ABSTRACT
In an attempt to explore the microbial content of functionally critical niches of the mouse gastrointestinal tract, we targeted molecular microbial diagnostics of the crypts that contain the intestinal stem cells, which account for epithelial regeneration. As current evidence indicates, the gut microbiota affects epithelial regeneration; bacteria that are likely to primarily participate in this essential step of the gut, microbiota cross talk, have been identified. We show in this article that only the cecal and colonic crypts harbor resident microbiota in the mouse and that regardless of the line and breeding origin of these mice, this bacterial population is unexpectedly dominated by aerobic genera. Interestingly, this microbiota resembles the restricted microbiota found in the midgut of invertebrates; thus, the presence of our so-called “crypt-specific core microbiota” (CSCM) in the mouse colon potentially reflects a coevolutionary process under selective conditions that can now be addressed. We suggest that CSCM could play both a protective and a homeostatic role within the colon. This article is setting the bases for such studies, particularly by providing a bona fide—and essentially cultivable—crypt microbiota of reference.
IMPORTANCE
Metagenomic typing of the whole-gut luminal microbiome was recently provided, revealing great opportunities for physiological and physiopathological analysis of the host-microbiota interface. On this basis, it appears increasingly important to analyze which niches of the gut exposed to a particular microbiota are of major functional importance, specifically focusing on the crypt, which accounts for permanent epithelial renewal, and to analyze how this microbiota compares to its luminal counterpart in composition and quantity. Crypt-specific core microbiotas may show themselves as important elements regarding crypt protection and homeostasis of its functions.
Introduction
The intestinal crypt contains stem cells (1, 2), and hence it is the site of epithelial restitution. It represents a rare situation in which a differentiating and proliferative epithelium is directly exposed to bacteria, both permanent symbionts and occasional pathogens. One can thus hypothesize that coevolution of mammals with their gut microbiota has led to a balance, protecting the crypt against microbial insults while maintaining a capacity to sense and integrate microbial signals to convert them into signals boosting epithelial regeneration (3, 4). In the small intestine, the crypt is robustly shielded against bacterial colonization by a combination of various effector defenses, including mucins (5) and antimicrobial molecules (6), which are largely produced by Paneth cells. In the colon, mucus is also largely produced, but the extraordinary density of the colonic microbiota (i.e., 1011 CFU/g of feces) compared to that of the small intestine (104 CFU/ml) and the absence of Paneth cells in the crypts may create a more permissive environment for a crypt microbiota to develop. This work explored the possibility that a particular microbiota (i.e., crypt-specific core microbiota [CSCM]) is selected to survive in the crypt environment particularly because of its adaptation to the niche environment. Such a CSCM may play a homeostatic role by acting as a gatekeeper, preventing the proliferation of more aggressive symbiotic microorganisms (i.e., pathobionts) (7) and pathogens, and by providing optimal signaling to the crypt and its environment. Metagenomic analysis of the fecal microbiome (8) alone cannot address this question, which requires a dedicated approach to explore this particular niche. Prior publications have shown the presence of bacteria in the colonic crypts of healthy rodents (i.e., mice and rats) and patients presenting with ulcerative colitis (9–12), although no attempt at identifying these microorganisms was made.
This work aimed to determine the presence of CSCM in the murine gut. Using a panel of histological and molecular biology approaches, we demonstrated that these bacteria colonize murine colonic crypts and more precisely that aerobic bacterial species are localized within this particular niche.
RESULTS
Bacteria colonize murine colonic crypts.
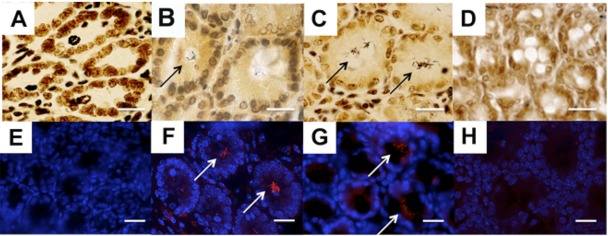
A critical aspect of our study was prevention of any potential contamination of the crypt content by components of the luminal microbiota. Throughout the entire work, all tissue blocks were oriented cautiously, and sectioning was carried out by starting at the peritoneal/muscular side and cutting to the luminal surface. In order to detect bacteria, we first examined the presence of bacterial bodies in the small intestinal and colonic crypts using the highly sensitive Warthin-Starry silver/nitrate-based staining method, which was successfully used to demonstrate the presence of helicoidal bacteria on the gastric surface of people suffering gastritis and peptic ulcers (13). This initial approach showed the consistent absence of bacteria in small intestinal crypts and in the crypts of the distal colon, though bacteria were present in almost 70% of crypts of the cecum and proximal colon (Fig. 1A to 1D). These data were confirmed by fluorescence in situ hybridization (FISH), using the pan-bacterial Eub338 probe (Fig. 1E to H). A positive hybridization signal was never observed using similar technical conditions with the non-Eub338 probe (see Fig. S1A in the supplemental material), nor with hybridization using the pan-bacterial Eub338 probe on colonic or luminal intestinal sections from germfree mice (see Fig. S1B).
