Abstract
In vitro, transcript elongation by RNA polymerase II is impeded by DNA sequences, DNA-bound proteins, and small ligands. Transcription elongation factor SII (TFIIS) assists RNA polymerase II to transcribe through these obstacles. There is however, little direct evidence that SII-responsive arrest sites function in living cells nor that SII facilitates readthrough in vivo. Saccharomyces cerevisiae strains lacking elongation factor SII and/or containing a point mutation in the second largest subunit of RNA polymerase II, which slows the enzyme’s RNA elongation rate, grow slowly and have defects in mRNA metabolism, particularly in the presence of nucleotide- depleting drugs. Here we have examined transcriptional induction in strains lacking SII or containing the slow polymerase mutation. Both mutants and a combined double mutant were defective in induction of GAL1 and ENA1. This was not due to an increase in mRNA degradation and was independent of any drug treatment, although treatment with the nucleotide-depleting drug 6-azauracil exacerbated the effect preferentially in the mutants. These data are consistent with mutants in the Elongator complex, which show slow inductive responses. When a potent in vitro arrest site was transcribed in these strains, there was no perceptible effect upon mRNA accumulation. These data suggest that an alternative elongation surveillance mechanism exists in vivo to overcome arrest.
Several factors have been identified that act as transcription elongation factors in vitro. One of the best studied is transcription elongation factor SII (also known as TFIIS), which facilitates readthrough of transcription arrest sites and other blocks to RNA polymerase II (pol II)1 elongation in vitro (1, 2). SII acts by binding to pol II and activating an intrinsic RNA cleavage activity within the enzyme that shortens the newly transcribed RNA and allows re-extension of the arrested transcript past the block to elongation (1, 2).
The transcription arrest process has been difficult to study in vivo, due in part to the rapid processing of primary transcripts and degradation of incomplete transcripts. Sensitivity of yeast to nucleotide-depleting drugs such as 6-azauracil (6AU) and mycophenolic acid has served as a phenotypic indicator of yeast with mutations in genes encoding pol II subunits and transcription elongation factors, including RPB1, RPB2, RPB6, RPB9, DST1, ELP1, ELP3, SPT4, SPT5, SPT6, SPT16, and RTF1 (3–13). This drug sensitivity is thought to result from stress upon the elongation machinery due to a reduction in the intracellular pools of nucleotides used for RNA synthesis (3, 8). It is well known that, in vitro, a low concentration of nucleotide substrates causes pol II to transcribe at a slower rate, become arrested more often, and therefore be dependent upon SII for efficient elongation (4, 14–17). Hence, upon drug treatment, yeast may become more dependent upon efficient transcript elongation by pol II. Nevertheless, not all 6AU-sensitive mutants carry mutations in genes that are obviously related to transcription elongation (18, 19), underscoring the fact that the mechanistic basis of 6AU sensitivity is not well understood.
Yeast deleted for ELP1 and ELP3, two genes encoding Elongator subunits, show delayed transcriptional induction in response to different environmental stimuli (9, 10). Inactivation of the elongation-related factor Spt5 when a conditional mutant strain is shifted to the nonpermissive temperature results in loss of some, but not all, pol II transcripts (11). In related studies, yeast with a disruption of the SII gene (dst1) or a point mutation in the second largest subunit of pol II that reduces elongation rate (rpb2–10), are sensitive to the nucleotide-depleting drugs 6AU and mycophenolic acid (4, 8, 20). These cells are compromised in their ability to induce transcription of the gene for IMP dehydrogenase, IMD2 (also known as PUR5) in response to 6AU treatment (12). As well, the rpb2–10 mutation results in cold sensitivity and inositol auxotrophy, the latter phenotype is attributed to the inability to transcribe INO1 (21). Cells lacking functional SII and containing the slow elongation mutation display a synergistic sensitivity to 6AU. They also have reduced poly(A+) mRNA levels and reduced transcription of a number of genes compared with wild-type (5). Relatively little is known about the spectrum of genes whose expression requires or is augmented by elongation factors in general and SII in particular. Cumulatively, these results suggest that one of the hallmarks of mutation in elongation factor genes is a slow transcriptional induction phenotype and a reduction in the efficiency of mRNA synthesis. Recently, a gene called SSM1 (suppressor of 6-azauracil sensitivity of the SII null mutant 1) was identified in a high copy suppressor screen of the 6AUs phenotype of a DST1 deletant (22). SSM1 expression is reduced in DST1 null yeast and is restored by SII expression (22). Specific sequences in SSM1 that arrest transcription in vitro could explain why this gene requires SII for its transcription (22). A pressing question is whether elongation factors contribute to the efficient transcription of many or most genes or if they are particularly important for the function of those genes with specific arrest sites.
