Abstract
Coffea arabica L. (arabica coffee), the only tetraploid species in the genus Coffea, represents the majority of the world's coffee production and has a significant contribution to Nicaragua's economy. The present paper was conducted to determine the genetic diversity of arabica coffee in Nicaragua for its conservation and breeding values. Twenty-six populations that represent eight varieties in Nicaragua were investigated using simple sequence repeat (SSR) markers. A total of 24 alleles were obtained from the 12 loci investigated across 260 individual plants. The total Nei's gene diversity (H T) and the within-population gene diversity (H S) were 0.35 and 0.29, respectively, which is comparable with that previously reported from other countries and regions. Among the varieties, the highest diversity was recorded in the variety Catimor. Analysis of variance (AMOVA) revealed that about 87% of the total genetic variation was found within populations and the remaining 13% differentiate the populations (F ST = 0.13; P < 0.001). The variation among the varieties was also significant. The genetic variation in Nicaraguan coffee is significant enough to be used in the breeding programs, and most of this variation can be conserved through ex situ conservation of a low number of populations from each variety.
1. Introduction
Coffea arabica L. (arabica coffee) is a self-fertile allotetraploid species that belongs to the genus Coffea in the family Rubiaceae [1, 2]. Out of the 103 species in the genus, arabica coffee is the only tetraploid species (2n = 4x = 44), the remaining species being diploid with 2n = 2x = 22 chromosomes [3]. Arabica coffee originated from a relatively recent hybridization between Coffea canephora (robusta coffee) and C. eugenioides or their ecotypes in the plateaus of Central Ethiopia [2, 4]. Coffee is mainly grown in tropical and subtropical regions and is an important cash crop in more than 60 countries in South and Central America, Asia, and Africa with an acreage of over 11 million ha [5].
Coffee production is an important economic activity in Central America and accounts for about 10% of the world coffee production [6]. In Nicaragua, large-scale coffee production was started in the 1850s, and since 1870, coffee is the main export crop [7, 8]. Most of the coffee production in the country comes from arabica coffee and the most cultivated varieties are Caturra, Catuai, Bourbon, and Typica [9]. More than 70% of Nicaraguan coffee is produced at elevations between 600 and 1500 m above sea level (asl) in the north central part of the country, where it is considered optimal for coffee production; and the rest is produced below 600 m asl in the south pacific region [10]. During 2010, the total production in the country was about 78 kilo tonnes [11].
Several studies have shown that the genetic diversity of arabica coffee is low when compared to that of robusta coffee [2, 12–19] due to its narrow genetic base associated with autogamy, evolutionary history, and domestication. This narrow genetic base has been reflected in different forms that include the lack of resistant genotypes to various pests and diseases (e.g., [20–22]). The genetic base of arabica coffee in the American content is even narrower, as it represents only a small subset of the genetic variations present within the arabica coffee gene pool [14, 20] and are more prone to various pests and diseases [20, 23, 24]. Thus, enhancement of its resistance to pests and diseases is becoming a crucial priority for economic and sustainable coffee production. This is being done through crossing arabica coffee with other coffee species, particularly robusta coffee [12, 21, 23, 25, 26] and through selection of genotypes of interest from the arabica coffee gene pool [27, 28].
Detecting and quantifying genetic variation in crop species is important for successful conservation of genetic resources and plant breeding. Molecular marker techniques, such as random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), and simple sequence repeats (SSR, also known as microsatellites) have been used for genetic diversity analysis in wild and cultivated coffee [13, 14, 16–19, 29–31]. However, there is little information on the genetic diversity of arabica coffee varieties in Nicaragua. Hence, the present study was conducted to estimate the genetic diversity and population genetic structure of arabica coffee in Nicaragua using SSR markers.
2. Materials and Methods
2.1. Plant Material and DNA Extraction
Coffee seeds from twenty-six populations representing eight arabica coffee varieties were used in this study. Each population was represented by ten individual coffee trees. Fresh coffee berries were collected between December 2009 and February 2010 from the main coffee growing provinces of Nicaragua (Table 1). The berries were dried up at room temperature and processed to obtain seeds. The seeds were then grown in pots in a greenhouse at a mean temperature of 28°C. Individually sampled leaf tissue from the plants grown in the greenhouse was placed in 2 mL Eppendorf microcentrifuge tubes and immediately frozen in liquid nitrogen and stored at −80°C until DNA extraction. After the frozen samples were milled using a Retsch MM400 shaker (Haan, Germany), DNA was extracted using a modified CTAB procedure, as described in Bekele et al. [32]. DNA quality and concentration was measured using a Nanodrop ND-1000 spectrophotometer (Saveen Werner, Sweden).
Table 1.
List of 26 arabica coffee populations used in this study representing eight coffee varieties in Nicaragua, and their geographic descriptions and SSR-based estimates of gene diversity (H Loci) and percent polymorphic loci (PPL).
| Population/variety | Province | Position | H Loci a | PPL* | Population/variety | Province | Position | H Loci a | PPL | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| B1 | Masatepe | N 11°54′ | W 86°08′ | 0.26 | 0.42 | CA1 | El Crucero | N 12°00′ | W 86°17′ | 0.24 | 0.42 |
| B2b | Jinotepe | N 11°50′ | W 86°12′ | 0.47 | 0.58 | CA2 | Masatepe | N 11°54′ | W 86°07′ | 0.24 | 0.42 |
| B3c | Dipilto | N 13°45′ | W 86°30′ | 0.26 | 0.42 | CA3e | Matagalpa | N 13°10′ | W 85°29′ | 0.24 | 0.33 |
| B4 | Jinotega | N 13°10′ | W 85°51′ | 0.25 | 0.42 | CA4 | Dipilto | N 13°45′ | W 86°47′ | 0.27 | 0.33 |
| B5 | Quilalí | N 13°36′ | W 85°56′ | 0.28 | 0.50 | CA5 | Sn Juan Río Coco | N 13°28′ | W 86°09′ | 0.27 | 0.33 |
| Bourbon (B) | 0.30 | 0.47 | Catuai amarillo (CM) | 0.25 | 0.35 | ||||||
| CT1 | Masatepe | N 11°53′ | W 86°08′ | 0.28 | 0.50 | CM1 | Dipilto | N 13°45′ | W 86°32′ | 0.32 | 0.50 |
| CT2 | Jinotepe | N 11°50′ | W 86°12′ | 0.40 | 0.58 | CM2 | Jinotega | N 13°13′ | W 85°54′ | 0.33 | 0.42 |
| CT3 | Matagalpa | N 13°10′ | W 85°45′ | 0.25 | 0.42 | CM3 | Quilalí | N 13°36′ | W 85°56′ | 0.43 | 0.58 |
| CT4 | Dipilto | N 13°45′ | W 86°30′ | 0.23 | 0.33 | CM4f | Sn Juan Río Coco | N 13°34′ | W 86°11′ | 0.40 | 0.58 |
| CT5 | Jinotega | N 13°13′ | W85°52′ | 0.32 | 0.58 | Catimor | 0.37 | 0.52 | |||
| CT6d | Quilalí | N 13°37′ | W 85°58′ | 0.27 | 0.42 | M | Dipilto | N 13°45′ | W 86°29′ | 0.24 | 0.33 |
| Caturra (CT) | 0.29 | 0.47 | Maracaturra | 0.24 | 0.33 | ||||||
| CR1 | Matagalpa | N 13°09′ | W 85°44′ | 0.25 | 0.33 | P | Jinotega | N 13°11′ | W 85°51′ | 0.30 | 0.50 |
| CR2 | Dipilto | N 13°45′ | W 86°32′ | 0.24 | 0.42 | Pacas | 0.30 | 0.50 | |||
| CR3 | Sn Juan Río Coco | N 13°29′ | W 86°10′ | 0.26 | 0.42 | PA | Dipilto | N 13°45′ | W 86°32′ | 0.27 | 0.50 |
| Catuai rojo (CR) | 0.25 | 0.39 | Pacamaras | 0.27 | 0.50 | ||||||
aCalculated based on the eight polymorphic loci; bpopulation B2 has a unique allele at the locus Cam35; cpopulation B3 has a unique allele at the locus Sat207; dpopulation CT6 has a unique allele at the locus SSR03; epopulation CA3 has a unique allele at the locus Sat207; fpopulation CM4 has a unique allele at the locus Sat207. Note: variety names and their corresponding mean genetic diversity values are shown in bold; populations listed above each variety belong to that variety; the overall mean H Loci and PPL across all populations and loci were 0.29 and 0.42, respectively.
