Abstract
For this special issue on 90 years of polarography, the following personal account describes how my early research in electrochemistry and polarography in the laboratory of Prof. Petr Zuman led to a major research effort in the determination of proteins for cancer detection and monitoring. It reviews the very recent history of nanoparticle labels and multiplexed detection in protein immunosensors. It then describes our journey of discovery that has led to ultrasensitive protein immunosensors achieved by combining nanostructured electrodes with particles labeled with up to ½ million enzymes that can detect down to as little as 1 fg mL−1 protein in diluted serum. Our most mature multiple protein detection system is a microfluidic device with eight sensors coated with 5-nm gold nanoparticles that uses off-line protein detection with heavily labeled magnetic particles. This approach has led to reliable sub pg mL−1 detection limits for multiple proteins, provides excellent correlation with referee ELISA methods, and is currently being used for validation of panels of biomarkers for oral and prostate cancer. The article ends with a section on future perspectives.
Keywords: electrochemistry, immunosensors, microfluidics, nanostructures, proteins
1. Introduction
It is a great honor for me to contribute this Personal Account article in the special issue on electrochemistry honoring the 90th anniversary of the discovery of polarography by Jaroslav Heyrovsky in 1922. Polarography was the very first quantitative electroanalytical technique, since prior to this time the irreproducibility of electrode surfaces led to serious problems in reproducibility. Heyrovsky’s genius was to use a dropping mercury electrode (DME), featuring renewal of a new, clean Hg surface every few seconds when the drop fell off and a new one began to form. Heyrovsky became Professor of Physical Chemistry at Charles University in Prague and went on to found and head the Polarographic Institute in Prague, which for many years was the international leader in polarographic research as well as in the development of new electrochemical instrumentation. The popularity of the DME has waned in recent years, due to the toxicity of mercury and our greatly improved understanding of how to reproducibly prepare solid electrode surfaces. However, the discovery of polarography facilitated extensive experience in quantitative experiments that extrapolated to solid electrodes, and that led directly to the wide variety of subsequent electroanalytical applications that we find now at the beginning of the 21st century.
This article deals with the development of electrochemical immunosensors that detect proteins as well as viruses and bacteria. These biosensors are part of a long list of electroanalytical methods designed to help solve biological problems. Examples include enzyme-based home-use electrochemical biosensors for measuring blood glucose by diabetic patients,1 biosensors that detect DNA for genetic analysis and protein biomarkers for diseases,2,3 and electrochemiluminescence (ECL) detectors for nucleic acids and proteins.4,5
It is personally rewarding to me to contribute to this issue as I am the scientific grandson of Jaroslav Heyrovsky, having earned my Ph.D. in Chemistry at Clarkson University in 1979 under the direction of Professor Petr Zuman, who was Heyrovsky’s Ph.D. student at Charles University. Petr Zuman also served as the head of the Organic Electrochemistry Division of Heyrovsky’s Polarographic Institute in the 1950s and 60s before moving briefly to the University of Birmingham in the UK, and then on to Clarkson in the US in 1970.
My graduate education with Prof. Zuman in the late 1970s involved mainly mechanistic organic electrochemistry. The skills I developed there in fundamental electrochemistry and instrumentation provided me with an excellent background for a later move into independent bioelectrochemical research at University of Connecticut. For many years prior to the present century, our work in Connecticut involved catalytic electrochemical decomposition of organic pollutant molecules in detergent dispersions6 as well as fundamental studies of protein film voltammetry.7 I first developed an interest in electrochemical immunosensors in 2001 during a sabbatical period in the laboratory of Prof. Malcolm Smyth at the Irish National Center for Sensor Research at Dublin City University. Our group has since translated this interest into a major research effort involving biologists, materials scientists, statisticians, and clinical researchers in the US and around the world. Major colleagues in this group include nanomaterials scientist Prof. Folios Papadimitrakopoulos at the University of Connecticut, and cancer biologists at the US National institutes of Health led by Dr. Silvio Gutkind and Dr. Vyomesh Patel. In the sections below, I begin with a brief history of electrochemical immunosensors, then provide an overview of the advantages of nanoparticles in these immunosensors. I then focus the remainder of the review on recent contributions of our laboratory to ultrasensitive detection of cancer biomarker proteins with nanoparticle-enhanced electrochemical immunosensors and immunoarrays.
Our research in this area focuses on developing electrochemical arrays for ultrasensitive detection of multiple biomarker measurements for cancer diagnostics. The US National Institutes of Health defines biomarkers as “molecules that can be objectively measured and evaluated as indicators of normal or disease processes and pharmacologic responses to therapeutic intervention”.8 Our research focuses on panels of biomarker proteins that can provide a “snapshot” of patient status with regard to the disease.13,14 Cancer detection and monitoring demands the ability to detect amounts of protein below the normal patient levels for the least abundant protein. It requires good sensitivity for all protein biomarkers at normal and elevated levels. Detection limits well below the pg mL−1 level and good sensitivity up to hundreds of ng mL−1 will be necessary for the broadest applicability. For example, prostate specific antigen (PSA), a clinical biomarker for prostate cancer, has a normal serum level of 0.5–3 ng mL−1, while elevated levels of 4–10 ng mL−1 indicate the possibility of prostate cancer.9 Interluekin-6 (IL-6) is a biomarker protein that is overexpressed in head and neck, prostate, lung, multiple myeloma and renal cancers.10 Its normal serum level is <6 pg mL−1 while levels in cancer patients can elevate to hundreds of pg mL−1 or more. Detection of both biomarkers in serum illustrates the fact that detection needs to be viable over a broad range of concentrations that are often different for different proteins. This is all the more important since single biomarkers usually have insufficient predictive ability, and devices that measure panels of 4–10 proteins will ultimately be needed for reliable cancer diagnostics.13,14
2. Development of Electrochemical Immunoassays and Immunosensors
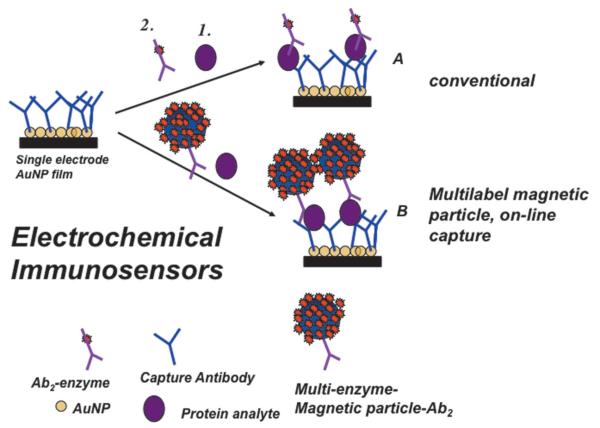
The basic principles of electrochemical immunoassays are illustrated by Figure 1, which includes some aspects of nanoparticle enhancement that we will discuss later. Basically, primary antibodies are attached to a solid substrate or an electrode to selectively capture the analyte protein from the sample. After washing, a secondary antibody with a label attached is added, and binds selectively to the protein on the surface. This label, shown as an enzyme in Figure 1, is then detected electrochemically.11-13 This general approach was adapted from enzyme-linked immunosorbent assays (ELISA), which have served as workhorse methods for detection of proteins for many years.14,15 ELISA uses optical detection of a colored enzyme reaction product and, while still widely used, has known limitations in multiplexing, analysis time, and sample size.