Here we have analyzed transcriptional induction in yeast bearing mutations in the elongation machinery. An advantage of this system is that the biochemical consequence of this mutation has been characterized as has the mechanism by which SII relieves arrest in vitro (2). The effect of 6AU on transcriptional responses, which is expected to slow elongation rates and promote arrest in vivo, was also assessed in wild-type and mutant cells. We find that gene induction is impaired in dst1 and rpb2–10 single mutants and the rpb2–10 dst1 double mutant in the absence of drug and that this effect is exacerbated in the presence of 6AU. Finally, we measured the efficiency of transcription through a strong, well-characterized in vitro arrest site for yeast pol II in the presence and absence of 6AU. Surprisingly, the arrest site was ineffective in reducing transcriptional output in any strain in either the presence or absence of drug. That arrest sites defined in vitro were not particularly effective in vivo may reflect the fact that this type of elongation impediment is not rate-limiting in vivo. SII would therefore seem to participate in gene expression in a somewhat more general manner than would be expected considering the well described arrest-relieving activity of SII in vitro. An alternative mechanism may exist that prevents arrest in living cells. Nevertheless, SII and an efficiently elongating pol II have general effects in augmenting transcription in vivo.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Medium
Strains used here are listed in Table I. Yeast transformation was performed as previously described (23). The growth medium used for ENA1 induction experiments was synthetic complete medium lacking uracil with glucose (SC-Ura + Glu), to which NaCl was added to a final concentration of 1 m. The growth medium for galactose induction experiments was SC-Ura with raffinose (2% w/v; SC-Ura + Raff) (24), to which galactose was added to a final concentration of 2% (w/v). In the glucose repression experiments, glucose was added to a final concentration of 2% (w/v).
Table I.
Strains used
| Strain | Genotype |
|---|---|
| DY103 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 |
| [pRP214 (RPB2 LEU2 CEN)] [pRS316 (URA3 CEN)] | |
| DY105 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 |
| [pRP2–10L (rpb2–10 LEU2 CEN)] [pRS316 (URA3 CEN)] | |
| DY106 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 |
| dst1::hisG [pRP214 (RPB2 LEU2 CEN)] [pRS316 (URA3 CEN)] | |
| DY108 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 |
| dst1::hisG [pRP2–10L (rpb2–10 LEU2 CEN)] [pRS316 (URA3 CEN)] | |
| DY500 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 |
| [pRP214 (RPB2 LEU2 CEN)] [pC1023 (URA3 2μ)] | |
| DY501 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 |
| dst1::hisG [pRP214 (RPB2 LEU2 CEN)] [pC1023 (URA3 2μ)] | |
| DY502 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 |
| [pRP2–10L (rpb2–10 LEU2 CEN)] [pC1023 (URA3 2μ)] | |
| DY503 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 |
| dst1::hisG [pRP2–10L (rpb2–10 LEU2 CEN)] | |
| [pC1023 (URA3 2μ)] | |
| DY510 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 |
| [pRP214 (RPB2 LEU2 CEN)] [pC1024 (URA3 2μ)] | |
| DY511 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 |
| dst1::hisG [pRP214 (RPB2 LEU2 CEN)] [pC1024 (URA3 2μ)] | |
| DY512 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 |
| [pRP2–10L (rpb2–10 LEU2 CEN)] [pC1024 (URA3 2μ)] | |
| DY513 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 |
| dst1::hisG [pRP2–10L (rpb2–10 LEU2 CEN)] [pC1024 (URA3 2μ)] | |
| DY520 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 |
| [pRP214 (RPB2 LEU2 CEN)] [pYes2 (URA3 2μ)] | |
| DY521 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 |
| dst1::hisG [pRP214 (RPB2 LEU2 CEN)] [pYes2 (URA3 2μ)] | |
| DY522 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 |
| [pRP2–10L (rpb2–10 LEU2 CEN)] [pYes2 (URA3 2μ)] | |
| DY523 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 |
| dst1::hisG [pRP2–10L (rpb2–10 LEU2 CEN)] [pYes2 (URA3 2μ)] |
Plasmids
The plasmids pC1023 and pC1024 were made by inserting a ~700-base pair fragment (XbaI/NdeI) containing two tandem Ia arrest sites from the plasmid pAdTerm2-2HH-HH (Reines et al., 1993) in the correct or inverse orientation, respectively, into the vector pYES2 (Stratagene, La Jolla, CA).