FIG 1 .
Bacteria reside in the murine proximal colonic crypts. Warthin-Starry staining (A to D) or FISH (E to H) with the pan-bacterial probe Eub338 at different levels of the intestine of C57BL/6 mice is shown: small intestine (A and E), cecum (B and F), proximal colon (C and G), or distal colon (D and H). Black and white arrows indicate the presence of bacteria. Scale bars, 10 µM.
A CSCM inhabits the colonic crypt.
Encouraged by the results of the histological data, we proceeded with molecular identification of the corresponding bacteria. We combined laser capture microdissection (LCM), DNA amplification with primers flanking the V5-V6 hypervariable regions of 16S rRNA encoding sequences, and 454 sequencing. This combination of techniques was used successfully in numerous studies (14–18). Moreover, we focused our attention on the most representative genera found, which were systematically validated by FISH using genus-specific probes. This combination allowed the confirmation of the existence of a restricted set of bacterial genera associated with murine colonic crypts. A pilot pyrosequencing experiment using a pool of microdissected crypts isolated from the proximal colon of 3 C57BL/6 mice, generating 450,975 reads, indicated that Acinetobacter was the predominant resident bacterial genus (see Fig. S2 in the supplemental material).
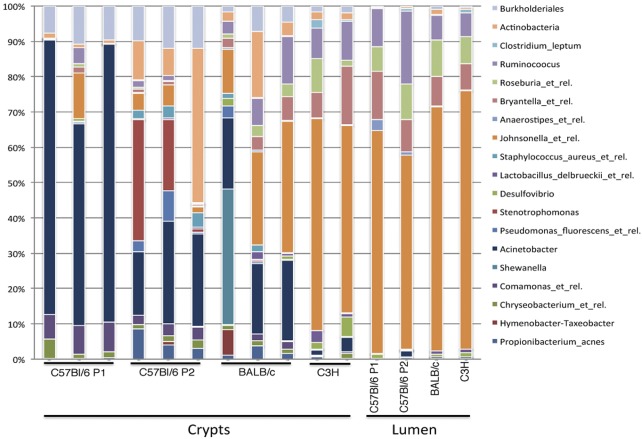
We then performed a multiplexed bar code pyrosequencing approach using various strains of mice from independent providers in order to confirm whether or not the Acinetobacter genus was common to all murine strains studied. We compared the relative abundances of major bacterial groups, as defined by Silva database taxonomy, present in the crypt and the luminal microbiota. Differences between microbiota were assessed using the principal coordinate analysis (PCoA) of Bray-Curtis distances, allowing the samples to be discriminated into two clusters: the crypt and the lumen, with one exception for the crypt samples of C3H/HeN mice (Fig. 2). Interestingly, the samples corresponding to the crypt of the C57BL/6 mice obtained from the two providers showed very close sequence similarity. To perform these analyses, we clustered sequences into species-level operational taxonomic units (OTUs) of 97% sequence similarity by the furthest-neighbor method, using the Mothur software program. The sequences were grouped into 45,926 OTUs belonging to various phylotypes, revealing subtle variations between samples. This experiment generated 906,262 reads, leading to 419,179 bacterial gene sequences from 11 crypt samples and 4 lumenal samples from colons (see Table S1 in the supplemental material). Fourteen bacterial phyla were detected, but most sequences could be assigned to five phyla: Firmicutes (73%), Beta- and Gammaproteobacteria (16%), Actinobacteria (3.5%), and Bacteroidetes (1.7%). Figure 3 illustrates the phylogenetic abundances of the most represented OTUs. Whereas members of the Bacteroidetes were rather poorly represented within both crypt and luminal samples, the Firmicutes represented the majority of luminal sequences (95.5%). Among members of the Firmicutes, the Clostridiales were associated with more than 97% of the sequences, and Johnsonella (or the Lachnospiraceae) was the most abundant bacterial group, with 84.3% of the sequences. With the exception of the crypts of C3H/HeN mice, where members of the Firmicutes reached 74% to 88% of the sequences, the Proteobacteria represented the most abundant sequences found in crypts (47.6%, versus 2.7% for the lumen). The Betaproteobacteria/Gammaproteobacteria subphyla comprised 43.8% of sequences, with a predominance of gammaproteobacteria, 32.5% in crypts versus 1.2% in luminal samples. The major bacterial groups identified were the Moraxellaceae (23.7%), with 23% of Acinetobacter spp. sequences in crypts versus 1.6% in the lumen of C57BL/6 and BALB/c mice. Shewanella spp. were also found in abundance in a single crypt sample of the BALB/c mice. This strict aerobic bacterium may also be selected in this particular ecological niche, but we cannot exclude a possible luminal contamination during LCM. The most abundant taxa of the beta branch, also observed in crypt samples, were the Burkholderiales (Comamonas, 3.2%) and the Xanthomonadales (Stenotrophomonas, 4.7%; for mice from Charles River Breeding facilities), two other groups of strictly aerobic bacteria.
