ABSTRACT
The Pseudomonas aeruginosa extracytoplasmic functioning (ECF) sigma factor σ22 is encoded by algT/algU and is inhibited by anti-sigma factor MucA. σ22 was originally discovered for its essential role in the expression of the exopolysaccharide alginate by mucoid strains associated with chronic pulmonary infection. However, σ22 is now known to also have a large regulon associated with the response to cell wall stress. Our recent transcriptome analysis identified 293 open reading frames (ORFs) in the σ22 stress stimulon that include genes for outer envelope biogenesis and remodeling, although most of the genes have undefined functions. To better understand the σ22-dependent stress response, mutants affected in 27 genes of the σ22 stimulon were examined and expression was studied with lacZ fusions. Mutants constructed in the 27 genes showed no major change in response to cell wall-acting antibiotics or growth at elevated temperatures nor in alginate production. The mutants were examined for their effects on the expression of the σ22-dependent promoter of the alginate biosynthetic operon (PalgD) as a measure of σ22 derepression from MucA. By testing PalgD expression under both planktonic and sessile growth conditions, 11 genes were found to play a role in the stress response that activates σ22. Some mutations caused an increase or a decrease in the response to cell wall stress. Interestingly, mutations in 7 of the 11 genes caused constitutive PalgD expression under nonstressed conditions and thus showed that these genes are involved in maintaining envelope homeostasis. Mutations in PA0062 and PA1324 showed constitutive PalgD expression during both the planktonic and the sessile modes of growth. However, the PA5178 mutation caused constitutive PalgD expression only during planktonic growth. In contrast, mutations in PA2717, PA0567, PA3040, and PA0920 caused constitutive PalgD expression only in the sessile/biofilm mode of growth. This provides evidence that the σ22 stimulon for cell envelope homeostasis overlaps with biofilm control mechanisms.
IMPORTANCE
During chronic lung infections, such as in cystic fibrosis patients, Pseudomonas aeruginosa produces the exopolysaccharide alginate and forms biofilms that shield the organisms from the immune response and increase resistance to antibiotics. Activation of alginate genes is under the control of an extracytoplasmic stress response system that releases an alternative sigma factor (σ22) in response to cell wall stress and then activates expression of a large regulon. In this study, a mutant analysis of 27 members of the regulon showed that 11 play a role in envelope homeostasis and affect the stress response system itself. Interestingly, some genes demonstrate effects only in either the planktonic (free-swimming) or the sessile (biofilm) mode of growth, which leads to persistence and antibiotic tolerance. The studies presented here provide an important initial step in dissecting the mechanisms that regulate a critical signal transduction pathway that impacts P. aeruginosa pathogenesis.
Introduction
Pseudomonas aeruginosa is a Gram-negative bacterium capable of thriving in diverse environmental niches. It is also an important opportunistic human pathogen and major contributor to chronic lung infections in patients with cystic fibrosis (CF). The ability of this organism to successfully colonize these different environments is often multifactorial. One survival strategy used by P. aeruginosa in the CF lung is to overproduce the exopolysaccharide alginate, which gives colonies a distinctive mucoid phenotype and is also a harbinger of increased morbidity and mortality for these patients (1). The alginate barrier in mucoid P. aeruginosa has been shown to provide protection against many common antibiotics (2) and to confer increased resistance to phagocytic killing and antibody-dependent bactericidal mechanisms (3–5). The ability of P. aeruginosa to form biofilms is another important factor in establishing chronic infections in the lungs of CF patients (6, 7). Mucoid P. aeruginosa in a biofilm is more resistant to killing by human leukocytes in the presence of gamma interferon than is its isogenic nonmucoid form, suggesting that alginate plays an important role in protecting mucoid P. aeruginosa biofilm bacteria from the human immune system (3).
The master regulator for alginate production in mucoid P. aeruginosa is the extracytoplasmic functioning (ECF) sigma factor, σ22 (8, 9). This 22-kDa alternative sigma factor, encoded by algT (also known as algU), is essential for expression of the algD promoter (PalgD), which controls the expression of the 12-gene alginate biosynthetic operon (algD-alg8-alg44-algKEGXLIJFA) (10–12). Three additional positive regulatory proteins under σ22 control are required for PalgD expression: the two-component response regulators AlgB and AlgR (13–15) and a small ribbon-helix-helix family DNA-binding protein, AmrZ (16).
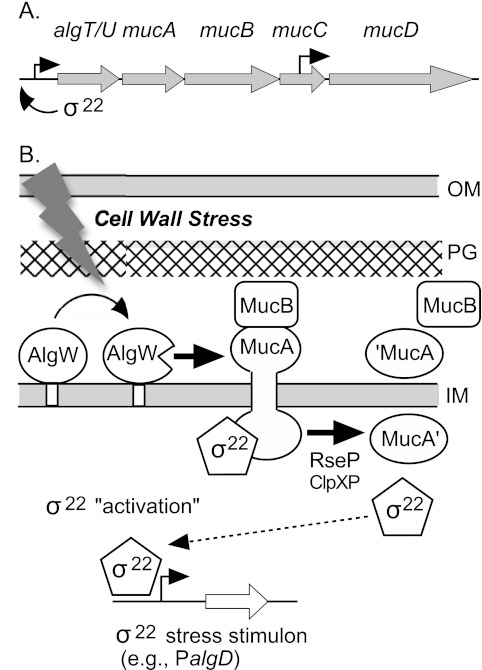
σ22 is encoded by the algT/U-mucABCD operon along with posttranslational regulatory proteins (Fig. 1A). MucA is an anti-sigma factor and primary inhibitor of σ22 activity; it acts by sequestering the sigma factor at the cytoplasmic membrane. Mutations in mucA are the most common cause of the constitutive expression of alginate observed in CF isolates of mucoid P. aeruginosa (17). MucA spans the inner membrane with the N-terminal cytoplasmic domain binding σ22 and the C-terminal periplasmic domain binding MucB (18, 19) (Fig. 1B). The formation of this macromolecular membrane complex results in diminished expression of σ22-regulated genes. The mucD gene encodes an HtrA/DegP-like periplasmic protease that apparently degrades peptide signals that lead to σ22 release and thus helps maintain the integrity of the σ22-MucAB complex. The mucD gene is also under the control of an internal promoter within mucC, a gene of unknown function (20).
FIG 1 .
(A) Map of the algT/U operon. Arrows indicate promoters. (B) Model for the “activation” of σ22 activity by regulated intramembrane proteolytic (RIP) degradation of the anti-sigma factor MucA. Under nonstress conditions, σ22 activity is low due to its sequestration at the inner membrane by the MucA-MucB complex. In response to cell wall stress (e.g., d-cycloserine), AlgW cleaves MucA in the C terminus and then RseP (YaeL) and ClpXP cleave the MucA N-terminal region, releasing σ22 from posttranslational repression. This allows σ22 to complex with core RNA polymerase, thus increasing transcription of target promoters in the σ22 stimulon, which includes PalgD of the alginate biosynthetic operon. Abbreviations: OM, outer membrane; PG, peptidoglycan; IM, inner membrane.
Liberation of σ22 from the inner membrane complex and the resultant increase in σ22-mediated transcription occur by regulated intramembrane proteolytic (RIP) destruction of MucA (Fig. 1B). Cell wall stress triggers the activation of this process, such as by exposure to cell wall-active antibiotics (e.g., d-cycloserine) or compounds that disrupt biological membranes (e.g., Tween and triclosan) or by overproduction of certain outer membrane proteins (e.g., MucE) (19, 21, 22). AlgW is an inner membrane endoserine protease that senses envelope stress conditions by binding sequence-specific polypeptide signal molecules at its PDZ domain, which relieves steric inhibition of the catalytic domain and allows AlgW to cleave MucA in the periplasmic domain (18). The initiating AlgW-dependent cleavage of MucA is followed by further degradation of the truncated anti-sigma polypeptide through the actions of RseP (YaeL) protease and several cytoplasmic ClpXP proteases, resulting in the ultimate release of σ22 so that it can complex with core RNA polymerase and direct transcription of its target genes (18, 21–23).
