Abstract
In this paper, we develop a powerful test for identifying SNP-sets that are predictive of survival with data from genome-wide association studies (GWAS). We first group typed SNPs into SNP-sets based on genomic features and then apply a score test to assess the overall effect of each SNP-set on the survival outcome through a kernel machine Cox regression framework. This approach uses genetic information from all SNPs in the SNP-set simultaneously and accounts for linkage disequilibrium (LD), leading to a powerful test with reduced degrees of freedom when the typed SNPs are in LD with each other. This type of test also has the advantage of capturing the potentially non-linear effects of the SNPs, SNP-SNP interactions (epistasis), and the joint effects of multiple causal variants. By simulating SNP data based on the LD structure of real genes from the HapMap project, we demonstrate that our proposed test is more powerful than the standard single SNP minimum p-value based test for association studies with censored survival outcomes. We illustrate the proposed test with a real data application.
Keywords: cox model, genetic studies, gene-based analysis, kernel machine, multi-locus test, score test, single nucleotide polymorphism
1 Introduction
There has been increasing interest in identifying single nucleotide polymorphisms (SNPs) that are associated with disease phenotypes. The ultimate goal of most of these association studies is to help uncover the biological mechanisms involved in the disease, which can subsequently lead to better understanding of the disease process and improved strategies for disease prevention and management [Hirschhorn, 2009]. Genome-wide association studies (GWAS) have become popular tools for discovering loci in the human genome that can give rise to disease susceptibility. A GWAS is a hypothesis free method wherein a large number of markers are genotyped in many samples to find genetic variations associated with a disease phenotype.
Most GWAS employ a case-control design and are concerned with identifying SNPs that affect disease susceptibility. On cancer phenotypes alone, more than 50 studies have been published to date [Ioannidis et al., 2010]. However, recently there has been increased interest in identifying genetic markers that characterize a patient's prognosis [Azzato et al., 2010; Huang et al., 2009; Pillas et al., 2010]. Prognostic markers are important for identifying patients with more aggressive disease who may need more aggressive or prioritized treatment to improve survival outcomes. In addition to potentially using these markers to tailor individual treatments, knowledge of these markers can help gain understanding into the biological processes that underlie disease progression. To identify prognostic markers, an investigator will typically employ prospective cohort design and collect information on baseline predictors and the time from baseline to the occurrence of a clinical outcome of interest. Examples of clinical event times include time to death or time to secondary tumor from diagnosis. GWAS of survival outcomes have become available for various diseases. For instance, Azzato et al. [2010] conducted a GWAS of survival after diagnosis of breast cancer. Pillas et al. [2010] performed a GWAS on time to first tooth eruption.
A typical GWAS design includes (at least) two stages. The first is the discovery stage which usually involves genotyping samples at a large number of SNPs using readily available commercial chips. Instead of genotyping each individual at every SNP in the genome, by utilizing the linkage disequilibrium (LD) and data from the completed HapMap project [International HapMap Consortium, 2005], only a smaller subset of SNPs (called typed SNPs) are genotyped. A selected number of top-ranked loci are then followed-up in independent replication samples at the subsequent replication stages. The region around the replicated typed SNP is then fine-mapped to examine the true disease locus. In the discovery stage, a single locus approach is most commonly used for statistical analysis. The typical analysis in this stage involves fitting a logistic regression model (for case-control study) or Cox proportional hazards model (for time-to-event outcome) to each SNP, often adjusting for non-genetic covariates.
Such a single SNP approach has several limitations. It may suffer from low power due to a large number of tests and small effect sizes of individual SNPs, likely resulting in a large number of false positives. Specifically, the true causal SNP is often not genotyped but is captured through LD with the typed SNPs. Since each typed SNP is likely to be in partial LD with the true causal SNP, the observed effect size might be smaller. Furthermore, multiple SNPs jointly affecting the disease phenotype via a complex structure (e.g. epistasis) will not be captured by the single SNP approach.
Few multi-locus tests for censored survival observations exist. The existing methods can be broadly classified into two categories: (i) individual-SNP based minimum p-value analysis (min test); (ii) haplotype-based tests. For multi-marker tests based on min p-value analysis, in order to test for the effect of a group of loci, one calculates the p-values for individual SNPs and corrects the minimum of these individual p-values to account for multiple comparison by estimating the effective number of SNPs [Cheverud, 2001; Moskvina and Schmidt, 2008; Nyholt, 2004] or via Monte Carlo methods [Lin, 2005]. As these tests rely heavily on the individual SNP approach, they do not fully utilize the correlation between the SNPs and focus only on combining p-values from the various tests. The second class of methods is haplotype-based analysis [Tregouet and Tiret, 2004]. Studies have shown that haplotype-based analysis have little advantage over single SNP analysis [Chapman et al., 2003; Roeder et al., 2005].
To overcome the difficulties in a single SNP approach, in this paper we apply a kernel machine regression based approach to jointly analyze multiple SNPs for association with censored survival outcomes. We first group SNPs into SNP-sets based on biological criteria such as genes or LD blocks, and then test for the overall joint effects of all SNPs in a SNP-set. Specifically, we use a kernel machine Cox regression framework and apply a score test to assess the joint effect of a SNP-set on the survival outcome. This approach utilizes genetic information from all SNPs in a SNP-set simultaneously, leading to a more powerful test with reduced degrees of freedom when the typed SNPs are in LD with each other. By employing non-linear kernels, this test can also capture the potentially non-linear and/or interaction effects of the SNPs.
The kernel-machine (KM) approach has been developed and applied for continuous and binary phenotypes to test for the pathway effects of gene expressions [Liu et al., 2007, 2008]. Kwee et al. [2008] applied linear KM to candidate gene studies with quantitative traits. Wu et al. [2010] applied logistic KM to case-control GWAS to test for the SNP-set effect. However, these methods are not applicable to survival data, which are subject to censoring and use the Cox proportional hazards model. Li and Luan [2003] proposed a kernel Cox regression model for modeling the effect of gene expression on survival, where they focused on estimating regression coefficients and prediction, and did not provide any inference procedures for the regression coefficient estimates of the gene expression level effects. Cai et al. [2011] extended their work by developing a KM based score test for assessing the pathway effect based on gene expression data on censored survival outcomes, where they considered the linear kernel for linear effects and the Gaussian kernel for interactions of gene expressions in a pathway.
Our work is an extension of Cai et al. [2011] in which we tailor their methodology for application to genotype data from a GWAS survival study. Specifically, we take into account the particular characteristics of SNPs in a GWAS study. For example, we consider scenarios where the true causal SNP may be untyped, as well as use kernels that are appropriate only for SNP data, e.g. the identity-by-state (IBS) kernel, for modeling SNP-SNP interactions. Note that the Gaussian kernel is suitable for continuous gene expression data but is not suitable for discrete SNP data. We perform an extensive simulation study to evaluate the performance of the proposed survival KM methods for testing for the SNP-set effect in GWAS. Our paper can also be viewed as an extension of the logistic KM method of Wu et al. [2010] where we apply the KM method to a prospective cohort GWAS in which the phenotype is a censored survival outcome modeled using the Cox proportional hazards model and the test is based on martingale residuals to incorporate censoring.
