasmy occurred, we focused on ESCs, representing the early epiblast lineage. We established three stable ESC lines (designated as Hetero-1, -2 and -3) from ICMs of heteroplasmic blastocysts. All three cell lines were male (XY) and expressed typical primate markers characteristic for pluripotent cells. Analysis of samples from early passage cells (p. 3–5) revealed that two ESC lines already contained predominantly alien mtDNA with heteroplasmy levels reaching 93% (Hetero-1) and 97.9% (Hetero-3). However, the third cell line, Hetero-2, carried mainly resident mtDNA with only 5% heteroplasmy for alien mtDNA. To investigate if ESCs represent heterogeneous population of cells, we subcloned Hetero-1 ESCs and analyzed heteroplasmy. Nine clones, each representing a progeny of a single cell, were assayed and six clones contained exclusively alien mtDNA with no traces of resident mtDNA (Table S4). Heteroplasmy in remaining three clones ranged from 90.7 to 92.9%.
Together, these results indicate that in contrast to cleaving embryos and blastocysts, average heteroplasmy levels in fetal lineages of the embryo proper have shifted more towards homoplasmy. Moreover, analysis of heteroplasmy in early passage ESCs suggests that this genetic shift occurs early during specification of the epiblast. Interestingly, mtDNA segregation occurred towards either alien or resident mtDNA, independently of the nuclear genetic background, indicating that nuclear-mitochondrial interactions do not favor selective amplification of the resident mtDNA or that this interaction has been conserved between the two sub species of macaques.
Heteroplasmy in the female germline
Intrigued by the dramatic segregation of mtDNA haplotypes during primate preimplantation development and the unusual shift seen in the somatic linage, we recovered ovaries from the female fetus and analyzed individual primordial oocytes. In contrast to nearly homoplasmic (towards alien mtDNA) fetal somatic tissues, analysis demonstrated that heteroplasmy levels varied significantly among individual oocytes. The levels of alien mtDNA in fifty-one oocytes ranged from 3.7% to 99.2% (Table 3). These findings demonstrate striking differences in mtDNA transmission between somatic and germline lineages.
Table 3.
Heteroplasmy in fetal F2 oocytes.
| Individual oocytes |
Alien mtDNA (%) |
Individual oocytes |
Alien mtDNA (%) |
|---|---|---|---|
| 1 | 99.2 | 27 | 65.2 |
| 2 | 95.5 | 28 | 64.2 |
| 3 | 95.4 | 29 | 63.9 |
| 4 | 90.1 | 30 | 62.9 |
| 5 | 88.1 | 31 | 61.8 |
| 6 | 87.3 | 32 | 53.1 |
| 7 | 86.3 | 33 | 52.2 |
| 8 | 86.2 | 34 | 50.9 |
| 9 | 84.6 | 35 | 50.3 |
| 10 | 83.4 | 36 | 49.0 |
| 11 | 82.3 | 37 | 47.7 |
| 12 | 81.6 | 38 | 46.5 |
| 13 | 80.3 | 39 | 43.4 |
| 14 | 79.9 | 40 | 33.1 |
| 15 | 79.3 | 41 | 29.1 |
| 16 | 78.3 | 42 | 24.0 |
| 17 | 75.5 | 43 | 20.4 |
| 18 | 75.1 | 44 | 20.2 |
| 19 | 74.2 | 45 | 18.1 |
| 20 | 74.1 | 46 | 14.6 |
| 21 | 72.9 | 47 | 12.1 |
| 22 | 72.6 | 48 | 8.9 |
| 23 | 71.1 | 49 | 4.9 |
| 24 | 68.2 | 50 | 3.9 |
| 25 | 67.9 | 51 | 3.7 |
| 26 | 67.5 |
MtDNA carryover in offspring produced by spindle transfer
We previously demonstrated that mtDNA can be efficiently replaced in mature monkey oocytes by spindle-chromosomal complex transfer (ST) resulting in healthy offspring carrying different nuclear and mitochondrial genomes (Tachibana et al., 2009). In clinical IVF settings, this approach offers a potentially reliable option to prevent mtDNA disease transmission from mothers to children. During ST procedure, a small amount of cytoplasm (1%), containing bound mtDNA, is co-transferred with chromosomes into recipient cytoplasm. We questioned if observed segregation and bottleneck could result in significant increase of carryover mtDNA in the somatic and germline cells of ST offspring.