We have analyzed the transcriptional profiles of genes upregulated by σ22 activation in response to cell wall stress (19). Exposing P. aeruginosa PAO1 to d-cycloserine, which directly targets peptidoglycan synthesis, results in the increased expression of 293 genes that are dependent on σ22 deregulation. Among those upregulated are the genes for alginate biosynthesis and its regulation. Other members of the σ22 stimulon include genes involved in peptidoglycan biosynthesis (mdoH, mrcB, and mpl), lipopolysaccharide (LPS) biosynthesis (wzz, rmlD, wbpH, and wbpD), and genes encoding proteins with known adaptive or protective functions, such as bacitracin resistance protein (BacA), chloroperoxidase (Cpo), and two mechanosensitive channels encoded by PA4394 and PA4614 (19). A number of lipoproteins that are σ22 dependent have predicted roles in outer membrane repair and/or maintenance. However, the large majority of genes in the σ22 stimulon encode hypothetical or conserved hypothetical proteins for which limited biological information is known. It has also been shown that a mutation in algT/algU, which disrupts the whole σ22 stimulon of 293 genes, also results in less robust (i.e., less shear-resistant) biofilms (24). However, it is unknown which genes of the stimulon are involved with biofilm formation.
In this study, we sought to better understand the σ22-mediated stress response by characterizing mutants with defects in specific σ22-dependent genes for potential roles in maintaining cell envelope integrity. This analysis revealed that several gene products play a role in envelope homeostasis and thus affect the stress response system itself. Further study revealed that some of these genes demonstrate this effect only in either the planktonic (free-swimming) or the sessile (biofilm) mode of growth.
RESULTS
Construction of σ22 stress stimulon mutants.
There are 293 known genes in the σ22 stress stimulon of strain PAO1 as determined by a transcriptome analysis of planktonically grown bacteria subjected to cell wall stress with d-cycloserine (19). In this study, we selected a set of undercharacterized genes within the σ22 stress stimulon (Table 1) for a mutant analysis to shed light on the output of this complex stress response system. These genes can be under the direct or indirect control of σ22; previous studies show that at least two of them (PA0059 and PA3819) have promoter regions with the σ22 consensus sequence (25). Transposon insertion mutants of PAO1 with sequence-defined insertions in genes of the σ22 stress stimulon were obtained from the University of Washington Genomics Resource Center (WGRC). To ensure a consistent PAO1 strain background, which is known to show some diversity among laboratory strains (26), each WGRC transposon-mutated allele was transduced into this laboratory’s reference PAO1 isolate (also known as PDO1). In all, 27 transduced transposon insertions were constructed for study and are shown in Table 1 along with their stress induction values from the microarray analyses (19). Each was tested by PCR to verify that the transposon was in the correct gene (see Materials and Methods).
TABLE 1 .
Verification that selected genes were σ22 dependent by using a lacZ fusion analysis of each promoter in algT (σ22-deficient) and mucA (anti-sigma-factor-deficient) backgrounds
| Gene | Gene product descriptiona | Stress induction (15/60 min)b | P-lacZ WT:algT:mucA activityc |
|---|---|---|---|
| PA0059 | OsmC, redox protein, osmotically induced | 9.9/8.9 | 156:15:450 |
| PA0062 | Hypothetical, predicted type II lipoprotein | 11.4/10.5 | ND |
| PA0460 | Hypothetical, predicted periplasmic | 18.9/7.7 | 1,569:116:5,161 |
| PA0567d | Proteolipid homolog, membrane integrity | 6.0/6.2 | 220:142:976 |
| PA0854 | FumC2, fumarate hydrase | 4.7/10.7 | ND |
| PA0919 | Hypothetical, predicted secretion usher | 3.0/<2 | Operon |
| PA0920d | Aminoacyl-phosphatidylglycerol synthase | 3.0/<2 | 3,264:2,825:5,239 |
| PA1243 | Predicted transcriptional regulator, PAS domain | 2.0/<2 | 87:30:307 |
| PA1323 | Hypothetical, DUF883 family | 19.8/15.9 | 269:13:1,278 |
| PA1324 | Predicted to bind/transport polysaccharides | 19.1/14.3 | Operon |
| PA2167 | Hypothetical, unclassified | <2/20.7 | 72:21:470 |
| PA2176 | Hypothetical, unclassified | <2/7.0 | 76:19:296 |
| PA2177d | Predicted sensor response hybrid, PAS domain | 2.2/<2 | 99:101:244 |
| PA2717 | Cpo, chloroperoxidase | <2/14.4 | ND |
| PA3040d | Hypothetical, DUF883 family | 7.1/6.2 | 1,352:1,073:4,148 |
| PA3459d | Predicted glutamine amidotransferase | 4.4/<2 | 402:415:839 |
| PA3691 | Hypothetical, predicted lipoprotein | 15.7/17.8 | 608:13:2,216 |
| PA3795d | Predicted oxidoreductase | 5.2/8.8 | 707:605:1,593 |
| PA3819 | SlyB homolog, outer membrane protein | 10.3/8.2 | 1,221:568:3,351 |
| PA4311 | Predicted glycosyltransferase | 4.0/6.9 | ND |
| PA4394 | Predicted mechanosensitive channel, McsS | 3.0/4.0 | 130:15:797 |
| PA4717d | Predicted periplasmic metalloprotease | 5.3/6.3 | 1,404:1,101:3,215 |
| PA5107 | Lipocalin Blc, outer membrane protein | 8.1/4.4 | Operon |
| PA5108 | Hypothetical, predicted lipoprotein | 7.5/3.0 | 714:387:3,732 |
| PA5178 | Hypothetical, LysM domain | 9.6/7.1 | 452:284:2,532 |
| PA5212 | Hypothetical, unclassified | 15.9/7.8 | 1,555:44:5,758 |
| PA5424d | Hypothetical, predicted inner membrane protein | 8.3/7.4 | 807:660:3,904 |
PAO1 gene names and descriptions were obtained from the Pseudomonas Genome Database (33) and are listed numerically. Sequence-defined transposon insertions in each gene were purchased from the University of Washington Genome Center. All were transduced into the PAO1/PDO1 reference isolate and verified by PCR analysis.
Stress induction shows data previously described (19) for the fold increase of the genes’ transcriptional activity expressed from the PAO1 chromosome when treated with d-cycloserine (400 µg/ml) for 15 or 60 min as determined by microarray analysis.
PAO1 (wild type [WT]), PDO-LS586 (algT), and PDO351 (mucA) strains containing each lacZ fusion plasmid were grown in L broth at 37°C with aeration, collected during logarithmic growth, and assayed for β-galactosidase activity. Data show each gene’s promoter activity (Miller units) when fused to lacZ (P-lacZ) when expressed in the wild type or in an algT or mucA mutant. “Operon” indicates that the gene is in an operon of a gene already tested. ND, not determined.
The mutant did not show the predicted σ22 stimulon phenotype until tested under cell wall stress conditions (see Table 2).
Verification of membership in the σ22 stress stimulon.