2.2. SSR-PCR
Twenty-five SSR primer-pairs were initially screened for good amplification, polymorphism, specificity to their target loci, and suitability of the allele size for multiplexing. This led to the selection of twelve primer-pairs for final analysis (Table 2). The forward primers of selected primer-pairs were fluorescently 5′-labeled with either 6FAM, VIC, NED, or PET fluorescent dyes. The reverse primers were PIG-tailed with “GCTTCT” to avoid a nontemplated addition of a single nucleotide by Taq DNA polymerase to the PCR product, as described in Ballard et al. [34].
Table 2.
List of primer-pairs used to amplify the SSR loci used in this study and their annealing temperature: the repeat motifs of the loci and observed fragment sizes of the alleles.
| Locus name | Repeat motif | Primer sequence | Annealing temperature | Observed allele size (bp) | |
|---|---|---|---|---|---|
| X°C | Y°C | ||||
| 471 a | CT | F: TTACCTCCCGGCCAGAC | 60 | 54 | 292, 318, 320 |
| R: CAGGAGACCAAGACCTTAGCA | |||||
| 838 a | AC | F: CCCGTTGCCATCCTTACTTA | 57 | 61 | 97 |
| R: ATACCCGATACATTTGGATACTCG | |||||
| CaM03 b | AC | F: CGCGCTTGCTCCCTCTGTCTCT | 68 | 62 | 188, 194 |
| R: TGGGGGAGGGGCGGTGTT | |||||
| CaM16 b | TC | F: AAGGCAGCTGAAGCGGGACAAA | 68 | 62 | 188, 194 |
| R: TGGGGAGAGCTGCAGTTGGAGG | |||||
| CaM35 b | TGGAAG | F: CGAGCTAGAATGGATGACTTGGTTGG | 65 | 59 | 207, 213 |
| R: ATACCCGATACATTTGGATACTCG | |||||
| CM5 c | CCT | F: GTAACCACCACCTCCTCTGC | 60 | 54 | 185, 188 |
| R:TGGAGGTAACGGAAGCTCTG | |||||
| DCM06 d | (T) (TTC) | F: GTAGTCGGTGGGCTTGTGTT | 60 | 54 | 213 |
| R: AACGCGGACTAATTGAGGAA | |||||
| Sat207 e | ng | F: GAAGCCGTTTCAAGCC | 57 | 51 | 82, 89, 93, 97 |
| R: CAATCTCTTTCCGATGCTCT | |||||
| Sat235 e | ng | F:TCGTTCTGTCATTAAATCGTCAA | 60 | 54 | 167 |
| R: GCAAATCATGAAAATAGTTGGTG | |||||
| SSR03 d | TC | F: GGACAAAACACCGCCCAAAATA | 62 | 56 | 142, 148 |
| R: AGCGAGACAGAGGAAGGGAATATT | |||||
| SSR06 d | AAAGG | F: CAGGCACAGAAGGAATGAAGAGC | 62 | 56 | 126 |
| R: TGGTGGTATGGAAAACAGGAAGG | |||||
| SSR09 d | GT | F: TTGGCTTTTGTCCCTCCTTCCTCTG | 62 | 56 | 124, 126, 130 |
| R: AGCCCATTTCCCTCTCATCATTTCAAG | |||||
The PCR reactions were carried out in a volume of 25 μL containing 25 ng genomic DNA, 0.3 μM forward and reverse primers, 2 mM MgCl2, 0.3 mM dNTPs, 1 U Taq DNA polymerase (Sigma, Germany), and 1 × PCR buffer (10 mM Tris-HCl, pH 8.3 and 50 mM KCl). The reactions were performed using the GeneAMP PCR system 9700 thermocycler using the following temperature profiles: initial denaturation at 95°C for 3 min, followed by six touchdown cycles of denaturation at 94°C for 30 sec, annealing at X-Y°C (−1°C/cycle) for 30 sec and extension at 72°C for 45 sec, and then 32 cycles of denaturation at 94°C for 30 sec, annealing at Y°C for 30 sec, and extension at 72°C for 45 sec, and a 20 min final extension step at 72°C. The annealing temperature (T a) was changed based on the melting temperature (T m) of each primer-pair (Table 1).
For each locus, amplification was confirmed by running 5 μL of the PCR products on 1.5% ethidium bromide containing agarose gels. The PCR products of the twelve primer-pairs were multiplexed into two panels, each of which containing six PCR products. In each panel, the size difference between the PCR products labeled with the same fluorescent dyes was at least 80 bp to avoid overlapping. The multiplex PCR products were then analyzed using an ABI Prism 3730 DNA Analyzer (Applied Biosystems) at Genomics Core Facility of the University of Gothenburg, Sweden.
2.3. Genotyping and Data Analysis
The allele peaks were visually inspected and then analyzed using PEAK SCANNER V1.0 software (Applied Biosystems) based on the internal Genescan-500 LIZ size standard. Each peak was considered as an allele at a codominant locus and the genotype of each individual at each locus was recorded. The Free Tree-Freeware program [35] was used to generate Nei's standard genetic distance and for cluster analysis and bootstrapping. TreeView (Win32) 1.6.6 program [36] was used to view the trees. Analysis of molecular variance (AMOVA) was conducted using Arlequin ver. 3.5-2 [37].
3. Results
3.1. Total and within-Population Genetic Variation
Out of the 12 loci analyzed, eight loci were polymorphic whereas only one allele was detected across the 260 individuals analyzed in each of the remaining four loci. The four monomorphic loci were 838, DCM06, Sat235, and SSR06 (Table 2). The overall gene diversity for each polymorphic locus varied from 0.01 (Cam35) to 0.55 (SSR09). In addition to SSR09, the other loci with a relatively high level of gene diversity were CM5, Sat207, and Cam03 with H T of 0.50, 0.50 and 0.54, respectively (Table 3).
Table 3.
