ABSTRACT
Bacterial pathogens must be able to both recognize suitable niches within the host for colonization and successfully compete with commensal flora for nutrients in order to establish infection. Ethanolamine (EA) is a major component of mammalian and bacterial membranes and is used by pathogens as a carbon and/or nitrogen source in the gastrointestinal tract. The deadly human pathogen enterohemorrhagic Escherichia coli O157:H7 (EHEC) uses EA in the intestine as a nitrogen source as a competitive advantage for colonization over the microbial flora. Here we show that EA is not only important for nitrogen metabolism but that it is also used as a signaling molecule in cell-to-cell signaling to activate virulence gene expression in EHEC. EA in concentrations that cannot promote growth as a nitrogen source can activate expression of EHEC’s repertoire of virulence genes. The EutR transcription factor, known to be the receptor of EA, is only partially responsible for this regulation, suggesting that yet another EA receptor exists. This important link of EA with metabolism, cell-to-cell signaling, and pathogenesis, highlights the fact that a fundamental means of communication within microbial communities relies on energy production and processing of metabolites. Here we show for the first time that bacterial pathogens not only exploit EA as a metabolite but also coopt EA as a signaling molecule to recognize the gastrointestinal environment and promote virulence expression.
IMPORTANCE
In order to successfully cause disease, a pathogen must be able to sense a host environment and modulate expression of its virulence genes as well as compete with the indigenous microbiota for nutrients. Ethanolamine (EA) is present in the large intestine due to the turnover of intestinal cells. Here, we show that the human pathogen Escherichia coli O157:H7, which causes bloody diarrhea and hemolytic-uremic syndrome, regulates virulence gene expression through EA metabolism and by responding to EA as a signal. These findings provide the first information directly linking EA with bacterial pathogenesis.
Introduction
To establish infection, bacterial pathogens must be able to sense the host environment, express genes essential for colonization and virulence, and compete with commensal flora for nutrients. Ethanolamine (EA) is a major component of both bacterial and mammalian cell membranes (1–3), and the rapid turnover and exfoliation of intestinal cells releases EA into the intestine (4, 5). The ability to use ethanolamine as a noncompetitive source of carbon and/or nitrogen may aid a pathogen in colonizing the host (6); however, it is currently not known whether EA directly influences a pathogen’s ability to recognize the host environment and/or coordinate expression of virulence genes.
Enterohemorrhagic Escherichia coli O157:H7 (EHEC) is a food-borne pathogen that causes bloody diarrhea and hemolytic-uremic syndrome (HUS) worldwide (7). In the intestines, EHEC causes the formation of attaching and effacing (AE) lesions that are characterized by the destruction of microvilli and rearrangement of the cytoskeleton to form a pedestal-like structure that cups the bacteria individually (8). Most of the genes involved in AE lesion formation are carried on the locus of enterocyte effacement (LEE) pathogenicity island (9). Additionally, EHEC produces Shiga toxin (Stx). Stx is responsible for the severe complications associated with diseases caused by EHEC, including hemorrhagic colitis, and HUS, which may be fatal (7). EHEC virulence gene regulation is complex and involves interkingdom signaling. EHEC carries genes that encode the sensor kinases QseC and QseE that recognize diverse environmental signals, including the host hormones epinephrine (EPI) and norepinephrine (NE), to coordinate expression of its virulence genes (10, 11). The transcriptional regulator QseA is also part of EHEC’s regulatory cascade through its direct interaction and activation of ler, the master regulator of the LEE genes (12) and of putative virulence genes (13). Although previous studies have suggested a role for EA in bacterial pathogenesis (reviewed in reference 14), we demonstrate for the first time a direct role for EA in virulence gene regulation of a bacterial pathogen. Furthermore, we show that EA stimulates virulence gene expression not only due to metabolism, but also through signaling when EA is present at concentrations that are insufficient to support growth.
RESULTS AND DISCUSSION
EA regulates virulence gene expression.
The ability to harvest nutrients is fundamental for the successful colonization of the gastrointestinal (GI) tract by a bacterium. Pathogens have to compete for nutrients with the members of the indigenous microbiota, which far outnumber them. Hence, one advantage for pathogens is to proficiently use nutrients to gain a competitive advantage over the microbiota. EA is an abundant source of carbon and nitrogen in the intestine and is used as a nitrogen source by EHEC in the GI tracts of mammals (15). Genes that encode components involved in EA metabolism have been identified in many members of the family Enterobacteriaceae (16), and organization of the eut (ethanolamine utilization) operon is similar in EHEC and Salmonella (17–19). The eut operon is comprised of 17 genes that code for components involved in transport and catabolism of EA, as well as homologs of carboxysome shell proteins (18, 20–24). The eut operon also contains genes that encode EutR, which activates expression of the entire eut (ethanolamine utilization) operon in response to EA and vitamin B12 (20, 22).
We first investigated the ability of EHEC strain 86-24 to grow on a modified minimal M9 medium (25) with EA (EA medium) as the sole carbon or nitrogen source. Our initial studies were conducted aerobically with 30 mM EA and 150 nM vitamin B12, conditions frequently used to study the EA metabolism in E. coli and Salmonella (20, 23, 24, 26–29). EHEC grew using EA as a nitrogen source, although its growth was slower than growth with NH4 or glutamine. EHEC strain 86-24 did not grow when EA served as the sole carbon source under aerobic or anaerobic conditions (data not shown). These results are consistent with EA metabolism in EHEC strain EDL933 for which it was previously reported that EHEC can use EA only as a nitrogen source and not as a carbon source (15).