Fig. 1.
Protocols for electrochemical immunosensors illustrated for a gold nanoparticle sensor using a single enzyme label (path A) or a multi-labeled nanoparticle (path B). The sensor features primary antibodies (Ab1) that capture the analyte protein from the sample. In the conventional approach (path A), an enzyme-labeled secondary antibody (Ab2) is added that binds to the analyte protein on the surface. In the high sensitivity approach (path B) a nanoparticle with antibodies and thousands of enzyme labels is added to bind to the analyte protein. Addition of an enzyme substrate and electrochemical detection provides signals proportional to the amount of protein in the sample.
Bill Heinemann and co-workers pioneered electrochemical immunosensing in the 1980s, utilizing sandwich immunoassays with enzyme label alkaline phosphatase that produces electroactive products. In his group’s approach, the capture antibodies in the sandwich immunoassay reside in a flow system upstream from the detector. Products formed from the enzyme label are transported by a chromatographic or fluidic system to the detecting electrode.16,17 Advances have included interdigitated electrodes to achieve high sensitivity,18 and incorporation into microfluidic devices.19 This team has achieved excellent detection limits (DLs) in the pg mL−1 to ng mL−1 range for small molecules and proteins.
Efforts in the 1990s began to be directed toward developing single analyte enzyme-linked electrochemical immunosensors by researchers including Malcolm Smyth, Gerry Guilbault and others.12,20-23 Here, the antibodies (Ab) are attached directly to the sensor electrode, and antigen capture, secondary antibody binding, and label detection are all done on the same surface. The sandwich format provides the most selective and sensitive assays (see Figure 1).
In addition to alkaline phosphatase, horseradish peroxidase (HRP) is a suitable label. When activated by hydrogen peroxide, the iron heme HRP enzyme is converted to an FeIV=O form that can be reduced on electrodes at low potentials.24 Many other types of labels have been used, including glucose oxidase, electroactive metal ions and complexes, nanoparticles,25,26 conductive polymers, and liposomes loaded with electroactive compounds.23 Savéant et al. analyzed electrochemical responses of enzyme labels in detail, and identified key immunosensor sensitivity factors.27-29
A critical issue in immunoassays is minimization of non-specific binding (NSB) of interfering species in samples such as serum or blood, as well as NSB of the labeled Ab2 that arises when this signal producing species is bound to non-antigen sites on the sensor. In labeled assays, non-enzyme Ab2 bound to sites other than the analyte protein still gives a signal, but it is not proportional to the analyte protein concentration. This can increase detection limits and degrade sensitivity. NSB is usually minimized by washing with a cocktail that includes casein or bovine serum albumin and detergents in NSB-blocking steps. The literature suggests that tailoring the sensor surface with appropriate chemical groups can also inhibit protein adsorption, and one of the most effective surfaces features polyethylene glycol (PEG) moieties.23,30,31 Such functionalized surfaces, although useful, may still permit small amounts of NSB that could significantly increase background in the pg to ng mL−1 analyte concentration ranges.
Multiplexing has also been achieved with electrochemical immunosensors. For example, iridium oxide sensor chips enabled simultaneous electrochemical immunoassays using alkaline phosphatase-labeled Ab2 and detection of product hydroquinone with DLs of 1–3 ng mL−1 for up to eight cancer biomarker proteins.32-34 Results showed good correlation with ELISA. Sensors coated with a composite DNA dendrimer/conducting polymer were used in a 16-sensor immunoelectrochemical array.35 Protein biomarkers for oral cancer (IL-8 and IL-1b) and IL-8mRNA were measured using HRP-labeled secondary antibodies in buffer with DL of 100–200 fg mL−1 for the proteins and 10 aM for the IL-8 mRNA. In saliva, DLs degraded to 4 fM IL-8mRNA and 7.4 pg mL−1 IL-8.36
Unlabeled electrochemical immunoassays can be achieved with impedance or capacitance sensors, field-effect transistors (FETs) or nanowire transistors.37,38 Ultrasensitive nanowire transistors require significant future development and cost reductions before they can compete with more accessible, lower-cost procedures. A less-sensitive nanoribbon sensor coupled with a microfluidic device that measures cancer biomarker proteins also shows promise.39
3. Nanoparticle-Based Electrochemical Immunosensors
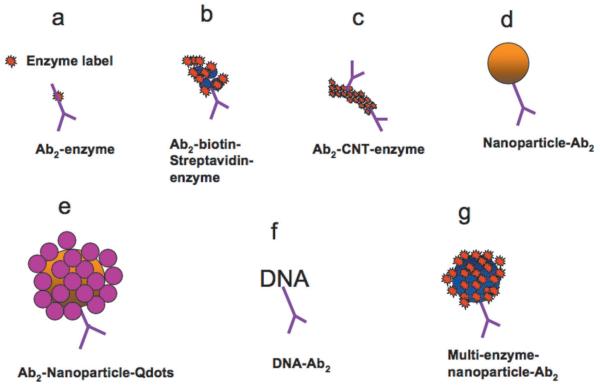
When I was a Ph.D. student in the 1970s, we in Petr Zuman’s group were members of the Institute of Colloid and Surface Science at Clarkson University. In those days, consistent fabrication of monodisperse gold or metal oxide particles with diameters under 20 nm would have been a multi-year effort. Since that time, simple, rapid methodologies for nanoparticle fabrication have emerged that have enabled many new applications. Ready availability of these particles has led to new opportunities for ultrasensitive approaches to electrochemical protein sensors.11,13,25,31,40-43 Sandwich immunosensor strategies using secondary antibody(Ab2)-nanoparticle bioconjugates have included dissolvable nanoparticles leading to electroactive ions, Ab2-nanoparticles with thousands of enzyme labels, and Ab2-nanoparticles with multiple redox probes (Figure 2).25,30,41,44-47 These approaches greatly enhance sensitivity by providing a large number of labels for each protein bound on the sensor surface.
Fig. 2.
Amplification strategies for electrochemical immunosensors using nanoparticles or other moieties attached to secondary antibody (Ab2).