To verify that the genes in Table 1 were indeed upregulated by σ22 activation, plasmids that contained each gene’s upstream promoter region transcriptionally fused to lacZ using broad-host-range vector pSS269 were constructed (27). All the reporter plasmids showed β-galactosidase activity in wild-type PAO1 (Table 1), indicating that their promoters had been cloned to form lacZ fusions. Compared to PAO1, the promoter activity of σ22 regulon members should show reduced expression in an algT mutant (PDO-LS586) devoid of σ22 and high expression in a mucA mutant (PDO351) lacking the anti-sigma factor for σ22. In general, these genes’ promoter reporters (P-lacZ) showed this predicted expression pattern, indicating that they were directly or indirectly under the control of σ22 (Table 1).
Many of the promoters fused to lacZ (P-lacZ) showed a dramatic reduction in β-galactosidase activity in the algT mutant (i.e., devoid of σ22) as expected, but some had only a modest reduction or none in the algT mutant (i.e., PA0567, PA0920, PA2177, PA3040, PA3459, PA3795, PA4717, and PA5424) (Table 1). Thus, their P-lacZ reporters were examined under conditions of cell wall stress in wild-type PAO1 and in an algT (PDO-LS586) mutant (Table 2). The results showed that all but one (PA0567) had a ≥5-fold increase in β-galactosidase activity in PAO1 as a result of exposure to d-cycloserine, which as predicted was not observed in the algT mutant. With PA0567-lacZ, β-galactosidase activity rose by only 2.3-fold following cell wall stress and then activity actually increased instead of decreasing in the algT mutant, suggesting that the regulation of PA0567 is more complex. Nevertheless, we kept this gene in our study because PA0567-lacZ transcriptional activity was markedly elevated in the mucA mutant (Table 1), indicating that this gene was under σ22 control.
TABLE 2 .
Confirmation of σ22-dependent gene expression during cell wall stress for selected promoters
| Promoter-lacZ fusiona | Fold increase in β-galactosidase activity due to d-cycloserine treatment |
|
|---|---|---|
| PAO1 | algT | |
| PA0567-lacZ | 2.3 | 10.9 |
| PA0920-lacZ | 5.6 | 2.6 |
| PA2177-lacZ | 14.2 | 2.3 |
| PA3040-lacZ | 10.0 | 2.2 |
| PA3459-lacZ | 9.0 | 2.4 |
| PA3795-lacZ | 11.4 | 3.3 |
| PA4717-lacZ | 8.8 | 2.8 |
| PA5424-lacZ | 19.2 | 1.7 |
Listed are promoter-lacZ fusions from Table 1 that did not yield the predicted phenotypes under unstressed conditions in algT and/or mucA mutant backgrounds. Here they were compared for β-galactosidase activity under cell wall stress conditions in PAO1 and an algT mutant by exposure to d-cycloserine. Bacteria were grown under routine lab conditions (L broth with aeration at 37°C) to an OD600 of 0.3 and treated with a sub-MIC level (400 µg/ml) of d-cycloserine for 60 min. The fold increase in β-galactosidase activity (Miller units) shown is a comparison to that of untreated control cultures. In PAO1 containing functional σ22, all 8 promoter fusions above showed an increase in transcriptional activity when exposed to d-cycloserine. None of these promoters, except PA0567-lacZ, showed high induction in the σ22 knockout, PAO1algT, indicating their dependence on σ22 for increased expression during cell wall stress.
Phenotypes of σ22 stimulon mutants.
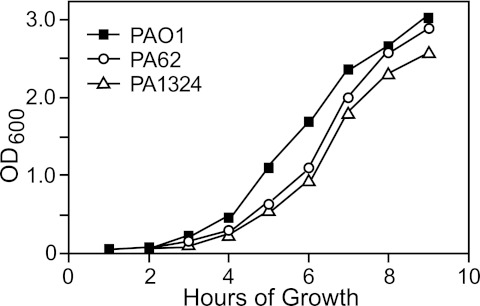
Because the genes of the σ22 stress stimulon were discovered by their upregulation following peptidoglycan damage, we looked at the mutants for changes in sensitivity to cell wall-inhibiting antibiotics (i.e., d-cycloserine, fosfomycin, and carbenicillin [Cb]); however, none showed an altered zone of inhibition compared to parent strain PAO1 in a standard disk diffusion assay (data not shown). Sensitivity to elevated growth temperatures is also a common phenotype of mutants defective in the ability to recover from stress, but the pattern of growth at 43°C in L broth with aeration showed no change from that of wild-type PAO1 (data not shown). However, when the growth temperature was raised to 45°C, the PA0062 and PA1324 mutants did show slightly lower growth rates than did PAO1 (Fig. 2). Because previous studies indicate a link between the σ22 stimulon and the regulation of biofilm formation (24), we looked for biofilm defects in the mutants with knockouts in genes of the σ22 stimulon. We examined microbial adherence to the walls of polystyrene tubes (24-h incubation at 37°C) and at the ability to form air-to-medium interface flocculation in statically grown culture (over 12 days at 25°C) using methods previously described (28). However, no obvious alteration from the wild-type PAO1 biofilm phenotypes could be observed with any of the mutants (data not shown).
FIG 2 .
Demonstration of a temperature-sensitive (Ts) growth phenotype. Mutants with Tn5-related (Tcr) insertions in genes of the σ22 stimulon were examined for growth defects compared to the parental strain, PAO1, in L broth at 45°C in a shaking water bath incubator. Among the 27 mutants examined, mutations in PA0062 or PA1324 caused a modest temperature-sensitive growth defect at 45°C. Shown is one of three experiments, all of which showed comparable results.
The σ22 stress stimulon genes are coregulated with those for alginate production, so we examined the possibility that some of the mutants might be altered in envelope homeostasis that would activate AlgW protease sufficiently to cause alginate biosynthesis. However, none showed a mucoid phenotype on agar plates, nor was production of alginate measurably above background PAO1 levels (data not shown). To determine if any of these mutations might cause a block in alginate production, the gene-specific transposon insertions (Tcr) were transduced into the chromosome of an isogenic mucoid strain, PDO351 (mucA::Gmr). However, all of these double mutants retained the mucoid phenotype and produced levels of alginate similar to that produced by PDO351 (data not shown), indicating that none of the genes under study was required for high-level alginate production.
Effect of mutations in the σ22 stimulon on PalgD induction in planktonic or sessile culture.
To determine whether any of the selected genes in the σ22 stress stimulon have a role in envelope homeostasis, we examined the mutants for effects on σ22 activation, which responds with high sensitivity to disturbances in the cell wall. Expression of PalgD, a well-characterized σ22-dependent promoter for an important virulence factor, was examined using a lacZ transcriptional fusion (PalgD-lacZ) as an indicator of the σ22 activity level under unstressed and stressed conditions. When wild-type PAO1 carried a PalgD-lacZ reporter plasmid (pLW149a) and was grown under planktonic conditions (i.e., L broth with aeration), it produced only a low level of β-galactosidase (13 ± 7 Miller units), but when stressed by exposure to d-cycloserine at a sub-MIC for 60 min, expression increased ~100-fold (1,161 ± 231 Miller units). Thus, when normalized to 100%, the planktonic PalgD-lacZ untreated/treated ratio of expression in PAO1 was typically ~1:100 (Table 3). This 1:100 ratio was then compared to the expression of PalgD-lacZ in the mutants of the σ22 stress stimulon under the same conditions.
TABLE 3 .