FIG 2 .
Unweighted Unifrac Bray-Curtis principal coordinate analysis shows dissimilarity in samples. Axis 1, percent variation explained (24.2%); axis 2, percent variation explained (17.17%). Squares and triangles represent crypt and luminal samples, respectively. Each murine strain is represented by a color code (C57BL/6 from provider 1, purple; C57BL/6 from provider 2, blue; BALB/c, green; C3H/HeN, orange).
FIG 3 .
Proportional abundances of the 20 most-represented OTUs in mice belonging to different strains at the crypt and luminal levels. These family-level phylogenetic relative contributions are based on 16S rRNA gene frequencies.
OTUs from Acinetobacter spp. were shared among all crypts, representing a possible common bacterial phylogroup with possible quantitative variations according to the mouse line studied (85% of the sequences in C57BL/6 mice and only 1.8% or 4.7% in C3H/HeN mice). However, in all cases, levels of Acinetobacter spp. in crypts were significantly higher than those observed in luminal samples (Fig. 3).
Despite individual and murine strain variations in crypt DNA composition, the presence of sequences belonging to the Acinetobacter species in all samples confirmed our pilot study. Results in previous metagenomic studies have suggested that the presence of 16S rRNA gene sequences could reflect residual bacterial DNA in autoclaved food and also possible annealing of bacterial universal primers to corn mitochondrial genes or to rice and wheat chloroplast rRNA genes (19, 20). In order to address this possibility, we performed quantitative reverse transcription-PCR (qRT-PCR) on DNA extracted from autoclaved chow, showing that bacterial DNA could be amplified, especially from the Firmicutes, but not from Acinetobacter spp.; in the latter case, the threshold cycle (CT) values obtained were similar to those obtained using water as a template (see Fig. S3 in the supplemental material).
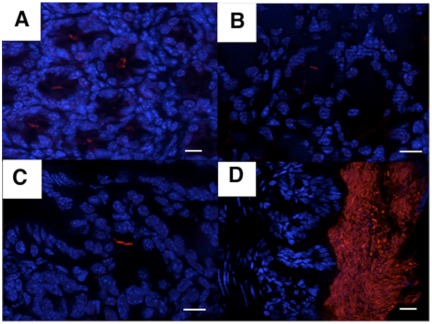
As stressed by Amann and Fuchs, “metagenomics cannot substitute for the information that is gained by visualizing the identity and activity of single microbial cells in situ” (21). Thus, using FISH with probes specific for bacterial families and/or genera (listed in Table S2 in the supplemental material), the presence of Acinetobacter spp. was unequivocally confirmed in crypt samples from different murine strains (more than 10% of the crypts were colonized by Acinetobacter, as visualized by FISH) (Fig. 4A to C), whereas members of the Firmicutes were localized in the lumen (Fig. 4D). These FISH experiments were also performed with fixed tissues, using the Carnoy reagent, a technique which guarantees maximal preservation of the mucus layer. Both methods provided identical results, indicating that the native spatial relationships between the bacteria and the host were maintained in the protocols used in our study and that we did not lose significant bacterial populations associated with the mucus in the colonic glands.
FIG 4 .