Nanostructured sensor surfaces can provide additional increases in sensitivity by providing high surface areas enabling attachment of a large number of capture antibodies,13,48 and by facilitating better access of protein analytes to these antibodies.49 For example, nanostructured immunosensors have been made in our laboratory using films of carbon nanotubes30,31,41 or gold nanoparticles,50 and in Shana Kelley’s lab by engineering highly nanostructured sensor surfaces by electrodepositing gold.49,51
Nanoparticle labels in immunoassays were first reported by Delequaire et al.52 The metals in the nanoparticles served as labels after dissolution of the particles. After the antibodies capture analyte proteins, Ab2-nanoparticles are added to bind to them, and an NSB blocking wash is done. Then, the nanoparticles are dissolved in acid to produce high concentrations of electroactive metal ions. With gold nanoparticle-Ab2 labels, Au3+ released by acid dissolution was detected by anodic stripping voltammetry to give a 3 pM DL for IgG in buffer. Joe Wang and his team further enhanced sensitivity25 by using cyclic accumulation of gold nanoparticles (AuNPs) and using gold particles to catalyze Ag precipitation, detecting the large amounts of metal ions released by stripping analysis. For example, Ag deposition provided a 0.5 ng mL−1 (22 pM) DL for cardiac troponin I.53 Multiple AuNPs were attached to larger Au spheres and used for Ag deposition.25 Similarly, magnetic particles were decorated with CdS quantum dots (Qdots), isolated magnetically, then dissolved for potentiometric stripping detection of Cd.25 Ag deposition also afforded ultrasensitive conductivity immunoassays of human IgG in buffer.54 A novel antibody-coated nanopore immunoassay was developed that filters blood and detects proteins utilizing gold nanoparticle labels, then depositing Ag. Detection relies on the rate of decreased diffusion of an electroactive probe through the pores. This method was used to detect IgGs including a cancer biomarker in whole blood.55 Other strategies include loading Ab2-nanoparticles or Ab2-polymer beads with electroactive labels such as ferrocene, and later releasing the labels for detection.25,26,30,40
Multiplexed protein assays using nanoparticle approaches were also developed.25 “Bar code” labeling of secondary antibodies with distinct nanoparticles having different electrochemical characteristics have been used. For example, different Ab2’s can be labeled with dissolvable nanoparticles or Qdots of different metals that give ions with different reduction potentials. For example, cadmium, zinc, copper, and lead sulfide Qdots were attached to four individual Ab2’s to measure the four different cognate proteins.56 The Qdots were then dissolved to yield four different metal ions, each tracking a different protein, and measured by electrochemical stripping analysis. Multiple-metal rods, spheres or alloy rods have also been used as multiplexing tools in immunoassays. Upon dissolution, these materials give a series of metal stripping peaks whose potentials and relative intensities are associated with individual analyte proteins.25
4. Enzyme-Nanoparticle Bioconjugates for Ultrasensitive Electrochemical Immunoassays
Nanoparticles can be coated with high loadings of enzymes and secondary antibodies to amplify signals in sandwich immunoassays. Joe Wang pioneered this approach for ultrasensitive detection of DNA and proteins.57 Joe’s group used multiwall carbon nanotubes (MWCNTs) derivatized with thousands of copies of alkaline phosphatase and secondary antibodies, and reported femptomolar detection levels for proteins in buffer. Alternate electrostatic layer-by-layer deposition of alkaline phosphatase with oppositely charged polyions on MWCNTs was used to make alternative detection particles and achieve a DL of ca. 70 aM for IgG in buffer.58
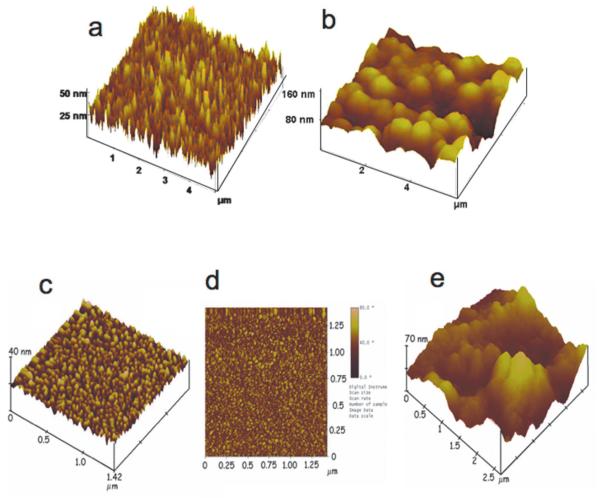
In collaboration with Bernard Munge of Salve Regina University, a former postdoctoral fellow with Wang, our research team began to use multi-enzyme labeled nanoparticles in immunosensor protocols for PSA, IL-6, and other prostate cancer biomarkers.13,31,44,41,59-61 Sensitivity was promoted further by using nanostructured electrodes featuring densely packed films of ca. 30 nm long upright single wall carbon nanotube (SWCNT) forests,62,63 or 5 nm glutathione-decorated AuNPs.50 These high-area sensor surfaces feature populations of carboxylate groups ready for attachment of large amounts of capture antibodies48 by amidization. AFM images of these surfaces illustrate their large areas and changes in surface morphology that result when capture antibodies are attached (Figure 3).
Fig. 3.
Atomic force microscope images of immunosensor platforms: (a) SWCNT forest on silicon; (b) SWCNT forest coated with chemically attached antibodies; (c) a PDDA/gold nanoparticle (AuNP) bilayer on smooth mica; (d) phase contrast image of the same PDDA/AuNP bilayer on mica; (e) anti-PSA antibodies attached onto carboxylate groups of the AuNP/PDDA bilayer. Images (a) and (b) reproduced with permission from reference [62]. Images (c), (d), and (e) reproduced with permission from reference [50].
We used nanostructured sensors coated with SWCNT forests or AuNP films for sandwich immunoassays for prostate cancer biomarker PSA.31,41,50,59 As in Figure 2a, conventional secondary antibodies (Ab2) conjugated with single or multiple HRPs, as well as carbon nanotubes (CNT) or magnetic particles conjugated with Ab2 and large copy numbers of HRPs (Figure 2c,g), were used for detection. When these heavily labeled detection particles64 replace HRP-Ab2 in immunoassays (cf. Figure 1), the large number of labels per captured protein analyte greatly enhances sensitivity.
We used rotating disk amperometry to measure immunosensor responses using H2O2 to activate the enzyme, and hydroquinone (HQ) to mediate the reduction of the resulting ferryloxy form of HRP (PFeIV=O). This approach produces a steady-state amperometric current proportional to concentration (see Figures 4 and 5). The chemistry of the detection is illustrated in Scheme 1.
Fig. 4.
