INTRODUCTION
In eukaryotic cells, the genomic DNA is etched with a number of chemical modifications, called epigenomic modifications (epi- modifications). These epi- modifications add an extra layer of information onto the genomic sequence, and enable it to encode a more complex program of gene regulation (Karlić et al., 2010; Maunakea et al., 2010a). Different epi- modifications affect how the DNA interacts with transcription factors, although many mechanisms remain unknown (Campos and Reinberg, 2009). Adding to the complexity, the genomes are far from being completely annotated on the functional level, making it necessary to first find regulatory genomic sequences before we can understand their complex regulatory roles.
Evolutionary comparisons provide a powerful tool to study genome functions. This became obvious when it was recognized that the majority of DNA can mutate freely without deleterious effects, while certain sequence elements are more constrained (Kimura, 1968). Leveraging this theory, researchers have inferred functional genomic segments by examining genomic sequence conservation (Hardison, 2003) and have identified human-specific regulatory DNA by looking for sequences with accelerated rates of evolutionary change (Pollard et al., 2006).
The successes in genomic comparisons beg the question: can we also use evolution to study the functions of the epigenome? To do so, the basic evolutionary properties of the epigenome must be established first, preferably in the contexts of both genomic and transcriptomic evolution. To explore relationships among evolutionary changes to the genome, the epigenome, and the transcriptome, several specific questions were of critical interest. First, evolutionary selection has left clear traces on the human genome (Ren, 2010); what are the traces of evolutionary selection on the human epigenome? Second, are evolutionary changes to the epigenome merely a consequence of genomic sequence changes or, rather, has the epigenome made the genome more or less susceptible to evolutionary selection? Third, the degree of gene expression conservation correlates poorly with the extent to which nonexonic sequences are conserved among vertebrates (Chan et al., 2009; Wilson et al., 2008); might this discrepancy be explained by the epigenome? Fourth, mammalian orthologous transcription factors (TF) often do not bind to orthologous DNA sequences (Jegga et al., 2008), as only ~5% of the Oct4 and Nanog binding sites occupy homologous sequences in human and mouse embryonic stem (ES) cells (Kunarso et al., 2010); do epigenetic modification enzymes apply the same types of modifications to orthologous sequences in mammals?
Among many types of epi- modifications (Tan et al., 2011), a subset is known to correlate with gene transcription. For example, DNA cytosine methylation (Cm) (Maunakea et al., 2010a), histone 3 lysine 27 tri-methylation (H3K27me3), and histone 3 lysine 9 tri-methylation (H3K9me3) may repress gene transcription, whereas histone 3 lysine 4 mono-, di-, and tri-methylation (H3K4me1/2/3), lysine 27 acetylation (H3K27ac), and lysine 36 tri-methylation (H3K36me3) are positively associated with transcription (Karlić et al., 2010). The roles of some epigenomic marks (epi- marks) remain controversial. For example, histone variant H2A.Z is generally assumed to be associated with active promoters because it anti-correlates with Cm in plants, insects, and fish (Zemach et al., 2010); consistently with this, H2A.Z is associated with active promoters in flies (Weber et al., 2010). However, H2A.Z is associated with inactive promoters in yeasts (Guillemette et al., 2005; Raisner et al., 2005). The role of H2A.Z has yet to be tested in mammals. Even for the epi- modifications whose roles are better established, they may have undiscovered functions.
The functions of many epi- modifications have so far only been evaluated individually, primarily due to the difficulty of assessing the functional significances of co-localized epi- marks. Any two epi- marks can co-localize in some genomic regions, but most of such co-localizations do not serve any regulatory functions. The best documented epi- marks co-localization is probably the bivalent domain (H3K27me3 + H3K4me3), which is hypothesized to be poised for activation during differentiation of embryonic stem cells (ESCs) (Mikkelsen et al., 2007). This hypothesized function was derived from comparing ESCs with other cell types. However, a mechanistic understanding of how bivalent domains regulate lineage-specific ESC differentiation is still lacking. We wish to provide a new approach to systematically examine the functions of epi- modifications, and more importantly, the functions of combinations of epi- marks. We propose to leverage the connection between evolutionary conservation and functional importance to achieve this goal.
Here we introduce ‘comparative epigenomics’ – interspecies comparison of epigenomes – as a novel approach for annotation of the regulatory sequences of the genome. We created a multi-species epigenomic dataset from pluripotent stem cells of humans, mice and pigs, which is comprised of genomic distributions of DNA methylation and eight histone modifications, the binding intensities of four transcription regulators (Nanog, Oct4, P300, Taf1), and transcribed RNA sequences. We first examined the co-evolution properties among the epigenome, the genome, and the transcriptome. Comparing epigenomic changes to genomic changes, we observed strong epigenomic conservation for both fast-evolving and slowly evolving DNA sequences, but not on neutrally evolving DNA sequences. These data suggest epigenomic conservation is not completely dictated by genomic sequences. On the other hand, interspecies epigenomic changes are linearly correlated with evolutionary changes of transcription factor binding and gene expression, suggesting that comparative epigenomics can directly reveal critical information on gene regulation. Based on these initial analyses, we set out to discover regulatory sequences by conserved co-localization of different epi- marks. To test the functions of these putative regulatory sequences, we developed a differentiation assay in which mouse embryonic stem cells were differentiated into mesendoderm cells. Our time-course ChIP-seq and RNA-seq data in this differentiation process confirmed the regulatory functions of all of the seven pairs of epi- marks identified by conserved co-localization. Thus, conserved co-localization is an efficient approach to identify functional epi- mark combinations from a large (combinatorial) number of random combinations of epi- marks. More importantly, comparative epigenomics reveals regulatory features of the genome that cannot be discerned from sequence comparison alone.