The partitioning of total gene diversity into within and among variety components for eight polymorphic SSR loci, and the number of alleles observed at each locus.
| Variety | HSSR03 | HCM5 | HCaM35 | HSat207 | HCaM16 | HCaM03 | H471 | HSSR09 | Overall |
|---|---|---|---|---|---|---|---|---|---|
| Bourbon | 0.136 | 0.500 | 0.036 | 0.501 | 0.075 | 0.516 | 0.166 | 0.500 | 0.304 |
| Catuai amarillo | 0.142 | 0.500 | 0.000 | 0.504 | 0.000 | 0.499 | 0.000 | 0.384 | 0.254 |
| Catimor | 0.241 | 0.500 | 0.000 | 0.506 | 0.224 | 0.538 | 0.387 | 0.556 | 0.369 |
| Catuai rojo | 0.048 | 0.500 | 0.000 | 0.500 | 0.000 | 0.498 | 0.000 | 0.433 | 0.247 |
| Caturra | 0.136 | 0.500 | 0.000 | 0.500 | 0.060 | 0.525 | 0.140 | 0.468 | 0.291 |
| Maracaturra | 0.000 | 0.500 | 0.000 | 0.500 | 0.000 | 0.420 | 0.000 | 0.480 | 0.238 |
| Pacas | 0.139 | 0.500 | 0.000 | 0.500 | 0.000 | 0.420 | 0.465 | 0.375 | 0.300 |
| Pacamaras | 0.049 | 0.500 | 0.000 | 0.500 | 0.180 | 0.480 | 0.000 | 0.480 | 0.274 |
| HS | 0.136 | 0.500 | 0.007 | 0.502 | 0.070 | 0.509 | 0.141 | 0.465 | 0.291 |
| HT | 0.229 | 0.500 | 0.008 | 0.503 | 0.186 | 0.536 | 0.310 | 0.550 | 0.353 |
| GST | 0.409 | 0.000 | 0.091 | 0.002 | 0.624 | 0.051 | 0.544 | 0.154 | 0.234 |
| NA | 2 | 2 | 2 | 4 | 2 | 2 | 3 | 3 | 2.5 (2.0)* |
*The mean number of alleles for the eight polymorphic loci and for all loci including monomorphic ones is 2.5 and 2.0, respectively.
The total gene diversity (H T) and the within-populations gene diversity (H S), estimated based on Nei's gene diversity [38], were 0.353 and 0.291, respectively (Table 3). The genetic diversity of each population (H Loci), which is the average gene diversity across the eight polymorphic loci, and the percent polymorphic loci (PPL) were also analyzed. H Loci ranged from 0.23 to 0.47, whereas %PL ranged from 0.33 to 0.58 (Table 1). At the variety level, the mean Nei's gene diversity ranged from 0.24 (variety Maracaturra) to 0.37 (variety Catimor) with corresponding lowest and highest %PL of 0.33 and 0.52. Variety Catimor showed the highest gene diversity in five of the eight polymorphic loci (Table 3). The overall mean gene diversity and %PL per population were 0.29 and 0.42, respectively.
Population-specific rare alleles, with frequencies ranging from 0.025 to 0.1, were detected in five of the 26 populations. An 89 bp allele unique to population B3 was detected at locus Sat207 (Figure 1) at a frequency of 0.025. Similarly, a 97 bp allele was detected in populations CA3 and CM5 at this locus at the same frequency. The other populations bearing unique and rare alleles were B4 and CT7. These populations had unique alleles at loci SSR03 (frequency = 0.1) and Cam35 (frequency = 0.025), respectively.
Figure 1.
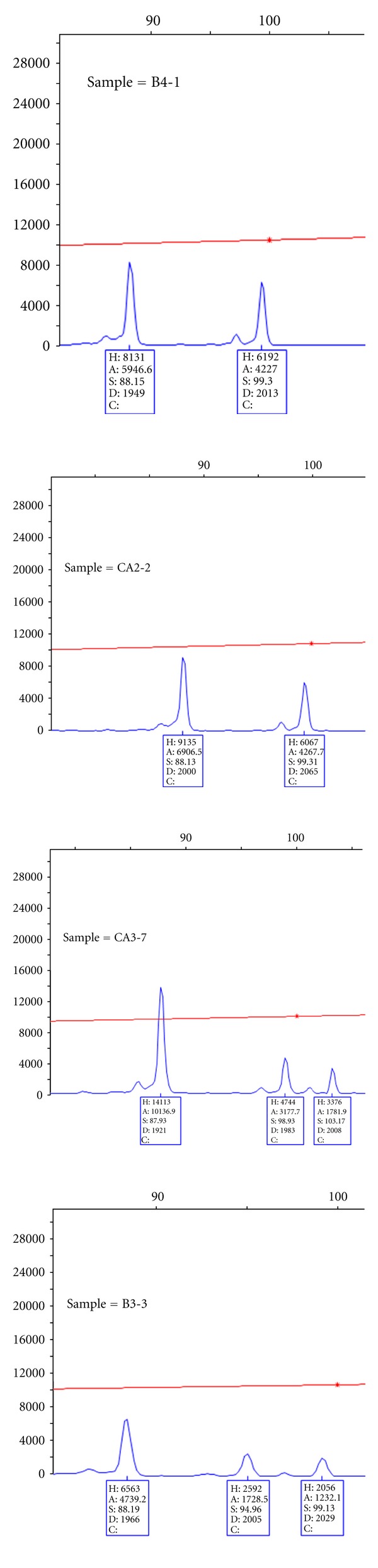
The electrophoretogram showing the alleles at the SSR locus Sat207.
3.2. Genetic Variation among Populations and Groups
The genetic differentiation of populations was estimated based on gene diversity (G ST; [39]) and AMOVA (F ST; [37]). The overall mean G ST and F ST were 0.23 and 0.13, respectively (Tables 3 and 4). The estimates of G ST varied from 0.00 (CM5) to 0.62 (Cam16) when calculated for each locus. The values of both G ST and F ST at locus CM5 were zero although each population has a gene diversity of 0.5. This is due to the fact that all individuals were heterozygous for the two alleles at this locus. The same is true for locus Sat207 except that few individuals in three of the 26 populations had additional rare alleles. At these two loci, both the total and within-population gene diversity was high but with no differentiation between the populations.
Table 4.
SSR-based AMOVA for the 26 populations of arabica coffee: (A) without grouping the populations, (B) by grouping the populations according to varieties, (C) by grouping the populations according to regions of origin, and (D) by grouping the populations according to province of origin.
| Groups | Sources of variation | Degrees of freedom | Variance components | Percentage of variation | Fixation indices | P-value |
|---|---|---|---|---|---|---|
| (A) without grouping populations | AP | 25 | Va = 0.194 | 13.50 | F ST = 0.130 | Va and F ST = 0.000 |
| WP | 494 | Vb = 1.25 | 86.50 | |||
| Total | 519 | 1.44 | ||||
|
| ||||||
| (B) Populations grouped by varieties |
AV | 7 | Va = 0.115 | 7.90 | F ST = 0.140 | Vc and F ST = 0.000 |
| APWV | 18 | Vb = 0.095 | 6.50 | F SC = 0.070 | Vb and F SC = 0.000 | |
| WP | 494 | Vc = 1.25 | 85.60 | F CT = 0.08 | Va and F CT = 0.023 | |
| Total | 519 | 1.46 | ||||
|
| ||||||
| (C) Populations grouped by regions of origin | AR | 1 | Va = −0.022 | −1.60 | F ST = 0.125 | Vc and F ST = 0.000 |
| APWR | 24 | Vb = 0.25 | 14.10 | F SC = 0.139 | Vb and F SC = 0.000 | |
| WP | 494 | Vc = 1.25 | 87.50 | F CT = −0.016 | Va and F CT = 0.812 | |
| Total | 519 | 1.43 | ||||
|
| ||||||
| (D) Populations grouped by province of origin | APr | 7 | Va = −0.002 | −0.14 | F ST = 0.134 | Vc and F ST = 0.000 |
| APWPr | 18 | Vb = 0.196 | 13.57 | F SC = 0.136 | Vb and F SC = 0.000 | |
| WP | 494 | Vc = 1.25 | 86.57 | F CT = −0.001 | Va and F CT = 0.419 | |
| Total | 519 | 1.44 | ||||
AP: among populations; WP: within populations; AV: among varieties; APWV: among populations within varieties; AR: among regions; APWR: among populations within regions; APr: among provinces; APWPr: among populations within provinces.