This paper makes three key contributions. First, we describe a powerful alternative using SNP-sets to the single SNP Cox model in analyzing whole genome data. Second, we illustrate the feasibility of employing a kernel machine framework in analyzing time-to-event outcome in GWAS, that takes into account biological information as well as allows for testing for multiple causal SNPs and epistasis. Finally, we demonstrate via numerical simulations that our approach has better performance than the standard single SNP based minimum p-value test (min test) when the typed SNPs are in LD with each other and with the true causal SNP. The remainder of this paper is organized as follows: we will first discuss how SNP-sets can be formed and then introduce the survival kernel machine method, before presenting simulation results. Lastly, we apply our method to a GWAS dataset to identify genes associated with lung cancer survival.
2 Forming SNP-sets
The motivation behind forming SNP-sets is two-fold. Firstly, it allows us to capture the joint effects of multiple SNPs and harness the LD between the SNPs in the SNP-set to increase test power. Secondly, it allows us to incorporate biological information on how SNPs may collectively affect the phenotype of interest, so the results have better biological interpretation. There are various ways to form SNP-sets, see Wu et al. [2010] for an overview. For example, one could form SNP-sets by including all the SNPs that are located near a gene. This could be done by taking all SNPs from the transcription start to end, and possibly include all SNPs that are upstream and downstream of a gene. A gene-based approach is useful in helping to identify genes that are associated with the disease. Since the true causal SNP is often not genotyped and is probably correlated with several SNPs in the SNP-set, by forming gene SNP-sets and using the kernel machine method, we can increase power by using the correlation between the SNPs in the SNP-set. Another way to form a SNP-set is by pathway, e.g. by including all SNPs that fall within a gene and including all genes in a biological pathway. The advantage of this approach is that if there are multiple causal SNPs falling within different regions associated with the disease, the kernel machine method would be able to capture this, leading to increased power. The drawback of forming SNP-sets using genes or biological pathways is that not all genes are known and some SNPs associated with a disease can be located in gene deserts. An alternative is to form SNP-sets based on LD blocks, recombination hot-spots, or using a sliding window approach. These methods allow complete coverage of the genome. For illustration purposes, we will form SNP-sets using the gene-based approach in this article.
3 Survival Kernel Machine Method
Consider a S × 1 covariate vector Zi containing the genotypes for the S SNPs in the SNP-set, and a R × 1 covariate vector Xi containing the R non-genetic covariates for individual i. For an additive effect of the allele, the genotype of each SNP is coded as 0, 1 or 2. Let Ti denote the survival time for individual i. Due to censoring, Ti is observable up to a bivariate vector (Ui, Δi), where Ui = min(Ti, Ci), Δi = I(Ti ≤ Ci), and Ci is the censoring time for the ith subject. We require the standard assumption that Ci is independent of Ti conditional on Zi and Xi. For a study with sample size n, the data consist of n independent and identically distributed copies of random vectors {(Ui, Δi, Zi, Xi), i = 1, ⋯, n}. Assume that survival time Ti is related to Zi and Xi through the Cox proportional hazards model [Cox, 1972]:
| (1) |
For example, specifying corresponds to the usual Cox proportional hazards model including only main effect terms for all the S SNPs in the SNP-set (while adjusting for the R non-genetic covariates). To allow for flexibility in modeling the effects of the SNPs in a SNP-set, we allow h(·) to be an arbitrary function generated by a given positive definite kernel function K(·, ·). To allow for the potential non-linear effects from the covariates, X may include non-linear bases of the original covariates such as polynomial or splines.
3.1 Kernels
A kernel function K(·, ·) implicitly specifies a functional space ℋK spanned by a particular set of orthogonal basis functions , where J is allowed to be infinite. By the representation theorem [Kimeldorf and Wahba, 1970], a properly regularized estimator of h(Z) based on the observed data can be written as both
| (2) |
and
| (3) |
where αi are the unknown parameters. The kernel function is a projection of the genotype data from the original space to a new space (spanned by the basis functions , in which h(·) is modeled linearly in this new space, as illustrated by equation (2). Intuitively, K(Zj, Zl) is a distance metric measuring the similarity between two individuals, the jth and lth subject, with respect to their genotype information in the SNP-set. A few popular choices of kernels K(·, ·) that can be used for SNP data are given below. The linear kernel given by:
| (4) |
implicitly specifies ℋK to be spanned by , which corresponds to the standard Cox model with main effects for all SNPs in the SNP-set, i.e. . The weighted linear kernel is similar to the linear kernel, except that weights can be incorporated to improve power.
| (5) |
For example, if we define the weights to be , where qs is the minor allele frequency (MAF) for the sth SNP in the SNP-set, this will cause rare variants to be given higher weights, while down weighting the common variants. Such a weight might be used to prevent information from SNPs with low MAF to be smoothed over by SNPs with high MAF.
The advantage of using the kernel machine method is that the basis functions are not always easily specified. With a variety of kernels to choose from, one can conveniently specify more complex models especially for high-dimensional data. Two such kernels are the identical by state (IBS) kernel and the weighted IBS kernel, which are defined using the number of alleles shared IBS by subjects j and l at the S typed SNPs. The IBS kernel is:
| (6) |
where IBS(Zj,s, Zl,s) is the number of alleles shared IBS (0, 1, or 2) by subjects j and l at SNP s in the SNP-set. The advantage of using the IBS kernel is that it allows for SNP-SNP interactions (epistasis). The weighted IBS kernel is similar to the IBS kernel, except that like the weighted linear kernel, it allows weights to be incorporated. Other choices of kernels could also be employed as long as the kernel function satisfies the requirements of Mercer's Theorem [Cristianini and Shawe-Taylor, 2000], which includes the condition that the kernel function is positive definite (eigenvalues must be positive). See Wessel and Schork [2006], Lin and Schaid [2009], Mukhopadhyay et al. [2010] for examples of additional kernels. The choice of a kernel specifies a metric with which the genetic distance between the two individuals are measured and will influence the power of the test.