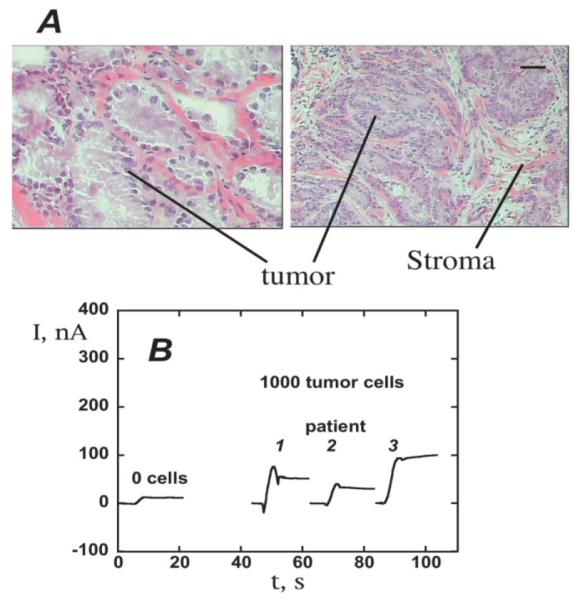
Measurement of PSA in prostate cancer cells using SWCNT forest immunosensors: (A) Micrograph of representative prostate cancer biopsy stained to reveal areas of tumor cells (scale bar 100 microns). Cells subsequently isolated by laser microdissection for PSA detection. (B) Sensor response at −0.3 V and 3000 rpm for tissue lysates (~1000 cells) in 10 μL buffer followed by 10 μL anti-PSA-CNT-HRP bioconjugate (11 pmol mL−1 in HRP) in 0.05% Tween-20. Reproduced with permission from reference [62].
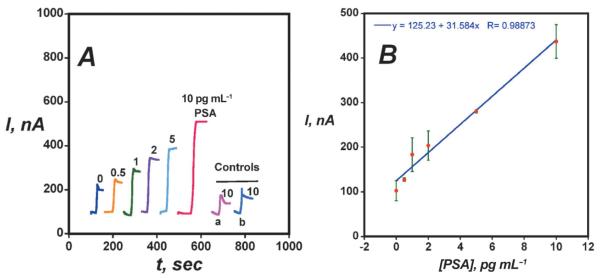
Fig. 5.
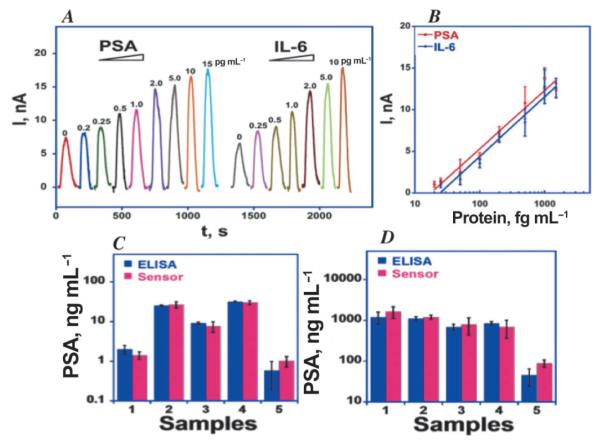
Amperometric responses for AuNP immunosensors at −0.3 V and 3000 rpm in buffer containing 1 mM hydroquinone after injecting 0.04 mM H2O2 to develop the signal (A) using Ab2-magnetic bead-HRP with 7500 labels/bead at PSA concentrations shown. Controls: (a) Immunosensors built on bare PG at 10 pg mL−1 PSA; (b) Immunosensors built on PDDA coated PG surface at 10 pg mL−1 PSA. (B) Influence of PSA concentration on steady state current for AuNP immunosensor using multi-label Ab2-magnetic bead-HRP. Reproduced with permission from reference [50].
Scheme 1.
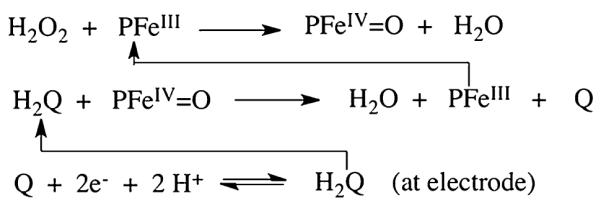
Detection of proteins using HRP (PFeIII) labels.
Immunosensors for PSA in serum using multi-labeled CNT-HRP-Ab2 particles using this approach gave detection limits (DL, 3 times the average noise level above the zero PSA control) of 4 pg mL−1 (150 fM).62 SWCNT forests provided 5 to 10-fold better sensitivity than immunosensors without nanotubes because of a large increase in density of Ab1 compared to that on a flat immunosensor, which was recently confirmed.48 SWCNT immunosensors were used to measure PSA levels in 1000 prostate cancer cells laser dissected from prostate tissue (Figure 4). These immunosensors were also used to obtain a 0.5 pg mL−1 DL for IL-6 released from a variety of oral cancer cells into cell growth media.61
AuNP-coated sensors were made by depositing 5 nm glutathione-decorated AuNPs onto a 0.5 nm polycation layer on pyrolytic graphite (PG). Glutathione terminates in carboxyl groups that provided functionality to attached capture antibodies to the AuNP layer. Sensitivity was further improved by using detection particles with Ab2 and ca.7500 HRPs attached onto 1 μm magnetic beads.50 Using these multiply labeled magnetic beads with AuNP platforms (Figure 5), the detection limit was 0.5 pg mL−1 (20 fM) PSA in serum, eight-fold better, and sensitivity was four-fold better than SWCNT forest immunosensors. Controls (a) and (b) in Figure 5A show that the dense AuNP platform also provides enhanced sensitivity over immunosensors without AuNPs.
SWCNT forest and AuNP immunosensors gave excellent correlations with ELISA results for determination of PSA in serum from human cancer patients.50,62 A four-electrode SWCNT forest array was used to detect PSA, IL-6, platelet factor-4, and prostate specific membrane antigen simultaneously.65 Since these four proteins are present in different concentration ranges in the serum, we combined multiply and single labeled antibodies to achieve the necessary individual sensitivities. Excellent correlations with single-protein ELISAs were obtained for samples from cancer patients and cancer-free controls (Figure 6).
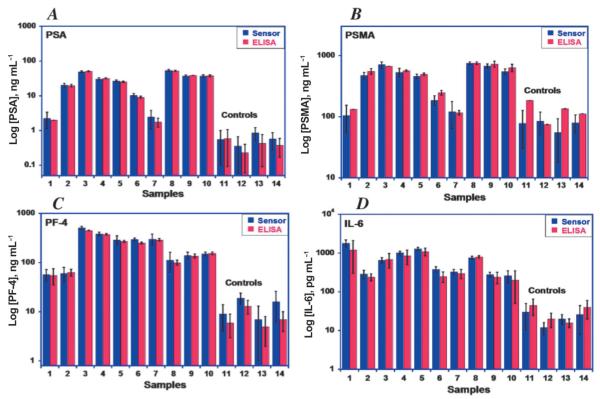
Fig. 6.