Overall, AMOVA revealed a highly significant genetic variation among populations (P < 0.0001; Table 4) accounting for 13.5% of the total variation. The differentiation among varieties was also significant (F CT = 0.08; P = 0.023) contributing 7.9% to the total genetic variation. The presence of rare alleles in four of the eight varieties contributed to the significant differentiation obtained. On the other hand, AMOVA revealed no significant variation among the two coffee growing regions and among the eight provinces (P > 0.4; Table 4). The pairwise AMOVA in the 26 populations revealed that each population was significantly differentiated from at least four populations. The most differentiated populations were CM2, CM3, and CM4, all of which belong to the variety Catimor. Pairwise F ST showed that CM2 and CM3 were significantly differentiated from each other as well as from all other populations. Population CM4 was significantly differentiated from all populations except from B2 (Table 5). Population CA5 was significantly differentiated from only four populations (B2, CM2, CM3, and CM4). At variety level, Catimor, Catuai rojo, and Pacas were differentiated from each other and all other varieties (Pairwise F ST; Table 6).
Table 5.
The Nei's standard genetic distance (above the diagonal) and the pairwise F ST (below the diagonal) between the 26 pairs of arabica coffee populations.
| B1 | B2 | B3 | B4 | B5 | CA1 | CA2 | CA3 | CA4 | CA5 | CM1 | CM2 | CM3 | CM4 | CR1 | CR2 | CR3 | CT1 | CT2 | CT3 | CT4 | CT5 | CT6 | M | P | PA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B1 | 0.065 | 0.006 | 0.005 | 0.005 | 0.013 | 0.013 | 0.014 | 0.001 | 0.001 | 0.010 | 0.376 | 0.255 | 0.093 | 0.001 | 0.006 | 0.000 | 0.002 | 0.018 | 0.006 | 0.012 | 0.008 | 0.002 | 0.006 | 0.015 | 0.004 | |
| B2 | + | 0.073 | 0.069 | 0.056 | 0.074 | 0.074 | 0.076 | 0.064 | 0.063 | 0.060 | 0.156 | 0.077 | 0.014 | 0.067 | 0.072 | 0.065 | 0.058 | 0.017 | 0.070 | 0.077 | 0.051 | 0.066 | 0.070 | 0.042 | 0.064 | |
| B3 | − | + | 0.002 | 0.003 | 0.004 | 0.005 | 0.005 | 0.004 | 0.007 | 0.010 | 0.392 | 0.275 | 0.112 | 0.009 | 0.023 | 0.004 | 0.004 | 0.023 | 0.002 | 0.004 | 0.006 | 0.003 | 0.006 | 0.030 | 0.004 | |
| B4 | − | + | − | 0.001 | 0.002 | 0.002 | 0.003 | 0.002 | 0.006 | 0.008 | 0.379 | 0.267 | 0.110 | 0.008 | 0.023 | 0.003 | 0.002 | 0.021 | 0.000 | 0.002 | 0.004 | 0.001 | 0.005 | 0.030 | 0.003 | |
| B5 | − | + | − | − | 0.003 | 0.003 | 0.004 | 0.002 | 0.005 | 0.008 | 0.350 | 0.242 | 0.092 | 0.008 | 0.021 | 0.003 | 0.001 | 0.014 | 0.001 | 0.003 | 0.003 | 0.001 | 0.005 | 0.024 | 0.003 | |
| CA1 | + | + | − | − | − | 0.000 | 0.000 | 0.007 | 0.013 | 0.011 | 0.373 | 0.272 | 0.124 | 0.017 | 0.037 | 0.010 | 0.007 | 0.026 | 0.002 | 0.001 | 0.006 | 0.004 | 0.009 | 0.043 | 0.007 | |
| CA2 | − | + | − | − | − | − | 0.000 | 0.006 | 0.013 | 0.011 | 0.373 | 0.272 | 0.124 | 0.017 | 0.036 | 0.009 | 0.006 | 0.026 | 0.001 | 0.001 | 0.006 | 0.004 | 0.009 | 0.042 | 0.006 | |
| CA3 | + | + | + | − | − | − | − | 0.008 | 0.015 | 0.012 | 0.375 | 0.275 | 0.127 | 0.019 | 0.040 | 0.011 | 0.008 | 0.028 | 0.002 | 0.001 | 0.006 | 0.005 | 0.010 | 0.045 | 0.007 | |
| CA4 | − | + | − | − | − | + | − | + | 0.001 | 0.007 | 0.364 | 0.252 | 0.098 | 0.003 | 0.012 | 0.001 | 0.000 | 0.017 | 0.003 | 0.007 | 0.004 | 0.000 | 0.005 | 0.020 | 0.003 | |
| CA5 | − | + | − | − | − | − | − | − | − | 0.008 | 0.360 | 0.246 | 0.091 | 0.001 | 0.007 | 0.001 | 0.002 | 0.017 | 0.007 | 0.013 | 0.007 | 0.003 | 0.005 | 0.013 | 0.004 | |
| CM1 | − | + | − | − | − | − | − | + | + | − | 0.297 | 0.209 | 0.100 | 0.011 | 0.024 | 0.009 | 0.008 | 0.019 | 0.008 | 0.011 | 0.002 | 0.007 | 0.006 | 0.026 | 0.003 | |
| CM2 | + | + | + | + | + | + | + | + | + | + | + | 0.023 | 0.191 | 0.379 | 0.379 | 0.376 | 0.353 | 0.248 | 0.378 | 0.389 | 0.306 | 0.369 | 0.369 | 0.311 | 0.345 | |
| CM3 | + | + | + | + | + | + | + | + | + | + | + | + | 0.092 | 0.255 | 0.250 | 0.257 | 0.243 | 0.147 | 0.267 | 0.280 | 0.213 | 0.257 | 0.259 | 0.196 | 0.240 | |
| CM4 | + | − | + | + | + | + | + | + | + | + | + | + | + | 0.090 | 0.085 | 0.096 | 0.091 | 0.036 | 0.112 | 0.124 | 0.089 | 0.103 | 0.101 | 0.044 | 0.100 | |
| CR1 | − | + | − | − | − | + | − | + | − | − | − | + | + | + | 0.004 | 0.001 | 0.003 | 0.019 | 0.009 | 0.016 | 0.010 | 0.005 | 0.006 | 0.012 | 0.005 | |
| CR2 | − | + | + | + | + | + | + | + | + | − | + | + | + | + | − | 0.009 | 0.013 | 0.026 | 0.025 | 0.037 | 0.024 | 0.016 | 0.020 | 0.011 | 0.018 | |
| CR3 | − | + | − | − | − | + | − | + | − | − | − | + | + | + | − | + | 0.001 | 0.018 | 0.004 | 0.009 | 0.006 | 0.001 | 0.005 | 0.018 | 0.003 | |
| CT1 | − | + | − | − | − | − | − | − | − | − | − | + | + | + | − | + | − | 0.015 | 0.003 | 0.007 | 0.004 | 0.001 | 0.005 | 0.019 | 0.003 | |
| CT2 | + | − | + | + | − | + | + | + | + | − | − | + | + | + | + | + | + | − | 0.021 | 0.027 | 0.013 | 0.018 | 0.020 | 0.014 | 0.018 | |
| CT3 | + | + | − | − | − | − | − | − | − | − | − | + | + | + | − | + | − | − | + | 0.001 | 0.003 | 0.001 | 0.005 | 0.032 | 0.003 | |
| CT4 | + | + | − | − | − | − | − | − | + | − | − | + | + | + | + | + | − | − | + | − | 0.006 | 0.004 | 0.007 | 0.042 | 0.006 | |
| CT5 | − | + | − | − | − | − | − | + | + | − | − | + | + | + | + | + | − | − | − | − | − | 0.004 | 0.004 | 0.023 | 0.002 | |
| CT6 | + | + | − | − | − | + | − | + | + | − | − | + | + | + | + | + | + | − | + | − | + | − | 0.004 | 0.024 | 0.002 | |
| M | − | + | − | − | − | + | − | + | + | − | − | + | + | + | − | + | − | − | + | − | − | − | + | 0.020 | 0.002 | |
| P | − | + | + | + | + | + | + | + | + | − | + | + | + | + | − | − | + | + | − | + | + | + | + | + | 0.022 | |
| PA | − | + | − | − | − | − | − | − | − | − | − | + | + | + | − | + | − | − | − | − | − | − | − | − | + |
+: Significant differentiation between the pair of populations (P < 0.05).