Acinetobacter and the Firmicutes are localized mainly in the crypt and lumen of the colon, respectively. Hybridization of an Alexa555-labeled Acinetobacter-specific probe on colonic sections from C57BL/6 (A), BALB/c (B), or C3H/HeN (C) or of an Alexa555-labeled Firmicutes-specific probe on colonic sections from C57BL/6 (D) is shown.
Ability of Acinetobacter to colonize colonic crypts.
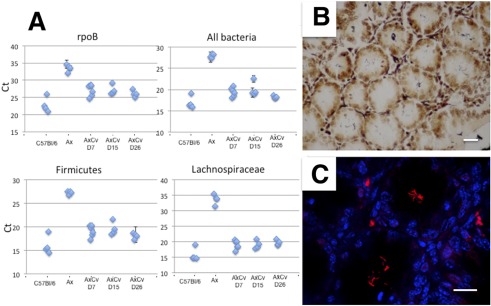
In order to confirm the tropism of Acinetobacter, germfree, adult, BALB/c female mice were colonized using a conventional microbiota originating from littermates, and the appearance of bacteria in the luminal content was monitored in feces at days 7, 15, and 26 postcolonization. As early as day 7 of colonization, bacterial DNA could be detected by qRT-PCR in the feces of colonized mice at a level equivalent to that for conventional mice (Fig. 5A). Firmicutes and Lachnospiraceae DNAs were amplified, whereas Acinetobacter DNA was never amplified from these fecal samples. Following FISH using an Acinetobacter-specific probe, we were unable to observe significant luminal staining at any time point. On the other hand, after 26 days of colonization, bacteria were also observed by silver staining in colonic crypts (Fig. 5B), and the presence of Acinetobacter spp. was clearly demonstrated by FISH (Fig. 5C), thus establishing its strong crypt tropism.
FIG 5 .
Acinetobacter colonizes the colonic crypt. (A) qRT-PCR amplification using phylum- and family-specific primers with DNA extracted from the feces of SPF C57BL/6 mice or from germfree BALB/c mice before and after 7, 15, and 26 days of colonization with conventional microbiota. (B and C) Warthin-Starry (B) or FISH using an Alexa555-labeled Acinetobacter-specific probe (C) on colonic crypt sections after 26 days of colonization of germfree BALB/c mice with a conventional microbiota. Scale bars, 10 µM.
DISCUSSION
Association of a limited number of specific bacteria in particular niches is not uncommon. Alcaligenes is an almost exclusive inhabitant of murine Peyer’s patches, and the segmented filamentous bacterium (SFB) dominates at the epithelial surface of these gut-associated lymphoid structures (22). Only 0.16% of the sequences of the crypt microbiota were associated with Alcaligenes in our study. FISH analysis has shown that 16% of proximal colonic crypts harbor bacteria (Clostridiales, Alphaproteobacteria, and Lactobacillaceae) (10). Helicobacter hepaticus was also shown to colonize the cecum and colonic crypts in mice (23); however, H. hepaticus was never detected in crypts from the murine strains used in this study by either qRT-PCR or FISH. Regarding the results obtained in the present study on luminal bacterial composition, our observations are not in total agreement with data from the literature on the microbiota found in the murine intestine. Indeed, it was described that the Firmicutes represent almost 50% of the gut microbiota, and the Actinobacteria and the Bacteroidetes represent around 20% each (19, 24). However, in the present study making use of LCM, we concentrated on a luminal area closely associated with the epithelium, and therefore, we addressed the composition of bacterial populations that are not necessarily representative of those found in intestinal lavage specimens or in metagenomic studies of fecal material. Our data do coincide with findings of a very recent study showing that the microbiota in close contact with the epithelium, named the interfold region, are composed mainly of members of the Lachnospiraceae, in contrast to the luminal area, named the digesta, where the Actinobacteria and Bacteroidetes were present (25).
The present study highlights the dominant presence of the aerobic bacterial genus Acinetobacter in the colonic crypt, a finding in accordance with our recent work showing the presence of oxygen at the apical surface of intestinal epithelial cells (26). This represents a possible mechanism of exclusion of strictly anaerobic, extremely oxygen-sensitive microorganisms.