Initially, we determined that isolated karyoplasts indeed carry bound mtDNA (mtDNA copy number in karyoplasts 3,538 ± 2,310 vs. in cytoplasts 576,948 ± 209,069; n=9) resulting on average a level of 0.6% heteroplasmy. We next produced a total of 102 oocytes by the ST technique of which 93 (91%) were successfully fertilized and 63 (62%) developed to blastocysts, confirming our previous conclusions that manipulated oocytes retain their high developmental potential. To select female blastocysts for embryo transfers, we performed a TE biopsy in expanded blastocysts and performed PCR-based gender analysis using size differences in the amplicons of the X- and Y-linked zinc finger protein genes (ZFX and ZFY) (Mitalipov et al., 2007). Of 32 biopsied blastocysts, we successfully amplified DNA and determined gender for 29 embryos. Both male and female embryos were equally represented among tested ST embryos (15 XX and 14 XY). Next, we generated two singleton pregnancies using preselected female blastocysts and recovered preterm fetuses at 135 days post embryo transfer.
We analyzed the extended heteroplasmy in various tissues and organs of these female fetuses as described above. As expected, mtDNA carryover from the spindle donor oocytes was very low or undetectable in sampled somatic tissues (Table S5). We next isolated ovaries from both ST fetuses and analyzed twenty-four individual oocytes. The majority of eggs displayed low or undetectable mtDNA heteroplasmy. However, two oocytes (one from each fetus) contained a substantial degree of mtDNA carryover with 16.2% and 14.1% heteroplasmy levels (Table 4). We carried out an independent ARMS-qPCR assay using different sets of primers and probes and corroborated these unusual results. Taken together, the data support our observations seen with heteroplasmic fetuses and imply that different mtDNA transmission mechanisms may exist for somatic and germline lineages.
Table 4.
mtDNA carryover in oocytes derived from ST offspring
| Individual oocytes | Karyoplast mtDNA (%) | |
|---|---|---|
| Fetus 1 | Fetus 2 | |
| 1 | ND | ND |
| 2 | ND | ND |
| 3 | ND | ND |
| 4 | 0.19 | ND |
| 5 | 0.45 | ND |
| 6 | 0.45 | ND |
| 7 | 0.53 | ND |
| 8 | 1.04 | ND |
| 9 | 1.17 | 0.46 |
| 10 | 1.46 | 5.26 |
| 11 | 2.72 | 5.53 |
| 12 | 14.15 | 16.24 |
The lower detection limit for mtDNA carryover by ARMS-qPCR was 0.15%.
ND, non-detectable.
DISCUSSION
Rapid intergenerational segregation of mtDNA is a poorly studied phenomenon despite its importance for understanding principles of the fundamental biology and management of clinical syndromes caused by mtDNA mutations. Such unpredictable intergenerational mtDNA fluctuations appear to be even more extreme and frequent in humans than in the mouse (Wonnapinij et al., 2010). To explain the segregation of mtDNA variants between generations, a genetic bottleneck hypothesis in the female germline has been proposed. According to this model, segregation and rapid shift of mtDNA occurs during oogenesis resulting in partitioning of mtDNA haplotypes between individual oocytes (Jenuth et al., 1996; Olivo et al., 1983).
Although, the existence of mtDNA segregation during early embryogenesis and subsequent shift from transiently heteroplasmic embryos to the homoplasmic offspring was hypothesized (Laipis et al., 1988), experimentally this was never demonstrated. We have now tested this hypothesis by generating heteroplasmic rhesus oocytes with equal mixture of two wild-type mtDNA haplotypes and followed the transmission of mtDNA to preimplantation embryos, fetuses and germ cells. Using a single cell analysis, we demonstrate rapid segregation of mtDNA variants between daughter blastomeres of cleaving preimplantation embryos, similar to that seen in germ cells, i.e. in individual oocytes (Figure 1). Intriguingly, three ESC lines and one fetus produced from these heteroplasmic embryos were nearly homoplasmic suggesting that return to the homoplasmic condition can occur during development of an individual organism from a zygote to birth, without actual passage of mtDNA through the germline (Figure 1). The fact that early passage ESCs derived from ICMs of heteroplasmic blastocysts have already shifted towards homoplasmy suggests that strong segregation and bottleneck occurs during early epiblast lineage specification rather than during later fetal development. Our study also supports the model that possibly few cells within epiblast progenitors give rise to the somatic cell lineage of embryo proper. This concept is supported by evidence in the mouse that whole somatic lineage of the embryo proper can be derived from just one founder epiblast cell (Wang and Jaenisch, 2004). Another interesting point is that only progenitors with low heteroplasmy (towards either resident or alien mtDNA) contributed to the embryo proper lineage while intermediate variants where lost. This observation supports the notion that the genetic bottleneck at this stage may not be random but rather preferentially selects homoplasmic conditions.