Northern Analysis
RNA was prepared by the hot phenol extraction method and quantitated by measuring absorbance at 260 nm (25). 15 µg of RNA per sample was run on a 1% agarose-formaldehyde gel and transferred to a Zeta-Probe GT nylon membrane (Bio-Rad, Hercules, CA). The filter was dried, baked at 80 °C under vacuum for 30 min, and cross-linked in a Stratalinker 1800 (Stratagene, La Jolla, CA). Filters were prehybridized for 3 h at 42 °C in 5× SSC, 5× Denhardt’s solution (25), 50% formamide, 1% SDS, and 100 µg/ml salmon sperm DNA. The filters were then hybridized with ~100 ng of 32P-labeled probes overnight at 42 °C, and washed twice at room temperature in 2× SSC/0.1% SDS for 5 min each, twice at room temperature in 0.2× SSC/0/1% SDS for 5 min each, and twice at 42 °C in 0.2× SSC/0.1% SDS for 15 min each. Additional washes (twice at 65 °C in 0.1× SSC/0.1% SDS for 15 min each) were performed for GAL1, SED1, and pC1023 and pC1024 Northern hybridizations. Filters were exposed to x-ray film and PhosphorImager screens. Quantitation was performed with a Fujifilm BAS1000 imaging system. The ENA1 probe was made using the PCR product amplified using the following primers: 5′-AGGTGCCGTTAACGATATCTGTTCTGA-3′ and 5′-CCATCTTAGTAGCAAACACTTGAATCG-3′, producing a 400-base pair probe. The SED1 probe was made using a SmaI/EcoRI fragment from the SED1 gene. The GAL1 probe was made using the PCR product amplified from yeast genomic DNA using the following primers: 5′-TCTCGCGAAGAATTCACAAGAGAC-3′ and 5′-GCTGCCCAATGCTGGTTTAGAGAC-3′, producing a 467-base pair fragment. The pC1023/pC1024 probe was made using the PCR product amplified from pYES2 using the following primers: 5′-AATAGGGACCTAGACTTCAGG-3′ and 5′-CTGCAGATATCCATCACACTG-3′, producing a 150-base pair fragment. The probes were radiolabeled with [α-32P]dATP to a specific activity of ≈108 cpm/µg using random hexamer primers and Klenow DNA polymerase.