RESULTS
Epigenomes of human, mouse, and pig pluripotent stem cells
To answer the above questions, we conducted a “comparative epigenomics” study (Mikkelsen et al., 2010) with a focus on evolution. We generated and compiled from published work the genomic distributions of nine epigenetic modifications including Cm, H2A.Z, H3K4me1/2/3, H3K9me3, H3K27me3, H3K27ac, H3K36me3, and the binding of four transcription regulators, P300, Taf1, Oct4, and Nanog, in pluripotent stem cells of humans, mice, and pigs (Sus scrofa) (West et al., 2010). Cm was assayed by both methylated DNA immunoprecipitation followed by sequencing (MeDIP-seq) and DNA digestion by methyl-sensitive restriction enzymes followed by sequencing (MRE-seq) (Maunakea et al., 2010b). Histone modifications and binding of transcription regulators were assayed with chromatin immunoprecipitation followed by sequencing (ChIP-seq). Gene expression was measured by RNA sequencing (RNA-seq) technology. Taken together, a total of 48 sequencing datasets were compiled, among which 29 datasets (73 billion bases) were generated from this study (GEO accession number: GSE36114) and the 19 other datasets (27 billion bases) were compiled from 3 published works (Chen et al., 2008; Goren et al., 2010; Lister et al., 2009) (Table S1 upper panel).
Comparative Epigenome Browser
To manage, visualize, and compare these data, we built an interactive data analysis system, the Comparative Epigenome Browser (http://sysbio.igb.uiuc.edu/cpbrowser). A novel analytical feature of this system is a side-by-side comparison of epi- modifications on orthologous genomic regions.
Interspecies epigenomic variation is greater than within-species variation
We compared interspecies and within-species epigenomic variations. Based on histone modifications, human and mouse cell lines cluster separately. Hierarchical clustering showed that two human embryonic stem cell (hESC) lines and three human induced pluripotent stem cell (hiPS) lines cluster tightly together, and mouse embryonic stem cell (mESC) and two mouse epiblast stem cell (mEpiSC) lines cluster together separately from the human cells (Figure S1A–F). The tight clustering of different sources of pluripotent cells within a species was based on comparison of all human-mouse orthologous genes, and the results were consistent for both the activation mark H3K4me3 and the repressive mark H3K27me3, regardless of the distance metric used (Figure S1C–D). These data indicate that interspecies epigenomic differences are greater than within-species differences. H3K4me3 and H3K27me3 are the only two marks that have been assayed in both naïve and primed pluripotent stem cells (Nichols and Smith, 2009) in a genome-wide manner. Lab-to-lab variation in ChIP-seq data is relatively small (Figure S1G) and thus unlikely to affect this result. Consistently, hierarchical clustering of gene expression data showed strong within-species similarities (Pearson correlation = 0.86 ± 0.19, Rank correlation = 0.91±0.09) and a clear interspecies difference (Pearson correlation = 0.36±0.03, Rank correlation = 0.60±0.04) (Figure S1E).
Traces of evolutionary conservation of the epigenome
The pronounced interspecies epigenomic differences provoked the question of whether there are any traces of conservation of the epigenome. We started by analyzing the epi- modification intensities for various features of the genome, including intergenic regions, promoters, exons, introns, and 5′ and 3′ untranslated regions (UTRs). The distribution of the intensities of each epi- modification on each type of genomic feature was summarized (box plots, Figure 1), and then these distributions were combined into an ‘epi- mark intensity’ distribution for all genomic regions for a species (any panel of 9 box plots, Figure 1). A non-parametric test with the null hypothesis that epi- mark intensity distributions are different across species generated a p-value for every epi- mark. These p-values ranged from 10−10 (Cm) to 10−2 (H3K4me3), indicating that the relative difference in epi- modification intensities on different genomic features is in general consistent across species, although the consistency levels vary from modification to modification. This pattern of conservation provides evolutionary support for the idea of using epigenomic data to predict functional noncoding genomic features (Ernst and Kellis, 2010).
Figure 1.
Interspecies conservation of epigenomic modifications. Each box plot represents the distribution of the normalized intensities of the indicated epi- modifications (e.g., Cm, upper-most row) in various genomic regions (e.g., 500bp upstream of genes, left-most column). Median, quartiles, maximum, and minimum intensity values are shown in each box plot (see insert). The assembly of 9 box plots shows the distribution of relative intensities of an epi- modification on different genomic regions in a species (e.g., Cm in human, upper-most and left-most panels). P-value: the support to conservation of each epi- modification, calculated from a non-parametric test comparing the data in the left, middle, and right panels.
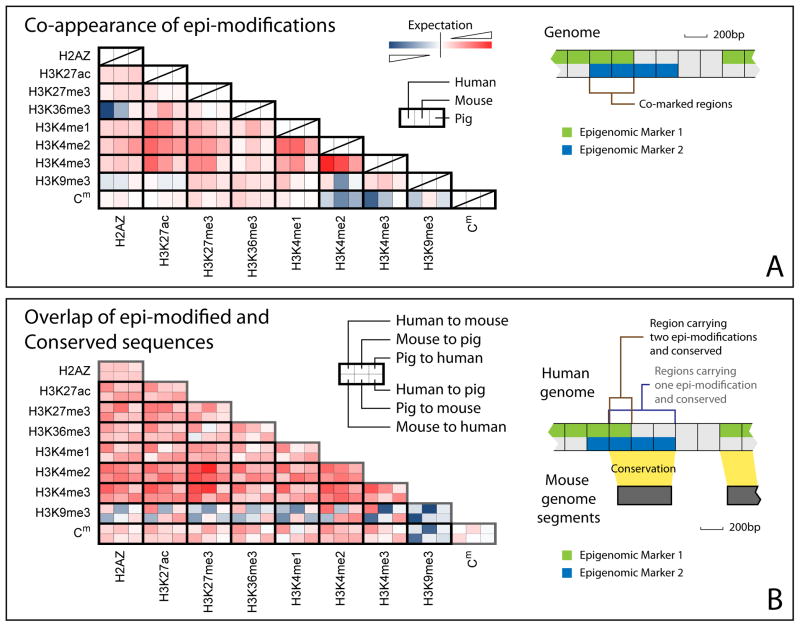
We then asked whether the co-occupancy of two epi- marks is conserved across species. In the human genome, non-randomly co-appearing epi- marks include H3K4me1/2/3 (null hypothesis: epi- marks appear independently in the genome, minimum odds ratio = 4.55, p-value < 10−20), H3K27ac and H3K4me1/2/3 (minimum odds ratio = 10.3, p-value < 10−20), as well as H3K27me3 and H3K4me1/2/3 (minimum odds ratio = 4.14, p-value < 10−20). Non-random, mutual-avoiding epi- modifications include Cm vs. H3K4me2/3 (odds ratio = 0.70, p-value < 10−20). These non-random co-occupancy patterns are conserved in mouse and swine genomes (Figures 2A, S2A–C). Even weak co-occupancy patterns between any two of H3K27me3, H3K36me3, and H3K4me1/2/3 are conserved in all three species (p-value < 10−9). In contrast to being antagonistic chromatin marks in plants, insects, and fish (Zemach et al., 2010; Zilberman et al., 2008), H2A.Z and Cm are not clearly anti-correlated in any of the three mammals (observed total length of co-marked regions ≥ expected total length of co-marked regions). Instead, H3K4me2/3 and H3K9me3 exhibit stronger anti-correlation to Cm (Figures 2A, S2A–C).