In contrast to relatively homogeneous fetal somatic tissues with low heteroplasmy levels, we found a wide range of heteroplasmic variants distributed in individual oocytes. These data support the existence of a secondary mtDNA bottleneck responsible for segregation of mtDNA variants in the female germline. Previous studies concluded that much of the mtDNA segregation occurs during oocyte development (Jenuth et al., 1996). However, based on our results, we cannot exclude possibility that this segregation takes place much earlier, in precursors of germ cells within the epiblast. This model postulates that the majority of cells in the ICM may contribute to the germline.
The biological mechanism underlying the mtDNA bottleneck is not clear yet. Earlier mouse studies suggested that the basis of the mtDNA bottleneck may be a significant reduction in mtDNA copy number per cell (about 200) during a specific point in oocyte development (Cree et al., 2008). However, recent evidence suggests that segregation occurs without a reduction of mtDNA contents but rather by preferential amplification of specific mtDNA variants (Cao et al., 2007). Neither of these models seems to be able to explain the mtDNA segregation between individual blastomeres in cleaving monkey embryos seen in our study. First, mtDNA copy numbers in blastomeres of cleaving embryos could be in the tens of thousands, well above of estimated minimum (≈200) to be effective for segregation. Secondly, there is no known replication of mtDNA during preimplantation development excluding the possibility of selective amplification. Although, a very short period of mtDNA synthesis immediately after fertilization has been described (McConnell and Petrie, 2004). Mitochondria are not passive organelles, but rather exist as dynamic and interacting structures maintained through balance of fusion and fission. Hence, the distribution of mitochondria and mtDNA to daughter cells during cell division seems to be regulated by mechanisms responsible for mitochondrial fusion and fission (Kashatus et al., 2011). It would be important to investigate the extent of mitochondrial dynamics in preimplantation embryos and their role in regulation of mtDNA segregation and bottleneck.
In our study, we utilized mtDNA differences between Indian- and Chinese-origin rhesus macaques to generate heteroplasmic embryos and offspring. It is believed that sequence polymorphisms between these two subpopulations of macaques are as great as those between some primate species (Smith, 2005). Transcription and replication of mtDNA is tightly controlled by the nuclear genome, suggesting that the resident mtDNA may have a replicative advantage over the alien mtDNA during development (Meirelles and Smith, 1998). However, segregation towards either resident or alien mtDNA haplotypes in fetuses or ESCs, irrespective of the nuclear genetic background seen in our study, either conflicts with this assumption or suggests that the nuclear control of mtDNA replication is highly conserved.
Our study also has far reaching clinical implications for genetic management of mtDNA diseases. Currently, PGD is actively pursued for monitoring mtDNA disorders by sampling one or two blastomeres and selecting embryos for transfer with low mtDNA mutation loads (Poulton and Bredenoord, 2010; Thorburn et al., 2009). However, based on our results, heteroplasmy in biopsied blastomeres from cleaving embryos may not be fully predictive of total mutation load in remaining blastomeres and thus in the embryo. Moreover, heteroplasmy levels in fetal tissues may change drastically compared to preimplantation embryos due observed bottleneck. In contrast, chorionic villus sampling (Poulton et al., 2010) could be more reliable assays based on observations that mtDNA segregation patterns between placenta and fetus were relatively low. It is worth to note that our study is based on heteroplasmy using wild-type mtDNA haplotypes. However, segregation patterns for pathogenic mutations in human embryos could be different (Marchington et al., 1997; Marchington et al., 1998; Steffann et al., 2006). Available clinical observations are often made on limited number of blastomeres or on arrested embryos (Monnot et al., 2011) suggesting that more extensive human studies for each mutation type are needed.