RESULTS
Induction of ENA1 Transcription Is Impaired in Elongation Mutants
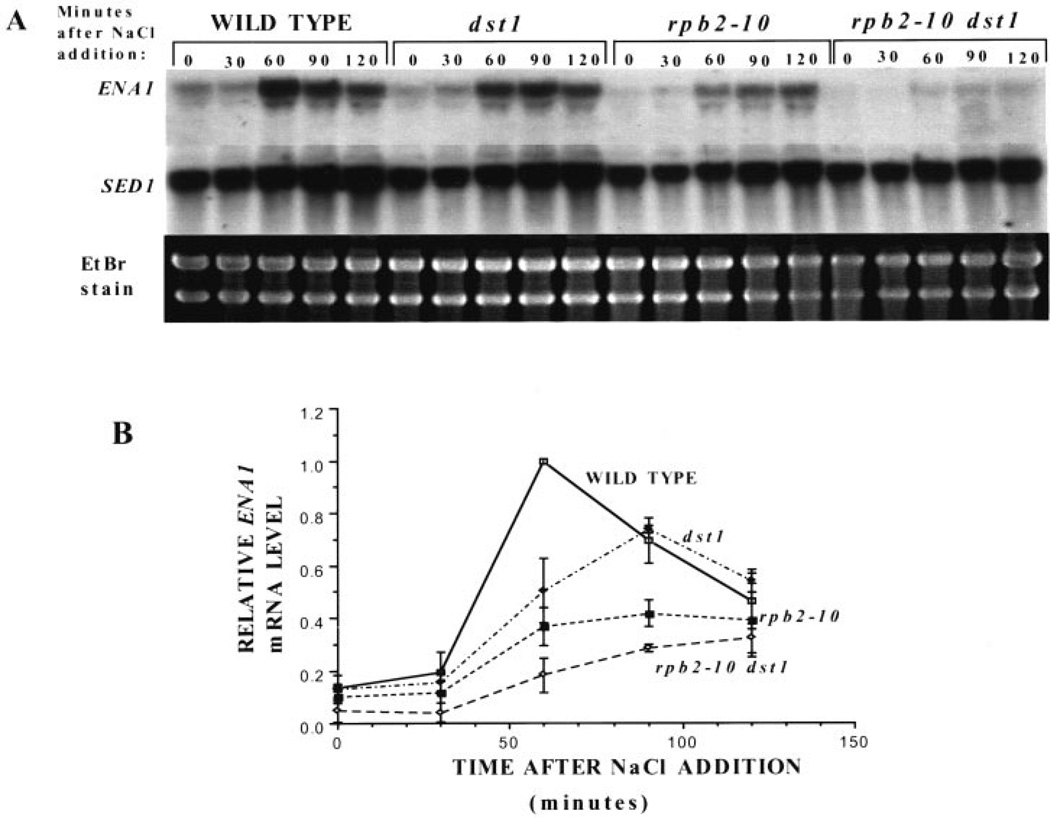
Deletion of the Elongator subunit genes ELP1 and ELP3 result in delayed transcriptional induction responses. To better understand the consequences of mutations that either slow pol II’s elongation rate (rpb2–10) or remove SII from the cell (dst1), we tested strains bearing either or both of these changes for their ability to induce transcription of the sodium-inducible ENA1 gene, which encodes a sodium pump (26). Cultures were challenged with 1 m NaCl, and RNA was collected for Northern analysis. A representative experiment is shown in Fig. 1A, and triplicate experiments are quantitated in Fig. 1B. Yeast with no SII (dst1) were slightly defective in their ability to induce ENA1 transcription relative to wild-type cells, showing a delay in reaching a maximal level of induction, which was below the wild-type yeast maxima. Yeast with a slowly elongating pol II (rpb2–10) had a similar, but more attenuated response than the dst1 disruptant. Cells with both of these mutations (rpb2–10 dst1) were quite severely defective in their ability to induce ENA1 transcription relative to wild-type cells.
Fig. 1. Induction of ENA1 in wild-type and mutant yeast strains.
A, RNA from DY103 (WILD TYPE), DY106 (dst1), DY105 (rpb2–10), and DY108 (rpb2–10 dst1) cells were taken at the time points indicated following addition of NaCl (1 m) to the medium. Blots were probed with a portion of the ENA1 gene, and results from three separate experiments were quantitated using a PhosphorImager. ENA1 transcript levels were corrected for background levels of the same area for each sample lane. These values were divided by the maximal level of ENA1 transcript present in wild-type at 60 min, and the means and standard deviations (error bars) were plotted (B).
