Abstract
How do the actions of individual genes contribute to the complex morphologies of animals and plants? How widespread are these genes taxonomically? How many genes are involved in the morphological differences observed between species, and can we identify them? To what extent can empirical data and theory be reconciled? We provide an overview of some recent attempts to answer these questions, answers that have taken us to the threshold of understanding the mechanistic basis and evolutionary factors that underlie morphological innovation.
Over the past two decades, developmental biologists have made great strides in understanding embryonic pattern formation at the genetic, molecular, and cellular levels. Much of this progress is because of the remarkable success of studies of pattern formation in model systems, such as the fruit fly Drosophila melanogaster. Identification of genes that play major roles in setting up the body plan, followed by the discovery that many of these genes are well conserved even between different phyla, has also led to a renaissance in the investigation of the links between evolution and development. Using data from model systems, we are beginning to explore the degree to which developmental pathways have been conserved or altered between various organisms. Insights gained will help us understand the evolutionary changes in the mechanisms of pattern formation and provide a molecular basis for analyzing the diversification of body morphologies and developmental mechanisms. Eventually, such studies may allow us to understand the nature of the mutations that provide selectively advantageous changes for an organism.
A number of comparative studies aimed at examining the evolution of body morphology have focused on a well characterized set of genes termed the homeotic (Hox) genes. The Hox genes are known to play a major role in specifying regional identity along the anterior–posterior axis of animals from a wide range of phyla (1). Their potential role in altering body plan patterning during evolution was recognized soon after their characterization (2). For example, because altering the regulation of the Hox gene Ultrabithorax (Ubx) transforms a normally two-winged fly into a four-winged mutant, it was thought that evolutionary changes in Ubx regulation might explain the difference between insects that normally have four wings versus those that normally have two (2). A comparison of Ubx expression in flies and butterflies (butterflies normally have four wings) revealed that in fact the difference does not seem to be at the level of Ubx regulation (3), but instead at the level of genes downstream of Ubx (4). Thus, despite their clear potential to alter body plans on mutation in Drosophila, it has been difficult to document actual evolutionary changes in arthropod body plans that can be attributed to alterations in the initial boundaries of Hox gene expression.
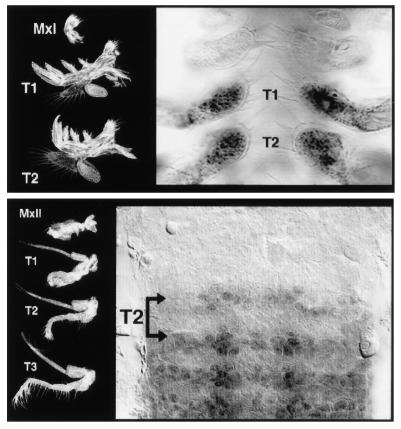
Recently, however, Averof and Patel (5) demonstrated a striking correlation within the crustaceans (lobsters, shrimp, crabs, etc.) between Hox gene expression and the evolution of their body morphology. By analyzing the expression of the Hox genes Ubx and abdominal-A (abd-A) in 13 crustacean species in 9 different orders, they showed that the initial embryonic anterior expression boundary of Ubx/abd-A can be used to predict where in the body plan the transition from the anterior feeding appendages to the distinctive posterior locomotory appendages occurs (Fig. 1). Depending on the species, this transition occurs anywhere from the first to the fourth thoracic segment. In a few instances, appendages with an intermediate morphology are found, and these are associated with segments showing intermediate levels and/or mosaic patterns of Ubx/abd-A expression during development.
Figure 1.