The utilization of EA as a nitrogen source during growth in the GI tract confers a competitive advantage to EHEC (15). However, it has never been investigated whether EA could also modulate virulence gene expression in bacterial pathogens. Hence, we compared virulence gene expression in EHEC cells grown with NH4 or EA. We first examined expression of the qseC and qseE genes that encode global regulators in EHEC (10, 11). Transcription of these genes was significantly increased in EA medium at early and mid-log growth phases compared to EHEC grown with NH4, and no significant differences in gene expression were measured at late log growth phase (Fig. 1A). The sensor kinases QseC and QseE play critical roles in EHEC gene expression by integrating environmental cues present in the human GI tract and activating virulence genes. In response to EPI, phosphate, or sulfate, QseE autophosphorylates and subsequently phosphorylates its cognate response regulator (RR) QseF that in turn promotes AE lesion formation and Stx production (11, 30). QseC senses EPI, NE, and the bacterial signaling molecule autoinducer-3 (AI-3) to increase phosphorylation (10, 31). QseC then phosphorylates its cognate RR QseB that activates flagella (32). QseC can also phosphorylate the response regulators KdpE, which activates ler, and QseF (33). These data suggest that EA metabolism induces expression of genes necessary for EHEC to integrate environmental signals and consequently activate virulence genes.
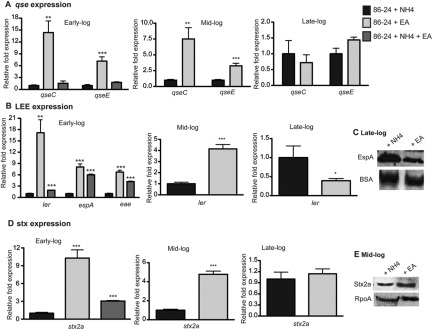
FIG 1 .
EA increases expression of EHEC virulence genes. (A) qRT-PCR of qseC and qseE of WT EHEC strain 86-24 grown in minimal medium containing glucose (glucose minimal medium) and either NH4, EA, or NH4 plus EA to early, mid-, and late log growth phases. (B) qRT-PCR of ler of WT EHEC grown in glucose minimal medium containing either NH4, EA, or NH4 plus EA to early, mid-, and late log phases. (C) Western blots of secreted proteins from WT EHEC grown to late log in glucose minimal medium containing either NH4 or EA probed with antisera against EspA from WT EHEC grown to late log. BSA, bovine serum albumin. (D) qRT-PCR of stx2a of WT EHEC grown in glucose minimal medium containing either NH4, EA, or NH4 plus EA to early, mid-, and late log. (E) Western blots of whole-cell lysates of WT EHEC grown in glucose minimal medium containing either NH4 or EA to mid log with antisera against Stx and RpoA (loading control). qRT-PCR expression values in panels A, B, and D are presented as relative values compared to the value for WT EHEC strain 86-24 grown with NH4. Values are means plus standard deviations (error bars) for 3 independent experiments. Values that are significantly different from the value for WT EHEC strain 86-24 grown with NH4 are indicated by asterisks as follows: *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
We also examined expression of ler (Fig. 1B). At early log phase, ler expression is increased more than 15-fold when EHEC is grown with EA as the sole nitrogen source and ler expression continued to be significantly increased at mid-log phase. At late log phase, ler transcription was decreased in minimal medium with EA only. The LEE encodes a type III secretion system (TSS) that injects bacterial effectors into host cells as well as the translocon of this system made up of EspA, EspB, and EspD that are secreted through the TTS. EHEC secreted proteins can be detected at late log phase; therefore, we examined EspA expression at this time point (Fig. 1C). Expression of EspA was decreased in cells grown with EA compared to cells grown with NH4. These data agreed with the ler transcription data.
Mortality due to EHEC-caused disease is most commonly associated with the production of Stx; thus, we examined how EA affected Stx expression (Fig. 1D and E). Expression of the stx2a gene that encodes Stx was most significantly increased during early and mid-log growth phases in EA medium at both the transcriptional and translational levels (Fig. 1D). At late log phase, no significant differences in expression were observed for EHEC grown in the NH4 or EA medium.
To confirm that the increase in gene expression was due to growth on EA, as opposed to NH4 causing a decrease in expression, we grew EHEC cells with glucose and both EA and NH4 as nitrogen sources and measured gene expression at early log phase. When cells were grown aerobically in the presence of alternative nitrogen sources, EA metabolism did not seem to play a role in qse expression, as no significant differences in expression were observed in EHEC cells grown with both EA and NH4 compared to EHEC cells grown with NH4 only (Fig. 1A). However, expression of the ler gene, which is carried on LEE, the espA gene, which is carried in LEE4, and the eae gene, which is carried in LEE5, as well as stx2a was significantly increased in EHEC cells grown with both EA and NH4 compared to EHEC cells grown with NH4 only (Fig. 1B and D). These data indicate that EA metabolism is important for assimilating environmental information by activating expression of major EHEC regulators and virulence factors. The greatest differences in gene expression were measured at early log growth phase, suggesting that EA metabolism and/or that EA recognition trigger induction of virulence genes that allows for rapid adaptation to the host environment.
Expression of the eut genes in EHEC grown with alternative N sources.
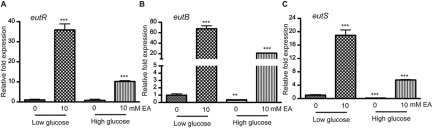
The GI tract contains several nitrogen sources available to commensal and pathogenic bacteria (34). To better understand expression of the genes encoding components involved in EA metabolism and their roles in EHEC gene regulation, we examined expression of eutR, eutB, which encodes the EA lyase large subunit (20, 21), and eutS, which encodes a carboxysome structural protein (18) in EHEC cells grown in Dulbecco’s modified Eagle medium (DMEM) with or without the addition of EA (Fig. 2A to C). DMEM contains glutamine as the major nitrogen source, which is metabolized by EHEC, and DMEM promotes expression of the LEE (35). These studies were conducted using both low- and high-glucose conditions (5.56 mM and 25 mM glucose, respectively), because a previous study showed that EutBC activity was decreased when E. coli was grown with glucose compared to glycerol, potentially due to catabolite repression (36). Our data indicated that expression of eutR, eutB, and eutS was very significantly increased in EHEC cells grown with EA compared to EHEC cells grown in the absence of EA under both low- and high-glucose conditions (Fig. 2A to C). Because expression of these genes was highest when EHEC cells were grown in the low-glucose DMEM, all subsequent experiments were performed using this medium. Additionally, expression of eut genes in EHEC cells grown with alternative nitrogen sources suggests that EHEC metabolizes EA even when alternative nitrogen sources are available. These data are consistent with previous work that showed that E. coli strain NCIB8114 metabolized EA when grown in the presence of both EA and NH4 (29).