Induction of GAL1 Transcription Is Impaired in Elongation Mutants
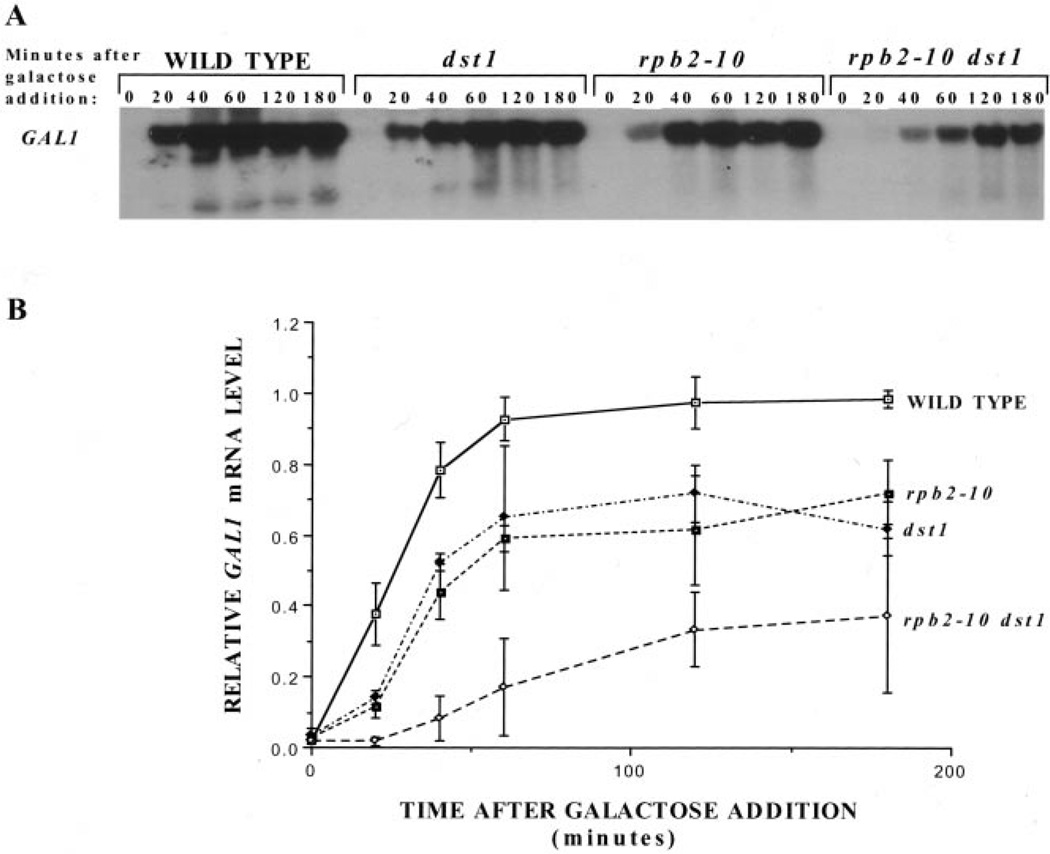
Another well-characterized induction system in yeast is that of galactose activation of GAL1 transcription. This response was examined in the elongation-defective strains described above. Galactose was added to cultures grown in raffinose, and RNA was subjected to Northern analysis. A representative experiment is shown in Fig. 2A and triplet experiments were plotted in Fig. 2B. Again we observed that yeast with mutations in the transcription elongation machinery (dst1, rpb2–10, and rpb2–10 dst1) were defective in their ability to induce transcription with respect to both the rate and extent of GAL1 mRNA synthesis. The double mutant strain (rpb2–10 dst1) was only able to induce GAL1 mRNA to ≈37% of the maximal wild-type level after 3 h of galactose exposure. Although the mutant strains were not able to attain wild-type levels of either ENA1 or GAL1 mRNA, they were able to grow on solid medium containing 1 m NaCl or galactose as their sole carbon source, respectively, suggesting that biologically significant amounts of gene product were ultimately produced in both cases (data not shown). Although these cells show reduced levels of total poly(A)+ mRNA (5), the induction defects shown here provide more direct evidence of a transcriptional defect in these cells, which is notable, because it is observed in the absence of any drug treatment.