−: No significant differentiation between the pair of populations (P > 0.05).
Table 6.
The Nei's standard genetic distance (above the diagonal) and the pairwise F ST (below the diagonal) between the eight arabica coffee varieties.
| Bourbon | Catuai amarillo | Catimor | Catuai rojo | Caturra | Maracaturra | Pacas | Pacamaras | |
|---|---|---|---|---|---|---|---|---|
| Bourbon | 0.004 | 0.091 | 0.007 | 0.001 | 0.007 | 0.017 | 0.004 | |
| Catuai amarillo | + | 0.121 | 0.011 | 0.001 | 0.005 | 0.030 | 0.003 | |
| Catimor | + | + | 0.117 | 0.102 | 0.119 | 0.083 | 0.108 | |
| Catuai rojo | + | + | + | 0.008 | 0.009 | 0.012 | 0.007 | |
| Caturra | − | − | + | + | 0.004 | 0.022 | 0.002 | |
| Maracaturra | − | − | + | + | − | 0.020 | 0.002 | |
| Pacas | + | + | + | + | + | + | 0.022 | |
| Pacamaras | − | − | + | + | − | − | + |
+: Significant differentiation between the pair of populations (P < 0.05).
−: No significant differentiation between the pair of populations (P > 0.05).
3.3. Genetic Distance and Cluster Analysis
The Nei's standard genetic distance between populations ranged from less than 0.001 (e.g., CA1 versus CA2) to 0.392 (B3 versus CM2) with the overall mean of 0.060 (Table 5). The genetic distance between the varieties ranged from 0.001 (Caturra versus Bourbon and Caturra versus Catuai amarillo) to 0.121 (Catimor versus Catuai amarillo) with the overall mean of 0.031 (Table 6). The Nei's genetic distance-based cluster analysis revealed five clusters supported by moderate-to-high bootstrap values. Cluster I contained two populations from the variety Catimor (CM2 and CM3) with a bootstrap support of 100%. The 98% bootstrap supported Cluster II contained three populations (CM4, B2, and CT2), which belong to three different varieties. Similarly, cluster III comprised two populations (CR2 and P) from the Catuai-rojo and Pacas varieties with a 62% bootstrap support. Populations CM1, CT5, M, and PA, each of which belongs to different varieties, were placed under Cluster IV with a 65% bootstrap support. Cluster V is the largest cluster comprising 15 populations that were subclustered into three groups. However, the bootstrap support for the subclusters was low. In all clusters except cluster I, populations from more than one variety were clustered together showing a poor clustering of populations according to their varieties (Figure 2). At the variety level, Maracaturra and Pacamaras formed cluster I, whereas Catuai Amarillo, Bourbon, and Caturra formed cluster II with a 94% bootstrap support (Figure 3). The remaining three varieties remained solitary.
Figure 2.
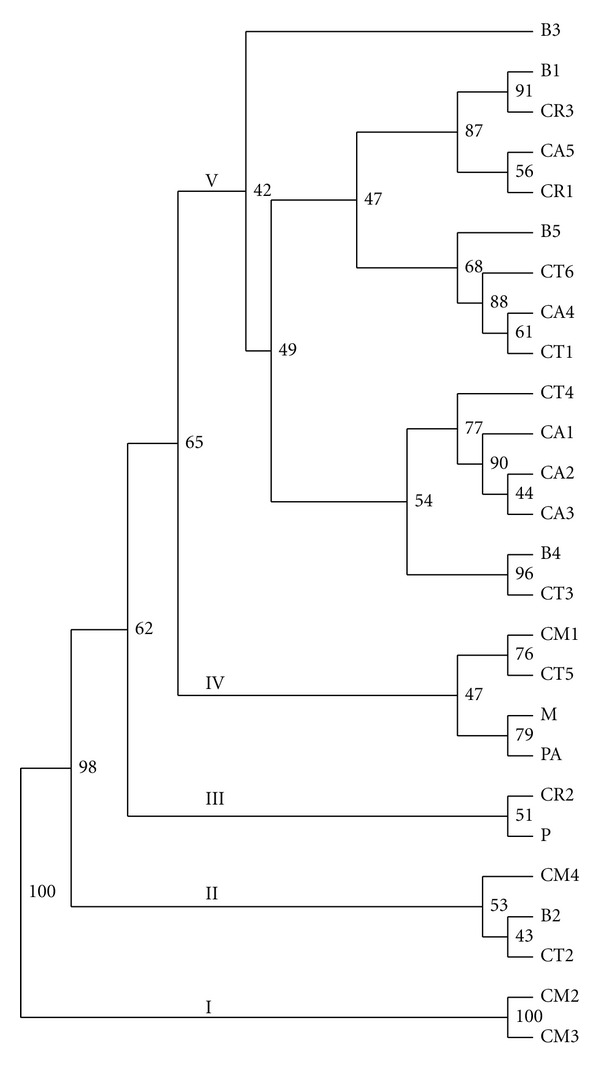
UPGMA phenogram for the 26 coffee populations based on Nei's standard genetic distance. Numbers in front of the branches are bootstrap values.
Figure 3.