Appendage morphology in crustaceans correlates with boundaries in Ubx/abd-A expression. (Upper) In Triops longicaudatus (tadpole shrimp), Ubx/abd-A expression begins in the first thoracic segment (T1), the position in the body where the transition from feeding to locomotory appendages occurs. (Lower) In Mysidium colombiae (opossum shrimp), Ubx/abd-A expression begins in T2 but is rather weak in this segment. Stronger expression is seen in T3 and all thoracic segments more posterior. The transition from the feeding type morphology to locomotory morphology occurs at T2, although T2 is somewhat intermediate between the morphology of T1 and T3. Mxl, morphology of typical feeding appendage seen in the head. [Reproduced with permission from ref. 5 (Copyright 1997, Macmillan Magazines)].
Given the widely documented role of Hox genes in specifying segmental identity in a number of organisms, Averof and Patel (5) suggest that the association between Hox gene expression and appendage morphology during crustacean evolution may be direct and causal, and thus that homeotic genes may play a role in the normal process of adaptive evolutionary change. Furthermore, the existence of segments with intermediate morphology associated with reduced levels or mosaic patterns of Hox gene expression suggests that these “homeotic” changes may occur gradually through the accumulation of mutations that slowly alter homeotic gene expression during arthropod evolution. For now, we do not know whether these changes in Ubx/abd-A expression are because of cis-regulatory changes in these genes themselves, or trans changes in one or more upstream regulators of these homeotic genes.
Furthermore, in studies of the role of Ubx/abd-A in the regulation of abdominal appendages in insects, a variety of sequential changes in this regulatory interaction have been found (6). It appears that in phylogenetically primitive insects (and crustaceans), neither Ubx nor abd-A represses limb formation; in phylogenetically intermediate insects, abd-A, but not Ubx, appears to repress limb formation; and in the most phylogenetically derived insects, such as Drosophila, both Ubx and abd-A repress limb formation.
We now turn to questions concerning the number of genes involved in differences between species and the sizes of their effects. Does the evolution of animal and plant form typically result, for instance, from the action of many genes of small effect or from a few genes of large effect? Historically, evolutionists have favored the first, polygenic view. Indeed, traditional quantitative genetic theory largely rests on the so-called infinitesimal assumption, i.e., the assumption that phenotypic change involves many factors of very small effect each.
Recent quantitative trait locus (QTL) analyses have, however, called this view into question. QTL analysis, a powerful fusion of molecular and quantitative genetics, allows genetic dissection of morphological differences between pairs of crossable taxa. By producing hybrids who carry random combinations of chromosome regions from two taxa (where species identity of regions is inferred from molecular markers), and by scoring the mean phenotype of each genotype, one can map, count, and estimate the effects of genes underlying the trait studied (7). Such analyses routinely reveal that morphological differences involve a modest number of chromosome regions of substantial effect. To date, QTL studies have focused on agriculturally important organisms. Recent studies by Doebley and colleagues (8, 9), for instance, have shown that morphological differences between maize and its ancestor, teosinte, may involve as few as five factors. Remarkably, differences in lateral branching pattern appear to reflect the action of a single gene, teosinte branched-1 (tb1) (9).
Given their history of strong artificial selection, crops may not, of course, be representative of more natural phenotypic differences that do not involve human intervention. But a small but growing body of evidence suggests that “natural adaptations” may also involve a modest number of genetic factors (10–12). In summary, it appears that the distribution of gene effects underlying morphological evolution may be highly leptokurtic: whereas many genes of small effect may be involved, a few factors of large effect might account for the lion’s share of phenotypic differences between taxa (9).
Can we account for such results theoretically? Although evolutionary biology possesses a rich and formidable body of mathematical theory, virtually all assumes that evolution reflects either the cumulative effects of many infinitesimal contributions (quantitative genetics) or changes at single loci (population genetics). Although not widely appreciated, it is clear that neither tradition allows prediction of phenotypic effects among factors found in QTL studies.