Comparison of PalgD induction in mutants of the σ22 stimulon when in the planktonic and sessile states of growth
| Phenotype | Strain/ mutation |
Activity of untreated planktonic PalgD-lacZ:activity of that treated with d-cycloserinea |
Sessile PalgD-cat treated with d-cycloserineb |
Putative role of gene product |
|
|---|---|---|---|---|---|
| Wild type | PAO1 | 1:100 | Ring | ||
| Alterations in stressed PalgD expression |
PA3459 |
1:67 |
Weaker ring |
Osmoprotectant |
|
| PA5424 | 1:52 | Weaker ring | Membrane protein | ||
| PA1243 | 1:153 | Ring | Sensor/regulator | ||
| PA5107 | 1:134 | Ring | Lipocalin | ||
| Growth-independent alterations in unstressed PalgD expression |
PA0062 |
6:131 (Ts) |
Lawn |
Lipoprotein |
|
| PA1324 | 1.7:151 (Ts) | Lawn | Polysaccharide binding | ||
| Planktonic growth-specific alterations in unstressed PalgD expression |
PA5178 | 7:215 | Ring | LysM, BON domains | |
| Sessile growth-specific alterations in unstressed PalgD expression |
PA0567 |
1:100 |
Lawn |
UPF0057 domain |
|
| PA3040 | 1:100 | Lawn | DUF883 family | ||
| PA2717 | 1:142 | Lawn | Chloroperoxidase | ||
| PA0920 | 1:55 | Lawn | A-PG synthasec |
PalgD-lacZ Miller units of β-galactosidase in PAO1 strains were 13 (±7) if untreated and 1,161 (±231) if treated with d-cycloserine. To be significant in the fold change, the ratio needed to be <0.5 or >1.5 for untreated cultures and <80 or >120 for treated cultures. When maximum activity was normalized to 100%, the untreated/treated ratio of PalgD-lacZ expression in PAO1 was typically 1:100. Ts indicates a temperature-sensitive growth defect at 45°C.
PalgD-cat activity in PAO1 strains was estimated on L agar plates containing chloramphenicol to prevent growth unless the cat fusion was activated by d-cycloserine placed in the center of the plate, which resulted in a ring of growth. In some mutants, a weaker ring of growth was observed. A lawn of growth indicated constitutive PalgD-lacZ expression under such sessile conditions.
A-PG, aminoacyl-phosphatidyl glycerol.
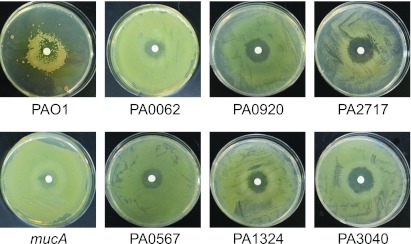
We also employed another tool for measuring PalgD induction, but under sessile conditions using a plate bioassay (22). Here, a low-copy-number reporter plasmid (pLW117, Gmr) has the algD promoter fused to a promoterless cat gene (PalgD-cat) such that growth occurs in the presence of chloramphenicol (Cm) when PalgD is activated. When a culture of PAO1(pLW117) was spread onto an L agar plate containing Cm (50 µg/ml) and d-cycloserine was spotted in the center of the plate, a ring of growth formed around the antibiotic due to the activation of the PalgD-cat fusion (Fig. 3, PAO1). When the PalgD-cat fusion was tested in a mucA mutant (PDO351), a lawn of growth on the Cm plate was observed because σ22 is constitutively active (Fig. 3, mucA); this lawn phenotype did not require stress for induction (data not shown). These PalgD-cat phenotypes allowed us to test σ22 activity under sessile (i.e., biofilm-like) conditions for comparison to the above PalgD-lacZ phenotypes under planktonic culture conditions.
FIG 3 .
Sessile growth assay for observing the effects of mutations in genes of the σ22 stimulon on the activation of the σ22-dependent promoter, PalgD. Shown are pictures of plates of L agar plus chloramphenicol (Cm) coated with mutant derivatives of PAO1 that hosted pLW117, a plasmid containing PalgD-cat, which confers Cmr and thus growth when activated. A 5-mm filter disk impregnated with d-cycloserine was placed in the center of the plate. With PAO1, exposure to d-cycloserine (10 µg) in the center of the plate produced a ring of Cmr growth after 3 days at 25°C. A mucA mutant (devoid of the σ22 anti-sigma factor) is shown as an example of σ22 derepression. The other plates show a lawn of growth with the mutants indicated, which occurred with or without d-cycloserine and indicated constitutive activation of PalgD-cat due to the mutation.
The PalgD-lacZ and PalgD-cat reporter plasmids (pLW149a and pLW117, respectively) were transferred to each of the mutant strains for comparison of the effects of planktonic and sessile conditions. When grown under the same planktonic conditions, most of them produced β-galactosidase at the same ~1:100 untreated/treated ratios, similar to the wild type. Likewise, in the sessile state of growth, most showed the same ring of growth as did the wild type upon exposure to d-cycloserine. However, there were 11 mutants that showed distinguishing phenotypes compared to the wild type and are presented in Table 3. Four mutants showed wild-type levels of PalgD-lacZ activity under nonstressed conditions but showed alterations in the response to cell wall stress. Two (PA3459 and PA5424) showed reduced responsiveness to stress in the planktonic assay; in the sessile assay, they also displayed a consistently weaker ring of growth (data not shown). Two other mutants (PA1243 and PA5107) showed an increased response to stress when in planktonic culture, although a normal ring of growth was seen in the plate assay rather than an enlarged ring, which might have been predicted.
Three mutants (PA0062, PA1324, and PA5178) had activity higher than that of the wild-type PalgD-lacZ under planktonic and unstressed conditions (Table 3), indicating that loss of this gene product caused σ22 activation even without stress. This suggests that the loss of these gene products affected envelope homeostasis such that AlgW-mediated degradation of MucA is increased. Two of these (PA0062 and PA1324) showed the same results under sessile-biofilm conditions when carrying PalgD-cat and produced a lawn (Fig. 3), which was consistent with elevated σ22 activation in the absence of stress. Both of these mutants also showed a temperature-sensitive phenotype at 45°C (Fig. 2). Interestingly, the PA5178 mutant produced a wild-type ring of growth when PalgD-cat was induced by d-cycloserine, rather than a lawn, indicating that its role in envelope homeostasis was observable only under planktonic conditions.
There were four other mutants (PA0567, PA3040, PA2717, and PA0920) with elevated σ22 activity that grew as a lawn when expressing algD-cat (Fig. 3), indicating constitutive PalgD-cat expression under unstressed sessile conditions. However, all showed normal PalgD-lacZ activity under unstressed planktonic conditions (Table 3). Thus, the roles of these σ22 stimulon gene products in maintaining envelope homeostasis were dependent on the sessile-biofilm state of growth.
DISCUSSION
The σ22 stimulon is a large stress response system in P. aeruginosa that includes the genes for the production of alginate, an important virulence factor; however, most of the gene products in the stimulon have undefined functions (19). Here we sought to better understand the σ22 stress response system by examining the expression of 27 undercharacterized genes of this stimulon and the phenotypes resulting from their mutations. Interestingly, none of the mutations of this stress response stimulon had a major effect on typical stress-related phenotypes like growth rate, temperature sensitivity, or alginate production.
We considered the recent observation that a mutation in algT/U, which disrupts the whole σ22 stimulon, results in less robust (i.e., less shear-resistant) biofilms (24). Biofilm growth of P. aeruginosa occurs on natural surfaces in the environment and during human infections, such as in the lungs of CF patients, causing chronic bronchopneumonia (6, 7). The development of a biofilm is initiated by planktonic (freely moving) bacteria that attach to a surface (become sessile) and form microcolonies that produce a polymeric matrix and become increasingly tolerant to antibiotics (29). When we compared the 293 genes in the σ22 stimulon (19) to those activated during biofilm development (30), we found that 59% of the σ22 stimulon genes were in common. Among the genes selected in our study, all but one (PA0919) are upregulated in PAO1 biofilms as determined by cluster analysis of whole-genome expression profiles of PAO1 transcriptomes derived from planktonic cultures that developed into mature biofilms (30). In addition, a recent screen of a strain PA14 transposon insertion library for biofilm defects revealed that mutants in 3 of the genes studied here (PA0854, PA1243, and PA4394) exhibit reduced biofilm formation as observed by microscopic analysis (31). Given this link between the σ22 stimulon and biofilm formation, we examined our σ22 stimulon mutants for biofilm defects. However, no obvious alteration from the wild-type biofilm phenotypes was observed using simple biofilm models. Instead, our studies focused on potential effects that the mutations might have on envelope homeostasis, as measured by σ22 activity on the alginate operon promoter, PalgD, during planktonic and sessile growth conditions.