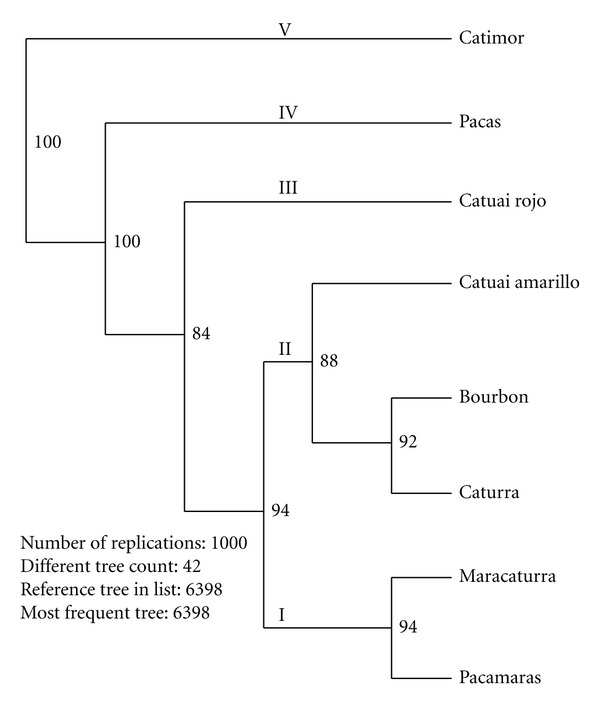
UPGMA phenogram for the eight coffee varieties based on Nei's standard genetic distance. Numbers in front of the branches are bootstrap values.
4. Discussion
4.1. The SSR Loci and Alleles
The difference in number of nucleotides between alleles obtained at the polymorphic loci in the present study indicates that the source of polymorphism was mainly the variation in number of repeat motifs of the SSRs. For example, CAM35 is a hexanucleotide repeat SSR [33], and the size of the alleles obtained in the present study was 207 bp and 213 bp. Similarly, the size of the two alleles of CM5, a trinucleotide repeat SSR locus [15], was 91 bp and 94 bp. However, the difference in size of the alleles observed at locus Sat207 appeared to be due to a combination of differences in the repeat motif and other types of variation, such as indels in the flanking sequences, as the difference in length between the alleles varied from three to four nucleotides (82 bp, 89 bp, 93 bp, 97 bp; Figure 1).
In addition to their application for analysis of genetic diversity, SSR markers have several other applications that include their use as markers for desirable traits. Among the SSRs used in the present study, Sat207 and Sat235 were reported to be tightly linked to locus Ck-1 that carries a major gene conferring resistance to the coffee berry disease (CBD) with Sat235 more closely linked to the gene than Sat207 [23]. CBD is a fungal disease caused by Colletotrichum kahawae that may cause severe damage in arabica coffee.
The SSR Sat235 was monomorphic across the 26 populations, and it is less likely that it can be a useful marker for genetic linkage analysis of Ck-1 in Nicaraguan coffee. On the other hand, Sat207 was polymorphic with two major alleles (ca 82 bp and 93 bp) and two rare alleles (ca 89 bp and 97 bp). Taking into consideration the amphidiploid nature of arabica coffee, it is most likely that the 82 bp allele on one hand and the other three alleles on the other hand originated from different progenitor genomes of arabica coffee (Figure 1).
If variation exists at the Ck-1 locus in Nicaraguan arabica coffee that gives resistance to CBD, the polymorphism detected at Sat207 is worth considering during the development of molecular markers linked to the resistance trait. Since arabica coffee is generally considered susceptible to CBD (e.g., [23, 40]), resistant genotypes should be rare and thus it would be interesting to evaluate the genotypes carrying the two rare alleles for resistance to this disease. Developing CBD-resistant arabica coffee varieties through identification of mutants is a method of choice, as it is simple and straight forward as compared to transferring resistance genes from other coffee species that requires crossing with donor genotypes followed by backcrossing to restore desirable traits. Considering that alleles of the same size at locus sat207 are identical by decent, it would also be interesting to compare the allele linked to resistance to CBD [23] with the allele introgressed to arabica coffee from robusta coffee [12], as this helps to assign the alleles to the two progenitor genomes.
Another interesting locus to discuss is CM5. Two alleles were detected at this locus and all the 260 individual plants studied were heterozygous for the two alleles. Baruah et al. [15] also identified only two alleles at this locus in arabica coffee. Several authors have reported a high cross-species transferability of SSR markers including the EST-SSRs within the genus Coffea (e.g., [15, 18, 41]). Given that arabica coffee is autogamous [3], the 100% heterozygosity obtained at this locus can only be explained by its amphidiploid nature. The two alleles should have been originated from different arabica coffee ancestral genomes. Baruah et al. [15] obtained 70% heterozygosity in arabica coffee at this locus, unlike the present study, suggesting that the two arabica coffee genomes of some genotypes carried the same alleles due to homoplasy. At this and other similar loci, fixed heterozygosity is the result when the two homoeologous loci are monomorphic and homozygous within the studied populations. The results clearly suggest the lack of recombination between the chromosomes of the two ancestral genomes due to the amphidiploid nature of arabica coffee.
Cubry et al. [19] obtained only two alleles per locus in the study that involved sixty SSR loci, and based on this they treated their data as diploid species data. However, a maximum of two alleles per locus is not always the case in arabica coffee, as shown in the present study. For example, three alleles were obtained at locus CAM03 in most individual plants analyzed. Three alleles per genotype were also observed at locus Sat207. These SSRs are reliable evidence that shows the presence of loci bearing nonrecombining alleles in arabica coffee representing the homoeologous loci from the two progenitor genomes. This supports the amphidiploid nature of the allotetraploid arabica coffee previously reported based on cytological evidence [2, 42].
4.2. The within- and among-Population Genetic Variation
The narrow genetic base of arabica coffee caused by rigorous selection during domestication and breeding has been reported by several authors (e.g., [14–16, 18, 19]). For example, Cubry et al. [19] reported a mean of 2.1 alleles per locus for arabica coffee, which was the lowest among the Coffea species they studied. This is comparable with 2.3 alleles/locus obtained in the present study. Similarly, Moncada and McCouch [16] reported a mean of 1.9 alleles per locus.
In the present study, the mean Nei's total (H T) and within-population (H S) gene diversity were estimated to be 0.35 and 0.29, respectively. The estimates for these parameters were 0.22 and 0.07, in that order, for the RAPD-based study of the Ethiopian arabica coffee by Anthony et al. [13]. Similarly, Cubry et al. [19] reported a mean gene diversity of 0.30 for the arabica coffee material they studied using SSR markers, whereas Aga et al. [43] reported an H T of 0.37 using ISSR markers in Ethiopian forest coffee. Thus, the level of genetic variation in Nicaraguan arabica coffee is comparable to that previously reported from several countries and regions. The presence of the major SSR alleles across all the populations in very high frequencies in the present study suggests a narrow gene pool of arabica coffee in Nicaragua in line with previous reports. This suggests some difficulties in finding genotypes bearing desirable traits, such as resistance to diseases and pests within the domesticated arabica coffee gene pool.