Figure 2.
Interspecies conservation of co-occupancy of different epi- modifications. (A) Log ratio between the number of genomic regions carrying two epi- modifications (shown as row and column names) and the expected number, calculated from a null model that the epi- modifications appear independently of each other (each small box, red: log ratio > 0, co-occupancy; blue: log ratio < 0, anti-co-occupancy). With a few exceptions, both positive and negative co-occupancies of any epi- marks are conserved across species, as seen in similar colors of the three consecutive boxes in a row. (B) Log ratio between the number of conserved regions carrying one (diagonal boxes) or two (non-diagonal boxes) epi- modifications and the expected number, calculated from a null model in which conserved regions and epi- modified regions appear independently. Conserved genomic regions are determined by six pair-wise comparisons, shown in six small boxes outlined with a darker edge. For example, the left-most upper box refers to the human genomic regions conserved in a human vs. mouse comparison. All genomic regions with epi- modifications except H3K9me3 were positively associated with conserved regions (red). H3K9me3 selectively marks non-conserved regions (blue). Bivalent domains (co-marked by repression mark H3K27me3 and activation mark H3K4me2/3) exhibited the strongest association with conserved regions. (See also figure S2)
Next we tested whether the genomic regions with one or a pair of epi- modifications are correlated with conserved genomic sequences. Genomic regions with all assayed epi- modifications except for H3K9me3 are correlated with sequence-conserved regions (odds ratio = 1.43, p-value < 10−20) (Figures 2B, S2D–F). With the exception of H3K9me3-marked regions, the genomic regions with two epi- modifications are also correlated with conserved sequences (p-value < 10−9), often with stronger correlations than single epi- modification regions.
If the epigenome is evolutionarily conserved, one would expect to see not only that epi- modification marked and conserved sequences are positively correlated, but also that orthologous sequences in two genomes share the same epi- modifications. To test this hypothesis, we categorized conserved regions of the human genome into three distinct sets, which are strongly, moderately, and weakly conserved in 46 vertebrate species, respectively (Siepel et al., 2005). In each set, we quantified epigenomic conservation by the ratio between the observed number of sequences whose orthologous sequences share the same epi- modification and the expected number of epi- sharing orthologous sequences (Figure S2K). H3K27me3, H3K36me3, H3K4me2, and H3K4me3 showed 3- to 20-fold increases in co-occupancy of human-mouse or human-pig orthologous sequences than would be expected by random chance (maximum p-value < 10−7). For H3K27me3, H3K36me3, H3K4me1/2/3, and Cm, as the level of sequence conservation increases, the chance of orthologous sequences sharing the same epi- modification also increases. Taken together, these data suggested that examinations of the co-occupancy of epi- marks, the correlation between epi- modification and conserved regions, and the co-occupancy of orthologous sequences may be viable approaches to assess epigenomic conservation.
The “U-shaped” correlation between epigenomic and genomic conservations
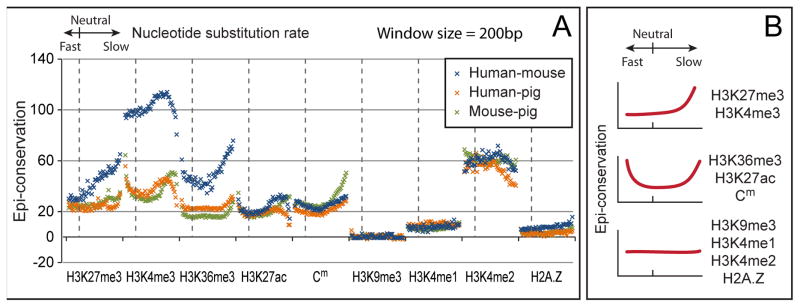
To globally examine the relationship between genomic evolution and epigenomic changes, we categorized the human genome into 50 distinct sets of sequence segments, and ordered these sets by nucleotide substitution rate (see Experimental procedures). We then identified the epigenomic conservation levels in every set (Figures 3A, S2G–J). The enhancer mark H3K27ac, gene-body mark H3K36me3, and Cm exhibited increased conservation in both the accelerated substitution rate (fast-changing) and reduced substitution rate (conserved) sets. H3K4me3 and H3K27me3 exhibited increased conservation in reduced substitution rate sets. In summary, a U-shaped correlation was observed between genomic selection and epigenomic conservation; the large portion of evolutionary neutral or near-neutral sequences exhibit a baseline epigenomic conservation level, forming the bottom of the U-shape; the sequences with accelerated or reduced substitution rates, respectively exhibit enhanced epigenomic conservation, making the two ends of the U-shape tilt upward (Figure 3B).
Figure 3.