3.2 Kernel Machine Score test for Censored Survival Outcomes
We are interested in testing the null hypothesis that a SNP-set, for example a gene, is not associated with the event time of interest after adjusting for covariates X. This corresponds to testing H0 : h(Z) = 0 under model (1). From equation (3), testing H0 is equivalent to testing . Assuming that α = (α1, …, αn)T follows an arbitrary distribution with mean zero and variance covariance matrix τ𝕂−, H0 is equivalent to testing H0 : τ = 0, where 𝕂 is the n × n matrix whose (i, j)th element is K(Zi, Zj) and 𝕂− is the generalized inverse of 𝕂. Under such a random effect framework, H0 can tested using a score statistic for the variance component, namely
| (7) |
where M̂ = (M̂1, ⋯, M̂n)⊤, is the estimated martingale residual for individual i under H0, Yi(t) = I(Ui ≥ t), γ̂ is the partial likelihood estimator of γ and is the Breslow’s estimator of under the null model of , and
As discussed in Cai et al. [2011], the test statistic Q under the null follows a mixture of chi-square distributions which can be approximated via resampling methods. To obtain a p-value for the test using resampling methods, Cai et al. [2011] derived asymptotic expansions of the score statistic as double integrated martingale processes, and then generated realizations of the score statistic under the null by approximating the distribution of the martingale processes via resampling. Intuitively, the score statistic determines the extent to which genetic similarities as described by the kernel is correlated to similarities in phenotype. This can be seen if we rewrite the first term in the score statistic
| (8) |
The term K(Zi, Zj) would be large if the ith and jth patients have similar genetic profiles, while the term M̂iM̂j would be large if they also have similar disease prognosis. Thus the term M̂iM̂jK(Zi, Zj) would be large if genotype similarities are correlated with phenotype similarities for the ith and jth patients. The second term of the score statistic is a centering term so that the score statistic Q has mean zero. Hence the score statistic describes whether patients with similar genetic profiles have similar disease prognosis.
4 Simulations
We conducted simulation studies to investigate the performance of the survival kernel machine based SNP-set test in a genome-wide association study, whereby SNP-sets are formed by including all SNPs located near a gene. For simplicity, no additional covariates were included. We generated SNP-sets based on the LD structure of a single gene using the program HAPGEN [Spencer et al., 2009]. The LD structure was derived from the CEU population in the HapMap project (build 35, release 21). For illustration purposes, we considered four different genes of varying sizes as summarized in Table i. The gene locations were obtained from the gene list used by PLINK [Purcell et al., 2007].
Table i.
Summary of Genes used in simulations.
| Gene | Chr | Start | End | HapMap SNPs | Typed SNPs |
|---|---|---|---|---|---|
| ASAH1 | 8 | 17958204 | 17986787 | 86 | 14 |
| FGFR2 | 10 | 123227833 | 123347962 | 232 | 35 |
| NAT2 | 8 | 18293034 | 18303003 | 22 | 4 |
| HLA-B | 6 | 31429627 | 31432968 | 6 | 2 |
For each gene, we simulated genotype data for all HapMap SNPs from the transcription start to the transcription end of the gene. A SNP is considered as a typed SNP if it is on the Illumina HumanHap550 array. These are the observed SNPs that make up the SNP-sets. In our simulations, we restricted testing to common variants, i.e. typed SNPs with MAF ≥ 0.05. We compared the kernel machine method to the single SNP based minimum p-value test (min test), which is calculated by fitting the standard Cox model to each SNP in the SNP-set individually and correcting the smallest p-value pmin for multiple testing by the effective number of SNPs Neff in the SNP-set, where Neff is estimated using the method developed by Moskvina and Schmidt [2008]:
| (9) |
For the kernel machine method, we considered three different kernels: (i) linear kernel, (ii) IBS kernel, and (iii) inverse root MAF weighted IBS kernel ( where qs =MAF). To obtain a p-value for a SNP-set, perturbations were used (see Cai et al., 2011 for details).
4.1 Size Simulations
To investigate the empirical size of the survival kernel machine SNP-set test, we conducted simulations in which survival times were simulated from the exponential distribution with constant hazard λ0(t) = 1. This corresponds to the null model:
| (10) |
Censoring times were generated from the exponential distribution with mean μC = 1 to yield about 25% censoring. For each dataset, only typed SNPs with MAF ≥ 0.05 were included in the testing procedure. The empirical size was computed using the proportion of datasets (out of 5000) that had p-value ≤ 0.05.
4.2 Power Simulations
To compare the power of the kernel machine SNP-set test to the single SNP based min test, we considered the case where there is only one causal variant and simulated survival time T under the linear Cox model:
| (11) |
As in the size simulations, censoring times were generated from the exponential distribution with mean μC = 1, giving approximately 20%–22% censoring. The causal SNP was allowed to vary across all HapMap SNPs (including both typed and untyped SNPs) one at a time, however, only typed common SNPs with MAF ≥ 0.05 were included in the testing procedure. We used β = 0.2. This setting closely mirrors the usual GWAS setting in which the true causal SNP is often not genotyped but is captured only through LD with the typed SNPs. The empirical power was calculated using the proportion of datasets (out of 500) that had p-value ≤ 0.05. Note that both the kernel machine SNP-set test and the min test evaluate the global null hypothesis that none of the SNPs in the SNP-set is associated with survival, i.e. there is no association between survival and a SNP-set, e.g. a gene.
4.3 Power Simulations in the presence of epistasis
To compare the power of different kernels in the presence of epistasis, we generated survival time from the model
| (12) |
We considered several scenarios using the ASAH1 gene. The first scenario is that the two causal SNPs were set as the untyped SNPs rs4377998 and rs6586684 which are in low LD with each other (R2 = 0.392). The typed SNP rs1049874 has high LD with rs4377998 (R2 = 0.836). The type SNP rs10112857 has high LD with rs6586684 (R2 = 0.819). The second scenario sets the two causal SNPs to be one untyped SNP rs4377998 and one typed SNP rs10112857. The third scenario sets the two causal SNPs to be one untyped SNP rs6586684 and one typed SNP rs1049874. The fourth scenario sets the two causal SNPs to both typed (rs10112857 and rs1049874).
We then repeated this experiment for another two untyped SNPs, rs12541181 and rs2427746, which are also in low LD with each other (R2 = 0.422). The SNP rs12541181 is in high LD with the typed SNP rs7830490 (R2 = 0.986), and rs2427746 is in high LD with the typed SNP rs12155668 (R2 = 0.953).
4.4 Size and Power Simulations using imputed SNPs
A common current practice in GWAS is to impute all the HapMap SNPs using the typed SNPs and HapMap data. For this set of simulations, we restricted our attention only to SNPs that segregate in the CEU panel, which consists of 62 HapMap SNPs and 13 typed SNPs for the ASAH1 gene. We first simulated genotype data for all 62 SNPs in ASAH1 gene using HAPGEN (LD structure for simulated genotypes was based on the CEU population). We then used only the 13 typed SNPs to impute for all 62 SNPs in the ASAH1 gene using the program MaCH [Li et al., 2009, 2010] and the CEU HapMap reference panel. The imputed dataset consisting of both typed and imputed SNPs (in the form of dosage) was then used for testing for the gene effect. Again we restricted testing to common variants with MAF ≥ 0.05. No filtering of the imputed SNPs was necessary as the median estimated squared correlation between imputed and true genotypes was ≥ 0.5 for all SNPs (a cut-off of 0.3 will flag most poorly imputed SNPs). For size simulations, we generated survival times as was done in Section 4.1. Additionally, we compared the power of the kernel machine SNP-set test (linear kernel) to the single SNP based min test, where there is only one causal variant and simulated survival time T under the linear Cox model:
| (13) |
We varied the causal SNP one at a time across each of the 62 HapMap SNPs. The effective number of SNPs Neff was estimated using the most likely genotypes using the Moskvina and Schmidt [2008] method. Note that the survival time was always simulated using the true genotype Zcausal, not the imputed genotype, but the testing procedure was applied to both typed and imputed SNPs.