In our follow up studies with the ST approach designed for efficient replacement of mtDNA in oocytes, the amount of mtDNA carryover was insignificant or undetectable in major organs and tissues of offspring. However, a few oocytes recovered from the ST female offspring carried mtDNA from spindle donor oocytes at levels reaching 16.24%. This was unexpected since carryover levels during ST procedure were estimated to be below 1%. While these heteroplasmy levels are below known mutation thresholds for phenotypic expression of many mitochondrial diseases, the possibility exists that mtDNA heteroplasmy may change in subsequent generations through the bottleneck. This observation also suggests that PGD selection for embryos carrying 30% or less mutation loads most likely will not eliminate the possibility of recurrence of mitochondrial diseases in subsequent generations.
EXPERIMENTAL PROCEDURES
Animals
Adult and fetal rhesus macaques were used in this study and all animal procedures were approved by the Institutional Animal Care and Use Committee at the Oregon National Primate Research Center.
Production of heteroplasmic and spindle transfer (ST) oocytes, embryos and offspring
Cumulus-oocyte complexes were collected from anesthetized animals by laparoscopic follicle aspirations and placed in Hepes-buffered TALP (modified Tyrode solution with albumin, lactate, and pyruvate) containing 0.3% bovine serum albumin (TH3) at 37°C. Mature MII oocytes were transferred into 30 µl manipulation droplet of TH3 medium supplemented with 5 µg/ml cytochalasin B on a glass-bottom micromanipulation dish (World Precision Instruments) and incubated at 37°C for 10–15 min. The dish was then mounted on inverted microscope (Olympus, Tokyo, Japan) equipped with micromanipulators (Narishige,Tokyo, Japan), laser objective (XYClone, Hamilton Thorne, Beverly, MA) and spindle imaging system (Oosight, CRi, Woburn, MA). Approximately 50% of the cytoplasm (without spindle) from each oocyte was aspirated into a cytoplast using a 40 µm (inner diameter) glass pipette and then gently expelled into a drop containing inactivated viral envelope from the Haemagglutinating virus of Japan (HVJ-E, Ishihara Sangyo Kaisha Ltd, Japan). A cytoplast was then placed under the zona pellucida of a second oocyte containing another half cytoplasm with a spindle (karyoplast) to induce fusion. Reconstructed oocytes were next fertilized by ICSI, placed into 4-well dishes (Nalge Nunc) containing embryo culture medium and cultured at 37°C in 6% CO2, 5% O2, and 89% N2 (Tachibana et al., 2010). Complete mitochondrial gene replacement by the ST procedure and embryo transfers were carried out as we previously described (Tachibana et al., 2010; Tachibana et al., 2009).
Sex determination in biopsied blastocysts
An expanded blastocyst was immobilized with holding pipette and a few TE cells were aspirated into a biopsy pipette (inner diameter 25–30 µm). The pipette was pulled gently away and biopsied TE portion was separated using a laser pulse. Biopsied TE cells were gently rinsed in TH3 medium and transferred into PCR tubes containing 5 µl of lysis buffer [20 mM Tris-HCl (p.H 8.0), 0.9% Tween-20, 0.9% Nonidet-P40 and 0.4 mg/ml Protenase-K]. Isolated DNA was amplified by PCR using size differences in the amplicons of the X- and Y-linked zinc finger protein genes (ZFX and ZFY) (Mitalipov et al., 2007). The PCR primers were: For 5’-ATT CCA GGC AGT ACC AAA CAG-3’ and Rev 5’-CCA TCA GGG CCA ATA ATT ATT-3’. PCR reactions were carried out in a 20 µl volume containing 5 µl DNA template, 0.1 µM of each primer and 15 ul of Platinum PCR SuperMix (Invitogen). PCR conditions were 40 cycles at 94/55/72°C for 30/30/60 sec (denaturation/annealing/extension). Amplicons were electrophoresed through 1.6% agarose gels stained with ethidium bromide and visualized on a UV transilluminator. Female (XX) samples produced 1149 bp fragment while male (XY) samples contained an additional 771 bp fragment (Figure S2).