Validation of accuracy of nanostructured four-sensor immunoarray for human serum samples showing correlation with results from ELISA for: (A) PSA, (B) prostate specific membrane antigen (PSMA), (C) platelet factor-4 (PF-4), (D) interleukin-6 (IL-6). Samples numbered 1-10 are from prostate cancer patients; samples 11–14 are controls from cancer-free individuals showing much lower protein levels.
Paramagnetic beads are very useful to aid in protein capture, separation, and transport in immunoassays. Paramagnetic labeled particles have a distinct advantage over non-magnetic particles since only a simple magnet is required to separate and manipulate them, and filtration and centrifugation are avoided.66 They can also be used to achieve magnetic position control in microfluidic devices. We have now attached secondary antibodies and 7,500 to 500,000 HRP labels to magnetic particles for immunosensor applications. The number of HRP units can be adjusted depending on the sensitivity needs for specific analyte proteins. Individual sensors have been developed that achieved DLs well below 1 pg mL−1 for PSA, IL-6, IL-8, and matrix metalloprotein-3 (MMP3).50,67-70 Our record, in collaboration with Bernard Munge, is an unprecedented DL of 1.0 fg mL−1 for IL-8 using magnetic bead bioconjugates with a half-million HRP units each. An added advantage is that tens of thousands of antibodies can be included on 1 μm beads, promoting binding of analyte proteins to the beads at concentrations well below what can be achieved with a single antibody.
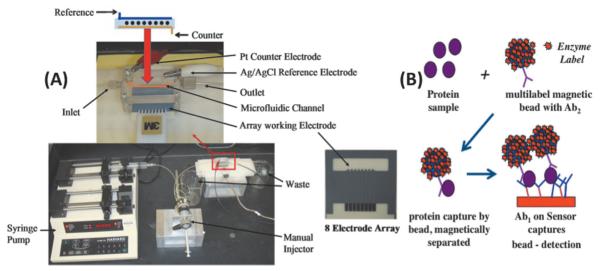
We recently coupled the immunosensor approaches described above into a microfluidic system featuring an eightbiosensor array. This simple microfluidic immunoassay system (Figure 7) features AuNP sensor electrodes, and is currently being used to validate biomarker panels. We used off-line protein capture by multiply labeled magnetic particles to achieve very low DLs of 10–300 fg mL−1 for proteins in serum. Other advantages include multiplexing capability, speed (ca. 1 hr per assay), and low cost.
Fig. 7.
Microfluidic protein array: (A) microfluidic array chip, pump (bottom left), and sample injector. The eight-sensor microfluidic device is in the red box, expanded at top with arrangement of eight nanostructured sensors (black dots), Pt counter and Ag/AgCl reference electrodes shown above. (B) Illustration of use of the array for immunoassays showing one sensing element and massively labeled (7,000–400,000 horseradish peroxidases) 1 mm magnetic particle with multiple secondary antibodies (Ab2) attached. The magnetic particle captures protein analytes from serum off-line, is washed and magnetically separated, then injected into the microfluidic device, and captured by antibodies (Ab1) on the elements of the sensor chip for amperometric measurement using H2O2 and hydroquinone mediator to activate the HRP label. Reproduced with permission from reference [66].
Figure 8 shows calibration data obtained for the simultaneous detection of PSA and IL-6 in diluted serum using the microfluidic system in Figure 7.72 After off-line capture of the proteins by labeled magnetic particles, these particles are separated and washed. They are then injected into the device via the injector sample loop, and the flow is stopped when they reach the measuring chamber. After 15 min to allow the particles to bind to the capture antibodies on the electrodes and suppression of NSB by the appropriate blocking cocktail, flow is resumed and a mixture of H2O2 and hydroquinone is injected to develop the signal. The device gives low noise peaks in the pg mL−1 range with negligible crosstalk72 (Figure 8). Excellent dynamic ranges and DLs of ca. 0.2 pg mL−1 were obtained for both proteins in assays that require only 2–5 μL of diluted serum. Even if not required to determine panel proteins, such very high sensitivity will allow sample dilution to further minimize non-specific binding of interferences.
Fig. 8.
Simultaneous calibration using eight-sensor microfluidic array with off-line analyte capture by multi-label HRP-MP-Ab2 particles using 200,000 HRP labels per particle for PSA and IL-6 in serum. Signals developed by injecting 1 mM hydroquinone mediator + 100 mM hydrogen peroxide as enzyme activator. (C) Simultaneous determinations by the array compared to individual ELISAs for PSA and IL-6 in patient serum: 1–4 are from prostate cancer patients; 5 is a cancer-free control. Reproduced with permission from reference [66].
We are developing several alternatives to the above approach for ultrasensitive multi-protein measurements. We recently reported a prototype multi-protein array using electrochemiluminescence (ECL) detection, which involves electrochemically generated light emission from a luminescent label. The label used is tris (2,2′-bipyridyl) ruthenium(II), [RuBPY], which initiates ECL when it reacts in a complex pathway with oxidized forms of sacrificial reductant tripropylamine.4,73 An advantage of ECL is that an electrode turns on the light, avoiding light sources and sophisticated optics required for conventional luminescence approaches.
We fabricated an immunosensor array on a conductive pyrolytic graphite chip (1 × 1 in.) that features upright single-wall carbon nanotube forests (SWCNTs) attached at the bottoms of 10 μL wells with polymer walls (Figure 9).74 Antibodies to capture the analyte proteins were attached to the carboxylate groups on the ends of the SWCNTs. This array requires only a single connection to a potentiostat to achieve ECL, avoiding the individually addressable sensor electrodes required above. Silica nanoparticles containing [Ru(bpy)3]2+ and secondary antibodies (RuBPY-silica-Ab2) were synthesized for highly sensitive two-analyte detection. Antibodies to PSA and IL-6 were attached to the same RuBPY-silica-Ab2 particle. A sandwich immunoassay protocol was used in which antibodies on the SWCNTs in the wells capture analyte proteins from 10 μL of sample. The 100 nm RuBPY-silica-Ab2 particles are added to bind to the captured proteins. Then, detection is initiated in the microwells by electrochemical oxidation of tripropylamine (TprA), which generates emission of ECL from [Ru(bpy)3]2+ in the 100 nm particles. ECL is measured in an open top electrochemical cell placed in a dark box with a charge-coupled device (CCD) camera above it (Figure 10). The hydrophobic polymer wall barriers confine reagents and sample in the wells, and enable simultaneous assays of different proteins in serum while avoiding cross-contamination. DLs in serum were 1 pg mL−1 for PSA and 0.25 pg mL−1 for IL-6.74 Measurements of PSA and IL-6 in the serum of prostate cancer patients agreed well with single-protein ELISA assays. These microwell ECL immunoarrays provide a simpler though slightly less sensitive approach to detecting multiple proteins than the microfluidic electrochemical assays described above. Both systems are under further development, and each is expected to have advantages in different diagnostic scenarios.