We expected that many of the genes in the σ22 stimulon would contribute to cell envelope homeostasis because their expression was upregulated during extracytoplasmic stress induced by exposure to d-cycloserine. This is an antibiotic that directly causes disruption of the peptidoglycan cell wall. Disruption of envelope homeostasis results in the activation of σ22, which can be measured by the expression of a σ22 promoter. One such σ22 promoter is PalgD, the promoter of the alginate operon. Like other characterized ECF sigma factors, such as σE in Escherichia coli (32), σ22 is known to be activated by stress conditions via signal RIP degradation of the anti-sigma factor that normally sequesters the sigma factor, thus releasing or “activating” sigma factor activity (Fig. 1). We have previously shown that in P. aeruginosa PAO1, σ22 activation is highly responsive to cell wall stress (19, 22). Indeed, our mutant analysis of 27 genes of the σ22 stimulon revealed that over one-third of them (i.e., 11 of 27) had effects on σ22 activation under conditions of stress or nonstress.
Four mutants that altered the ability of P. aeruginosa to respond to cell wall stress with σ22 activation were found. Mutations in PA3459 and PA5424 reduced PalgD responsiveness to cell wall stress under planktonic and sessile conditions. This suggests that their gene products are required for full activation of the stress response system involved in σ22 control. PA3459 is the first gene of a three-gene operon (33) that putatively encodes functions involved in the synthesis of a cytoplasmic osmoprotectant, N-acetyl-glutaminyl-glutamine amide (NAGGN) and is upregulated in response to osmotic stress (34). Thus, loss of this osmoprotectant resulted in a reduced ability to respond to cell wall stress in both planktonic and sessile states of growth. PA5424 encodes a small (81-amino-acid) conserved hypothetical protein predicted to contain 3 transmembrane helices and to localize to the inner membrane. Its closest homolog is the hypothetical protein YeaQ in E. coli, sharing 61% similarity, but for which little else is known.
Mutants with defects in PA1243 and PA5107 showed elevated expression during cell wall stress that could be measured during planktonic growth. PA1243 is predicted to encode a sensor/response regulator hybrid protein, located in the cytoplasmic membrane. It contains both a histidine kinase domain and a sensory domain (with a PAS motif). Thus, PA1243 may directly control a subset of genes in the σ22 stimulon under conditions recognized by its sensor domain. Interestingly, it was reported that a transposon insertion in PA1243 of strain PA14 exhibits reduced biofilm formation (31). PA5107 is predicted to encode a bacterial lipocalin (Blc), which is an outer membrane lipoprotein. Blc in E. coli is known to be expressed under cell envelope stress conditions caused by high osmolarity (35). Recent reports indicate that E. coli Blc is a dimer with a binding preference for lysophospholipids, which suggests a role for this protein in the storage/transport of lipids important for membrane biogenesis and repair (36).
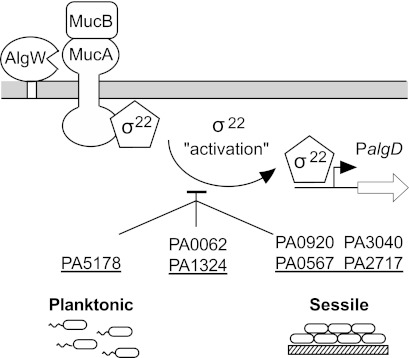
Of particular interest to this study were the 7 mutants that demonstrated elevated σ22 activation even in the absence of stress: PA0062, PA1324, PA5178, PA2717, PA0567, PA3040, and PA0920. This suggests that these gene products are important for maintaining envelope homeostasis and/or preventing AlgW from spontaneously activating the σ22 stress response (Fig. 4). Mutations in PA0062 and PA1324 caused higher expression of PalgD with and without stress induction, and this was observed under planktonic and sessile conditions. Interestingly, mutations in PA0062 or PA1324 also caused mild temperature-sensitive growth defects at 45°C. PA0062 encodes a hypothetical uncharacterized protein that is probably a lipoprotein. The PA1324 open reading frame (ORF) is classified in the Pseudomonas Genome Database (33) as an unknown hypothetical protein with a predicted type II lipoprotein signal. Recently, a nuclear magnetic resonance (NMR) structure of the PA1324 protein predicted a pre-albumin-like fold, and based on ligand screening studies, it is postulated to be involved in the binding and/or transport of polysaccharides (37).
FIG 4 .
Illustration of the roles of specific gene products in the σ22 stimulon on the “activation” of σ22 activity (i.e., by release from the MucA-MucB inhibitory complex). This was determined by examining the effect of mutations on envelope homeostasis, which leads to activation of PalgD. Mutations showed their effects when P. aeruginosa grew in the planktonic state of growth (left), the sessile/biofilm mode (right), or both (center).
The PA5178 mutant showed higher constitutive and induced expression of PalgD only during planktonic growth, while induced and uninduced PalgD expression appeared normal under sessile conditions. Thus, PA5178 plays a role in envelope homeostasis primarily during the planktonic mode of growth. PA5178 encodes an unclassified hypothetical protein that is predicted to contain one LysM domain, which is a widely distributed peptidoglycan-binding domain (38), suggesting that it acts directly on the cell wall. It also has a BON domain, which is a putative phospholipid-binding region (39). A proteomic analysis of P. aeruginosa outer membranes shows that the cellular concentration of this protein is increased with increasing resistance to several antibiotics, including ampicillin (40).
The rest of the mutants (PA0920, PA0567, PA3040, and PA2717) showed defects in unstressed envelope homeostasis that could be observed only in the sessile state of growth (Fig. 4). Mutations in PA0567 and PA3040 had no effect on PalgD expression and induction during planktonic growth but caused constitutive PalgD expression under sessile growth. Thus, the effects due to the loss of these gene products were severe enough to trigger the σ22 stress response only in adherent cultures. PA0567 encodes a small, 52-amino-acid, hydrophobic peptide with 2 predicted transmembrane helices. It shares 78% similarity with the proteolipid YqaE of E. coli, and both are members of the Pfam domain UPF0057. UPF0057 proteolipids have been characterized in yeast and plants as stress-responsive proteins that help maintain membrane integrity under environmental conditions of high salinity and low temperatures (41). PA3040 encodes an ortholog of the DUF883 family of membrane proteins. It is the first gene of a three-gene operon expressing hypothetical proteins. Interestingly, another member of the DUF883 protein family is PA1323, which was also included in this study of σ22 stimulon members, although its inactivation did not lead to any apparent alteration in σ22 activity.
PA2717 is predicted to encode a cytoplasmic chloroperoxidase (Cpo). The PA2717 mutant was normal for unstressed PalgD activity under planktonic conditions, but elevated PalgD expression was seen under planktonic stressed conditions. It formed a lawn in the plate assay, indicating constitutive PalgD activity under sessile conditions, suggesting that it normally attenuates σ22 activation in the absence of stress.