Global comparison of genomic and epigenomic conservations. (A) The human genome was categorized into 50 distinct sets by nucleotide substitution rates (x-axis). These sets were ordered from the fastest changing (1st), to neutral (17th), and to slowest changing (50th). Epi- conservation levels by human-mouse (green) and human-pig (orange) comparisons are plotted on the y-axis. Similarly, the mouse genome was categorized into 50 sets, and the epi- conservation levels in a mouse-pig comparison were plotted (blue). (B) Schematic representations of the correlations between sequence selection and epi- conservation. Some epi- marks exhibit a U-shaped correlation, while others can be represented by the right half or the flat bottom of the U-curve. (See also Figure S2)
The increased conservation levels for H3K27ac, H3K36me3, and Cm in fast-changing sequences indicate that epigenomic conservation is not completely determined by interspecies sequence similarity. To further test this hypothesis, we directly correlated epigenomic conservation with interspecies sequence similarity. This is a different test from the vertebrate conservation analysis (Figure 3A) because the nucleotide substitution rate estimated from 46 vertebrates does not necessarily correlate with pairwise sequence similarity between two species (Prabhakar et al., 2008). Except for H3K9me3, pairwise comparisons among human, mouse, and pig genomes consistently rule out a direct correlation between sequence similarity and epigenomic conservation (p-value < 10−20) (Figure S2L). These data indicate that either pairwise alignment is not sufficient to detect epigenomic conservation, or epigenomic conservation is not a simple consequence of sequence similarity. Therefore, we hypothesized that either (1) some epi- modifications may directly facilitate nucleotide substitution or (2) some conserved epi- modifications may buffer negative selective pressure, providing the genome greater freedom to change. Consistent with the former hypothesis, Cm is more mutagenic than C (Coulondre et al., 1978); the C→T change occurs more frequently than other changes between human and chimpanzee genomes, and such a change depends on local GC content (Jiang and Zhao, 2006). Because mechanisms that associate histone modifications with DNA mutations remain unidentified, we explored the plausibility of the latter hypothesis.
To buffer sequence changes from negative selective pressure, the epigenome must buffer genomic changes from generating phenotypic outcomes through, for example, concomitant transcriptome changes. To explore this possibility, we started by asking whether the same combination of epi- modifications is predictive of gene expression in every species. In each species, we used a linear regression model to fit the expression value of every gene to the nine measured epi- mark intensities in its promoter and used a model selection procedure to choose the epi- modifications that are predictive of gene expression. With only four epi- mark intensity values, the expression of every gene can be predicted in each species (largest p-value < 10−16) (Figure S3A). The models did not overfit (Figure S3D), and the epi- marks to expression predictive power matches 52.7%–81.3% of using one RNA-seq dataset to predict another (as measured by R2, Figure S3C). The epi- modifications predictive of gene expression levels were almost identical among humans, mice, and pigs, including H3K4me3, H3K36me3, H3K27me3, and H3K27ac (Figure S3B). The only exception was that in pigs, H3K9me3 replaced H3K27ac in the final model, due to a large correlation (0.91) between H3K4me3 and H3K27ac data in pigs. These data show that gene expression can be predicted by a conserved set of epi- marks, reiterating the idea that epigenomic conservation can be used to study gene regulation.
Interspecies epigenomic changes are predictive of interspecies changes of gene expression and transcription factor binding
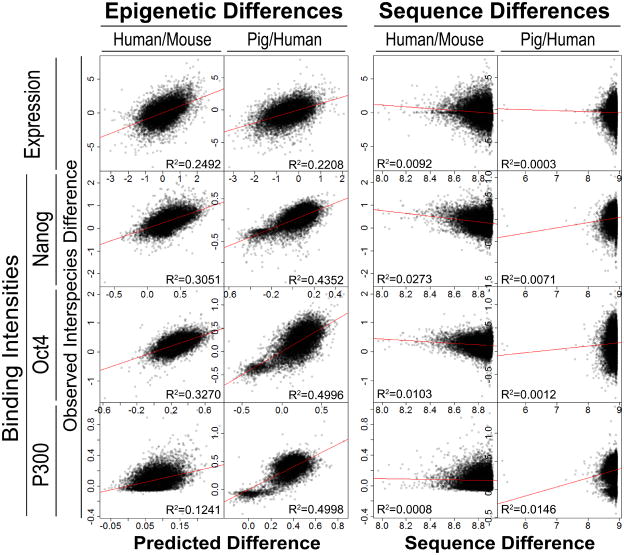
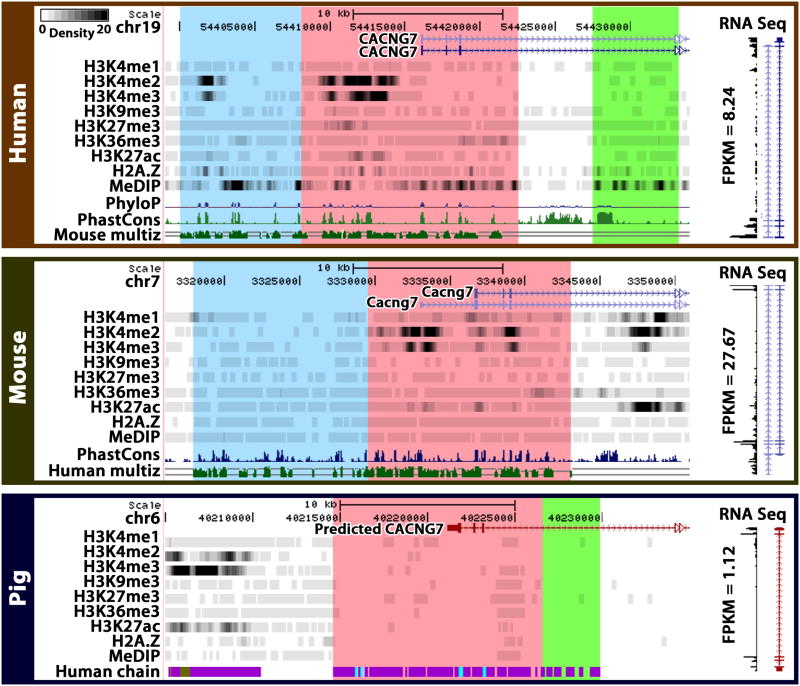
We then asked whether interspecies epigenomic changes are correlated to transcriptomic changes. The interspecies differences of epi- modification intensities are predictive of interspecies gene expression differences (p-value < 10−16) (Figure 4). In a control experiment, when interspecies epi- mark intensity differences were considered, the original epi- modification intensities in each species did not contribute to further explain gene expression difference (Table S2). This implies that the epigenomic information associated with changes of gene expression between species is distinct from the epigenomic information associated with gene expression variation within a single species. For example, the human RNA processing gene DDX17 (FPKM=70.89) expressed higher than the human AVPI1 (FPKM=2.99), a gene related to MAPK activity. Although this within-species expression difference can be predicted by human epi- mark intensities, the interspecies differences of expression for these genes cannot (Figure S3E). However, the moderate decrease of DDX17 expression (from humans to mice) and the 438% increase of AVPI1 expression were associated with interspecies changes of epi- modifications. In contrast, published cross-species analysis of tissue expression data found no identifiable sequence-to-expression correlation in vertebrates (Chan et al., 2009). Similarly, we could not find any apparent correlation between interspecies sequence difference and expression difference using a simple model (Figure 4). Take the CACNG7 (calcium channel, voltage-dependent, gamma subunit 7) gene as an example: its 12k bp upstream sequences are conserved in humans, mice and pigs, whereas its expression levels are high in humans and mice, but low in pigs. However, marks of active promoters H3K4me2/3 are conserved in humans and mice, but not in pigs, providing a correlation for the interspecies expression differences with epi- marks (Figure 5).