Interestingly, Acinetobacter has been found associated with the gut microbiota, even among invertebrates, which have a very restricted microbiota in their midgut compared to that of mammals. Acinetobacter sequences were found in Drosophila melanogaster, mosquito, and tsetse fly midguts (27–30). It was even shown that Acinetobacter could be beneficial for fly larva development (31). The presence of this particular gammaproteobacterium throughout many different species, from invertebrates to vertebrates, may have emerged during the selective pressure of a coevolutionary history whose rules have yet to be deciphered. Acinetobacter, alone or in combination with the other species characterized here in smaller amounts (such as Comamonas and Stenotrophomonas), constitutes a CSCM that may participate in the exclusion of pathobionts and in the maintenance of local homeostasis that is essential for proper epithelial regeneration. We are currently analyzing, as a model for the whole CSCM, the degree to which Acinetobacter pathogen-associated molecular patterns (PAMPs) and metabolic by-products may account for crypt homeostasis. Acinetobacter spp. are widely present in nature and are generally considered nonpathogenic (32). However, several species are prone to acquiring resistance to multiple antibiotics (33), and those emerging from an often-uncharacterized source can therefore cause severe nosocomial infections in hospitalized patients (34).
Next steps include confirmation of the existence of a similar CSCM in the human colon and assessment of its loss to a possible profit for other microbial populations in the context of inflammatory bowel diseases (IBDs) and colonic cancer. This would raise a novel concept of focal eubiosis/dysbiosis, the reality of which needs now to be experimentally addressed in vitro and in vivo.
MATERIALS AND METHODS
Mice.
Six- to eight-week-old C57BL/6 mice from Elevage Janvier (provider 1) and Charles River (provider 2) and C3H/HeN and BALB/c mice from Elevage Janvier were used in this study. BALB/c germfree mice were from the Pasteur Institute animal facilities. All mice were kept under specific-pathogen-free (SPF) conditions, and all animal experiments were approved by the committee on animal experimentation of the Institut Pasteur and by the French Ministry of Agriculture (agreement no. 75-305).
Colonization of germfree mice with conventional microbiota.
Female germfree BALB/c mice were colonized with a conventional microbiota by housing them in cages containing feces of SPF mice. DNA from murine feces was isolated using the Qiagen DNA stool isolation kit according to the manufacturer’s instructions. Bacterial colonization was monitored by qRT-PCR using bacterial DNA of colonized germfree mice at days 7, 15, and 26. Intestinal tissues from 4 mice were removed for histological staining at days 0, 15, and 26.
Histological processing and staining of tissue samples.
Intestinal tissues were embedded in OCT compound 4583 (Sakura), frozen in isopentane, cooled with dry ice, and stored at −80°C. Frozen blocks were cut with a thickness of 8 µm using a CM 3050S cryostat (Leica), and sections were collected on Superfrost plus slides (VWR) and stored at −20°C. Warthin-Starry staining was performed as previously described (35, 36). Slides were examined under an Eclipse E800 microscope (Nikon) equipped with a charge-coupled-device (CDD) camera, and images were processed with the Eclipse Net software program.
Fluorescence in situ hybridization (FISH).
Frozen sections were rehydrated in phosphate-buffered saline (PBS), covered with a solution of lysozyme at 10 mg/ml in PBS during 10 min at 37°C, and washed twice with PBS. After 30 min of incubation in hybridization buffer (20 mM Tris-HCl [pH 8], 0.9 M NaCl, 0.01% SDS, 30% formamide), slides were incubated overnight in hybridization buffer containing 20 nM of the fluorescent probes at 42°C. After washing twice in 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), slides were covered for 10 s with 4′,6-diamidino-2-phenylindole (DAPI) (0.125 µg/ml in PBS), washed in PBS, and mounted in ProLong gold antifade reagent (Invitrogen). The 16S rRNA-targeted oligonucleotide probes used in this study are listed in Table S2 in the supplemental material. The probes were covalently linked with fluorescein isothiocyanate (FITC) or Alexa 555 at their 5′ ends. Slides were examined under an Olympus BX50 microscope equipped with a CCD camera, and images were processed using the MetaVue software program or under a Widefield ApoTome inverted microscope (Zeiss) using the Axovision software program.
Laser microdissection.