4.3. The Arabica Coffee Varieties in Nicaragua
The cluster analysis of the SSR data for the 26 populations revealed that, in most cases, the clustering pattern of the populations was not in line with their varietal classification. The principal coordinate analysis (PCoA) of the 260 individual plants (data not shown) revealed the presence of divergent genotypes in populations B2, B3, CM3, CT2, CT5, and PA, which partly explains the poor clustering of populations according to their variety of origin. Given that arabica coffee is an autogamous species; such a poor clustering pattern of populations according to variety of origin is somewhat unexpected. However, the processes through which these varieties were developed may partly explain the lack of a clear differentiation between the populations of the different varieties. For example, the variety Caturra was developed from mutant genotypes of the variety Bourbon (http://www.coffeeresearch.org/coffee/varietals.htm). A significant differentiation between these two varieties may not be expected due to the relatively short time elapsed since the development of the variety Bourbon, especially at selectively neutral loci that include most of the SSRs used in the present study. Similarly, Catuai was the result of a cross between Mundo Novo and Caturra and thus there may not be a clear genetic differentiation between the Caturra and Catuai varieties at this stage. A relatively close relationship between the Bourbon, Caturra, and Catuai varieties can be observed from Figure 3. However, other possible factors, such as some degree of gene flow between varieties through cross pollination, might have also contributed to the population genetic structure obtained.
Among the eight coffee varieties we studied, the highest genetic diversity was recorded in the variety Catimor (GD = 0.37 and PPL = 0.52). This variety is interesting not only because of its high genetic diversity but also because two of its populations (CM2 and CM3) were significantly differentiated from all the other populations (Table 5). At locus 471, the alleles recorded in CM2 and CM3 were different from those in the other populations, excluding CM4. The relatively high diversity in Catimor can be partly explained by the fact that it was the result of a cross between the variety Caturra of arabica coffee and the Timor hybrid, which is a natural hybrid between arabica and robusta coffee [21, 23]. Robusta coffee has been reported to have a relatively high genetic diversity compared to arabica coffee in several studies (e.g., [16, 19]). Thus, a wise use of the genetic diversity in the locally adapted populations of the variety Catimor in coffee breeding programs in Nicaragua is very important. The presence of allelic variation at several loci in the other coffee varieties suggests the significance of using the existing genetic variation in these varieties in the hybrid breeding program to develop superior and improved varieties.
Root-knot Nematodes (RKN) of the genus Meloidogyne cause major damage in coffee worldwide, and coffee breeding for durable resistance to RKN is now a major goal in coffee producing countries [28]. Meloidogyne exigua and M. incognita are known RKN attacking arabica coffee in Nicaragua [44]. The best method to reduce the damage caused by RKN in coffee is through developing resistant varieties. Bertrand et al. [45] reported resistant arabica coffee genotypes to M. arabicida in Costa Rica, which suggests that identification of resistant genotypes to M. exigua and M. incognita from arabica coffee in Nicaragua may be possible. In addition, the resistance gene Mex-1 identified in robusta coffee and successfully introgressed into arabica coffee by Noir et al. [25] suggests the need to give attention to arabica coffee varieties developed through hybridization of the two cultivated Coffea species, such as the variety Catimor for pest and disease resistance.
Overall, the level of genetic diversity of arabica coffee in Nicaragua is generally low and is comparable to that previously reported for arabica coffee from other countries and regions. Therefore, it should be promoted through crossing with other closely related species such as robusta coffee. In addition, the presence of rare alleles in some populations suggests the need to explore such populations in order to identify mutants bearing desirable traits. The significant differentiation between most Nicaraguan arabica coffee varieties suggests that varieties grown in the country should be analyzed for resistance/tolerance to major biotic and abiotic stresses. On the other hand, the absence of a significant differentiation between the coffee populations based on regions of origin suggests that germplasm collecting missions should prioritize the representation of coffee varieties over coffee growing regions in the country.
Acknowledgments
The authors wish to thank the Swedish International Development Agency (Sida) for financing this work. They are grateful to Anna Zborowska for her assistance in the laboratory work. None of the authors has financial relations with any of the commercial identities mentioned in the paper.
References
- 1.Raina SN, Mukai Y, Yamamoto M. In situ hybridization identifies the diploid progenitor species of Coffea arabica (Rubiaceae) Theoretical and Applied Genetics. 1998;97(8):1204–1209. [Google Scholar]
- 2.Lashermes P, Combes MC, Robert J, et al. Molecular characterisation and origin of the Coffea arabica L. Genome. Molecular and General Genetics. 1999;261(2):259–266. doi: 10.1007/s004380050965. [DOI] [PubMed] [Google Scholar]
- 3.Davis AP, Govaerts R, Bridson DM, Stoffelen P. An annotated taxonomic conspectus of the genus Coffea (Rubiaceae) Botanical Journal of the Linnean Society. 2006;152(4):465–512. [Google Scholar]
- 4.Tesfaye K, Borsch T, Govers K, Bekele E. Characterization of Coffea chloroplast microsatellites and evidence for the recent divergence of C. arabica and C. eugenioides chloroplast genomes. Genome. 2007;50(12):1112–1129. doi: 10.1139/G07-088. [DOI] [PubMed] [Google Scholar]
- 5.Waller JM, Bigger M, Hillocks RA. Coffee Pests, Diseases and Their Management. Nosworthy Way Oxfordshire , UK: CABI; 2007. Coffee as a crop and commodity; pp. 1–17. [Google Scholar]
- 6.MIFIC (Ministerio de Fomento, Industria y Comercio) El Café en Nicaragua. Dirección de Políticas Externas (DPCE) Departamento de Análisis Económico, Managua, Nicaragua, pp. 47, 2005.
- 7.Kuhl AE. Nicaragua y su café. Primera edición, Hispamer, Managua, Nicaragua, pp. 376, 2004.
- 8.Beuchelt TD, Zeller M. Profits and poverty: certification’s troubled link for Nicaragua’s organic and fairtrade coffee producers. Ecological Economics. 2011;70(7):1316–1324. [Google Scholar]
- 9.Rivas C. El café en Nicaragua: Análisis y descripción del comportamiento del rubro. Banco Central de Nicaragua, Managua, Nicaragua, pp. 45, 2008.
- 10.Guharay F, Monterrey J, Momnteroso D, Staver C. Manejo integrado de plagas en el cultivo del café. CATIE, Managua, Nicaragua, pp. 272, 2000.
- 11.ICO. 2011, http://www.ico.org/trade_statistics.asp.