Figure 4.
Correlations among evolutionary changes of epi- modification intensities, gene expression levels, TF binding intensities, and genomic sequences. (Left panel) Evolutionary changes of epi- modification intensities are predictive of gene expression changes and TF binding intensity changes. X-axis: predicted gene expression or TF binding intensity changes with a linear model of interspecies epi- intensity changes; Y-axis: observed interspecies changes. (Right panel) Scatter plots between interspecies gene expression difference (y-axis) and promoter sequence difference (x-axis). For every orthologous gene pair, sequence difference was measured by log(m) − log(n), where m is the maximum log blastn score of all orthologous promoters (4k bp centered at TSS), and n is the blastn score of the orthologous promoter pair under consideration. R2: square of the sample correlation coefficient. (See also figure S3 and table S2)
Figure 5.
An example of correlated interspecies epi- and gene expression changes. The genomic and epigenomic neighborhoods of CACNG7 (calcium channel, voltage-dependent, gamma subunit 7) in three species are displayed by the Comparative Epigenome Browser. The orthologous regions determined by the liftOver program are shaded in the same color. Densities of ChIP-seq counts and MeDIP-seq counts are plotted in gray scale. ~12kb upstream sequences of the gene are conserved (pink). H3K4me2 and H3K4me3 are present and conserved in upstream regions in humans and mice, coinciding with conserved expression of the gene (RNA-seq data drawn vertically on the right). The conserved pig upstream sequence (in pink) is devoid of H3K4me2/3 marks, coinciding with a much lower expression level of the pig gene. (See also figure S3)
One possible mechanism to explain how transcriptomic evolutionary changes might relate to epigenomic changes is that epigenomic changes may influence TF binding intensities. In each species, epi- mark intensities are strongly correlated with the total binding intensities of all assayed TFs (Figure S4A–C). As a representative TF, Nanog’s in vivo binding sites are surrounded by cell type-specific epigenomic patterns (Figure S4D). The evolutionary changes of epi- mark intensities are predictive of binding intensity changes of Oct4 (p-value < 10−22), Nanog (p-value < 10−22), and P300 (p-value < 10−22) (Figure 4, left panel). The weak association between interspecies sequence changes and TF binding changes (Figure 4, right panel) is related to the finding that very few Oct4 and Nanog proteins bind to orthologous sequences (Kunarso et al., 2010). These data are in line with the hypothesis that the epigenome may be a buffer in the mapping of genome sequence to TF binding to gene expression to phenotypic outcome, stabilizing gene expression patterns in the face of sequence changes. Such a process would alleviate evolutionary selection pressure on sequence changes, by masking genetic changes from immediate phenotypic changes. These data do not rule out the idea that genetic changes can introduce coding changes and therefore can be evolutionarily selected.
Evolutionarily conserved co-localization of different epigenomic marks defines several classes of cis-regulatory sequences
We set out to test the functions of evolutionarily conserved epi- mark combinations (Figure 2, Table 1) using a cell differentiation assay. During ESC differentiation, the directions of epigenomic changes and expression changes of nearby genes were expected to reflect the function of an epi- mark combination (Rada-Iglesias et al., 2011; Zentner et al., 2011). We differentiated mESCs into mesendoderm cells (Tada et al., 2005; Yasunaga et al., 2005), a lineage in which the dynamic changes of the epigenome have not been examined. On the 6th day of differentiation, almost all cells expressed the mesendoderm protein Goosecoid (GSC) and endoderm protein Sox17 (Figure S5A), and exhibited typical mesendoderm morphology (Figure S5B). Pluripotency genes Oct4, Sox2, and Nanog were down-regulated, and mesendoderm markers GSC and Chordin and endoderm markers Foxa2, Sox17, Lim1, and Hnf4 were up-regulated (Figure S5C). We mapped the epigenomes and the transcriptomes of Day 4 and Day 6 differentiated cells using ChIP-seq and RNA-seq, adding a total of 18 datasets (38 billion bases) to our overall dataset (Table S1 lower panel, Figure S6).
Table 1.
Most conserved co-marks for each epi- modification. Comments in the “Biological inference” column only relate to mammalian species.