DNA extraction from oocytes and blastomeres
Zona pellucida from heteroplasmic oocytes and embryos was removed using brief (20 to 30 sec) exposure to 0.5% of pronase (Sigma Aldrich). Blastomeres from cleaving embryos were disaggregated by brief exposure to trypsin-EDTA and each blastomere was individually placed into a 0.2 ml PCR tube. ICM and TE cells from heteroplasmic blastocysts were isolated by immunosurgery as described previously (Mitalipov et al., 2006). In brief, zona-free blastocysts were incubated in anti monkey whole serum (Sigma Aldrich) for 30 min at 37°C, washed three times with TH3 and transferred into guinea pig complement (Sigma) for 30 min. Blastocysts were gently pipetted with a small bore pipette to disperse TE cells. Separated TE and ICM cells were then transferred into PCR tubes. Genomic DNA was extracted from singles blastomeres and from tissues using the NucleoSpin® Tissue XS Kit (Macherey-Nagel) and the Puregene DNA Purification Kit (Qiagen) according to the manufacturer’s instructions, respectively. NanoDrop 2000 spectrophotometer (NanoDrop Technologies) was used for DNA mass determination.
Heteroplasmy quantification by amplification refractory mutation system (ARMS)-qPCR
ARMS-qPCR was performed to measure heteroplasmy levels in single blastomeres and various tissues (Burgstaller et al., 2007). This method is based on qPCR quantification of less abundant mtDNA variants (alien mtDNA as default) and normalization to the total mtDNA content using discriminative and consensus assays. The delayed amplification from the non-target sequence limiting the dynamic range of ARMS-qPCR, was controlled with a homogenous/ homoplasmic template carrying the alternative sequence under study. Hydrolysis probes (Sigma-Aldrich or Integrated DNA Technologies in case of ZEN double quenched probes) and qPCR primers were designed to differentiate between alien and resident mtDNAs. The non-discriminative and discriminative assays were targeted to the 12S ribosomal RNA gene and to the control region, respectively, and were labeled with TET and FAM reporter fluorophores to facilitate also multiplexed measurement. The Rotor-Gene Multiplex PCR Kit (Qiagen) and the HOT FIREPol® DNA polymerase chemistry (Solis BioDyne) were used for multiplex and singleplex ARMS-qPCR, respectively, and run in duplicate 15 µL reactions. Oligonucleotide concentrations were 100 or 250 nM of each primer and 150 nM of each hydrolysis probe. From each experimental genomic DNA (1 to 4 ng) a 1:8 dilution was run to control for inhibition. The ABI 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) in combination with the SDS software versions 2.3 or 2.4 was used for qPCR measurement. Quantification cycle (Cq) values were determined using manual baseline adjustment. A standard curve was calculated based on four 8-fold serial dilutions plus last 4-fold dilution. The percentage of alien mtDNA in relation to the total mtDNA content was calculated by the equation 100 × quantity minor sequence / quantity total mtDNA. If the alien mtDNA was more abundant, the resident mtDNA was measured to determine the heteroplasmy level.
Measurements of mtDNA copy number
Karyoplasts and cytoplasts were transferred into 0.2 ml PCR tubes containing 10 µl of lysis buffer [50 mM Tris-HCl, (p.H 8.5), 1 mM EDTA, 0.5% tween-20 and 200 µg/mL Proteinase–K]. Samples were lysed at 55°C for 2 h, and incubated at 95°C for 10 min to inactivate Proteinase K. The cellular DNA extract was finally diluted in 30 µl H2O. In the case of cytoplast, an additional 1:10 dilution was performed (final dilution of 1:400). A mtDNA of 161 bp PCR products (nucleotides 3153 to 3313 in GenBank: AY 612638.1) was PCR amplified from 100 ng monkey skin fibroblast DNA using the primers 5’-ACC ACA CAT TCC ACC CGA AAA and 5’-ATG CTA TGG CGG CTA ATG TGG. The amplicon was purified from agarose gel using the QIAquick Gel Extraction Kit (Qiagen). DNA mass determined on a NanoDrop spectrophotometer (NanoDrop Technologies) was converted to DNA copy number using a web calculator (www.uri.edu/research/gsc/resources/cndna.html). Five 10-fold serial dilutions (106 to 102 copies in H2O) were prepared from the standard stock and stored at −20°C until use as standards in regression curve analysis. The 20 µl PCR contained 1× SYBR Green Master Mix (Applied Biosystems), 1 µM of each primer and 1 µl of template (standard DNA, 1:40 dilution of a karyoplast, or 1:400 dilution of a cytoplast). The iCycler (Bio-Rad) was used for qPCR monitoring in this data set. Following initial denaturation at 94°C for 5 min, amplification was performed over 40 cycles of 94/58/72°C for 30/30/45 sec, and completed with a final elongation step of 72°C for 10 min. A melting curve was analyzed to check the specificity of the PCR product. Three technical replicates of duplicate measurement were performed from each sample. The starting copy number of an experimental sample was concluded from the standard curve and adjusted for the dilution factor.