5 Simulation Results
The empirical Type 1 error rates at the nominal level of α = 0.05 are given in Table ii. The kernel machine SNP-set test had an appropriate size for all the three kernels and all genes considered. As a benchmark, we also report the empirical Type 1 error rate for the single SNP-based min test, which also gave the correct size.
Table ii.
Empirical Type 1 error rates at nominal level of α = 0.05.
| Gene | IBS Kernel | Weighted IBS Kernel | Linear Kernel | min test |
|---|---|---|---|---|
| ASAH1 | 0.0542 | 0.0528 | 0.0546 | 0.0486 |
| FGFR2 | 0.0460 | 0.0458 | 0.0466 | 0.0462 |
| NAT2 | 0.0500 | 0.0502 | 0.0476 | 0.0416 |
| HLA-B | 0.0578 | 0.0572 | 0.0576 | 0.0550 |
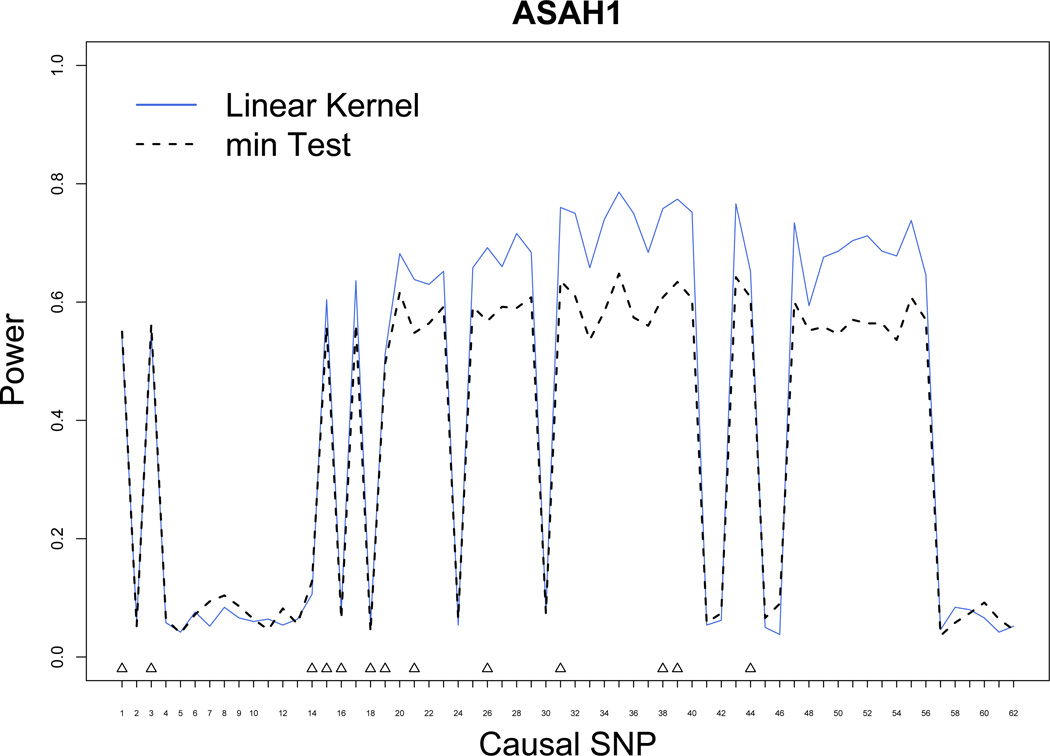
Figure 1 plots the empirical power of the kernel machine SNP-set test (using the linear kernel) and the min test for the ASAH1 gene. The ASAH1 gene is on 8p22-p21.3 and consists of 86 HapMap SNPs and 14 typed SNPs on the Illumina HumanHap550 array. The SNP-set is formed by including only these 14 typed SNPs. In Figure 1, we allowed each of the 86 HapMap SNPs to be the true casual SNP one-at-a-time and computed the power of both the survival kernel machine SNP-set test and the min test. The ASAH1 gene region is made up of SNPs with a good correlation structure, in which the typed SNPs are typically in good LD with each other and with the causal SNPs. To understand better when the kernel machine method performs better than the min test, for each casual SNP, we calculated the median R2 (a commonly used LD measure) of the causal SNP with the typed SNPs using the program Haploview [Barrett et al., 2005]. From Figure 2, we can see that when the median R2 of the causal SNP with the typed SNPs is moderate, the kernel machine method always has higher power than the min test. Not surprisingly, when the median R2 of the causal SNP with the typed SNPs is low, both methods have low and comparable power at levels around the Type 1 error rate of 0.05. In Figure 3, we also plot the power of the IBS and weighted IBS kernel. The plot shows that even when the survival time is simulated assuming a linear model, a scenario where the linear kernel and min test are optimized, the IBS and weighted IBS kernel suffer from only a slight power loss.
Figure 1.
Power Simulations for ASAH1. Blue solid line: power for Kernel Regression method. Black dashed line: power for min test. Typed SNPs are indicated with upright triangles.
Figure 2.
Kernel machine method has higher power than the min test when median R2 of the causal SNP with the typed SNPs is sufficiently high - Power Simulations for ASAH1. The causal SNPs on the x-axis are sorted by median R2 with the typed SNPs. Typed SNPs are indicated with upright triangles at the bottom. Blue solid line: power for Kernel Regression method. Black dashed line: power for min test. Red dotted line: minor allele frequency of causal SNP. Purple dotted and dashed line: median R2 of causal SNP with the typed SNPs.
Figure 3.
Power Simulations for ASAH1. Blue solid line: power for Linear Kernel. Red dotted line: Power for IBS kernel. Purple dotted and dashed line: Power for weighted IBS kernel. Black dashed line: power for min test. Typed SNPs are indicated with upright triangles.