| Epi- mark | Most conserved co-marks | Putative function | Biological inference |
|---|---|---|---|
| H2A.Z | H3K4me2/3 | Poised promoter | Negatively associated with gene activation in ES cells and during ESC differentiation |
| H3K27ac | H3K4me1/2/3 | Active enhancer (me1/me2) or promoter (me2/me3) | H3K27ac marks promoters as well |
| H3K27me3 | H3K4me1/2/3 | poised enhancer (me1/me2) or bivalent promoter (me2/me3) | Poised enhancers regulate as many genes as bivalent promoters do |
| H3K36me3 | H3K27ac, H3K4me1 | Active enhancer | Not correlated with H3K27me3 May be a neglected mark of active enhancers |
| H3K4me1 | H3K27me3, H3K4me2 | Poised enhancer | |
| H3K4me2 | H3K4me3 | Active or poised regulatory regions | |
| H3K4me3 | H3K4me2 | Active or poised regulatory regions | |
| H3K9me3 | Repressed region | Negatively correlated with sequence conservation | |
| Cm | Cm either only mildly influences gene regulation, or influences in a way that is independent from histone modifications |
Seven pairs of epi- marks were identified as conserved co-modifications in pluripotent stem cells, namely H2A.Z+H3K4me2/3, H3K27ac+H3K4me1/2, H3K27ac+H3K4me2/3, H3K27me3+H3K4me1/2, H3K27me3+H3K4me2/3, H3K36me3+H3K27ac, and H3K36me3+H3K4me1 (Table 1). The bivalent domain (H3K27me3+H3K4me2/3) was the most conserved epi- mark combination among all 36 pairs of modifications (Figure 2B), lending credence to the approach of using epigenomic comparison for identifying gene regulatory regions. To illustrate how our time-course experiment can reveal the functions of an epi- mark combination, we examined how bivalent domains regulate the early stages of mESC differentiation in a lineage-specific manner. It turned out that not all bivalent domains behave the same. Four subclasses of bivalent domains with different dynamic behaviors were discovered. On Day 6, the majority of sequences either retained both marks (40.7% of the sequences) or lost H3K27me3 while retaining H3K4me2/3 (36.7%). These sequences were preferentially located near transcription start sites (TSS) (Figure S7A, I and III). As expected, the genes whose promoters lost H3K27me3 but retained H3K4me2/3 exhibited higher expression (red line, Figure 6A) than those that kept both marks (purple line), which in turn were higher than those that kept H3K27me3 but lost H3K4me2/3 (green line). These data indicate that there are subclasses of bivalent promoters, which may be activated, still-poised, repressed, or suffer a loss of both marks during mesendoderm formation. We further examined the functions of genes regulated by each subclass of bivalent promoters. The genes with activated promoters are enriched for Gene Ontology (GO) terms of ‘TGFβ and receptor binding’ (p-value < 10−17), ‘mesoderm formation’ (p-value < 10−13), and ‘positive regulation of BMP pathway’ (p-value < 10−9), consistent with the mesendoderm differentiation process. For example, the bivalent promoters of mesendoderm marker gene GSC and mesoderm regulatory gene BMP7 were activated (Figure S6). On the other hand, ‘Neuron fate commitment’ was enriched in both the still-poised (p-value < 10−118) and the repressed (p-value < 10−13) subclasses. These data reveal an intricate coordination among distinct subclasses of bivalent promoters that facilitates lineage-specific differentiation.
Figure 6.
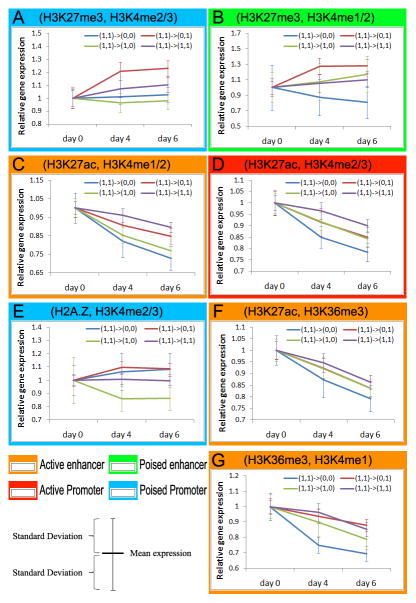
Epi- changes and gene expression changes during differentiation. Each panel represents a set of genomic regions associated with a pair of epi- marks. Each set of regions is categorized into four subclasses, i.e., kept both marks during differentiation (1,1→1,1), lost the first mark (1,1→0,1), lost the second mark (1,1→1,0), and lost both marks (1,1→0,0). For example, the red line (1,1→0,1) in Panel A (H3K27me3, H3K4me2/3) represents sequences with loss of H3K27me3 (the first sign changes from 1 to 0) and retention of H3K4me2/3 (the second sign stays at 1). Relative gene expression values of the nearest genes to the co-marked regions are plotted on the y-axis. (See also figure S5–7 and table S1)
We expected the conserved co-modifications H3K27me3+H3K4me1/2 to mark poised enhancers (Bernstein et al., 2006; Rada-Iglesias et al., 2011). On Day 6 of differentiation, 31.8% of H3K27me3+H3K4me1/2 marked regions removed repression mark H3K27me3 and kept activation mark H3K4me1/2. The genes next to this subset of hypothetically activated enhancers exhibited increased expression on Day 4 and Day 6 (red line, Figure 6B), even greater than the expression of genes associated with other dynamic epigenomic patterns (purple, green, and blue lines). Assuming an enhancer may regulate a target gene within 50k bp, we estimated that about 4,618 genes could be regulated by poised enhancers, on the same order as the number of genes (~5,194) regulated by bivalent promoters. In addition, conserved H3K27ac+H3K4me1/2 and H3K27ac+H3K4me2/3 marked active enhancers and promoters as expected (Figure 6C–D).
The most conserved co-mark of H2A.Z was H3K4me2/3. H2A.Z is a variant of H2A and is required for early mammalian development (Faast et al., 2001). Despite the usual assumption that H2A.Z is associated with active gene expression in multicellular organisms (Weber et al., 2010; Zemach et al., 2010), we found H2A.Z not to be positively associated with gene expression levels in mESCs (Pearson correlation = −0.0066, Figure S7D). This is consistent with the lack of global anti-correlation of H2A.Z and Cm in all three mammals (Figure 2A). Thus, H2A.Z could be a repressor mark in mammals, and H2A.Z+H3K4me2/3 could mark poised promoters, rather than active promoters as is generally assumed. Indeed, (H2A.Z+, H3K4me2/3-) promoters were less active than (H2A.Z-, H3K4me2/3-) promoters, and (H2A.Z+, H3K4me2/3+) promoters were less active than (H2A.Z-, H3K4me2/3+) (Figure S7C). More importantly, during differentiation, the (H2A.Z+, H3K4me2/3+) promoters in ESCs that lost the H2A.Z mark became more active (red line, Figure 6E), and those that lost H3K4me2/3 but kept H2A.Z were down-regulated (green line, Figure 6E). Thus, we propose that H2A.Z is a repressor mark in mammalian pluripotent stem cells and that H2A.Z+H3K4me3 marks a novel class of poised promoters.