FIG 2 .
Expression of the eut operon genes eutR (A), eutB (B), and eutS (C) is induced in cells grown with EA in both low- and high-glucose media. qRT-PCR expression values are presented as relative values compared to WT EHEC strain 86-24 grown in low-glucose DMEM without EA. Error bars represent the standard deviations for 3 independent experiments. **, P < 0.005; ***, P < 0.0005.
EHEC carries genes that encode two sensors that respond to EA.
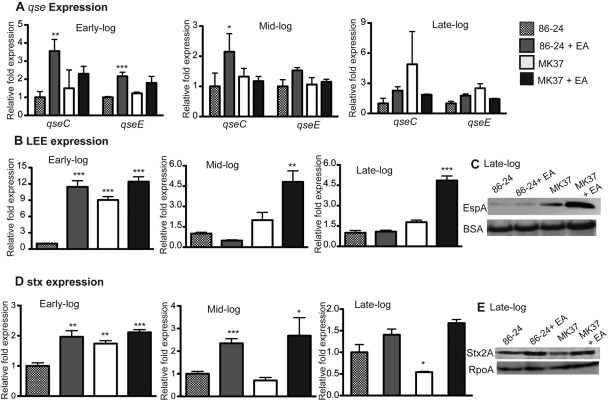
To better elucidate the role of EA in virulence gene regulation, we constructed an isogenic eutR mutant in EHEC (strain MK37). The eutR mutant did not grow in minimal medium with EA and vitamin B12, whereas growth in minimal medium was restored upon complementation with eutR expressed on a plasmid (strain MK41; data not shown). Subsequently, we examined expression of the qse, ler, and stx2a genes in wild-type (WT) EHEC strain 86-24 and the eutR mutant MK37 grown aerobically in Dulbecco’s modified Eagle medium with or without the addition of EA at early, mid-, and late log growth stages. At early log phase, expression of the global signaling genes qseC and qseE was significantly increased when EA was added to the medium, and this effect was slightly diminished at mid-log phase. At late log phase, no significant differences were observed in qseC or qseE expression in EHEC cells grown in DMEM or DMEM plus EA (Fig. 3A). These data are consistent with previous experiments that examined qse expression in EHEC cells grown in minimal medium with both EA and NH4 (Fig. 1A).
FIG 3 .
EA virulence gene regulation is both EutR dependent and independent. (A) qRT-PCR of qseC and qseE of WT EHEC strain 86-24 and the eutR mutant strain MK37 grown in DMEM or DMEM plus EA to early, mid-, and late log. (B) qRT-PCR of ler of WT EHEC strain 86-24 and the eutR mutant strain MK37 grown in DMEM or DMEM plus EA to early, mid-, and late log. (C) Western blots of secreted proteins from WT EHEC strain 86-24 and the eutR mutant strain MK37 grown to late log in DMEM or DMEM plus EA probed with antisera against EspA from WT EHEC. (D) qRT-PCR of stx2a of WT EHEC strain 86-24 and the eutR mutant strain MK37 grown in DMEM or DMEM plus EA to early, mid-, and late log. (E) Western blots of whole-cell lysates of WT EHEC strain 86-24 and the eutR mutant strain MK37 grown to late log in DMEM or DMEM plus EA with antisera against Stx and RpoA. RT-PCR expression values in panels A, B, and D are presented as relative values compared to WT EHEC strain 86-24 grown without EA. Error bars represent the standard deviations for 3 independent experiments. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Next, we measured expression of the LEE genes (Fig. 3B and C). During growth at early log phase, ler expression in WT EHEC was increased over 10-fold when EA was added to the medium. At mid- and late log phases, the addition of EA did not significantly affect the level of ler transcription. Expression of ler in the eutR mutant was increased compared to WT at early log phase, and this expression diminished throughout growth, suggesting that under aerobic conditions at early log phase, EutR represses LEE activation. The addition of EA to eutR mutant did not affect ler expression at early log phase. At mid- and late log phases, the addition of EA to the eutR mutant strain significantly increased ler expression compared to the eutR mutant grown without EA. Western blot analysis confirmed the transcription data, as EspA secretion was increased in the eutR mutant grown with EA at late log phase (Fig. 3C).
The pattern of Stx expression was similar to the pattern of ler expression (Fig. 3D). Addition of EA to WT cells significantly increased stx2a expression at early and mid-log growth phases. No significant differences in stx2a expression were observed at late log phase in WT cells and WT cells grown with EA. Expression of stx2a in the eutR mutant is increased slightly at early log phase compared to WT cells, but expression decreased to WT levels during mid- and late-log growth phases. Addition of EA to the eutR mutant did not affect transcription of stx2a at early log phase; however, stx2a expression was significantly increased when the eutR mutant was grown with EA at mid- and late log phases at both transcriptional and translational levels (Fig. 3D and E).
These results are interesting for several reasons. Gene expression in WT cells grown in DMEM plus EA was similar to expression when cells were grown in minimal medium with EA. The most significant expression differences occurred at early log phase, highlighting the importance of EA in adaptation to the GI tract. Furthermore, DMEM contains additional nitrogen sources; thus, EA gene regulation may not be completely dependent upon EA metabolism. Additionally, in the LEE and Stx analyses, no significant differences in expression in the eutR mutant grown with or without EA were observed at early log phase, but significant differences were observed at later time points. The fact that the eutR mutant is unresponsive to EA at early log phase suggests that EutR is the major sensor coordinating EA-related gene expression during this growth phase and that EHEC carries genes that encode an additional sensor that enables the eutR mutant to respond to EA at later growth phases.
EA-dependent virulence gene regulation is independent of EA metabolism.