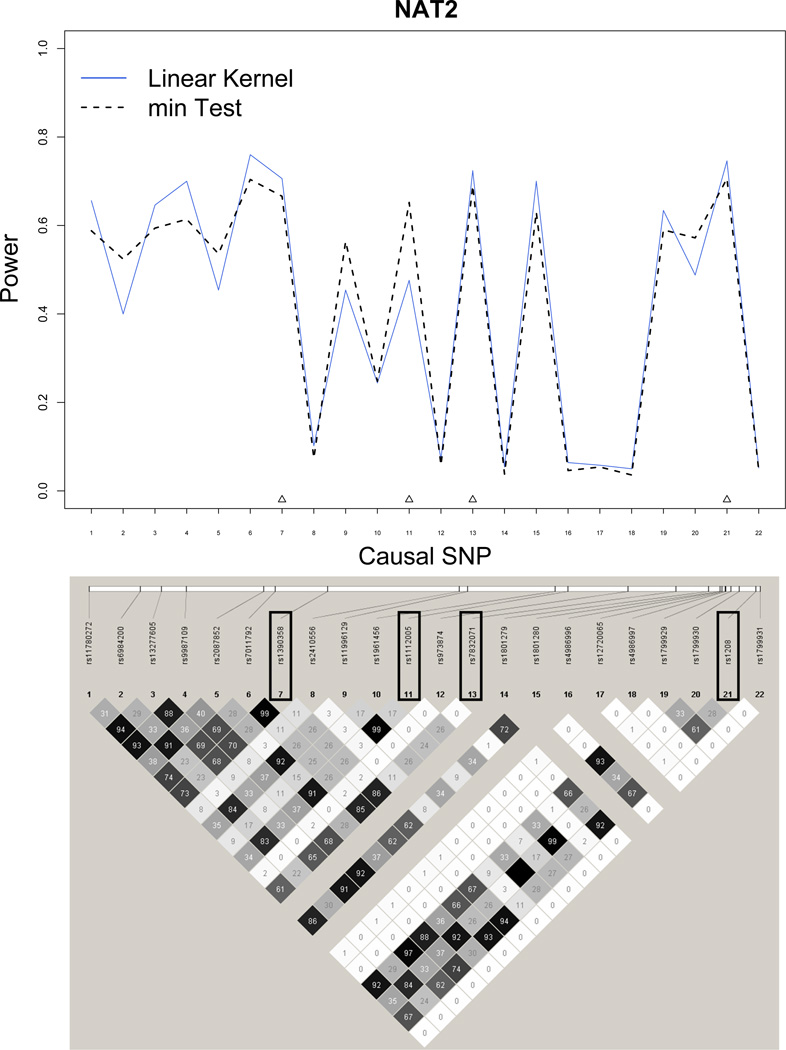
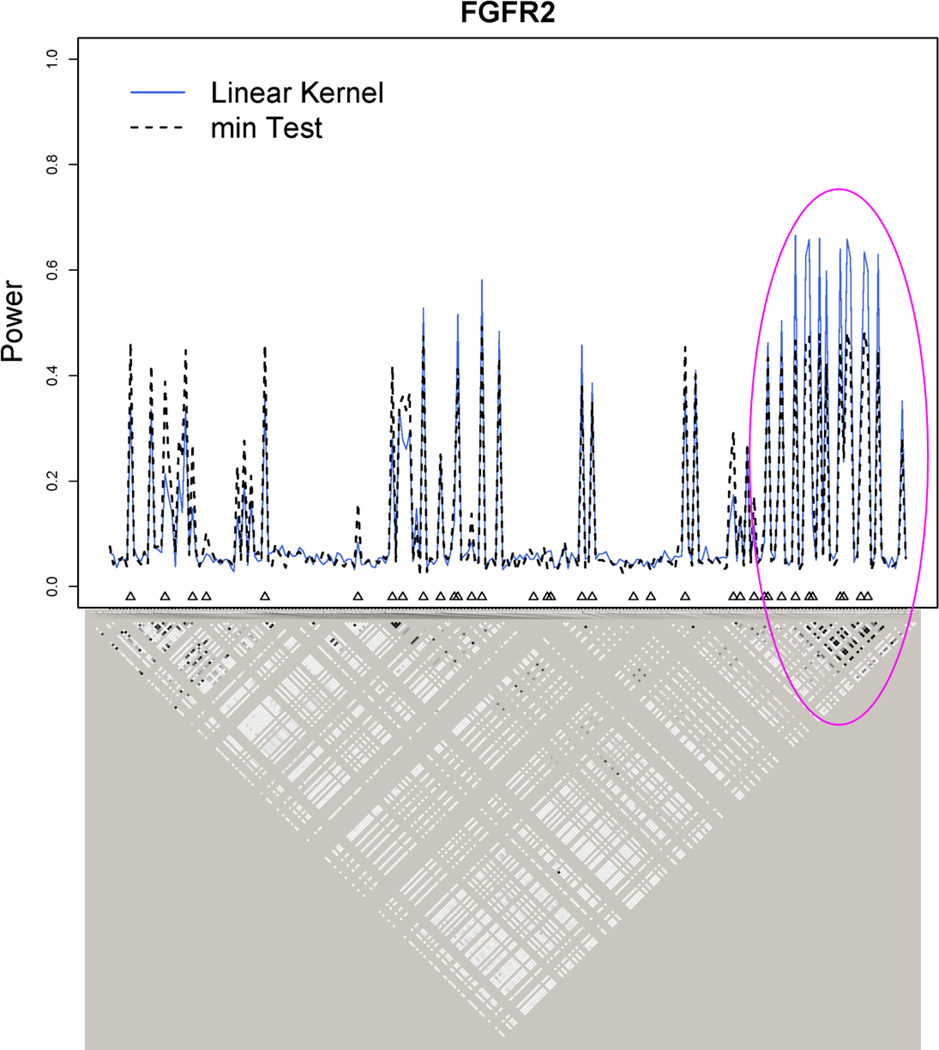
We also investigated the conditions under which the min test has higher power than the kernel machine method using the NAT2 gene. NAT2 lies on 8p22 and has 22 HapMap SNPs and only 4 typed SNPs from the transcription start to end of the gene. The 4 typed SNPs are rs1390358, rs1112005, rs7832071 and rs1208 and are labeled as SNPs 7, 11, 13, 21 in Figure 4. The power of the kernel machine SNP-set test is higher than the min test for most SNPs. The power of the min test is higher than that of the kernel machine method at SNPs 2, 5, 9, 11, 20. A close examination of the LD plot will reveal that SNPs 2 (R2 = 0.83), 5 (R2 = 0.91), 9 (R2 = 0.99), 20 (R2 = 0.99) are in high LD with SNP 11 which is a typed SNP. Hence SNPs 2, 5, 9 and 20 will have similar results as SNP 11. Additionally, SNP 11 is also in weak LD with the remainder typed SNPs (R2 = 0.26 − 0.27). Hence when there is only a single true causal SNP that is typed and tested (or one in high LD with it is typed and tested) but not in LD with other typed SNPs, the min test might give higher power than the kernel machine method. However, such a setting is unlikely to happen frequently in an actual GWAS. In contrast, SNPs 7, 13 and 21 are in good LD with each other and when each of these SNPs are the true causal SNPs, the power of the kernel machine method is higher than the min test. For a much larger gene like FGFR2 (Figure 5), in regions of high LD, the kernel machine method again outperforms the min test. In regions of weak LD, the two methods have comparable power.