H3K36me3, H3K27ac, and H3K4me1/2 exhibited pairwise conservation. While H3K4me1 and H3K27ac were previously associated with enhancers, H3K36me3 has been typically regarded as a mark for actively transcribed regions. The conserved co-localization of H3K36me3 with H3K27ac and H3K4me1/2 tempted us to explore H3K36me3 as an enhancer mark as well. Consistent with this thought, active enhancers make transcripts (eRNA) (Ren, 2010); H3K36me3 could be associated with any transcribed regions including active enhancers. If this hypothesis holds, we would predict that H3K36me3 should avoid overlapping with bivalent (poised) enhancers. Indeed, the epi- mark that has the least co-localization with H3K27me3 is H3K36me3 (Figure 2A). During differentiation, the genes near (not overlapping with) sequences that lost H3K36me3 and H3K27ac (or H3K4me1) exhibited lower expression than those with one mark lost, which in turn exhibited lower expression than those that retained two marks (Figure 6F–G). Thus, we propose that H3K36me3, when co-appearing with H3K27ac or H3K4me1/2, is a mark of active enhancers. In summary, it is powerful to use epi- mark combinations to annotate regulatory sequences and to form hypotheses about their functions. The difficulties of having too many epi- mark combinations and not knowing how to distinguish random vs. functional co-localizations can be overcome by using evolutionary conservation.
DISCUSSION
Many of the functional regions in the human genome have been identified by comparative genomic approaches, based on evolutionary principles. Here we provide a view of the evolutionary properties of the mammalian epigenome and illustrate co-evolutionary relationships among genomes, transcriptomes, and epigenomes. These results show how “comparative epigenomics,” an emerging field that studies evolutionary patterns of epigenomes, can use epigenomic information to functionally annotate genomes.
We compared interspecies epigenomic changes to both genomic and transcriptomic changes. Our data show that the degree of epigenomic conservation is not always correlated with the degree of genomic conservation, but that epigenomic conservation can yield additional information to genomic conservation. More importantly, the conservation levels of epigenomes are indicative of the conservation levels of gene expression, further illustrating that epigenomic comparison can shed new light on regulatory functions of the genome.
Evolution appears to have left traces on mammalian epigenomes, and one identifiable trace is in the combination of epi- marks. Some combinations co-appear (co-localize) in a conserved manner. The conservation of co-localized epi- marks is much stronger than the conservation level of each epi- mark, thus making the combinations computationally identifiable. We used a stem cell differentiation assay to test the regulatory functions of the conserved epi- mark combinations. These tests confirmed the regulatory functions of all (7 out of 7) conserved epi- mark combinations, including novel combinations and novel regulatory roles, suggesting that interspecies comparison can efficiently distinguish functional co-localization of epi- marks from non-functional combinations. This highlights an efficient approach to identify functional epi- mark combinations from a large (combinatorial) number of candidate combinations.
Our studies also reveal the phenomenon that the conservation levels of three epi- marks increase for genomic regions with accelerated sequence changes (related to positive selection (Sabeti et al., 2006)). This is a surprising finding because we expected the epigenomic conservation to be weak at locations where the genomic conservation is also weak. This suggests that epigenomic conservation may be used in conjunction with sequence comparison to identify positively selected regions to reveal functional sequences that make humans unique. Finally, the correlated evolutionary changes of the epigenome, the transcriptome, and TF binding suggest the functional importance of the epigenome in mammalian transcription networks (TNs). This may explain the limited successes in human TN reconstruction using only the information of DNA sequence motifs and gene expression, which were sufficient for reconstruction of yeast TNs (Segal et al., 2003).
EXPERIMENTAL PROCEDURES
Processing sequencing data
ChIP-seq and RNA-seq data were mapped to genome assemblies hg19, mm9 and susScr2 using Bowtie software. MeDIP-seq and MRE-seq were analyzed using methods described in (Maunakea et al., 2010b). Epi- modification intensities of a 200bp region were estimated based on the overlapping ChIP-seq reads in this region. A 200bp long sliding window was used to scan every genome to compute TF binding and epi- modification intensities. All analyses were repeated with 100bp and 300bp sliding windows, as well as a 200bp window with a different conservation threshold (Figure S2A–F).
Global analysis of genomic and epigenomic conservations. (1) Categorizing the human genome by rate of sequence change
The human genome was divided into 15 million 200bp segments. A PhyloP score was computed for every base from 46 vertebrate genomes (Cooper et al., 2005; Pollard et al., 2009), and an average PhyloP score was computed for each 200bp segment. These genomic segments were put into 50 equal-sized sets with increasing average PhyloP scores. The first set with the smallest PhyloP scores are the fastest-changing sequences. The last set with the largest PhyloP scores are the most conserved. (2) Quantifying epigenomic conservation. In the human-mouse comparison, a 200bp human genomic segment was determined to be epigenomicly conserved if the mouse orthologous sequence was marked by the same epi- modification. For each set of genomic segments, the average number of epigenomicly conserved segments was calculated. This average number was then divided by its expectation to obtain a ratio. The expectation was derived from an independence model in which epigenomic conservation was assumed to be independent of sequence conservation. The ratio was used as a quantitative measure of epigenomic conservation for each sequence set. Human-pig and mouse-pig comparisons were done similarly. All analyses were repeated with 100 bp and 300 bp segment sizes, as well as 200bp segments with a different conservation threshold (Figure S2G–J). This conservation threshold was only used to determine the orthologous sequences on which epigenomic data should be retrieved for estimating epigenomic conservation levels. To account for gene conversion, transposon, and multigene families, all “one-to-many” alignable sequences between any two species were removed from this analysis. Whether any sequence has a one-to-one or one-to-many alignment to another species was determined by processing UCSC pairwise alignment chain files.
Conservation level and sequence identity
Sequence conservation level was estimated by Phastcons (Siepel et al., 2005) scores computed by the UCSC Genome Browser group using 46 vertebrate genomes. The human genome were put into strongly, moderately and weakly conserved groups with Phastcons scores in [1, 1], [0.5, 1) and [0.1, 0.5), respectively. Human-mouse and human-pig orthologous regions were identified by the liftOver results stored in the UCSC Genome Browser. Sequence identities of these liftOver regions were estimated based on the proportion of matched nucleotides in local alignments to the total length of the two sequences.