Statistical analysis
Statistical analysis of mtDNA segregation was performed using Statview Software (SAS Institute, Inc.). In brief, the coefficient of variation (CV) and range (differences between maximum and minimum values in blastomeres within the same embryo) were determined by one-way ANOVA followed by a Tukey’s post hoc test. Data are presented as mean±s.d. In all analyses, P < 0.05 was considered as the level for statistical significance.
Highlights.
-
✓
Rapid segregation of heteroplasmic mtDNA in blastomeres of preimplantation embryos.
-
✓
Shift toward homoplasmy in somatic tissues of offspring produced from heteroplasmic oocytes.
-
✓
Heteroplasmic mtDNA was maintained in oocytes recovered from these offspring.
-
✓
Return to homoplasmy can occur during prenatal development, without a passage through the germline.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the Division of Animal Resources, Surgical Team, Assisted Reproductive Technologies & Embryonic Stem Cell, Endocrine Technology, and Molecular & Cellular Biology Cores at the Oregon National Primate Research Center for providing expertise and services that contributed to this project. We are grateful to Keith Masterson, Maidina Tuohetahuntila, Erin Wolff, Rebecca Tippner-Hedges, Ying Li, Monica Fraenkel and Dario Melguizo Sanchis for their technical support and Jinhee Kim for help with illustrative materials. We are indebted to Dr. Richard Stouffer for consulting, helpful discussions and critical reading of the manuscript. This study was supported by funds from Oregon National Primate Research Center, the Center for Women’s Health Circle of Giving and grants from the National Institutes of Health HD057121, HD059946, HD063276, HD047675, EY021214, HD018185 and RR000163.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information including two figures and five tables can be found with this article online.
REFERENCES
- Ashley MV, Laipis PJ, Hauswirth WW. Rapid segregation of heteroplasmic bovine mitochondria. Nucleic Acids Res. 1989;17:7325–7331. doi: 10.1093/nar/17.18.7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgstaller JP, Schinogl P, Dinnyes A, Muller M, Steinborn R. Mitochondrial DNA heteroplasmy in ovine fetuses and sheep cloned by somatic cell nuclear transfer. BMC Dev. Biol. 2007;7:141. doi: 10.1186/1471-213X-7-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Shitara H, Horii T, Nagao Y, Imai H, Abe K, Hara T, Hayashi J, Yonekawa H. The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells. Nat. Genet. 2007;39:386–390. doi: 10.1038/ng1970. [DOI] [PubMed] [Google Scholar]
- Craven L, Tuppen HA, Greggains GD, Harbottle SJ, Murphy JL, Cree LM, Murdoch AP, Chinnery PF, Taylor RW, Lightowlers RN, et al. Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature. 2010;465:82–85. doi: 10.1038/nature08958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cree LM, Samuels DC, de Sousa Lopes SC, Rajasimha HK, Wonnapinij P, Mann JR, Dahl HH, Chinnery PF. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat. Genet. 2008;40:249–254. doi: 10.1038/ng.2007.63. [DOI] [PubMed] [Google Scholar]
- Dean NL, Battersby BJ, Ao A, Gosden RG, Tan SL, Shoubridge EA, Molnar MJ. Prospect of preimplantation genetic diagnosis for heritable mitochondrial DNA diseases. Mol. Hum. Reprod. 2003;9:631–638. doi: 10.1093/molehr/gag077. [DOI] [PubMed] [Google Scholar]