- 12.Lashermes P, Combes MC, Ansaldi C, Gichuru E, Noir S. Analysis of alien introgression in coffee tree (Coffea arabica L.) Molecular Breeding. 2011;27(2):223–232. [Google Scholar]
- 13.Anthony F, Bertrand B, Quiros O, et al. Genetic diversity of wild coffee (Coffea arabica L.) using molecular markers. Euphytica. 2001;118(1):53–65. [Google Scholar]
- 14.Anthony F, Combes MC, Astorga C, Bertrand B, Graziosi G, Lashermes P. The origin of cultivated Coffea arabica L. varieties revealed by AFLP and SSR markers. Theoretical and Applied Genetics. 2002;104(5):894–900. doi: 10.1007/s00122-001-0798-8. [DOI] [PubMed] [Google Scholar]
- 15.Baruah A, Naik V, Hendre PS, Rajkumar R, Rajendrakumar P, Aggarwal RK. Isolation and characterization of nine microsatellite markers from Coffea arabica L., showing wide cross-species amplifications. Molecular Ecology Notes. 2003;3(4):647–650. [Google Scholar]
- 16.Moncada P, McCouch S. Simple sequence repeat diversity in diploid and tetraploid Coffea species. Genome. 2004;47(3):501–509. doi: 10.1139/g03-129. [DOI] [PubMed] [Google Scholar]
- 17.Silvestrini M, Junqueira MG, Favarin AC, et al. Genetic diversity and structure of Ethiopian, Yemen and Brazilian Coffea arabica L. accessions using microsatellites markers. Genetic Resources and Crop Evolution. 2007;54(6):1367–1379. [Google Scholar]
- 18.Aggarwal RK, Hendre PS, Varshney RK, Bhat PR, Krishnakumar V, Singh L. Identification, characterization and utilization of EST-derived genic microsatellite markers for genome analyses of coffee and related species. Theoretical and Applied Genetics. 2007;114(2):359–372. doi: 10.1007/s00122-006-0440-x. [DOI] [PubMed] [Google Scholar]
- 19.Cubry P, Musoli P, Legnaté H, et al. Diversity in coffee assessed with SSR markers: Structure of the genus Coffea and perspectives for breeding. Genome. 2008;51(1):50–63. doi: 10.1139/g07-096. [DOI] [PubMed] [Google Scholar]
- 20.Bertrand B, Peña Durán MX, Anzueto F, et al. Genetic study of Coffea canephora coffee tree resistance to Meloidogyne incognita nematodes in Guatemala and Meloidogyne sp. nematodes in El Salvador for selection of rootstock varieties in Central America. Euphytica. 2000;113(2):79–86. [Google Scholar]
- 21.Bertrand B, Anthony F, Lashermes P. Breeding for resistance to Meloidogyne exigua in Coffea arabica by introgression of resistance genes of Coffea canephora. Plant Pathology. 2001;50(5):637–643. [Google Scholar]
- 22.Villain L, Aribi J, Réversat G, Anthony F. A high-throughput method for early screening of coffee (Coffea spp.) genotypes for resistance to root-knot nematodes (Meloidogyne spp.) European Journal of Plant Pathology. 2010;128(4):451–458. [Google Scholar]
- 23.Gichuru EK, Agwanda CO, Combes MC, et al. Identification of molecular markers linked to a gene conferring resistance to coffee berry disease (Colletotrichum kahawae) in Coffea arabica . Plant Pathology. 2008;57(6):1117–1124. [Google Scholar]
- 24.Tscharntke T, Clough Y, Bhagwat SA, et al. Multifunctional shade-tree management in tropical agroforestry landscapes—a review. Journal of Applied Ecology. 2011;48(3):619–629. [Google Scholar]
- 25.Noir S, Anthony F, Bertrand B, Combes MC, Lashermes P. Identification of a major gene (Mex-1) from Coffea canephora conferring resistance to Meloidogyne exigua in Coffea arabica . Plant Pathology. 2003;52(1):97–103. [Google Scholar]
- 26.Alpizar E, Etienne H, Bertrand B. Intermediate resistance to Meloidogyne exigua root-knot nematode in Coffea arabica . Crop Protection. 2007;26(7):903–910. [Google Scholar]
- 27.Leroy T, Ribeyre F, Bertrand B, et al. Genetics of coffee quality. Brazilian Journal of Plant Physiology. 2006;18(1):229–242. [Google Scholar]
- 28.Albuquerque EVS, Carneiro RMDG, Costa PM, et al. Resistance to Meloidogyne incognita expresses a hypersensitive-like response in Coffea arabica . European Journal of Plant Pathology. 2010;127(3):365–373. [Google Scholar]
- 29.Aga E. Molecular genetic diversity study of forest coffee tree (Coffea arabica L.) populations in Ethiopia: implications for conservation and breeding. Swedish University of Agricultural Sciences, Acta Universitatis Agriculturae Sueciae; 2005. pp. 1652–6880, 2005:79, Ph.D. thesis. [Google Scholar]
- 30.Maluf MP, Silvestrini M, de C. Ruggiero LM, Filho OG, Colombo CA. Genetic diversity of cultivated Coffea arabica inbred lines assessed by RAPD, AFLP and SSR markers systems. Scientia Agricola. 2005;62(4):366–373. [Google Scholar]
- 31.Vieira ESN, von Pinho EVdeRV, Carvalho MGG, Esselink DG, Vosman B. Development of microsatellite markers for identifying Brazilian Coffea arabica varieties. Genetics and Molecular Biology. 2010;33(3):507–514. doi: 10.1590/S1415-47572010005000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bekele E, Geleta M, Dagne K, et al. Molecular phylogeny of genus Guizotia (Asteraceae) using DNA sequences derived from ITS. Genetic Resources and Crop Evolution. 2007;54(7):1419–1427. [Google Scholar]
- 33.Hendre PS, Phanindranath R, Annapurna V, Lalremruata A, Aggarwal RK. Development of new genomic microsatellite markers from robusta coffee (Coffea canephora Pierre ex A. Froehner) showing broad cross-species transferability and utility in genetic studies. BMC Plant Biology. 2008;8, article 51 doi: 10.1186/1471-2229-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ballard LW, Adams PS, Bao Y, et al. Strategies for genotyping: effectiveness of tailing primers to increase accuracy in short tandem repeat determinations. Journal of Biomolecular Techniques. 2002;13(1):20–29. [PMC free article] [PubMed] [Google Scholar]
- 35.Pavlícek A, Hrdá S, Flegr J. FreeTree—freeware program for construction of phylogenetic trees on the basis of distance data and Bootstrap/Jackknife analysis of the tree robustness, application in the RAPD analysis of genus frenkelia. Folia Biologica. 1999;45(3):97–99. [PubMed] [Google Scholar]
- 36.page RDM. TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12(4):357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 37.Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 38.Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89(3):583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nei M. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the United States of America. 1973;70(12):3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hein L, Gatzweiler F. The economic value of coffee (Coffea arabica) genetic resources. Ecological Economics. 2006;60(1):176–185. [Google Scholar]
- 41.Poncet V, Rondeau M, Tranchant C, et al. SSR mining in coffee tree EST databases: Potential use of EST-SSRs as markers for the Coffea genus. Molecular Genetics and Genomics. 2006;276(5):436–449. doi: 10.1007/s00438-006-0153-5. [DOI] [PubMed] [Google Scholar]
- 42.Clarindo WR, Carvalho CR. First Coffea arabica karyogram showing that this species is a true allotetraploid. Plant Systematics and Evolution. 2008;274(3-4):237–241. [Google Scholar]
- 43.Aga E, Bekele E, Bryngelsson T. Inter-simple sequence repeat (ISSR) variation in forest coffee trees (Coffea arabica L.) populations from Ethiopia. Genetica. 2005;124(2):213–221. doi: 10.1007/s10709-005-1484-6. [DOI] [PubMed] [Google Scholar]
- 44.Herrera I, Bryngelsson T, Monzón A, Geleta M. Identification of coffee root-knot nematodes based on perineal pattern, SCAR markers and nuclear ribosomal DNA sequences. Nematologia Mediterranea. 2011;39(2):101–110. [Google Scholar]
- 45.Bertrand B, Ramirez G, Topart P, Anthony F. Resistance of cultivated coffee (Coffea arabica and C. canephora) trees to corky-root caused by Meloidoqyne arabicida and Fusarium oxysporum, under controlled and field conditions. Crop Protection. 2002;21(9):713–719. [Google Scholar]