Recent work suggests, however, that such predictions are possible. These new studies take advantage of a mathematical idealization of evolution first offered by Fisher (13). Under this idealization, organisms are viewed as comprising n independent characters, with fitness falling off from the optimum at the same rate for all traits. Fisher used this model to calculate the probability that mutations of a given phenotypic size would be favorable, showing that, whereas small mutations have a good chance of being advantageous, larger ones suffer a rapidly decreasing probability. Fisher thus arrived at his now famous conclusion that small mutations are the stuff of adaptation. It was only much later realized, however, that Fisher had neglected an important aspect of adaptation—the probability of fixation; to contribute to adaptation, mutations must not only be favorable but must escape random loss when rare. Noting that the probability of such escape increases with the phenotypic size of a mutation, Kimura (14) argued that mutations of intermediate size are the most likely to underlie adaptation.
Unfortunately, Kimura’s analysis was also incomplete, because after some change in the environment, evolution toward a new morphological optimum might involve many evolutionary steps (substitutions), not one, as Kimura tacitly assumed. Thus, the distribution of greatest biological interest concerns phenotypic effects among factors fixed when summing over an entire “adaptive walk” to an optimum. This distribution—and not Kimura’s—roughly corresponds to the one glimpsed in QTL analysis.
This distribution was recently derived by Orr (15), who showed that (i) it is approximately exponential, unlike those of Fisher and Kimura; (ii) this result is remarkably robust to changes in the distribution of mutational effects, as long as small mutations are more common than large; and (iii) the expected size of the largest factor fixed during adaptation is quite large. This work thus provides a heuristic expectation for the results of QTL studies. Although such analyses remain in their infancy, this work suggests that evolutionary theory might provide surprisingly simple, robust predictions about the genetics of morphological change. Equally important, they suggest that the traditional infinitesimal view of phenotypic change is deeply flawed and provide population genetic support for the search for genes of large effect underlying morphological change.
References
- 1.Manak J R, Scott M P. Development Suppl. 1994. pp. 61–71. [PubMed] [Google Scholar]
- 2.Lewis E B. Nature (London) 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 3.Carroll S, Gates J, Keys D, Paddock S W, Panganiban G F, Selegue J, Williams J A. Science. 1994;265:109–114. doi: 10.1126/science.7912449. [DOI] [PubMed] [Google Scholar]
- 4.Weatherbee S D, Halder G, Kim J, Hudson A, Carroll S. Genes Dev. 1998;12:1474–1482. doi: 10.1101/gad.12.10.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Averof M, Patel N H. Nature (London) 1997;388:682–686. doi: 10.1038/41786. [DOI] [PubMed] [Google Scholar]
- 6.Palopoli M F, Patel N H. Curr Biol. 1998;8:587–590. doi: 10.1016/s0960-9822(98)70228-3. [DOI] [PubMed] [Google Scholar]
- 7.Tanksley S D. Annu Rev Genet. 1993;27:205–233. doi: 10.1146/annurev.ge.27.120193.001225. [DOI] [PubMed] [Google Scholar]
- 8.Doebley J, Stec A. Genetics. 1991;129:285–295. doi: 10.1093/genetics/129.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doebley J, Wang R-L. Cold Spring Harbor Symp Quant Biol. 1997;22:361–367. [PubMed] [Google Scholar]
- 10.Bradshaw H D, Otto K G, Frewen B E, McKay J K, Schemske D W. Genetics. 1998;149:367–382. doi: 10.1093/genetics/149.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones C D. Genetics. 1998;149:1899–1908. doi: 10.1093/genetics/149.4.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orr H A, Irving S. Evolution. 1997;51:1877–1885. doi: 10.1111/j.1558-5646.1997.tb05110.x. [DOI] [PubMed] [Google Scholar]
- 13.Fisher R A. The Genetical Theory of Natural Selection. Oxford: Oxford Univ. Press; 1930. [Google Scholar]
- 14.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1983. [Google Scholar]
- 15.Orr H A. Evolution. 1998;52:935–949. doi: 10.1111/j.1558-5646.1998.tb01823.x. [DOI] [PubMed] [Google Scholar]