Frozen sections were thawed and briefly stained with histogen (MDS Analytical Technologies), containing RnaseOut recombinant RNase inhibitor, washed in RNase-free water supplemented with ProtectRNA (Sigma-Aldrich), and dehydrated in ethanol (once in 70% for 30 s, twice in 95% for 1 min, and twice in 100% for 2 min) and in xylene (two baths for 5 min) before being air-dried. Slides were then transferred into a Veritas laser capture microdissector (MDS Analytical Technologies), microdissected, and captured on Capture Macro LCM caps (MDS Analytical Technologies). DNA was extracted using the PicoPure DNA extraction kit (MDS Analytical Technologies) after an incubation for 10 min at room temperature with lysozyme (10 mg/ml in PBS; Sigma-Aldrich). DNAs were stored at −20°C.
Quantitative RT-PCR.
Diluted DNAs extracted from the LCM crypt or luminal part of the proximal colon were used as a template for quantitative RT-PCR using specific primers (400 nM) for phyla and/or bacterial families in a 15-µl final volume containing Sybr green master mix (Roche) using the Applied 7900 thermocycler. Cycling conditions were as follows: initial denaturation step at 95°C for 10 min, with 40 cycles of denaturation at 95°C for 45 s, annealing at 50°C for 45 s, and elongation at 72°C for 60 s for amplification of rpoB DNA, or initial denaturation step at 95°C for 10 min, with 40 cycles of denaturation at 95°C for 45 s and annealing at 60°C for 45 s for all other primers. The list of all primers used in this study is provided in Table S2 in the supplemental material.
Pyrosequencing. (i) Pilot experiment.
DNAs extracted from microdissected crypts of proximal colon from 3 C57BL/6 mice were amplified independently using the primers 786F (5′ GATTAGATACCCTGGTAG 3′) and 1100R (5′ AGGGTTGCGCTCGTTG 3′) to obtain a 334-bp amplicon of the 16S rRNA gene encompassing the variable regions V5 and V6. PCR experiments were carried out in a 50-µl final volume with 2 µl of DNA as a template, using a 9700 thermocycler GeneAmp PCR system (Applied Biosystems). Cycling conditions were as follows: initial denaturation step at 94°C for 6 min, 37 cycles of denaturation at 94°C for 45 s, annealing at 57°C for 45 s, and elongation at 72°C for 75 s, and a final extension step at 72°C for 7 min. To avoid PCR biases, 4 PCR products of each sample were pooled, run, and extracted from a 1.3% agarose gel using the QIAquick gel extraction kit (Qiagen). After quantification, all extracted amplicons were equimolarly pooled. Pyrosequencing was performed using a Roche FLX Titanium genome sequencer at Beckman Genomics, producing a total of 450,975 reads.
(ii) Bar-coded pyrosequencing.
In order to carry out a multiplex sequencing approach, a bar code sequence (molecular identifier [MID]) was added at the 5′ ends of all primers during oligonucleotide synthesis. This bar code is a sequence of 10 additional nucleotides specific for each sample. The titanium sequencing adaptors A and B were added just before sequencing. PCR experiments were performed as described above.
For each mouse line, 2 or 3 samples of crypts and one sample of luminal content were amplified after LCM experiments and DNA extractions, and PCR products were purified as above. After a quantification step, all purified amplicons were equimolarly pooled. Multiplex pyrosequencing was performed using a Roche FLX Titanium genome sequencer at Beckman Genomics, producing a total of 906,262 reads.
(iii) Data analysis.
Sequences were processed and analyzed using the Mothur software program, V1.16.0 (37). Pyrosequencing reads of length >200 bp containing a correct primer sequence, showing an average quality above 25 without ambiguous base, and containing no more than 8 bp homopolymer, were extracted and cured from primer/bar code sequence. For the multiplexing, 63.6% of sequences were assigned to samples by examining the 10-nucleotide (nt) bar code. Identical sequences were then binned, and the 507,003 representative unique sequences were kept for the analysis. Trimmed fasta sequences (spanning variable regions V5 and V6) were aligned using the Silva-derived reference alignment. Sequences that did not align over the same span of nucleotide positions were removed. In order to reduce the number of sequences to a computationally manageable level, we filtered out all redundant sequences and merged rare sequences which differed from larger ones by one mismatch (using the precluster command in Mothur), ending with 189,967 sequences before clustering of phylotypes.
The presence of chimeras was checked using a fast method based on the ChimeraSlayer implementation (http://www.mothur.org/wiki/Chimera.slayer) (38). Chimera analysis showed that 7% of the 189,967 unique sequences were potential chimeric sequences and were removed before clustering and statistical analysis of the 176,508 unique sequences. Rarefaction curves and Simpson diversity indexes were calculated.