- Giles RE, Blanc H, Cann HM, Wallace DC. Maternal inheritance of human mitochondrial DNA. Proc. Natl. Acad. Sci. USA. 1980;77:6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauswirth WW, Clayton DA. Length heterogeneity of a conserved displacement-loop sequence in human mitochondrial DNA. Nucleic Acids Res. 1985;13:8093–8104. doi: 10.1093/nar/13.22.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauswirth WW, Laipis PJ. Mitochondrial DNA polymorphism in a maternal lineage of Holstein cows. Proc. Natl. Acad. Sci. USA. 1982;79:4686–4690. doi: 10.1073/pnas.79.15.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauswirth WW, Laipis PJ. Transmission genetics of mammalian mitochondria: A molecular model and experimental evidence. In: Quagliariello E, editor. Achievement and Perspectives in Mitochondrial Research. Vol. II. Amsterdam: Elsevier Biomedical; 1985. pp. 49–59. [Google Scholar]
- Hauswirth WW, Van de Walle MJ, Laipis PJ, Olivo PD. Heterogeneous mitochondrial DNA D-loop sequences in bovine tissue. Cell. 1984;37:1001–1007. doi: 10.1016/0092-8674(84)90434-3. [DOI] [PubMed] [Google Scholar]
- Hudson B, Vinograd J. Catenated circular DNA molecules in HeLa cell mitochondria. Nature. 1967;216:647–652. doi: 10.1038/216647a0. [DOI] [PubMed] [Google Scholar]
- Jenuth JP, Peterson AC, Fu K, Shoubridge EA. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat. Genet. 1996;14:146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- Kashatus DF, Lim KH, Brady DC, Pershing NL, Cox AD, Counter CM. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat. Cell Biol. 2011;13:1108–1115. doi: 10.1038/ncb2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laipis PJ, Van de Walle MJ, Hauswirth WW. Unequal partitioning of bovine mitochondrial genotypes among siblings. Proc. Natl. Acad. Sci. USA. 1988;85:8107–8110. doi: 10.1073/pnas.85.21.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnane AW, Marzuki S, Ozawa T, Tanaka M. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet. 1989;1:642–645. doi: 10.1016/s0140-6736(89)92145-4. [DOI] [PubMed] [Google Scholar]
- Marchington DR, Hartshorne GM, Barlow D, Poulton J. Homopolymeric tract heteroplasmy in mtDNA from tissues and single oocytes: support for a genetic bottleneck. Am. J. Hum. Genet. 1997;60:408–416. [PMC free article] [PubMed] [Google Scholar]
- Marchington DR, Macaulay V, Hartshorne GM, Barlow D, Poulton J. Evidence from human oocytes for a genetic bottleneck in an mtDNA disease. Am. J. Hum. Genet. 1998;63:769–775. doi: 10.1086/302009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PA, Matheson EC, Hall AG, Lightowlers RN. Mismatch repair activity in mammalian mitochondria. Nucleic Acids Res. 2003;31:1052–1058. doi: 10.1093/nar/gkg167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JM, Petrie L. Mitochondrial DNA turnover occurs during preimplantation development and can be modulated by environmental factors. Reprod. Biomed. Online. 2004;9:418–424. doi: 10.1016/s1472-6483(10)61277-1. [DOI] [PubMed] [Google Scholar]
- Meirelles FV, Smith LC. Mitochondrial genotype segregation during preimplantation development in mouse heteroplasmic embryos. Genetics. 1998;148:877–883. doi: 10.1093/genetics/148.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels GS, Hauswirth WW, Laipis PJ. Mitochondrial DNA copy number in bovine oocytes and somatic cells. Dev. Biol. 1982;94:246–251. doi: 10.1016/0012-1606(82)90088-4. [DOI] [PubMed] [Google Scholar]
- Mitalipov S, Kuo HC, Byrne J, Clepper L, Meisner L, Johnson J, Zeier R, Wolf D. Isolation and characterization of novel rhesus monkey embryonic stem cell lines. Stem Cells. 2006;24:2177–2186. doi: 10.1634/stemcells.2006-0125. [DOI] [PubMed] [Google Scholar]
- Mitalipov SM, Zhou Q, Byrne JA, Ji WZ, Norgren RB, Wolf DP. Reprogramming following somatic cell nuclear transfer in primates is dependent upon nuclear remodeling. Hum. Reprod. 2007;22:2232–2242. doi: 10.1093/humrep/dem136. [DOI] [PubMed] [Google Scholar]