The mutation in PA0920 was especially interesting in that under planktonic conditions PalgD expression was normal if unstressed but showed lower responsiveness (i.e., ~50% PalgD-lacZ activity) upon cell wall stress. However, the PA0920 mutation had the opposite effect under sessile growth and caused constitutive PalgD expression. PA0920 has recently been shown to encode an integral inner membrane enzyme required for the production of 2′-alanyl-phosphatidylglycerol in P. aeruginosa membranes (42). A PA0920 mutant analysis showed that alanyl-phosphatidylglycerol in P. aeruginosa PAO1 membranes confers increased resistance to the toxic effects of the heavy metal Cr3+, the osmolyte sodium lactate, the cationic peptide protamine sulfate, and cefsulodin, a β-lactam that impedes cross-linking of peptidoglycan. PA0920 is induced under acidic growth conditions, and here we show that it is also induced 5-fold by d-cycloserine as part of the σ22 stimulon. Changing the amount of alanyl-phosphatidylglycerol in P. aeruginosa membranes is a mechanism to control the fluidity and permeability of the cellular membranes, which are essential to maintaining envelope homeostasis (42). Here we also observed that loss of alanyl-phosphatidylglycerol in PAO1 membranes resulted in a disruption of envelope homeostasis leading to σ22 activation, but only under sessile/biofilm conditions.
In conclusion, we have found σ22-dependent genes whose loss resulted in altered stress responsiveness or constitutive activation of the σ22 sensory system that monitors cell envelope homeostasis. The effects were often dependent upon whether P. aeruginosa was in a planktonic or a sessile state of growth, suggesting an overlap with biofilm control mechanisms. Continued studies on the role of σ22 stimulon genes may reveal other links to the biofilm mode of growth and potential solutions for disrupting biofilms that often lead to antibiotic tolerance and persistence during infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains of P. aeruginosa used in this study are shown in Table 4. The PAO1 parent strain used in this study, also known as PDO1 (22), is a spontaneous Cms isolate of the original Cmr PAO1 strain obtained from B. W. Holloway (43). E. coli DH5α was used for routine plasmid manipulations. Bacteria were routinely cultured in L broth (10 g tryptone, 5 g yeast extract, and 5 g NaCl each per liter) or on L agar. LPIA plates were a 1:1 mix of L agar and Pseudomonas isolation agar (Difco) and were used to counterselect against E. coli following conjugal plasmid transfers to P. aeruginosa. Antibiotics (Sigma) were used at the concentrations indicated: gentamicin (Gm), 20 µg/ml and 100 µg/ml for E. coli and P. aeruginosa, respectively; tetracycline (Tc), 20 µg/ml and 60 µg/ml for E. coli and P. aeruginosa, respectively; kanamycin (Km), 30 µg/ml, and ampicillin (Ap), 100 µg/ml, both for E. coli; and carbenicillin (Cb), 100 µg/ml for P. aeruginosa.
TABLE 4 .
P. aeruginosa strains and plasmids used in this studya
| Strain or plasmid | Genotype/phenotype | Source or reference |
|---|---|---|
| P. aeruginosa | ||
| PAO1/PDO1 | Wild type, Cms | This laboratory |
| PDO-LS586 | algT::aacCI | 47 |
| PDO351 | mucA::aacCI (Gmr) Alg+ | 22 |
| PDO358 | algW::aacCIΩ | 22 |
| PDO-PA0059 | PDO1, PA0059::Tn (Tcr) (transduced) | This study |
| PDO-PA0062 | PDO1, PA0062::Tn (Tcr) (transduced) | This study |
| PDO-PA0460 | PDO1, PA0460::Tn (Tcr) (transduced) | This study |
| PDO-PA0567 | PDO1, PA0567::Tn (Tcr) (transduced) | This study |
| PDO-PA0854 | PDO1, PA0854::Tn (Tcr) (transduced) | This study |
| PDO-PA0919 | PDO1, PA0919::Tn (Tcr) (transduced) | This study |
| PDO-PA0920 | PDO1, PA0920::Tn (Tcr) (transduced) | This study |
| PDO-PA1243 | PDO1, PA1243::Tn (Tcr) (transduced) | This study |
| PDO-PA1323 | PDO1, PA1323::Tn (Tcr) (transduced) | This study |
| PDO-PA1324 | PDO1, PA1324::Tn (Tcr) (transduced) | This study |
| PDO-PA2167 | PDO1, PA2167::Tn (Tcr) (transduced) | This study |
| PDO-PA2176 | PDO1, PA2176::Tn (Tcr) (transduced) | This study |
| PDO-PA2177 | PDO1, PA2177::Tn (Tcr) (transduced) | This study |
| PDO-PA2717 | PDO1, PA2717::Tn (Tcr) (transduced) | This study |
| PDO-PA3040 | PDO1, PA3040::Tn (Tcr) (transduced) | This study |
| PDO-PA3459 | PDO1, PA3459::Tn (Tcr) (transduced) | This study |
| PDO-PA3691 | PDO1, PA3691::Tn (Tcr) (transduced) | This study |
| PDO-PA3795 | PDO1, PA3795::Tn (Tcr) (transduced) | This study |
| PDO-PA3819 | PDO1, PA3819::Tn (Tcr) (transduced) | This study |
| PDO-PA4311 | PDO1, PA4311::Tn (Tcr) (transduced) | This study |
| PDO-PA4394 | PDO1, PA4394::Tn (Tcr) (transduced) | This study |
| PDO-PA4717 | PDO1, PA4717::Tn (Tcr) (transduced) | This study |
| PDO-PA5107 | PDO1, PA5107::Tn (Tcr) (transduced) | This study |
| PDO-PA5108 | PDO1, PA5108::Tn (Tcr) (transduced) | This study |
| PDO-PA5178 | PDO1, PA5178::Tn (Tcr) (transduced) | This study |
| PDO-PA5212 | PDO1, PA5212::Tn (Tcr) (transduced) | This study |
| PDO-PA5424 | PDO1, PA5424::Tn (Tcr) (transduced) | This study |
| Plasmids | ||
| pKK61 | pCP19 (oriVRK2, Tcr) PalgD-cat | 22 |
| pLW117 | pKK61 aacC1(Gmr) PalgD-cat | This study |
| pLW127 | pSS269, PA0059osmC(−239/+50)-lacZ | 19 |
| pLW148 | pSS269, PA5178(−501/+50)-lacZ | This study |
| pLW149a | pSS269, PA3540 algD(−925/+50)-lacZ | 19 |
| pLW150 | pSS269, PA4394(−255/+60)-lacZ | This study |
| pLW152 | pSS269, PA3819(−660/+70)-lacZ | This study |
| pLW155 | pSS269, PA3691(−372/+59)-lacZ | 19 |
| pLW166 | pSS269, PA5108(−163/+70)-lacZ | 19 |
| pLW168 | pSS269, PA4717(−1130/+50)-lacZ | 19 |
| pLW179 | pSS269, PA1323(−450/+50)-lacZ | This study |
| pLW182 | pSS269, PA2176(−450/+50)-lacZ | This study |
| pLW183 | pSS269, PA2177(−450/+50)-lacZ | This study |
| pLW185 | pSS269, PA2167(−500/+50)-lacZ | This study |
| pLW186 | pSS269, PA0460(−500/+50)-lacZ | This study |
| pLW187 | pSS269, PA0567(−500/+50)-lacZ | This study |
| pLW188 | pSS269, PA0920(−500/+50)-lacZ | This study |
| pLW189 | pSS269, PA3040(−500/+50)-lacZ | This study |
| pLW190 | pSS269, PA3459(−500/+50)-lacZ | This study |
| pLW191 | pSS269, PA3795(−500/+50)-lacZ | This study |
| pLW194 | pSS269, PA5212(−500/+50)-lacZ | This study |
| pLW195 | pSS269, PA5424(−500/+50)-lacZ | This study |
| pRK2013 | ColE1-Tra(RK2)+ Kmr | 48 |
| pSS269 | pSS223 lacZ SF Apr | 27 |
All P. aeruginosa strains were derived from PDO1, a spontaneous Cmr isolate of strain PAO1 previously described (28). Abbreviations: Gmr, aacCI-encoded gentamicin resistance; Apr, ampicillin/carbenicillin resistance; Kmr, kanamycin resistance; Alg+, mucoid due to alginate overproduction; lacZ, β-galactosidase reporter in transcriptional fusions; SF, stabilization fragment for replication in P. aeruginosa. Numbers in parentheses before lacZ indicate the promoter region in base pairs relative to the start of translation (+1) of the open reading frame used to make the transcriptional reporter.