Differences between the samples were analyzed by comparing the community structures (Bray-Curtis index) in principal coordinate analysis (PCoA) and by comparing the community diversity using a phylogenic approach with the weighted and unweighted versions of Unifrac (39). When PCoA using Bray, Curtis, Jaccard, Sorensen, or Yue and Clayton distances were compared, the PCoA graphs obtained showed similar results, i.e., lumen samples were distantly related to crypt samples.
SUPPLEMENTAL MATERIAL
(A) FISH using the “no-bacteria” probe on different sections of the intestine of C57BL/6 mice. (B) FISH with the pan-bacterial probe Eub338 at the proximal colon and the lumen of germfree C3H/HeN mice. Download Figure S1, PDF file, 1.5 MB.
Pie chart showing the percentages of phylotypes (genus level) observed in three murine colonic crypt samples (pilot experiment). Download Figure S2, PDF file, 0.1 MB.
qRT-PCR experiments with DNA extracted from the autoclaved chow using phylum- and family-specific primers. Download Figure S3, PDF file, 0.1 MB.
Number of reads after multiplex pyrosequencing of murine crypt and luminal samples.
Sequences of primers and probes used in this study.
Relative contributions of OTUs at 97% similarity in the different samples resulting from bar-coded pyrosequencing.
ACKNOWLEDGMENTS
This work was supported in part by ERC program HOMEOEPITH (no. 232798) to P.J.S. and by grants from Danone-Research and Yakult.
We thank B. Regnault for her expertise in FISH experiments, M. Huerre, G. Eberl, P. Glaser, and I. Mozer for helpful discussions, B. Néron, N. Joly, and C. Maufrais for the installation of Bio-informatics tools (ARB and Mothur) on the central server of Institut Pasteur, M. Bérard, E. Maranghi, T. Angélique, and J. Perez for the availability of germfree mice in the Institut Pasteur animal facilities, and E. Arena and B. Sperandio for critical reading of the manuscript.
T.P. and P.J.S. designed research; T.P. and C.M. performed research; T.P., C.D., L.F., C.C., G.G., and P.J.S. analyzed data; and T.P. and P.J.S. wrote the article.
Footnotes
Citation Pédron T, et al. 2012. A crypt-specific core microbiota resides in the mouse colon. mBio 3(3):e00116-12. doi:10.1128/mBio.00116-12.
REFERENCES
- 1. Sato T, et al. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459:262–265 [DOI] [PubMed] [Google Scholar]
- 2. Fuchs E. 2009. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell 137:811–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. 2004. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118:229–241 [DOI] [PubMed] [Google Scholar]
- 4. Davis CP, Mulcahy D, Takeuchi A, Savage DC. 1972. Location and description of spiral-shaped microorganisms in the normal rat cecum. Infect. Immun. 6:184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McGuckin MA, Lindén SK, Sutton P, Florin TH. 2011. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 9:265–278 [DOI] [PubMed] [Google Scholar]
- 6. Bevins CL, Salzman NH. 2011. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9:356–368 [DOI] [PubMed] [Google Scholar]
- 7. Chow J, Mazmanian SK. 2010. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe 7:265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qin J, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sokol H, et al. 2010. Crypt abscess-associated microbiota in inflammatory bowel disease and acute self-limited colitis. World J. Gastroenterol. 16:583–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. 2005. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 43:3380–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamamoto K, et al. 2009. Histoplanimetrical study on the spatial relationship of distribution of indigenous bacteria with mucosal lymphatic follicles in alimentary tract of rat. J. Vet. Med. Sci. 71:621–630 [DOI] [PubMed] [Google Scholar]
- 12. Macfarlane S, Furrie E, Cummings JH, Macfarlane GT. 2004. Chemotaxonomic analysis of bacterial populations colonizing the rectal mucosa in patients with ulcerative colitis. Clin. Infect. Dis. 38:1690–1699 [DOI] [PubMed] [Google Scholar]
- 13. Marshall BJ, Warren JR. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 323:1311–1315 [DOI] [PubMed] [Google Scholar]
- 14. Mølbak L, Klitgaard K, Jensen TK, Fossi M, Boye M. 2006. Identification of a novel, invasive, not-yet-cultivated Treponema sp. in the large intestine of pigs by PCR amplification of the 16S rRNA gene. J. Clin. Microbiol. 44:4537–4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Hertogh G, et al. 2006. Validation of 16S rDNA sequencing in microdissected bowel biopsies from Crohn’s disease patients to assess bacterial flora diversity. J. Pathol. 209:532–539 [DOI] [PubMed] [Google Scholar]