Fig. 2. Galactose induction of GAL1 in wild-type and mutant yeast strains.
A, Northern analysis was carried out on RNA from DY103 (WILD TYPE), DY106 (dst1), DY105 (rpb2–10), and DY108 (rpb2–10 dst1) cells at various times following galactose addition. Blots were probed for GAL1 mRNA, and results from three separate experiments were quantitated using a PhosphorImager. GAL1 transcript levels were corrected for background levels of the same area for each sample lane. These values were divided by the maximal level of GAL1 transcript present in wild-type cells at 120 min, and the means and standard deviations (error bars) were plotted (B).
6-Azauracil Treatment Impairs the Ability of Elongation-defective Yeast to Induce GAL1 Transcription
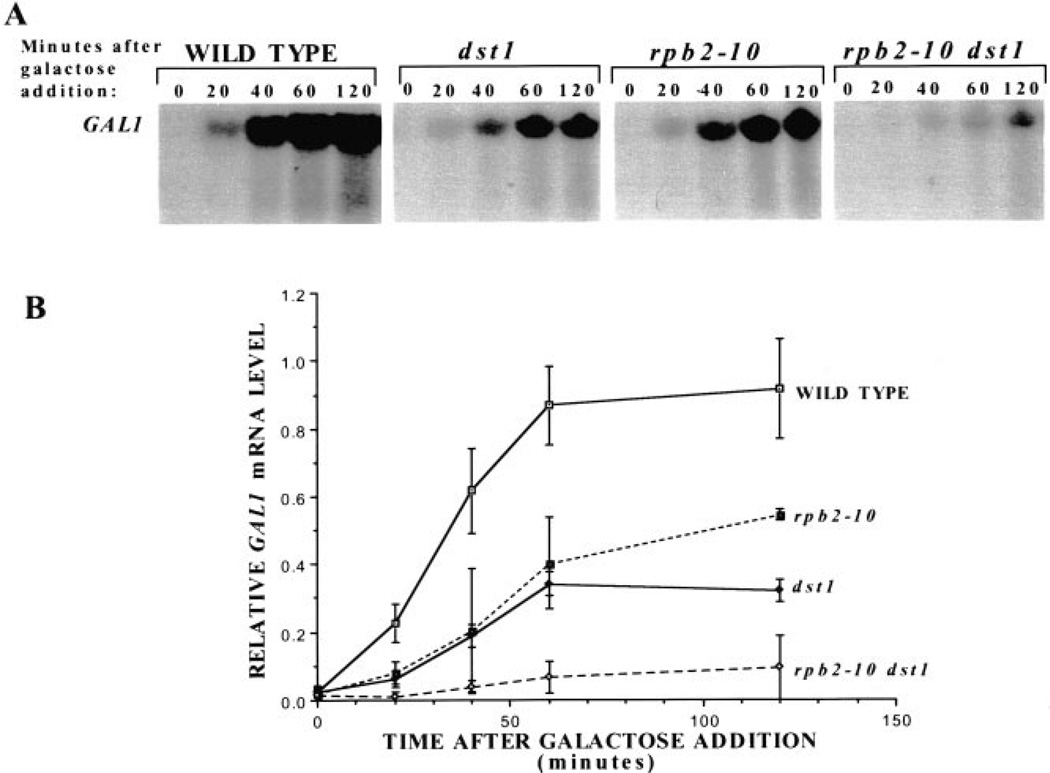
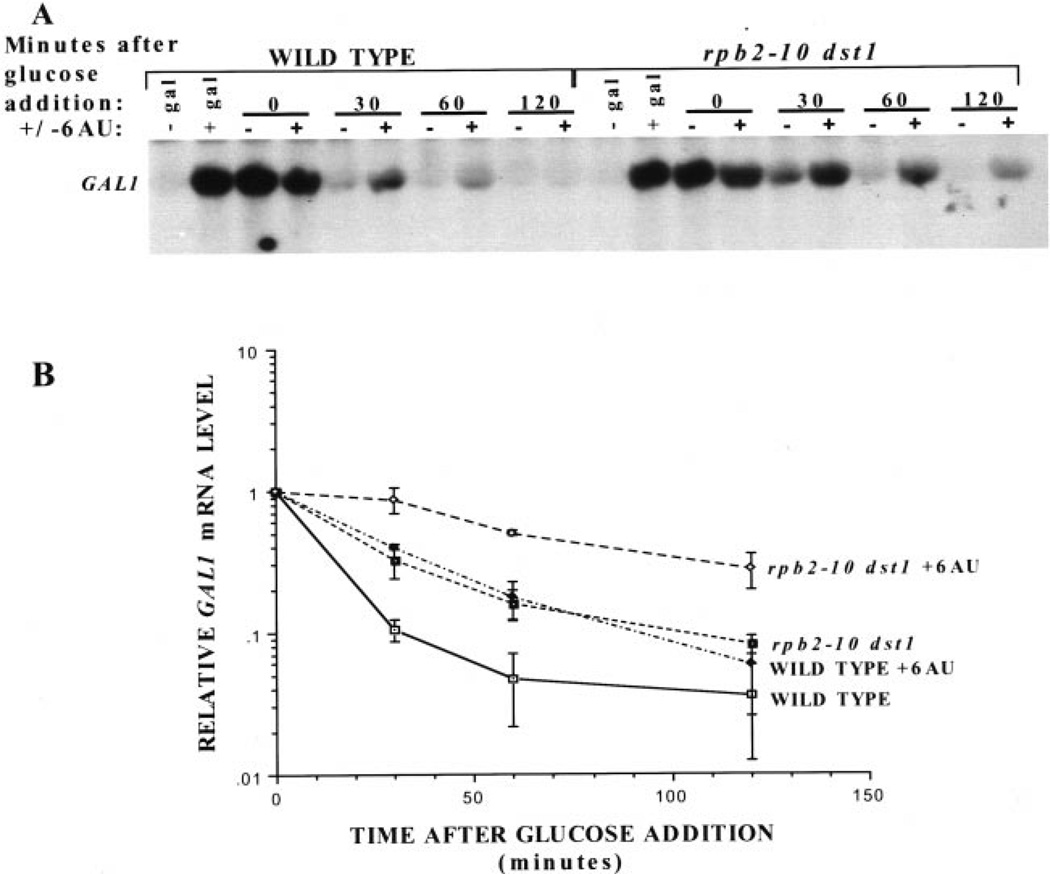
6-Azauracil (6AU) is thought to decrease the elongation rate of transcribing pol II in vivo because of its ability to inhibit the enzymes that generate nucleoside triphosphates, thereby producing effects similar to defects in the elongation machinery (3, 8). To determine whether 6AU would phenocopy the effect of the rpb2–10 and/or dst1 mutations upon GAL1 induction, yeast were treated with 6AU for 30 min, galactose was added, and RNA was prepared for Northern analysis (Fig. 3A). The single and double mutant strains were significantly impaired in both the rate and extent of induction in the presence of drug (Fig. 2 versus Fig. 3). In the absence of 6AU, the single mutants achieved 72% of wild-type GAL1 mRNA levels (Fig. 2B) versus 35–55% in the presence of drug (Fig. 3B). The rpb2–10 dst1 double mutant achieved maximal induction ≈37% of wild-type in the absence of drug (Fig. 2B) and 10% in its presence (Fig. 3B). These results provide additional evidence that 6AU affects transcription, and it does so in a manner similar to mutations in the elongation machinery, consistent with the idea that it slows transcription elongation in vivo subsequent to nucleotide depletion.
Fig. 3. The effect of 6AU upon galactose induction of GAL1 in wild-type and mutant yeast strains.