Mutant strain construction.
Mutants of P. aeruginosa PAO1 with sequence-defined transposon (Tn5-derived, Tcr) insertions of Tn5-lacZ or Tn5-phoA were purchased from the University of Washington Genomics Resource Center (WGRC, http://www.gs.washington.edu/labs/manoil/libraryindex.htm). Such Tn5-marked alleles were transduced into this laboratory’s PAO1 reference isolate (PDO1) with selection for tetracycline resistance (Tcr). Plate lysates with the generalized transducing phage F116L (43) were made with each WGRC mutant by incubating 10 ml F116L lysate at various 1:10 dilutions with 0.1 ml of an overnight L broth culture containing 10 mM MgSO4 and 5 mM CaCl2, which was then incubated at room temperature for 10 min to permit adsorption. Molten top agar at 50°C (3 ml L broth with 0.7% agar) was added to each tube and immediately poured onto the surface of an L agar plate. After 24 h at 37°C, 3 ml of L broth was added to a plate showing confluent plaques, the top agar was extracted with a sterile spreader, and the mix was centrifuged to remove the agar and bacterial cells. The supernatant was passed through a 0.45-µm filter to remove any residual bacteria. For transductions, 0.1 ml of a transducing lysate was mixed with 0.2 ml of the P. aeruginosa host strain grown overnight in L broth, which was incubated at room temperature for 15 min and was then shaken at 225 rpm at 37°C for 1 h; the culture was then plated onto L agar containing selective antibiotics and incubated at 37°C until colonies were observed. All transduced transposon insertions were examined by PCR to verify that each insertion was in the correct gene. For this, primers specific for Tn5-lacZ (LacZ148, GGGTAACGCCAGGGTTTTCC) or Tn5-phoA (PhoA138, CGGGTGCAGTAATATCGCCCT) and primers specific for the 5′ and 3′ ends of the gene of interest using sequences obtained from the Pseudomonas Genome Database (33) were used in standard PCRs with Taq polymerase; the products were observed using agarose gel electrophoresis for size analysis. To examine the effect of such mutations on the mucoid phenotype, Tn5 (Tcr)-mutated alleles were transduced into PDO351 (i.e., mucA::Gm). There were some F116L lysates of WGRC Tn5 mutants (i.e., PA0062, PA567, PA920, PA1323, PA1324, PA2717, PA3040, PA3819, PA4717, PA5107, and PA5424) where transductants were rare, in which case the mucA::aacCI (Gmr) allele from PDO351 was transduced into verified PAO1/PDO1::Tn5 (Tcr) mutants instead. Information on domains in proteins of interest was found in the Pseudomonas Genome Database (33) found at http://www.pseudomonas.com.
Disk diffusion assay.
The relative level of resistance to antibiotics by mutants was compared to that of the wild type by a standard disk diffusion assay. A sample (0.1 ml) of culture in the early logarithmic phase was spread onto an L agar plate, and then a 5-mm paper disk impregnated with the antibiotic was placed in the center to permit radial diffusion. After 24 h of incubation at 37°C, the diameter of the ring of growth inhibition was measured.
Assay for alginate.
P. aeruginosa strains were grown on L agar plates at 37°C for 48 h, and then the growth was resuspended in saline. Bacterial cells were removed by centrifugation, and then the supernatant was tested for alginate (which is composed of two uronic acids) using the carbazole-spectrophotometric method for the assay of uronic acids described by Knutson and Jeanes (44). Alginic acid from Macrocystis pyrifera (Sigma) was used as the standard.
Promoter-lacZ reporter plasmids.
High-fidelity Pfu Turbo (Stratagene) was used to amplify DNA by PCR with all primers custom synthesized by Eurofins MWG Operon using sequences available in the Pseudomonas Genome Database (33). Promoter regions obtained by PCR amplification included DNA ~0.5 kb upstream of each selected ORF, which was cloned into the broad-host-range transcriptional lacZ reporter pSS269 (27). Reporter plasmids were transferred from E. coli to P. aeruginosa strains by triparental mating as described elsewhere (27) using helper plasmid pRK2013 to mobilize the oriT-containing plasmids to P. aeruginosa with selection on LPIA plates containing an appropriate selective antibiotic. It was observed that reporter plasmids in some Tn5 mutants (i.e., PA0062, PA0920, PA1243, PA1324, PA2177, and PA3819) were somewhat unstable, and so fresh conjugations of reporter plasmids into these mutants were followed immediately by β-galactosidase assays in order to obtain consistent results. Transcriptional activity of reporter constructs was determined by measuring β-galactosidase activity, reported in Miller units, from promoter-lacZ fusions. For assays verifying σ22 dependence, strains were grown in L broth to an optical density at 600 nm (OD600) of 1.0, at which time 0.1-ml aliquots were assayed. To determine promoter activation in response to cell wall stress induced by d-cycloserine, strains were grown in 25 ml L broth at 37°C to an OD600 of 0.3; cultures were split in half, d-cycloserine was added to one half (400 µg/ml), and then both were incubated for an additional 60 min, at which time 0.1-ml samples of untreated and treated cultures were assayed for β-galactosidase activity.
Bioassay for PalgD induction on a solid substrate.
Plasmid pKK61 with an algD-cat fusion in low-copy-number vector pLAFR1 (Tcr) was previously described (45). Because the mutants in this study were Tcr due to the transposon insertions, a Gmr derivative of pKK61 named pLW117 was constructed to permit selection of the plasmid transferred to the mutant strains. pLW117 was constructed by cloning an aacCIΩ (Gmr) cartridge (46) into the HindIII site of pKK61. Bioassays of PAO1/PDO1::Tn strains (Cms) carrying pLW117 were performed as previously described (22). Briefly, expression of algD-cat was tested by incubating cultures to an OD600 of 1.2, at which time a 1.5-ml sample was centrifuged, and the bacterial cell pellet was resuspended in 10 mM MgSO4 to an OD600 of 0.2. A 25-µl sample of this cell suspension was spread onto an L agar plate containing chloramphenicol at 50 µg/ml. A 5-mm filter disk impregnated with 1 mg d-cycloserine (i.e., 10 µl of a 100-mg/ml stock solution) was placed in the center of the plate. The test plates were then incubated at 25°C for 3 days and examined for a ring of Cmr growth around the disk, indicating induced expression of algD-cat.
Biofilm assays.
Static L broth cultures of PAO1 and mutant derivatives were compared for their abilities to adhere to the walls of polystyrene tubes after a 24-h incubation at 37°C as previously described (28). Also, the ability to form an air-to-medium interface flocculation during static-growth culture at 25°C was performed as previously described (28), and the flocculation was examined daily over a 12-day period.
ACKNOWLEDGMENTS
We gratefully acknowledge the VCU Nucleic Acids Core Facility for assistance in the microarray analyses.
This work was supported by Public Health Service grant AI-19146 from the National Institute of Allergy and Infectious Diseases (D.E.O.) and in part by Veterans Administration medical research grant I01BX000477 (D.E.O.).
Footnotes
Citation Wood LF, Ohman DE. 2012. Identification of genes in the σ22 regulon of Pseudomonas aeruginosa required for cell envelope homeostasis in either the planktonic or the sessile mode of growth. mBio 3(3):e00094-12. doi:10.1128/mBio.00094-12.