Fig. 9.
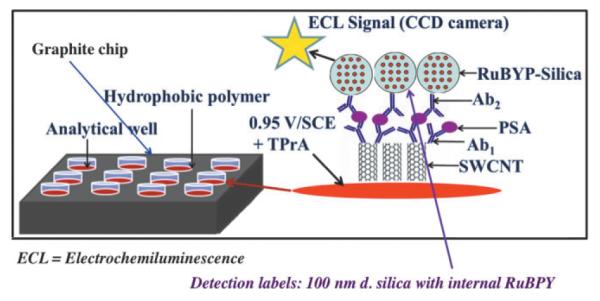
Representation of ECL immunoarray showing wells on a 1 × 1 in. pyrolytic graphite chip. SWCNT forests are surrounded by hydrophobic polymer (white) to make microwells on the chip. with SWCNT forests decorated with primary antibodies in the bottom. RuBPY-silica detection nanoparticles with cognate secondary antibodies are added and bind to the captured protein analytes. The chip is placed in an open top electrochemical cell, 0.95 V vs. SCE is applied, and ECL is detected by a CCD camera in a dark box. Reproduced with permission from reference [74].
Fig. 10.
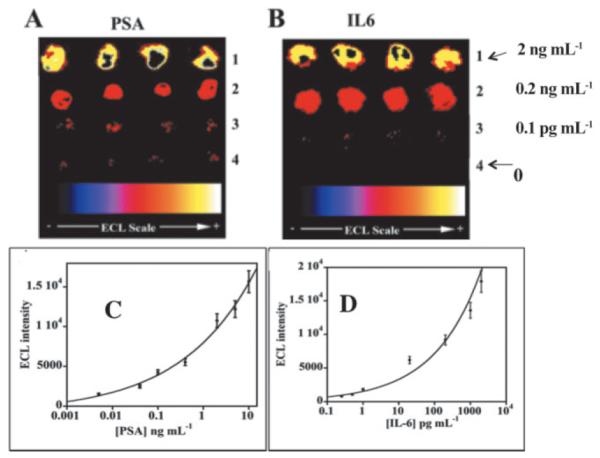
Detection of PSA and IL-6 in calf serum using ECL microwell arrays at 0.95 V vs. Ag/AgCl in the presence of 0.05% Tween 20 + 0.05% Triton-X 100 + 100 mM TPrA in 0.2 M phosphate buffer (pH 7.5). CCD images of ECL immunoarrays for (A) PSA at (1) 10 ng mL−1, (2) 0.4 ng mL−1, (3) 1 pg mL−1, (4) 0 pg mL−1 (control). (B) IL-6 at (1) 2 ng mL−1, (2) 0.2 ng mL−1, (3) 0.1 pg mL−1, (4) 0 pg mL−1 (control). Reproduced with permission from reference [74].
5. Summary and Future Perspectives
This article describes our recent progress in the development of ultrasensitive multiplexed electrochemical devices to detect biomarker proteins aimed for use in clinical cancer diagnostics. There is a great need for such devices since current clinical practice often involves single biomarkers with low predictive power, qualitative physical examinations, and biopsies coupled with pathological identification of the cancer. Certainly, measurement of a reliable panel of multiple biomarker proteins from a blood sample would be an enormous advance for cancer detection, for therapy monitoring, and for patient comfort and stress reduction.
Once devices are available that fit our requirements, what panels do we measure? The choice of these panels is not trivial. Reliability of the panels chosen needs to be validated quantitatively by studies on large numbers of patient samples, and panels need to be developed that have a high degree of reliability. We are currently planning validation studies of several biomarker protein panels that are designed to not only detect a particular kind of cancer, but to also predict the most likely subtype. These panels seem to be very promising for therapy guidance, but their predictive value needs to be determined rigorously.
From a bioanalytical science point of view, we have learned in our research that combining nanostructured electrodes with magnetic particle-antibody-HRP multi-label particles provides ultrahigh sensitivity, accuracy, flexibility, and low cost. The advantages of combining multiplexed immunoassays and microfluidics are realized in semi-automation, improved analysis speed, decrease of sample size into the low μL range, and higher signal/noise ratios. Further automation of immunoassays in more advanced microfluidic systems are underway as this article was being written.
Finally, in keeping with the theme of this special issue, our studies are built upon the solid foundation of early research discoveries in polarography, polarographic instrumentation and electrochemical enzyme catalysis that was initiated and pursued in Heyrovsky’s Polarographic Institute. Our sincere thanks are due to Professors Heyrovsky and Zuman and their early research colleagues for the immense and valuable legacy they have left us.
Acknowledgements
The author’s work described herein and the preparation of this review were supported by PHS grants ES013557 from NIEHS/NIH and EB014586 from NIBIB/NIH. The author thanks collaborators and research students named in joint publications for their excellent contributions to the project, without which progress would not have been possible.
Biography

James F. Rusling was awarded the B.Sc. in Chemistry from Drexel University in 1969, and Ph.D. from Clarkson University in 1979. He is Professor of Chemistry at University of Connecticut, Professor of Cell Biology at University of Connecticut Health Center, and adjunct Professor of Chemistry at National University of Ireland in Galway. His current research includes development of new cancer diagnostic tools measuring biomarker proteins, electrochemical and mass spectrometric arrays for toxicity screening, and fundamental bioelectrochemistry. He has authored more than 300 research papers and several books, and is a musician interested in Irish and American folk styles.