Figure 4.
min test can have higher power than the kernel machine method when the true causal SNP (or one in high LD with it) is typed and tested and not in LD with other typed SNPs - Power Simulations for NAT2. Blue solid line: power for Kernel Regression method. Black dashed line: power for min test. Typed SNPs are indicated with upright triangles.
Figure 5.
Kernel machine method outperforms the min test in regions of high LD - Power Simulations for FGFR2. Blue solid line: power for Kernel Regression method. Black dashed line: power for min test. Typed SNPs are indicated with upright triangles.
To illustrate how using MAF as weights in the weighted IBS kernel can upweight rare alleles, we present the empirical power for the HLA-B gene in Table iii. HLA-B gene is a part of a family of genes known as the human leukocyte antigen (HLA) complex and lies on 6p21.3. There are only 6 HapMap SNPs and 2 typed SNPs (rs1058026 and rs2523608) in the SNP-set. In Table iii, we simulated each of the 6 SNPs as the causal SNP. Since survival time was simulated by assuming a linear genetic effect, one would generally expect the power of the weighted IBS kernel to be no greater than that of the linear kernel, which corresponds to a Cox model with linear genetic effects of the SNPs. Of the two typed SNPs, rs1058026 and rs2523608, the former (0.118) has a lower MAF than the latter (0.478). When the SNP with the lower MAF was the causal SNP (rs1058026), the power of the weighted IBS kernel (0.358) was much greater than that of the linear kernel (0.214) and the IBS kernel (0.274). This is due to incorporating the information on the importance of rare variants into the kernel function. On the other hand, when the SNP with higher MAF was the causal SNP (rs2523608), the weighted IBS kernel (0.634) had a lower power than the IBS kernel (0.712) as the effect of the more common allele which is also the causal SNP was downweighted by the weighted IBS kernel.
Table iii.
Empirical power of the KM test and the min test for HLA-B gene: Weighted IBS kernel can lead to increased power if causal variants are uncommon.
| MAF | IBS Kernel | Weighted IBS Kernel | Linear Kernel | min test | |
|---|---|---|---|---|---|
| rs1058026 | 0.118 | 0.274 | 0.358 | 0.214 | 0.318 |
| rs2770 | 0.493 | 0.170 | 0.168 | 0.170 | 0.168 |
| rs3819299 | 0.018 | 0.056 | 0.060 | 0.070 | 0.064 |
| rs3819294 | 0.083 | 0.086 | 0.090 | 0.090 | 0.084 |
| rs2523608 | 0.478 | 0.712 | 0.634 | 0.782 | 0.708 |
| rs7769258 | 0.000 | 0.044 | 0.036 | 0.048 | 0.042 |
Table iv shows the power for testing for the ASAH1 gene effect using the different kernels in the presence of epistasis. In the cases studied, the IBS kernel always has the highest power, and the weighted IBS kernel has similar power. Both the linear kernel and min test suffer from substantial power loss, which is not surprising given that they mis-specify the model. These results are consistent when the causal SNPs are typed, and when the causal SNPs are untyped and are in good LDs with the typed SNPs.
Table iv.
Empirical Power in the presence of epistasis. The column labeled “LD” gives the R2 between the two causal SNPs. The results show that IBS kernel is useful for detecting epistasis.
| Causal SNP 1 | Causal SNP 2 | LD | IBS | weighted IBS | Linear | min test |
|---|---|---|---|---|---|---|
| rs4377998 (untyped) | rs6586684 (untyped) | 0.392 | 0.538 | 0.532 | 0.122 | 0.102 |
| rs4377998 (untyped) | rs10112857 (typed) | 0.539 | 0.588 | 0.588 | 0.168 | 0.18 |
| rs1049874 (typed) | rs6586684 (untyped) | 0.563 | 0.574 | 0.548 | 0.122 | 0.11 |
| rs1049874 (typed) | rs10112857 (typed) | 0.468 | 0.618 | 0.596 | 0.186 | 0.192 |
| rs12541181 (untyped) | rs2427746 (untyped) | 0.422 | 0.926 | 0.914 | 0.688 | 0.766 |
| rs12541181 (untyped) | rs12155668 (typed) | 0.461 | 0.94 | 0.918 | 0.668 | 0.738 |
| rs7830490 (typed) | rs2427746 (untyped) | 0.409 | 0.928 | 0.916 | 0.704 | 0.764 |
| rs7830490 (typed) | rs12155668 (typed) | 0.447 | 0.952 | 0.938 | 0.678 | 0.74 |
For simulations using imputed data, the empirical Type 1 error rates at the nominal level of α = 0.05 were 0.0566 and 0.0404 for the linear kernel and min test respectively. Figure 6 shows the empirical power of the kernel machine SNP-set test using the linear kernel and the min test for the overall effect of the ASAH1 gene based on imputed data. The conclusions are similar to before, specifically for a gene with a good LD structure like ASAH1 gene, the kernel machine SNP-set test outperforms the min test.
Figure 6.
Power Simulations for ASAH1 using both typed and imputed SNPs. Blue solid line: power for Kernel Regression method. Black dashed line: power for min test. Typed SNPs are indicated with upright triangles.
6 Data Analysis
To illustrate the feasibility of our approach on real data, we applied our method to a prospective GWAS identifying genetic markers associated with the overall survival of non-small-cell lung cancer (NSCLC) patients [Huang et al., 2009]. The study consists of two patient cohorts recruited from either the Massachusetts General Hospital (MGH) in Boston, USA or the National Institute of Occupational Health in Oslo, Norway. DNA extracted from the tumor tissues of the patients were genotyped using the Affymetrix 250K Nsp GeneChip (262,264 SNPs). After quality control filtering, there were a total of 149037 SNPs. To identify genes that are associated with NSCLC overall survival, we applied the kernel machine SNP-set testing procedures. For each of the 149037 SNPs, we first imputed the missing genotypes for the missing individuals for each SNP by using the minor allele frequency of the SNP and assuming Hardy-Weinberg equilibrium for the two cohorts separately. The two datasets were then combined. We then group the SNPs into SNP-sets based on genes and apply the kernel machine method. In addition to adjusting for study cohort, similar to Huang et al. [2009], we adjusted for five additional covariates including age (in continuous scale), sex, clinical stage (as ordinal categories), cell type (squamous cell carcinoma vs. adenocarcinoma) and smoking pack-years (in continuous scale). 185 patients (96 from the MGH cohort and 89 from the Norway cohort) which were successfully genotyped and had complete information in the five covariates were used in the analysis. There were a total of 96 events out of the 185 patients.