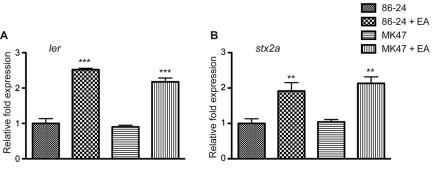
To determine whether EA metabolism was responsible for modulation of EHEC virulence gene regulation, we constructed a mutation in the eutB gene that encodes the ammonia lyase large subunit (strain MK47) (20, 21). Strain MK47 was unable to grown in minimal medium with EA as the sole nitrogen source; therefore, we examined expression of ler and stx2a at early log growth phase in DMEM with or without the addition of EA (Fig. 4A and B). Similar expression levels were measured in WT EHEC strain 86-24 and the eutB mutant when grown in the absence of EA, and the addition of EA significantly increased expression similarly in both strains, indicating that EA regulation of ler and stx2a does not depend upon the metabolism of EA. These data also suggest that EutR plays additional regulatory roles in EHEC beyond promoting expression of the eut operon.
FIG 4 .
EA virulence gene regulation is independent of EA metabolism at early log growth phase. (A) qRT-PCR of ler of WT EHEC strain 86-24 and the eutB mutant strain MK47 grown in DMEM or DMEM plus EA. (B) qRT-PCR of stx2a of WT EHEC strain 86-24 and the eutB mutant strain MK47 grown in DMEM or DMEM plus EA. qRT-PCR expression values are presented as relative values compared to WT EHEC strain 86-24 grown without EA. Error bars represent the standard deviations for 3 independent experiments. **, P < 0.005; ***, P < 0.0005.
EA is a signal that activates gene expression.
The GI tract is an anaerobic environment; therefore, we performed microarray analyses on WT EHEC grown anaerobically for 6 h in DMEM or DMEM plus 15 mM EA to gain better insight into EA gene regulation under conditions that are more similar to the host environment (NCBI GEO database accession no. GSE34046). The microarray data indicated that the most significant number of altered probes occurred when EHEC cells were grown with EA; more than 1,400 probes were significantly increased, and approximately 80 probes were significantly decreased, indicating that EA plays an important role in promoting gene expression in EHEC. Within these probes, the genes that are carried in the eut operon were the most significantly increased. These data further indicate that EHEC metabolizes EA even when in the presence of alternative nitrogen sources, and thus, EA may serve as an important nutrient for host colonization. Genes encoding components involved in vitamin B12 transport were also significantly increased. Global regulators, including QseC, QseE, and QseA, were increased in cells grown with EA as well as genes carried on LEE1 to LEE5.
To confirm the array data and compare gene expression in cells grown aerobically versus anaerobically, we performed quantitative reverse transcription-PCR (qRT-PCR) on the qse genes identified in the array, ler, and stx2a genes in EHEC cells grown in pure culture or in coculture with Bacteroides thetaiotaomicron, a predominant member of the GI tract (37). The pattern of gene expression in EHEC cells grown in pure culture was similar to that of cocultured cells (see Fig. S1 in the supplemental material). We also investigated whether EA is a signal for EHEC gene expression. Signaling molecules function at low concentrations and are not involved in primary metabolism (38); therefore, we grew WT EHEC and the eutR mutant in DMEM with EA concentrations ranging from 25 µM to 15 mM and compared gene expression to cells grown without added EA (Fig. 5A to E).
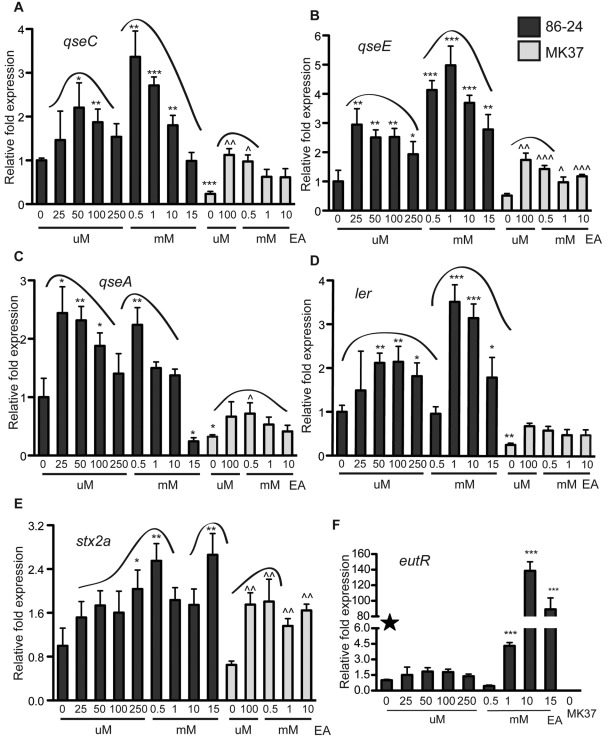
FIG 5 .
EA gene regulation in anaerobic conditions. (A) qRT-PCR of qseC in WT EHEC strain 86-24 and the eutR mutant strain MK37 in DMEM or DMEM plus EA. (B) qRT-PCR of qseE in WT EHEC strain 86-24 and the eutR mutant strain MK37 in DMEM or DMEM plus EA. (C) qRT-PCR of qseA in WT EHEC strain 86-24 and the eutR mutant strain MK37 in DMEM or DMEM plus EA. (D) qRT-PCR of ler in WT EHEC strain 86-24 and the eutR mutant strain MK37 in DMEM or DMEM plus EA. (E) qRT-PCR of stx2a in WT EHEC and eutR mutant strain MK37 in DMEM or DMEM plus EA. (F) qRT-PCR of eutR in WT EHEC grown in DMEM or DMEM plus EA and the eutR mutant strain MK37 grown in DMEM. The concentrations of EA (micromolar and millimolar) are shown on the x axes. qRT-PCR expression values are presented as relative values compared to WT EHEC strain 86-24 grown without EA. Error bars represent the standard deviations for 3 independent experiments. Statistical significance is indicated by symbols as follows. The asterisks indicate significance in gene expression between WT EHEC cells (0 µM EA), WT EHEC cells grown with EA, and the eutR mutant strain (0 µM EA): *, P < 0.05; **, P < 0.005; ***, P < 0.0005. The carets indicate significance in gene expression between the eutR mutant (0 µM EA) and eutR mutant cells grown with EA: ^, P < 0.05; ^^, P < 0.005; ^^^, P < 0.0005.