REFERENCES
- 1. Lyczak JB, Cannon CL, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051–1060 [DOI] [PubMed] [Google Scholar]
- 2. Govan JR, Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Leid JG, et al. 2005. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 175:7512–7518 [DOI] [PubMed] [Google Scholar]
- 4. Pier GB, Coleman F, Grout M, Franklin M, Ohman DE. 2001. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect. Immun. 69:1895–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schwarzmann S, Boring JR., III 1971. Antiphagocytic effect of slime from a mucoid strain of Pseudomonas aeruginosa. Infect. Immun. 3:762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bjarnsholt T, et al. 2009. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44:547–558 [DOI] [PubMed] [Google Scholar]
- 7. Moreau-Marquis S, Stanton BA, O’Toole GA. 2008. Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm. Pharmacol. Ther. 21:595–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jain S, Ohman DE. 2004. Alginate biosynthesis, p 53–81 In Ramos JL, Pseudomonas, vol 3: biosynthesis of macromolecules and molecular metabolism Kluwer Academic/Plenum Publishers, New York, NY. [Google Scholar]
- 9. Ramsey DM, Wozniak DJ. 2005. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol. Microbiol. 56:309–322 [DOI] [PubMed] [Google Scholar]
- 10. Chitnis CE, Ohman DE. 1993. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol. Microbiol. 8:583–590 [DOI] [PubMed] [Google Scholar]
- 11. Deretic V, et al. 1994. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. J. Bacteriol. 176:2773–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DeVries CA, Ohman DE. 1994. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternative sigma factor, and shows evidence for autoregulation. J. Bacteriol. 176:6677–6687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leech AJ, Sprinkle A, Wood L, Wozniak DJ, Ohman DE. 2008. The NtrC family regulator AlgB, which controls alginate biosynthesis in mucoid Pseudomonas aeruginosa, binds directly to the algD promoter. J. Bacteriol. 190:581–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lizewski SE, et al. 2004. Identification of AlgR-regulated genes in Pseudomonas aeruginosa by use of microarray analysis. J. Bacteriol. 186:5672–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wozniak DJ, Ohman DE. 1994. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J. Bacteriol. 176:6007–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baynham PJ, Ramsey DM, Gvozdyev BV, Cordonnier EM, Wozniak DJ. 2006. The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili. J. Bacteriol. 188:132–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martin DW, et al. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 90:8377–8381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cezairliyan BO, Sauer RT. 2009. Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB. Mol. Microbiol. 72:368–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wood LF, Ohman DE. 2009. Use of cell wall stress to characterize sigma 22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol. Microbiol. 72:183–201 [DOI] [PubMed] [Google Scholar]
- 20. Wood LF, Ohman DE. 2006. Independent regulation of MucD, an HtrA-like protease in Pseudomonas aeruginosa, and the role of its proteolytic motif in alginate gene regulation. J. Bacteriol. 188:3134–3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qiu D, Eisinger VM, Rowen DW, Yu HD. 2007. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 104:8107–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wood LF, Leech AJ, Ohman DE. 2006. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: roles of sigma (AlgT) and the AlgW and Prc proteases. Mol. Microbiol. 62:412–426 [DOI] [PubMed] [Google Scholar]
- 23. Qiu D, Eisinger VM, Head NE, Pier GB, Yu HD. 2008. ClpXP proteases positively regulate alginate overexpression and mucoid conversion in Pseudomonas aeruginosa. Microbiology 154:2119–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bazire A, et al. 2010. The sigma factor AlgU plays a key role in formation of robust biofilms by nonmucoid Pseudomonas aeruginosa. J. Bacteriol. 192:3001–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Firoved AM, Boucher JC, Deretic V. 2002. Global genomic analysis of AlgU (sigma(E))-dependent promoters (sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J. Bacteriol. 184:1057–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klockgether J, et al. 2010. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J. Bacteriol. 192:1113–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Suh SJ, Silo-Suh LA, Ohman DE. 2004. Development of tools for the genetic manipulation of Pseudomonas aeruginosa. J. Microbiol. Methods 58:203–212 [DOI] [PubMed] [Google Scholar]
- 28. Friedman L, Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675–690 [DOI] [PubMed] [Google Scholar]
- 29. Monds RD, O’Toole GA. 2009. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 17:73–87 [DOI] [PubMed] [Google Scholar]
- 30. Waite RD, et al. 2006. Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genomics 7:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Müsken M, Di Fiore S, Dötsch A, Fischer R, Häussler S. 2010. Genetic determinants of Pseudomonas aeruginosa biofilm establishment. Microbiology 156:431–441 [DOI] [PubMed] [Google Scholar]
- 32. Ades SE. 2008. Regulation by destruction: design of the sigmaE envelope stress response. Curr. Opin. Microbiol. 11:535–540 [DOI] [PubMed] [Google Scholar]
- 33. Winsor GL, et al. 2009. Pseudomonas genome database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 37:D483–D488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aspedon A, Palmer K, Whiteley M. 2006. Microarray analysis of the osmotic stress response in Pseudomonas aeruginosa. J. Bacteriol. 188:2721–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bishop RE. 2000. The bacterial lipocalins. Biochim. Biophys. Acta 1482:73–83 [DOI] [PubMed] [Google Scholar]
- 36. Campanacci V, Bishop RE, Blangy S, Tegoni M, Cambillau C. 2006. The membrane bound bacterial lipocalin Blc is a functional dimer with binding preference for lysophospholipids. FEBS Lett. 580:4877–4883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mercier KA, et al. 2009. Structure and function of Pseudomonas aeruginosa protein PA1324 (21-170). Protein Sci. 18:606–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Buist G, Steen A, Kok J, Kuipers OP. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 68:838–847 [DOI] [PubMed] [Google Scholar]
- 39. Yeats C, Bateman A. 2003. The BON domain: a putative membrane-binding domain. Trends Biochem. Sci. 28:352–355 [DOI] [PubMed] [Google Scholar]
- 40. Peng X, et al. 2005. Proteomic analysis of the sarcosine-insoluble outer membrane fraction of Pseudomonas aeruginosa responding to ampicilin, kanamycin, and tetracycline resistance. J. Proteome Res. 4:2257–2265 [DOI] [PubMed] [Google Scholar]
- 41. Koike M, et al. 2005. Molecular characterization of a cold-induced plasma membrane protein gene from wheat. Mol. Genet. Genomics 274:445–453 [DOI] [PubMed] [Google Scholar]
- 42. Klein S, et al. 2009. Adaptation of Pseudomonas aeruginosa to various conditions includes tRNA-dependent formation of alanyl-phosphatidylglycerol. Mol. Microbiol. 71:551–565 [DOI] [PubMed] [Google Scholar]
- 43. Holloway BW. 1969. Genetics of Pseudomonas. Bacteriol. Rev. 33:419–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Knutson CA, Jeanes A. 1968. A new modification of the carbazole analysis: application to heteropolysaccharides. Anal. Biochem. 24:470–481 [DOI] [PubMed] [Google Scholar]
- 45. Wozniak DJ, Ohman DE. 1991. Pseudomonas aeruginosa AlgB, a two-component response regulator of the NtrC family, is required for algD transcription. J. Bacteriol. 173:1406–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schweizer HP. 1993. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques 15:831–833 [PubMed] [Google Scholar]
- 47. Silo-Suh L, Suh SJ, Sokol PA, Ohman DE. 2002. A simple alfalfa seedling infection model for Pseudomonas aeruginosa strains associated with cystic fibrosis shows AlgT (sigma-22) and RhlR contribute to pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 99:15699–15704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]