We did the analysis using the linear, IBS and weighted IBS kernel (using inverse root MAF as weights). For comparison, we also report results from the min test, where we corrected the most significant p-value by the no. of SNPs using a Bonferroni correction. We restricted testing to SNP-sets that consists of at least two SNPs, giving a total of 6667 SNP-sets or genes. The top ten genes from each of the tests and the p-values obtained are shown in Table v. All four tests identified the same top gene. The top 10 genes identified from the IBS and weighted IBS kernels are largely identical, which is not surprising since testing was restricted only to common variants. The linear kernel gave a slightly different set of top 10 genes, but the top 10 genes identified from the linear kernel were generally highly ranked using the other two kernels. Likewise, the top 10 genes from IBS and weighted IBS kernels were also highly ranked by the linear kernel. In contrast, the top genes from the min test are quite different from the top genes identified from the kernel machine SNP-set tests. Using the Bonferroni correction (cut-off= 0.05/6667 = 7.5 × 10−6), none of the genes is significant. This is likely due to the small samples of the study. More research is needed in order to identify prognostic markers associated with NSCLC survival and validate these findings.
Table v.
Top 10 genes identified from linear kernel, IBS kernel, weighted IBS (wIBS) kernel and min test respectively.
| Gene | Chr | # SNPs | p-values | Rank | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Linear | IBS | wIBS | min test | Linear | IBS | wIBS | min test | |||
| DPY19L3 | 19 | 2 | 3.34e−05 | 2.07e−05 | 2.29e−05 | 6.86e−05 | 1 | 1 | 1 | 1 |
| KALRN | 3 | 53 | 1.06e−03 | 2.80e−04 | 2.40e−04 | 1.73e−02 | 12 | 2 | 2 | 180 |
| XKR4 | 8 | 46 | 2.06e−03 | 3.40e−04 | 5.10e−04 | 8.68e−03 | 17 | 3 | 6 | 102 |
| MARCH10 | 17 | 4 | 2.70e−04 | 5.10e−04 | 5.60e−04 | 1.04e−03 | 2 | 4 | 7 | 13 |
| ERI1 | 8 | 2 | 1.03e−03 | 5.30e−04 | 4.50e−04 | 3.06e−03 | 11 | 5 | 4 | 48 |
| RARB | 3 | 8 | 4.20e−04 | 5.30e−04 | 3.80e−04 | 4.99e−03 | 3 | 5 | 3 | 66 |
| ZNF230 | 19 | 2 | 1.19e−03 | 5.90e−04 | 1.09e−03 | 2.14e−03 | 14 | 7 | 14 | 34 |
| ZNF644 | 1 | 4 | 6.76e−03 | 6.30e−04 | 7.80e−04 | 8.50e−04 | 48 | 8 | 8 | 12 |
| TMEM106B | 7 | 5 | 6.40e−04 | 7.40e−04 | 4.90e−04 | 1.83e−03 | 4 | 9 | 5 | 27 |
| TNS4 | 17 | 2 | 1.11e−03 | 9.60e−04 | 9.20e−04 | 1.30e−03 | 13 | 10 | 10 | 18 |
| ADAM7 | 8 | 3 | 2.28e−03 | 1.01e−03 | 8.60e−04 | 7.58e−04 | 19 | 11 | 9 | 11 |
| PID1 | 2 | 26 | 6.50e−04 | 1.11e−03 | 1.51e−03 | 3.22e−03 | 5 | 12 | 16 | 50 |
| FNDC1 | 6 | 20 | 9.80e−04 | 1.23e−03 | 2.91e−03 | 4.20e−04 | 10 | 13 | 30 | 6 |
| C8orf37 | 8 | 2 | 8.10e−04 | 1.25e−03 | 1.05e−03 | 4.83e−04 | 7 | 14 | 13 | 8 |
| PITPNB | 22 | 2 | 8.30e−04 | 1.27e−03 | 1.01e−03 | 1.79e−03 | 9 | 15 | 12 | 26 |
| LYPD6 | 2 | 10 | 8.20e−04 | 2.20e−03 | 3.43e−03 | 1.99e−02 | 8 | 19 | 37 | 205 |
| PTCH1 | 9 | 6 | 7.00e−04 | 2.27e−03 | 1.19e−02 | 1.22e−03 | 6 | 21 | 86 | 17 |
| CASS4 | 20 | 2 | 1.14e−02 | 3.37e−03 | 9.20e−04 | 7.30e−04 | 78 | 33 | 10 | 10 |
| CYFIP1 | 15 | 9 | 8.86e−03 | 8.35e−03 | 2.44e−03 | 3.40e−04 | 62 | 61 | 25 | 4 |
| LAMA4 | 6 | 9 | 4.33e−02 | 1.87e−02 | 6.10e−03 | 4.20e−04 | 347 | 137 | 51 | 7 |
| MAP1B | 5 | 2 | 1.73e−02 | 1.88e−02 | 6.38e−03 | 5.87e−04 | 132 | 140 | 53 | 9 |
| SLC6A6 | 3 | 2 | 1.86e−02 | 2.01e−02 | 6.46e−03 | 3.14e−04 | 140 | 152 | 54 | 3 |
| EYA2 | 20 | 37 | 1.23e−02 | 3.45e−02 | 5.44e−02 | 2.49e−04 | 84 | 279 | 439 | 2 |
| A2BP1 | 16 | 257 | 1.52e−01 | 9.30e−02 | 5.92e−02 | 3.80e−04 | 1197 | 730 | 476 | 5 |
7 Discussions
In this article, we developed a more powerful alternative to the min test by accounting for LD among the typed SNPs. The kernel machine SNP-set test improves power by effectively utilizing the LD of the SNPs in the SNP-set and their correlation with untyped causal variants. Our approach offers a few practical advantages. Firstly, by grouping SNPs into SNP-sets based on some biological criteria, the results obtained are more easily interpretable. Secondly, our method is a multi-locus test that allows for multiple causal SNPs acting jointly. Thirdly, the kernel machine allows for easy adjustment of non-genetic covariates. Additionally, the kernel machine approach allows flexibility in modeling epistastic effects of SNPs, for example using the IBS kernel, without having to specify the functional form of the model. We considered in this paper both the linear kernel for the linear SNP effects and the IBS kernel to allow for SNP-SNP interactions in the SNP-set. Statistically choosing which kernel to use using the data is in fact a model selection problem [Liu et al., 2007], which is currently an active area of challenging statistical research with many open questions. More research is needed. Our experience suggests that the IBS kernel is a robust choice, in that it suffers a little loss in power when the effect of the SNP is linear, but is useful when the effects of the SNPs are more complex or when epistasis is present. However, the linear kernel is easier and faster to compute and works well if there is no strong evidence of epistatic effects.