Expression of these genes displayed several trends. Micromolar EA concentrations significantly increased expression of all genes examined compared to WT cells grown without EA. Expression was significantly decreased in the eutR mutant grown in DMEM without EA compared to WT cells. With the exception of ler, addition of EA to the eutR mutant significantly increased gene expression relative to the eutR mutant grown without EA. Gene expression profiles showed two peaks, one in the micromolar EA range and another in the millimolar EA range. We will discuss these points individually.
Expression of qseE and qseA was significantly increased when EHEC was grown with 25 µM EA, whereas the addition of 50 µM EA significantly increased qseC and ler expression, and 250 µM significantly increased stx2a expression. Since micromolar EA concentrations induce expression of these genes but do not support growth (data not shown), these data suggest that EA, under low concentrations, is a signal for gene regulation, independent of metabolism.
Expression of the qse, ler, and stx2a genes was significantly downregulated in the eutR mutant strain compared to the WT strain. These data indicate that EutR plays a positive role in virulence gene expression under anaerobic conditions, as these results are different from what was seen under aerobic conditions. Perhaps how EHEC gains energy, either by aerobic respiration or by fermentation, influences EutR gene regulation. EA regulates the expression of QseC and QseE both of which integrate environmental signals present in the GI tract; thus, it is possible that EA plays a global role in integrating multiple cues, including O2, to optimize timing of virulence gene expression.
Transcription of the qse and stx2a genes was significantly increased with the addition of 100 µM EA to the eutR mutant compared to eutR mutant grown without EA. These data indicate that EHEC carries a gene that encodes a second EA sensor that plays a role in regulation of these genes that act independently of EutR. Addition of EA did not induce differences in ler gene expression, suggesting that EutR is the major regulator involved in EA regulation of ler.
The two peaks of expression observed for all the genes examined further suggest that two EA sensors exist, one that responds to low concentrations in which EA is used as a signal, and a second sensor that requires higher EA concentrations that is most likely involved in EA metabolism. To test this hypothesis, we measured eutR transcription in EHEC grown with various concentrations of EA (Fig. 5F). The data revealed that micromolar EA concentrations were not sufficient to significantly increase eutR expression compared to WT cells grown without EA. However, the addition of 1 to 15 mM EA greatly increased eutR expression. These data are not surprising, because it would be energetically expensive to upregulate the genes involved in EA metabolism, including those that encode the carboxysome shell proteins (18). However, these data are intriguing. On the basis of the results of the DMEM time course experiments (Fig. 3), we hypothesized that EutR was the major sensor of EA at early log phase and that a second sensor was responsible for EA gene regulation at later growth phases. EA concentrations would be highest at earlier time points, during which EutR is expressed and involved in EA-related gene expression and metabolism. As EA is consumed, regulation by the second sensor that responds to EA as a signal becomes apparent. Expression of qseC, qseE, qseA and stx2a in the eutR mutant increased when cells were grown with 100 µM EA, highlighting that the second sensor responds to low EA concentrations.
To determine the global role of EA signaling-related gene regulation in EHEC, we also performed microarray analyses and compared gene expression of EHEC cells grown in the absence of EA to EHEC cells grown with 50 µM EA. These data revealed that nearly 200 probes were significantly increased and about 700 were mildly increased, whereas 50 probes were significantly decreased in cells grown with micromolar EA concentrations. The results of these analyses indicate that EA signaling is important in EHEC gene regulation but that EutR/metabolism-based regulation is more extensive. Current studies are being undertaken to identify the second EA sensor and to more clearly elucidate the roles of EutR and the second EA sensor in EHEC virulence gene expression.
EA influences timing of AE lesion formation.
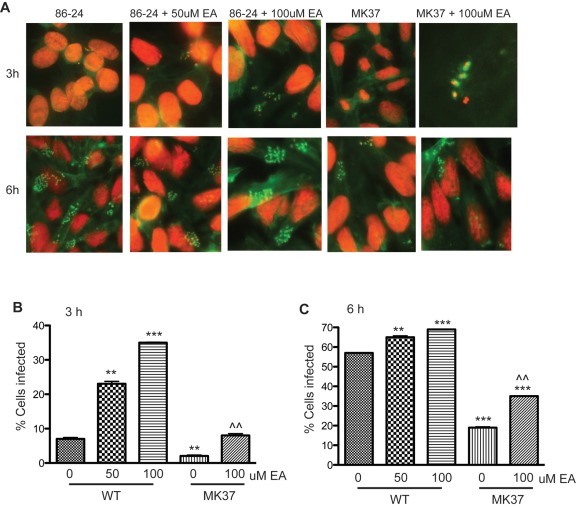
EA activates expression of the LEE when grown anaerobically in vitro; thus, we used the fluorescein actin staining (FAS) test to confirm the transcription data and determine how EA affects formation of AE lesions on epithelial cells. In this assay, the WT EHEC and eutR mutant strains were grown anaerobically in the presence of HeLa cells with 0, 50, or 100 µM EA. The cells were examined after 3 and 6 h of incubation. When 50 or 100 µM of EA was added to DMEM, WT EHEC formed more pedestals and infected significantly more cells at 3 h than cells grown without EA (Fig. 6A and B). At 6 h, the numbers of pedestals observed from WT EHEC cells grown with and without EA were similar; however, significantly more HeLa cells were infected in WT EHEC cells grown with 50 or 100 µM EA compared to EHEC cells grown in the absence of EA (Fig. 6C).
FIG 6 .
Detection of AE lesion formation using the FAS test on HeLa cells. (A) WT EHEC strain 86-24 and the eutR mutant strain MK37 in the presence and absence of EA at 3 h (top row) and 6 h (bottom row). The HeLa cell actin cytoskeleton (green) and the bacteria and HeLa cell nuclei (red) are shown. Pedestals are observed as bright green structures that are typically associated with bacterial cells. The cells were viewed at a magnification of ×640. The micromolar concentrations of EA are shown. (B and C) Percentage of infected HeLa cells after 3 h (B) or 6 h (C) of incubation. Error bars represent the standard deviations of 2 independent experiments. Statistical significance is indicated by symbols as follows. The asterisks indicate significance in gene expression between WT (0 µM EA), WT cells grown with EA, and the eutR mutant strain (0 µM EA): **, P < 0.005; ***, P < 0.0005. The carets indicate significance in gene expression between the eutR mutant cells (0 µM EA) and eutR mutant cells grown with EA; ^^, P < 0.005.