REFERENCES
- [1].Ramsay G, editor. Commercial Biosensors. Wiley; New York: 1998. [Google Scholar]
- [2].Palaček E, Scheller F, Wang J, editors. Electrochemistry of Nucleic Acids and Proteins—Towards Electrochemical Sensors for Genomics and Proteomics. Elsevier; Amsterdam: 2005. [Google Scholar]
- [3].Davis J, editor. Engineering the Bioelectronic Interface. Royal Soc. Chem.; Cambridge, UK: 2009. [Google Scholar]
- [4].Debad JB, Glezer EN, Leland JK, Sigal GB, Wholstadter J, Leland JK. In: Electrogenerated Chemiluminescence. Bard AJ, editor. Marcel Dekker; New York: 2004. pp. 359–396. [Google Scholar]
- [5].Forster RJ, Bertoncello P, Keyes TA. Annu. Rev. Anal. Chem. 2009;2:359–385. doi: 10.1146/annurev-anchem-060908-155305. [DOI] [PubMed] [Google Scholar]
- [6].Rusling JF. In: Encyclopedia of Electrochemistry, Vol. 2, Interfacial Kinetics and Mass Transport. Calvo E, editor. Marcel Dekker; 2003. pp. 418–439. [Google Scholar]
- [7].Rusling JF, Zhang Z. In: Biomolecular Films. Rusling JF, editor. Marcel Dekker; New York: 2003. pp. 1–64. [Google Scholar]
- [8].Atkinson AJ, Colburn WA, DeGruttola VG, DeMets DL, Downing GJ, Hoth DF, Oates JA, Peck CC, Schooley RT, Spilker BA, Woodcock J, Zeger SL. Clin. Pharmacol. Ther. 2001;69:89–95. [Google Scholar]
- [9].Ward MA, Catto JWF, Hamdy FC. Ann. Clin. Biochem. 2001;38:633–651. doi: 10.1258/0004563011901055. [DOI] [PubMed] [Google Scholar]
- [10].Riedel F, Zaiss I, Herzog D, Götte K, Naim R, Hörman K. Anticancer Res. 2005;25:2761–2766. [PubMed] [Google Scholar]
- [11].Wang J. Biosens. Bioelectron. 2006;21:1887–1892. doi: 10.1016/j.bios.2005.10.027. [DOI] [PubMed] [Google Scholar]
- [12].Warsinke A, Stocklein W, Leupold E, Micheel E, Scheller FW. Perspectives in Bioanalysis. Vol. 1. Elsevier B. V.; Amsterdam: 2007. [Google Scholar]
- [13].Rusling JF, Kumar CV, Patel V, Gutkind JS. Analyst. 2010;135:2496–2511. doi: 10.1039/c0an00204f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kingsmore SF. Nature Rev. Drug Discovery. 2006;5:310–320. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Williams TI, Toups KL, Saggese DA, Kalli KR, Cliby WA, Muddiman DC. J. Proteome Res. 2007;6:2936–2962. doi: 10.1021/pr070041v. [DOI] [PubMed] [Google Scholar]
- [16].Vijayawardhana CA, Halsall HB, Heineman WR. In: Electroanalytical Methods for Biological Materials. Chambers JQ, Bratjer-Toth A, editors. Marcel Dekker; New York: 2002. pp. 195–231. [Google Scholar]
- [17].Ronkainen-Matsuno NJ, Thomas JH, Halsall HB, Heineman WR. TrAC, Trends Anal. Chem. 2002;21:213–225. [Google Scholar]
- [18].Ronkainen NJ, Halsall HB, Heineman WR. Chem. Soc. Rev. 2010;39:1747–1763. doi: 10.1039/b714449k. [DOI] [PubMed] [Google Scholar]
- [19].Bange A, Halsall HB, Heineman WR. Biosens. Bioelectron. 2005;20:2488–2503. doi: 10.1016/j.bios.2004.10.016. [DOI] [PubMed] [Google Scholar]
- [20].Lu B, Smyth MR, O’Kennedy R. Anal. Chim. Acta. 1996;331:97–102. [Google Scholar]
- [21].Carter RM, Poli MA, Pesavento M, Sibley DET, Lubrano GJ, Guilbault GG. Immunomethods. 1993;3:128–133. [Google Scholar]
- [22].Warsinke A, Benkert A, Scheller FW. Fresenius J. Anal. Chem. 2000;366:622–634. doi: 10.1007/s002160051557. [DOI] [PubMed] [Google Scholar]
- [23].Yakovleva J, Emneus J. In: Handbook of Bioelectrochemistry. Bartlett PN, editor. John Wiley; New York: 2008. pp. 377–410. [Google Scholar]
- [24].Ruzgas T, Lindgren A, Gorton L, Hecht H-J, Reichelt J, Bilitewski U. In: Electroanalytical Methods for Biological Materials. Chambers JQ, Brajter-Toth A, editors. Marcel Dekker; New York: 2002. pp. 233–254. [Google Scholar]
- [25].Wang J. Electroanalysis. 2007;19:769–776. [Google Scholar]
- [26].Luo X, Morrin A, Killard AJ, Smyth MR. Electroanalysis. 2006;18:319–326. [Google Scholar]
- [27].Bourdillon C, Demaille C, Moiroux J, Savéant J-M. J. Am. Chem. Soc. 1999;121:2401–2408. [Google Scholar]
- [28].Limoges B, Marchal D, Mavré F, Savéant J-M, Schöllhorn B. J. Am. Chem. Soc. 2008;103:7259–7275. doi: 10.1021/ja7102845. [DOI] [PubMed] [Google Scholar]
- [29].Limoges B, Marchal D, Mavré F, Savéant J-M. J. Am. Chem. Soc. 2008;103:7276–7285. doi: 10.1021/ja7102873. [DOI] [PubMed] [Google Scholar]
- [30].Veetil JV, Ye K. Biotechnol. Prog. 2007;23:517–531. doi: 10.1021/bp0602395. [DOI] [PubMed] [Google Scholar]
- [31].Kim S-N, Rusling JF, Papadimitrakopolous F. Adv. Mater. 2007;19:3214–3228. doi: 10.1002/adma.200700665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wilson MS. Anal. Chem. 2005;77:1496–1502. doi: 10.1021/ac0485278. [DOI] [PubMed] [Google Scholar]
- [33].Wilson MS, Nie W. Anal. Chem. 2006;78:2507–2513. doi: 10.1021/ac0518452. [DOI] [PubMed] [Google Scholar]
- [34].Wilson MS, Nie W. Anal. Chem. 2006;78:6476–6483. doi: 10.1021/ac060843u. [DOI] [PubMed] [Google Scholar]
- [35].Wei F, Liao W, Xu Z, Yang Y, Wong DT, Ho C-M. Small. 2009;5:1784–1790. doi: 10.1002/smll.200900369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wei F, Patel P, Liao W, Chaudhry K, Zhang L, Arellano-Garcia M, Hu S, Elashoff D, Zhou H, Shukla S, Shah F, Ho C-M, Wong DT. Clin. Cancer Res. 2009;15:4446–4452. doi: 10.1158/1078-0432.CCR-09-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Patolsky F, Zheng G, Lieber CM. Anal. Chem. 2006;78:4260–4269. doi: 10.1021/ac069419j. [DOI] [PubMed] [Google Scholar]
- [38].Tkac J, Davis JJ. In: Engineering the Bioelectronic Interface. Davis JJ, editor. Royal Soc. Chem. UK; 2009. pp. 193–224. [Google Scholar]