- 16. Baker GC, Smith JJ, Cowan DA. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541–555 [DOI] [PubMed] [Google Scholar]
- 17. Veiga P, et al. 2010. Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc. Natl. Acad. Sci. U. S. A. 107:18132–18137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stecher B, et al. 2010. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 6:e1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hill DA, et al. 2010. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 3:148–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scupham AJ, et al. 2006. Abundant and diverse fungal microbiota in the murine intestine. Appl. Environ. Microbiol. 72:793–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Amann R, Fuchs BM. 2008. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 6:339–348 [DOI] [PubMed] [Google Scholar]
- 22. Obata T, et al. 2010. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proc. Natl. Acad. Sci. U. S. A. 107:7419–7424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Petnicki-Ocwieja T, et al. 2009. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc. Natl. Acad. Sci. U. S. A. 106:15813–15818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murphy EF, et al. 2010. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut 59:1635–1642 [DOI] [PubMed] [Google Scholar]
- 25. Nava GM, Friedrichsen HJ, Stappenbeck TS. 2011. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 5:627–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marteyn B, et al. 2010. Modulation of Shigella virulence in response to available oxygen in vivo. Nature 465:355–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Corby-Harris V, et al. 2007. Geographical distribution and diversity of bacteria associated with natural populations of Drosophila melanogaster. Appl. Environ. Microbiol. 73:3470–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pidiyar VJ, Jangid K, Patole MS, Shouche YS. 2004. Studies on cultured and uncultured microbiota of wild Culex quinquefasciatus mosquito midgut based on 16s ribosomal RNA gene analysis. Am. J. Trop. Med. Hyg. 70:597–603 [PubMed] [Google Scholar]
- 29. Zouache K, et al. 2011. Bacterial diversity of field-caught mosquitoes, aedes albopictus and Aedes aegypti, from different geographic regions of Madagascar. FEMS Microbiol. Ecol. 75:377–389 [DOI] [PubMed] [Google Scholar]
- 30. Geiger A, et al. 2009. First isolation of Enterobacter, Enterococcus, and Acinetobacter spp. as inhabitants of the tsetse fly (Glossina palpalis palpalis) midgut. Infect. Genet. Evol. 9:1364–1370 [DOI] [PubMed] [Google Scholar]
- 31. Lysyk TJ, et al. 1999. Rearing stable fly larvae (Diptera: Muscidae) on an egg yolk medium. J. Med. Entomol. 36:382–388 [DOI] [PubMed] [Google Scholar]
- 32. Fournier PE, Richet H. 2006. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin. Infect. Dis. 42:692–699 [DOI] [PubMed] [Google Scholar]
- 33. Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939–951 [DOI] [PubMed] [Google Scholar]
- 34. Castle M, Tenney JH, Weinstein MP, Eickhoff TC. 1978. Outbreak of a multiply resistant Acinetobacter in a surgical intensive care unit: epidemiology and control. Heart Lung 7:641–644 [PubMed] [Google Scholar]
- 35. Warthin AS, Chronister AC. 1920. A more rapid and improved method of demonstrating Spirochetes in tissues (Warthin and Starry’s cover-glass method). Am. J. Syph. 4:97–103 [Google Scholar]
- 36. Field AS, Marriott DJ, Hing MC. 1993. The Warthin-Starry stain in the diagnosis of small intestinal microsporidiosis in HIV-infected patients. Folia Parasitol. 40:261–266 [PubMed] [Google Scholar]
- 37. Schloss PD, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haas BJ, et al. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21:494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) FISH using the “no-bacteria” probe on different sections of the intestine of C57BL/6 mice. (B) FISH with the pan-bacterial probe Eub338 at the proximal colon and the lumen of germfree C3H/HeN mice. Download Figure S1, PDF file, 1.5 MB.
Pie chart showing the percentages of phylotypes (genus level) observed in three murine colonic crypt samples (pilot experiment). Download Figure S2, PDF file, 0.1 MB.
qRT-PCR experiments with DNA extracted from the autoclaved chow using phylum- and family-specific primers. Download Figure S3, PDF file, 0.1 MB.
Number of reads after multiplex pyrosequencing of murine crypt and luminal samples.
Sequences of primers and probes used in this study.
Relative contributions of OTUs at 97% similarity in the different samples resulting from bar-coded pyrosequencing.