We also examined the eutR mutant for its ability to form AE lesions (Fig. 6A to C). At 3 h, hardly any pedestals were observed. At 6 h, the eutR mutant formed fewer pedestals than the WT did, again suggesting that EutR activates the LEE under anaerobic conditions. The numbers of pedestals observed in the eutR mutant strain grown with and without EA were similar; however, the percentage of cells infected was higher when the eutR mutant was grown with EA at both 3 and 6 h (Fig. 6B and C). These data are consistent with the transcriptomic data that EA positively regulates LEE expression under anaerobic conditions and indicate that EA is an important signal, acting at micromolar concentrations to influence the timing of LEE expression. Although the addition of EA to DMEM did not significantly alter ler expression in the eutR mutant based on qRT-PCR (Fig. 5D), the addition of EA did increase the eutR mutant strain’s ability to infect a higher percentage of cells. These data are consistent with the ler transcription data, as pedestal formation is dependent on the LEE; however, the higher percentage of infected cells in the eutR mutant grown with EA could be due to the upregulation of other adhesins which EA may promote EHEC attachment (39).
Conclusions.
This study takes the first steps toward understanding the complex roles of EA in metabolism, cell-to-cell signaling, and virulence. Although EA metabolism influences gene expression, EHEC has coopted this nutrient for use as a signal, allowing the integration of multiple environmental cues and ultimately expression of genes involved in pathogenesis. The specific mechanisms of EA gene regulation are not fully understood, and additional studies will provide insights into bacterial pathogenesis and cell-to-cell signaling, including microbial-host interactions.
MATERIALS AND METHODS
Strains, plasmids, and recombinant DNA.
WT EHEC strain 86-24 (40) and its isogenic mutants as well as B. thetaiotaomicron strain VPI-5482 (ATCC) were used in this study. EHEC strains were grown at 37°C in low-glucose DMEM (catalog no. 11885; Invitrogen), high-glucose DMEM (catalog no. 11965; Invitrogen), LB, or M9 minimal medium. M9 minimal medium was made according to reference 25, except that no nitrogen source was added to the minimal salts. The nitrogen sources used were EA (ethanolamine hydrochloride; Sigma) at the concentrations listed in the text, NH4Cl (20 mM), and glutamine (30 mM). Vitamin B12 (cyanocobalamin; Sigma) was added at a final concentration of 150 nM whenever the media were supplemented with EA. The following antibiotics and concentrations were added: ampicillin, 100 µg/ml; streptomycin, 50 µg/ml; and kanamycin, 50 µg/ml. Recombinant DNA and molecular biology techniques were performed as previously described (25). All oligonucleotide primers are listed in Table 1. The mutant strains MK37 and MK47 were constructed using λ-Red as previously described (41) with primers eutR_LRF and eutR_LRR and eutB_LRF and eutB_LRR, respectively. To create the nonpolar mutants MK37 and MK47, the chloramphenicol cassettes were resolved using pCP20 (41). Strain MK37 was complemented with plasmid pMK47 to create strain MK41. Plasmid pMK47 was constructed from the PCR product of primer set MK260F and MK261R using EHEC as a template, and this product was digested with SacI and XbaI and inserted into pBAD33. Expression of pMK47 was induced by adding 0.2% arabinose to the growth medium. B. thetaiotaomicron was grown anaerobically at 37°C in TYG medium (42). The anaerobic conditions were prepared by filling a container with medium and then incubating the culture in an atmosphere of 5% CO2.
TABLE 1 .
Oligonucleotide primers used in this study
| Primera | Sequence | Function |
|---|---|---|
| eutR_LRF |
TAACTCCCTCACCCCCATTCCCG
CATCCGCTGATGCAACGTCAACGACGGCT GTGTAGGCTGGAGCTGCTTC |
Isogenic mutant construction |
| eutR_LRR |
ATCATGAAAAAGACCCGTACAGCC
AATTTGCACCATCTTTATCATGAACCCT CATATGAATATCCTCCTTAG |
Isogenic mutant construction |
| eutB_LRF |
CCGCGTCATCAGAAGAACAGTGAC
GGATCGCCCGCCCGTTTGGTCAGGCGAC GTGTAGGCTGGAGCTGCTTC |
Isogenic mutant construction |
| eutB_LRR |
CTTATGAAACTAAAGACCACATTG
TTCGGCAATGTATATCAGTTTAAGGATG CATATGAATATCCTCCTTAG |
Isogenic mutant construction |
| qseB-F | CGGTGATCCTGGATTTAACCTT | qRT-PCR |
| qseB-R | GCTGACCTTTTTCTCGCCATT | qRT-PCR |
| qseC-F | AATGGGAATACCGTGAAGACAT | qRT-PCR |
| qseC-R | CCAACCACGGGATCAATTG | qRT-PCR |
| qseE-F | ACAATCCCTGGCAATGCTTAA | qRT-PCR |
| qseE-R | GAAGCCACCAGCGAAAAGG | qRT-PCR |
| ler-F | CGACCAGGTCTGCCC | qRT-PCR |
| ler-R | GCGCGGAACTCATC | qRT-PCR |
| espA-F | TCAGAATCGCAGCCTGAAAA | qRT-PCR |
| espA-R | CGAAGGATGAGGTGGTTAAGCT | qRT-PCR |
| eae-F | GCTGGCCCTTGGTTTGATCA | qRT-PCR |
| eae-R | GCGGAGATGACTTCAGCACTT | qRT-PCR |
| stx2a-F | ACCCCACCGGGCAGTT | qRT-PCR |
| stx2a-R | GGTCAAAACGCGCCTGATA | qRT-PCR |
| qseA-F | GGCAAGGGAGATTTGTGACTAAT | qRT-PCR |
| qseA-R | GGCACCCGCCGTTAGC | qRT-PCR |
| rpoA-F | GCGCTCATCTTCTTCCGAAT | |
| rpoA-R | CGCGGTCGTGGTTATGTG | |
| eutRcompF | CTCGAGATGCCGCCACTGGTACGCTG | Construction of pMK47 |
| eutRcompR | CCCGGGCAGCTCTTTTCTGCTGGGAC | Construction of pMK47 |
The final letter in the primer name indicates the orientation: F, forward; R, reverse.