- [39].Stern E, Vacic A, Rajan NK, Criscione JM, Park J, Ilic BR, Mooney DJ, Reed MA, Fahmy Tarek M. Nat. Nanotech. 2009;5:138–142. doi: 10.1038/nnano.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang J. Small. 2005;1:1036–1043. doi: 10.1002/smll.200500214. [DOI] [PubMed] [Google Scholar]
- [41].Rusling JF, Yu X, Munge BS, Kim SN, Papadimitrakopoulos F. In: in Engineering the Bioelectronic Interface. Davis J, editor. Royal Soc. Chem.; UK: 2009. pp. 94–118. [Google Scholar]
- [42].Yánez-Sedeno Paloma, Riu Jordi, Pingarrón JM, Rius FX. Trends Anal. Chem. 2010;29:939–953. [Google Scholar]
- [43].Willner I, Willner B. NanoLett. 2010;10:3805–3815. doi: 10.1021/nl102083j. [DOI] [PubMed] [Google Scholar]
- [44].Zhang H, Zhao Q, Li X-F, Le XC. Analyst. 2007;132:724–737. doi: 10.1039/b704256f. [DOI] [PubMed] [Google Scholar]
- [45].de la Escosura-Muñiz A, Parolo C, Merkoçi A. Mater. Today. 2010;13:24–34. [Google Scholar]
- [46].Merkoçi A. Biosens. Bioelectron. 2010;26:1164–1177. doi: 10.1016/j.bios.2010.07.028. [DOI] [PubMed] [Google Scholar]
- [47].Perfézou M, Turnerbc A, Merkoçi A. Chem. Soc. Rev. 2011 in press. DOI: 10.1039/c1cs15134g. [Google Scholar]
- [48].Malhotra R, Papadimitrakopoulos F, Rusling JF. Langmuir. 2010;26:15050–15056. doi: 10.1021/la102306z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Das J, Kelley SO. Anal. Chem. 2011;83:1167–1172. doi: 10.1021/ac102917f. [DOI] [PubMed] [Google Scholar]
- [50].Mani V, Chikkaveeraiah BV, Patel V, Gutkind JS, Rusling JF. ACS Nano. 2009;3:585–594. doi: 10.1021/nn800863w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Soleymani L, Fang Z, Sargent EH, Kelley SO. Nature Nanotech. 2009;4:844–848. doi: 10.1038/nnano.2009.276. [DOI] [PubMed] [Google Scholar]
- [52].Dequaire M, Degrand C, Limoges B. Anal. Chem. 2000;72:5521. doi: 10.1021/ac000781m. [DOI] [PubMed] [Google Scholar]
- [53].Guo H, He N, Ge S, Yang D, Zhang J. Talanta. 2005;68:61. doi: 10.1016/j.talanta.2005.04.067. [DOI] [PubMed] [Google Scholar]
- [54].Velev OD, Kaler EW. Langmuir. 1999;15:3693. [Google Scholar]
- [55].de la Escosura-Muñiz A, Merkoçi A. Small. 2011;7:675–682. doi: 10.1002/smll.201002349. [DOI] [PubMed] [Google Scholar]
- [56].Liu G, Wang J, Kim J, Jan M, Collins G. Anal. Chem. 2004;76:7126–7130. doi: 10.1021/ac049107l. [DOI] [PubMed] [Google Scholar]
- [57].Wang J, Liu G, Jan MR. J. Am. Chem. Soc. 2004;126:3010–3011. doi: 10.1021/ja031723w. [DOI] [PubMed] [Google Scholar]
- [58].Munge B, Liu G, Collins G, Wang J. Anal. Chem. 2005;77:4662–4666. doi: 10.1021/ac050132g. [DOI] [PubMed] [Google Scholar]
- [59].Rusling JF, Sotzing G, Papadimitrakopoulos F. Bioelectrochem. 2009;76:189–194. doi: 10.1016/j.bioelechem.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Munge BS, Krause CE, Malhotra R, Patel V, Gutkind JS, Rusling JF. Electrochem. Comm. 2009;11:1009–1012. doi: 10.1016/j.elecom.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Malhotra R, Patel V, Vaqué JP, Gutkind JS, Rusling JF. Anal. Chem. 2010;82:3118–3123. doi: 10.1021/ac902802b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Yu X, Kim S-N, Papadimitrakopoulos F, Rusling JF. Molecular Biosys. 2005;1:70–78. doi: 10.1039/b502124c. [DOI] [PubMed] [Google Scholar]
- [63].Yu X, Munge B, Patel V, Jensen G, Bhirde A, Gong J, Kim S, Gillespie J, Gutkind S, Papadimitrakopolous F, Rusling JF. J. Am. Chem. Soc. 2006;128:11199–11205. doi: 10.1021/ja062117e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Jensen GC, Yu X, Munge B, Bhirde A, Gong JD, Kim SN, Papadimitrakopoulos F, Rusling JF. J. Nanosci. Nanotechnol. 2009;9:249–255. doi: 10.1166/jnn.2008.J016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Chikkaveeraih BV, Bhirde A, Malhotra R, Patel V, Gutkind JS, Rusling JF. Anal. Chem. 2009;81:9129–9134. doi: 10.1021/ac9018022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Mani V, Chikkaveeraiah BV, Rusling JF. Curr. Opinion Medical Diagnos. 2011;5:381–391. doi: 10.1517/17530059.2011.607161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Munge BS, Coffey AL, Doucette JM, Somba BK, Malhotra R, Patel V, Gutkind JS, Rusling JF. Angew. Chem. Int. Ed. 2011;50:7915–7918. doi: 10.1002/anie.201102941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Munge BS, Fisher J, Millord LN, Krause CE, Dowd RS, Rusling JF. Analyst. 2010;135:1345–1350. doi: 10.1039/c0an00028k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Rusling JF, Sotzing G, Papadimitrakopoulos F. Bioelectrochem. 2009;76:189–194. doi: 10.1016/j.bioelechem.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Malhotra R, Patel V, Vaqué JP, Gutkind JS, Rusling JF. Anal. Chem. 2010;82:3118–3123. doi: 10.1021/ac902802b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Munge BS, Krause CE, Malhotra R, Patel V, Gutkind JS, Rusling JF. Electrochem. Comm. 2009;11:1009–1012. doi: 10.1016/j.elecom.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Chikkaveeraiah BV, Mani V, Patel V, Gutkind JS, Rusling JF. Biosens. Bioelectron. 2011;26:4477–4483. doi: 10.1016/j.bios.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Forster RJ, Bertoncello P, Keyes TE. Annu. Rev. Anal. Chem. 2009;2:359–385. doi: 10.1146/annurev-anchem-060908-155305. [DOI] [PubMed] [Google Scholar]
- [74].Sardesai NP, Barron JC, Rusling JF. Anal. Chem. 2011;83:6698–6703. doi: 10.1021/ac201292q. [DOI] [PMC free article] [PubMed] [Google Scholar]