- Monnot S, Gigarel N, Samuels DC, Burlet P, Hesters L, Frydman N, Frydman R, Kerbrat V, Funalot B, Martinovic J, et al. Segregation of mtDNA throughout human embryofetal development: m.3243A>G as a model system. Hum. Mutat. 2011;32:116–125. doi: 10.1002/humu.21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivo PD, Van de Walle MJ, Laipis PJ, Hauswirth WW. Nucleotide sequence evidence for rapid genotypic shifts in the bovine mitochondrial DNA D-loop. Nature. 1983;306:400–402. doi: 10.1038/306400a0. [DOI] [PubMed] [Google Scholar]
- Poulton J, Bredenoord AL. 174th ENMC International Workshop: Applying pre-implantation genetic diagnosis to mtDNA diseases: Implications of scientific advances 19–21 March 2010, Naarden, The Netherlands. Neuromuscular Disorders. 2010;20:559–563. doi: 10.1016/j.nmd.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Poulton J, Chiaratti MR, Meirelles FV, Kennedy S, Wells D, Holt IJ. Transmission of mitochondrial DNA diseases and ways to prevent them. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001066. e1001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DG. Genetic characterization of Indian-origin and Chinese-origin rhesus macaques (Macaca mulatta) Comp. Med. 2005;55:227–230. [PubMed] [Google Scholar]
- Steffann J, Frydman N, Gigarel N, Burlet P, Ray PF, Fanchin R, Feyereisen E, Kerbrat V, Tachdjian G, Bonnefont JP, et al. Analysis of mtDNA variant segregation during early human embryonic development: a tool for successful NARP preimplantation diagnosis. J. Med. Genet. 2006;43:244–247. doi: 10.1136/jmg.2005.032326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinborn R, Schinogl P, Zakhartchenko V, Achmann R, Schernthaner W, Stojkovic M, Wolf E, Muller M, Brem G. Mitochondrial DNA heteroplasmy in cloned cattle produced by fetal and adult cell cloning. Nat. Genet. 2000;25:255–257. doi: 10.1038/77000. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sparman M, Mitalipov S. Chromosome transfer in mature oocytes. Nat. Protoc. 2010;5:1138–1147. doi: 10.1038/nprot.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sparman M, Sritanaudomchai H, Ma H, Clepper L, Woodward J, Li Y, Ramsey C, Kolotushkina O, Mitalipov S. Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature. 2009;461:367–372. doi: 10.1038/nature08368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima H, Sueoka K, Moon SY, Nakabayashi A, Sakurai T, Murakoshi Y, Watanabe H, Iwata S, Hashiba T, Kato S, et al. The development of novel quantification assay for mitochondrial DNA heteroplasmy aimed at preimplantation genetic diagnosis of Leigh encephalopathy. J. Assist. Reprod. Genet. 2007;24:227–232. doi: 10.1007/s10815-007-9114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn D, Wilton L, Stock-Myer S. Healthy baby girl born following pre-implantation Genetic diagnosis for mitochondrial DNA m.8993t>g Mutation. Mol. Genet. Metab. 2009;98:5–6. [Google Scholar]
- Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim. Biophys Acta. 2010;1797:113–128. doi: 10.1016/j.bbabio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Wai T, Teoli D, Shoubridge EA. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat. Genet. 2008;40:1484–1488. doi: 10.1038/ng.258. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial DNA mutations in disease and aging. Environ. Mol. Mutagen. 2010;51:440–450. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- Wang Z, Jaenisch R. At most three ES cells contribute to the somatic lineages of chimeric mice and of mice produced by ES-tetraploid complementation. Dev. Biol. 2004;275:192–201. doi: 10.1016/j.ydbio.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Wonnapinij P, Chinnery PF, Samuels DC. Previous estimates of mitochondrial DNA mutation level variance did not account for sampling error: comparing the mtDNA genetic bottleneck in mice and humans. Am. J. Hum. Genet. 2010;86:540–550. doi: 10.1016/j.ajhg.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.