RNA extraction and qRT-PCR.
Cultures of strains 86-24, MK37, MK41, and MK47 were grown aerobically in LB medium at 37°C overnight and then diluted 1:100 in DMEM and grown at 37°C. RNA from three biological replicate cultures of each strain/condition was extracted at the early exponential growth phase (optical density at 600 nm [OD600] of 0.2), mid-exponential growth phase (OD600 of 0.5), late exponential growth phase (OD600 of 1.0), or 6 h anaerobically (to mimic FAS conditions) using the RiboPure Bacteria RNA isolation kit (Ambion). In coculture experiments, EHEC was diluted 1:100, whereas B. thetaiotaomicron was diluted 9:100 into low-glucose DMEM. The primers used in the real-time qPCR assays were designed using Primer Express v1.5 (Applied Biosystems) (Table 1). The amplification efficiency and template specificity of each of the primer pairs were validated, and reaction mixtures were prepared as described previously (43). qRT-PCR was performed in a one-step reaction using an ABI 7500 sequence detection system (Applied Biosystems).
Data were collected using the ABI Sequence Detection 1.2 software (Applied Biosystems). All data were normalized to the levels of rpoA and analyzed using the comparative cycle threshold (CT) method (44). The expression levels of the target genes under the various conditions were compared using the relative quantification method (44). Real-time data are expressed as the changes in expression levels compared to the WT levels. Statistical significance was determined by Student’s t test, and a P value of ≤0.05 was considered significant.
Microarray.
Affymetrix 2.0 E. coli gene arrays were used to compare gene expression of strain 86-24 grown anaerobically with 5% CO2 in DMEM, DMEM plus 15 mM EA, or DMEM plus 50 µM EA at 37°C for 6 h. The RNA processing, labeling, hybridization, and slide-scanning procedures were performed as described by the manufacturer (Affymetrix). The array data analyses were performed as described previously (45). The output from scanning a single replicate of the Affymetrix GeneChip E. coli Genome 2.0 array for each of the biological conditions was obtained using GCOS v 1.4 according to the manufacturer’s instructions. Data were normalized using Robust Multiarray analyses (46, 47), and the resulting data were compared to determine features whose expression was increased or decreased in response to EA. Custom analysis scripts were written in Perl in order to sort the Affymetrix output data and complete the multiple array analyses. The microarray data have been deposited in the National Center of Biotechnology Information Gene Expression Omnibus database (accession no. GSE34046).
FAS assay.
Fluorescein actin staining (FAS) assays were performed as previously described (48). Briefly, OVN bacterial cultures were grown aerobically in LB at 37°C and then diluted 1:100 to infect HeLa cells. HeLa cells were grown on glass coverslips for 3 or 6 h at 37°C and 5% CO2. The coverslips were then washed, permeabilized with 0.2% Triton X, and treated with fluorescein isothiocyanate-labeled phalloidin to visualize actin accumulation. Propidium iodide was added to stain the bacteria.
SDS-PAGE and immunoblotting.
Secreted proteins from strains 86-24 and MK37 were harvested as previously described (49). Whole-cell lysates were prepared from strains grown in DMEM to mid- or late-log growth. SDS-PAGE and immunoblotting were completed as previously described (25). Preparations were probed by Western blot analysis using polyclonal antisera to EspA or monoclonal antisera against Stx2A (Santa Cruz Biotechnology) and RpoA (Neoclone).
SUPPLEMENTAL MATERIAL
Incubation of WT EHEC with B. thetaiotaomicron does not alter EA-dependent gene regulation. qRT-PCR of ler (A), qseC (B), and stx2a (C) in WT EHEC strain 86-24 grown with 0, 50 µM, or 10 mM EA in the presence or absence of B. thetaiotaomicron. qRT-PCR expression values are presented as relative values compared to the value for WT EHEC strain 86-24 grown without EA. *, P < 0.05; **, P < 0.005; ***, P < 0.0005. Download Figure S1, PDF file, 0.5 MB.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant AI053067, the Ellison Medical Foundation, the Burroughs Wellcome Fund, and a NIH Ruth L. Kirschstein Fellowship F32AI80115 to M.M.K.
The contents of this article are solely the responsibility of the authors and do not represent the official views of the NIH NIAID.