Linear regression
In each species, the expression level of every gene is regressed to the 8 epi- modification intensities on the upstream sequence of the gene. A forward stepwise model selection procedure was used to select the epi- modifications that were predictive. In evolutionary analysis, the interspecies gene expression differences were regressed to epi- modification differences. The same forward selection procedure was applied.
Control for hotspots of aberrant epigenomic reprogramming
All analyses were repeated with and without the genomic regions with epigenetic differences between iPS cells and ESCs (Table S3). The differences in computed statistics were all below 0.1% of the standard deviations of these statistics, making no changes to the order of computed p-values.
In vitro differentiation of mESCs
Undifferentiated E14 mESCs were cultured under feeder-free conditions. Guided differentiation was carried out as previously described (Tada et al., 2005). Briefly, 2×105 cells were seeded on Collagen IV-coated 10cm dishes (BD, 08-774-33) in serum-free medium ESF-B (Itochu Corporation) supplemented with 0.1% BSA, 50mM 2Me, and 10ng/ml Activin A. Medium was changed every day.
Immunofluorescence staining
10,000 to 20,000 cells were seeded on Collagen IV coated 35mm dishes (Ibidi 45074) with the same medium as in guided differentiation. Cells were fixed with 4% Paraformaldehyde. Primary antibodies for GSC (Origene, TA500087) and Sox17 (Millipore 09–038) were incubated at the same time for 2 hours at 37°C. Secondary antibodies, Goat anti-Mouse conj Alexa 568 (Invitrogen A-11031) and Goat Anti Rabbit conj Alexa 488 (Invitrogen A-11034), were each subsequently incubated for 2 hours at 37°C. DNA was stained by Hoechst 33342 (Invitrogen, H3570) for 15 minutes at room temperature. Images were taken by a Zeiss LSM 700 microscope.
Supplementary Material
Acknowledgments
This work was supported by NIH DP2-OD007417, NSF DBI 09-60583, and Sloan Research Fellowship to S.Z. and NIH 5U01ES017154, March of Dimes Foundation, Edward Jr. Mallinckrodt Foundation, DDRCC P30DK52574, and Siteman Cancer Center P50CA134254 to T.W. The authors thank Wei Huang, Darina McDee, Drs. Lisa Stubbs, Tetsuya Tanaka, Phillip Newmark, Rex Gaskins, Gene Robinson, David Clayton, Nigel Goldenfeld, Fei Wang, and Taekjip Ha for useful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: Annotating Chromatin. Annual Review of Genetics. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Chan E, Quon G, Chua G, Babak T, Trochesset M, Zirngibl R, Aubin J, Ratcliffe M, Wilde A, Brudno M, et al. Conservation of core gene expression in vertebrate tissues. Journal of Biology. 2009;8:33. doi: 10.1186/jbiol130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978;274:775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotech. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faast R, Thonglairoam V, Schulz TC, Beall J, Wells JR, Taylor H, Matthaei K, Rathjen PD, Tremethick DJ, Lyons I. Histone variant H2A.Z is required for early mammalian development. Curr Biol. 2001;11:1183–1187. doi: 10.1016/s0960-9822(01)00329-3. [DOI] [PubMed] [Google Scholar]
- Goren A, Ozsolak F, Shoresh N, Ku M, Adli M, Hart C, Gymrek M, Zuk O, Regev A, Milos PM, et al. Chromatin profiling by directly sequencing small quantities of immunoprecipitated DNA. Nat Meth. 2010;7:47–49. doi: 10.1038/nmeth.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC. Comparative genomics. PLoS Biol. 2003;1:E58. doi: 10.1371/journal.pbio.0000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegga AG, Inga A, Menendez D, Aronow BJ, Resnick MA. Functional evolution of the p53 regulatory network through its target response elements. Proc Natl Acad Sci U S A. 2008;105:944–949. doi: 10.1073/pnas.0704694105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Zhao Z. Directionality of point mutation and 5-methylcytosine deamination rates in the chimpanzee genome. BMC Genomics. 2006;7:316. doi: 10.1186/1471-2164-7-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlić R, Chung HR, Lasserre J, Vlahovićek K, Vingron M. Histone modification levels are predictive for gene expression. Proceedings of the National Academy of Sciences. 2010;107:2926–2931. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Kunarso G, Chia N, Jeyakani J, Hwang C, Lu X, Chan Y, Ng H, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nature Genetics. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010a;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D/’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010b;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. Comparative Epigenomic Analysis of Murine and Human Adipogenesis. Cell. 2010;143:156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2009;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, Siepel A, Pedersen JS, Bejerano G, Baertsch R, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2:e168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar S, Visel A, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Morrison H, Fitzpatrick DR, Afzal V, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B. Transcription: Enhancers make non-coding RNA. Nature. 2010;465:173–174. doi: 10.1038/465173a. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- Segal E, Shapira M, Regev A, Pe’er D, Botstein D, Koller D, Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nature Genetics. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada S, Era T, Furusawa C, Sakurai H, Nishikawa S, Kinoshita M, Nakao K, Chiba T. Characterization of mesendoderm: a diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development. 2005;132:4363–4374. doi: 10.1242/dev.02005. [DOI] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CM, Henikoff JG, Henikoff S. H2A.Z nucleosomes enriched over active genes are homotypic. Nat Struct Mol Biol. 2010;17:1500–1507. doi: 10.1038/nsmb.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West FD, Terlouw SL, Kwon DJ, Mumaw JL, Dhara SK, Hasneen K, Dobrinsky JR, Stice SL. Porcine Induced Pluripotent Stem Cells Produce Chimeric Offspring. Stem Cells and Development 2010 doi: 10.1089/scd.2009.0458. -Not available-, ahead of print. [DOI] [PubMed] [Google Scholar]
- Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VL, Fisher EM, Tavare S, Odom DT. Species-specific transcription in mice carrying human chromosome 21. Science. 2008;322:434–438. doi: 10.1126/science.1160930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunaga M, Tada S, Torikai-Nishikawa S, Nakano Y, Okada M, Jakt LM, Nishikawa S, Chiba T, Era T. Induction and monitoring of definitive and visceral endoderm differentiation of mouse ES cells. Nat Biotechnol. 2005;23:1542–1550. doi: 10.1038/nbt1167. [DOI] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.