If there is only one causal SNP that is genotyped and is uncorrelated with all the other typed SNPs, the min test may be more powerful, but such scenarios are uncommon, given the increasing number of SNPs that are genotyped in GWAS. Alternatively, one can also use a more powerful omnibus test, by taking the minimum p-value of the kernel machine SNP-set test and the min test, to cover both scenarios.
When a SNP-set associated with disease prognosis has been identified, there is often interest in identifying the causal variant(s). One could apply variable selection methods to the typed SNPs in the SNP-set to identify more “promising” SNPs, especially if there are many SNPs in the SNP-set. However such an approach is unsatisfactory since the true causal variant(s) is unlikely to have been genotyped, and the typed SNPs may all only be in partial LD with the causal variant(s). Thus to identify the causal variant(s), one would have to sequence the entire region. Alternatively, we need statistical methods that can effectively utilize known LD patterns in the human genome and the partial LD between typed SNPs and untyped causal variants to infer causal variant(s).
Acknowledgments
This research is supported by National Institutes of Health grants R37 CA076404 (X.L.), P01 CA134294 (X.L.), R01 CA092824 (D.C.C), R01 CA074386 (D.C.C), P50 CA090578 (D.C.C), P42 ES016454 (X.L and D.C.C), P30 ES00002 (X.L. and D.C.C), R01 GM079330 (T.C.) and National Science Foundation grant DMS 0854970 (T.C.).
References
- Azzato E, Pharoah P, Harrington P, Easton D, Greenberg D, et al. A genome-wide association study of prognosis in breast cancer. Cancer Epidemiology, Biomarkers and Prevention. 2010;19:1140–1143. doi: 10.1158/1055-9965.EPI-10-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J, Fry B, Maller J, Daly M. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Cai T, Tonini G, Lin X. Kernel machine approach to testing the significance of multiple genetic markers for risk prediction. Biometrics. 2011 doi: 10.1111/j.1541-0420.2010.01544.x. no. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J, Cooper J, Todd J, Clayton D. Detecting disease associations due to linkage disequilibrium using haplotype tags: a class of tests and the determinants of statistical power. Human Heredity. 2003;56:1831. doi: 10.1159/000073729. [DOI] [PubMed] [Google Scholar]
- Cheverud J. A simple correction for multiple comparisons in interval mapping genome scans. Heredity. 2001;87:52–58. doi: 10.1046/j.1365-2540.2001.00901.x. [DOI] [PubMed] [Google Scholar]
- Cristianini N, Shawe-Taylor J. An introduction to support vector machines. Cambridge University Press; 2000. [Google Scholar]
- Cox D. Regression models and life-tables (with discussion) Journal of the Royal Statistical Society, Series B. 1972;34:187–220. [Google Scholar]
- Hirschhorn J. Genomewide association studies - illuminating biologic pathways. New England Journal of Medicine. 2009;360:1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- Huang Y, Heist R, Chirieac L, Lin X, Skaug V, et al. Genome-wide analysis of survival in early-stage non-small-cell lung cancer. Journal of Clinical Oncology. 2009;27:2660–2667. doi: 10.1200/JCO.2008.18.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis J, Castaldi P, Evangelou E. A compendium of genome-wide associations for cancer: critical synopsis and reappraisal. Journal of the National Cancer Institute. 2010;102:846–858. doi: 10.1093/jnci/djq173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimeldorf G, Wahba G. A correspondence between bayesian estimation on stochastic processes and smoothing by splines. The Annals of Mathematical Statistics. 1970;41:495–502. [Google Scholar]
- Kwee L, Liu D, Lin X, Ghosh D, Epstein M. A powerful and exible multilocus association test for quantitative traits. American Journal of Human Genetics. 2008;82:386–397. doi: 10.1016/j.ajhg.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Luan Y. Kernel Cox Regression Models for Linking Gene Expression Profiles to Censored Survival Data. Pacific Symposium on Biocomputing. 2003;8:65–76. [PubMed] [Google Scholar]
- Li Y, Willer C, Ding J, Scheet P, Abecasis G. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiology. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype Imputation. Annual Review of Genomics and Human Genetics. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D. An efficient Monte Carlo approach to assessing statistical significance in genomic studies. Bioinformatics. 2005;21:781–787. doi: 10.1093/bioinformatics/bti053. [DOI] [PubMed] [Google Scholar]
- Lin W, Schaid D. Power comparisons between similarity-based multilocus association methods, logistic regression, and score tests for haplotypes. Genetic Epidemiology. 2009;33:183–197. doi: 10.1002/gepi.20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Lin X, Ghosh D. Semiparametric regression of multi-dimensional genomic pathway data: least square kernel machines and linear mixed models. Biometrics. 2007;63:1079–1088. doi: 10.1111/j.1541-0420.2007.00799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Ghosh D, Lin X. Estimation and testing for the effect of a genetic pathway on a disease outcome using logistic kernel machine regression via logistic mixed models. BMC Bioinformatics. 2008;9:292. doi: 10.1186/1471-2105-9-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskvina V, Schmidt K. On multiple-testing correction in genome-wide association studies. Genetic Epidemiology. 2008;32:567–573. doi: 10.1002/gepi.20331. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay I, Feingold E, Weeks D, Thalamuthu A. Association tests using kernel-based measures of multi-locus genotype similarity between individuals. Genetic Epidemiology. 2010;34:213–221. doi: 10.1002/gepi.20451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholt D. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. American Journal of Human Genetics. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillas D, Hoggart C, Evans D, OReilly P, Sipila K, et al. Genome-wide association study reveals multiple loci associated with primary tooth development during infancy. PLoS Genet. 2010;6(2) doi: 10.1371/journal.pgen.1000856. e1000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, et al. PLINK: a tool set for whole-genome association and population-based linkage analysis. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder K, Bacanu S, Sonpar V, Zhang X, Devlin B. Analysis of single-locus tests to detect gene/disease associations. Genetic Epidemiology. 2005;28:207–219. doi: 10.1002/gepi.20050. [DOI] [PubMed] [Google Scholar]
- Spencer C, Su Z, Donnelly P, Marchini J. Designing genome-wide association studies: Sample size, power, imputation, and the choice of genotyping chip. PLoS Genetics. 2009;5 doi: 10.1371/journal.pgen.1000477. e1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregouet D, Tiret L. Cox proportional hazards survival regression in haplotype-based association analysis using the Stochastic-EM algorithm. European Journal of Human Genetics. 2004;12 doi: 10.1038/sj.ejhg.5201238. 971974. [DOI] [PubMed] [Google Scholar]
- Wessel J, Schork N. Generalized genomic distance-based regression methodology for multilocus association analysis. American Journal of Human Genetics. 2006;79:792–806. doi: 10.1086/508346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Kraft P, Epstein M, Taylor D, Chanock S, Hunter D, Lin X. Powerful SNP-set analysis for case-control genome-wide association studies. American Journal of Human Genetics. 2010;86:929–942. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]