Footnotes
Citation Kendall MM, Gruber CC, Parker CT, Sperandio V. 2012. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. mBio 3(3):e00050-12. doi:10.1128/mBio.00050-12
REFERENCES
- 1. Bakovic M, Fullerton MD, Michel V. 2007. Metabolic and molecular aspects of ethanolamine phospholipid biosynthesis: the role of CTP:phosphoethanolamine cytidylyl-transferase (Pcyt2). Biochem. Cell Biol. 85:283–300 [DOI] [PubMed] [Google Scholar]
- 2. Dowhan W. 1997. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu. Rev. Biochem. 66:199–232 [DOI] [PubMed] [Google Scholar]
- 3. Dowhan W. 1997. Phosphatidylserine decarboxylases: pyruvoyl-dependent enzymes from bacteria to mammals. Methods Enzymol. 280:81–88 [DOI] [PubMed] [Google Scholar]
- 4. Cotton PB. 1972. Non-dietary lipid in the intestinal lumen. Gut 13:675–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Snoeck V, Goddeeris B, Cox E. 2005. The role of enterocytes in the intestinal barrier function and antigen uptake. Microbes Infect. 7:997–1004 [DOI] [PubMed] [Google Scholar]
- 6. Thiennimitr P, et al. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. 108:17480–17485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Karmali MA, Petric M, Lim C, Fleming PC, Steele BT. 1983. Escherichia coli cytotoxin, haemolytic-uraemic syndrome, and haemorrhagic colitis. Lancet ii:1299–1300 [DOI] [PubMed] [Google Scholar]
- 8. Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 9. McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 92:1664–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. 2006. The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U. S. A. 103:10420–10425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reading NC, et al. 2007. A novel two-component signaling system that activates transcription of an enterohemorrhagic Escherichia coli (EHEC) effector involved in remodeling of host actin. J. Bacteriol. 189:2468–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296–306 [DOI] [PubMed] [Google Scholar]
- 13. Kendall MM, Rasko DA, Sperandio V. 2010. The LysR-type regulator QseA regulates both characterized and putative virulence genes in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 76:1306-1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garsin DA. 2010. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat. Rev. Microbiol. 8:290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bertin Y, et al. 2011. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ. Microbiol. 13:365–377 [DOI] [PubMed] [Google Scholar]
- 16. Tsoy O, Ravcheev D, Mushegian A. 2009. Comparative genomics of ethanolamine utilization. J. Bacteriol. 191:7157–7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hayashi T, et al. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11–22 [DOI] [PubMed] [Google Scholar]
- 18. Kofoid E, Rappleye C, Stojiljkovic I, Roth J. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 181:5317–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Perna NT, et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533 [DOI] [PubMed] [Google Scholar]
- 20. Roof DM, Roth JR. 1988. Ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 170:3855–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roof DM, Roth JR. 1989. Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 171:3316–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roof DM, Roth JR. 1992. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J. Bacteriol. 174:6634–6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sheppard DE, Penrod JT, Bobik T, Kofoid E, Roth JR. 2004. Evidence that a B12-adenosyl transferase is encoded within the ethanolamine operon of Salmonella enterica. J. Bacteriol. 186:7635–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stojiljkovic I, Baümler AJ, Heffron F. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177:1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor: Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26. Jones PW, Turner JM. 1984. Interrelationships between the enzymes of ethanolamine metabolism in Escherichia coli. J. Gen. Microbiol. 130:299–308 [DOI] [PubMed] [Google Scholar]
- 27. Jones PW, Turner JM. 1984. A model for the common control of enzymes of ethanolamine catabolism in Escherichia coli. J. Gen. Microbiol. 130:849–860 [DOI] [PubMed] [Google Scholar]
- 28. Penrod JT, Mace CC, Roth JR. 2004. A pH-sensitive function and phenotype: evidence that EutH facilitates diffusion of uncharged ethanolamine in Salmonella enterica. J. Bacteriol. 186:6885–6890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scarlett FA, Turner JM. 1976. Microbial metabolism of amino alcohols. Ethanolamine catabolism mediated by coenzyme B12-dependent ethanolamine ammonia-lyase in Escherichia coli and Klebsiella aerogenes. J. Gen. Microbiol. 95:173–176 [DOI] [PubMed] [Google Scholar]
- 30. Reading NC, Rasko DA, Torres AG, Sperandio V. 2009. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 106:5889–5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. U. S. A. 100:8951–8956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sperandio V, Torres AG, Kaper JB. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43:809–821 [DOI] [PubMed] [Google Scholar]
- 33. Hughes DT, Clarke MB, Yamamoto K, Rasko D, Sperandio AV. 2009. The QseC adrenergic signaling cascade in enterohemorrhagic E. coli (EHEC). PLoS Pathog. 5:e10000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wrong O. 1978. Nitrogen metabolism in the gut. Am. J. Clin. Nutr. 3:1587–1593 [DOI] [PubMed] [Google Scholar]
- 35. Ebel F, Deibel C, Kresse AU, Guzmán CA, Chakraborty T. 1996. Temperature- and medium-dependent secretion of proteins by Shiga toxin-producing Escherichia coli. Infect. Immun. 64:4472–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blackwell CM, Scarlett FA, Turner JM. 1977. Microbial metabolism of amino alcohols. Control of formation and stability of partially purified ethanolamine ammonia-lyase in Escherichia coli. J. Gen. Microbiol. 98:133–139 [DOI] [PubMed] [Google Scholar]
- 37. Moore WE, Holdeman LV. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Keller L, Surette MG. 2006. Communication in bacteria: an ecological and evolutionary perspective. Nat. Rev. Microbiol. 4:250–258 [DOI] [PubMed] [Google Scholar]
- 39. Barnett-Foster D, et al. 1999. Phosphatidylethanolamine recognition promotes enteropathogenic E. coli and enterohemorrhagic E. coli host cell attachment. Microb. Pathog. 27:289–301 [DOI] [PubMed] [Google Scholar]
- 40. Griffin PM, et al. 1988. Illnesses associated with Escherichia coli. Ann. Intern. Med. 109:705-712 [DOI] [PubMed] [Google Scholar]
- 41. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Holdeman LV, Cato ED, Moore WE. 1977. Anaerobe laboratory manual. Virginia: Polytechnic Institute and State University Anaerobe Laboratory, Blacksburg, VA [Google Scholar]
- 43. Walters M, Sperandio V. 2006. Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect. Immun. 74:544–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Biosystems. Applied. 1997. ABI Prism 7700 sequence detection system: user bulletin #2. Applied Biosystems, Foster City, CA [Google Scholar]
- 45. Kendall MM, Rasko DA, Sperandio V. 2007. Global effects of the cell-to-cell signaling molecules autoinducer-2, autoinducer-3, and epinephrine in a luxS mutant of enterohemorrhagic Escherichia coli. Infect. Immun. 75:4875–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193 [DOI] [PubMed] [Google Scholar]
- 47. Irizarry RA, et al. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264 [DOI] [PubMed] [Google Scholar]
- 48. Knutton S, Baldwin T, Williams PH, McNeish AS. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jarvis KG, et al. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. U. S. A. 92:7996–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Incubation of WT EHEC with B. thetaiotaomicron does not alter EA-dependent gene regulation. qRT-PCR of ler (A), qseC (B), and stx2a (C) in WT EHEC strain 86-24 grown with 0, 50 µM, or 10 mM EA in the presence or absence of B. thetaiotaomicron. qRT-PCR expression values are presented as relative values compared to the value for WT EHEC strain 86-24 grown without EA. *, P < 0.05; **, P < 0.005; ***, P < 0.0005. Download Figure S1, PDF file, 0.5 MB.