A, Northern analysis was carried out on RNA from DY103 (WILD TYPE), DY106 (dst1), DY105 (rpb2–10), and DY108(rpb2–10 dst1) cells treated with 6AU (75 µg/ml) for 30 min before galactose induction. RNA was prepared at the indicated times, and the blot was probed for GAL1 mRNA. Results from three separate experiments were quantitated using a PhosphorImager. GAL1 transcript levels were corrected for background levels of the same area in each sample lane. These values were divided by the maximal level of GAL1 transcript in wild-type cells at 120 min, and the means and standard deviations (error bars) were plotted (B).
The Loss of GAL1 mRNA Levels after Glucose Shutoff Is Prolonged in Elongation Mutants
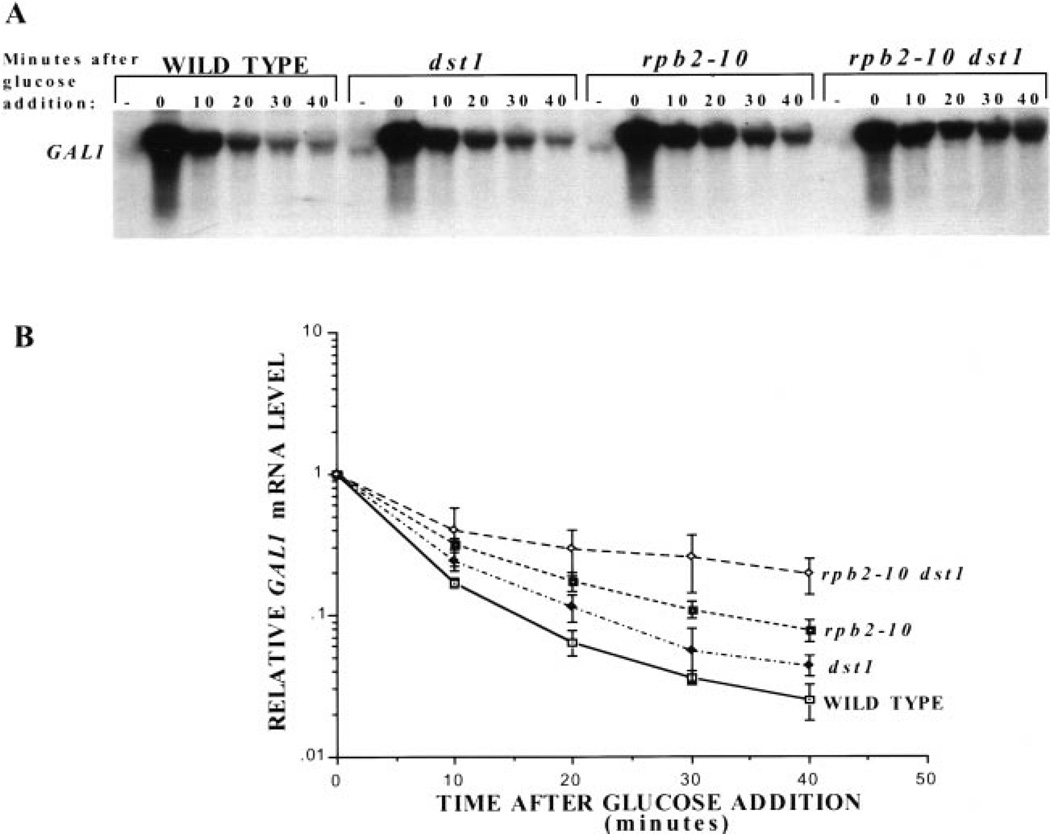
The reduced amount of GAL1 transcript observed in these mutant strains could be attributed to an alteration at one of several steps, including the initiation of transcription of GAL1, elongation through GAL1, mRNA export, or degradation of the transcript. To test if an increase in the level of mRNA degradation could account for the reduced level of GAL1 mRNA in the mutant strains, we used glucose to repress GAL1 initiation. This repression mechanism acts by preventing the recruitment of TBP and pol II to the gene and results in cessation of transcriptional initiation (27–29). GAL1 transcription was induced for 2 h with galactose then repressed by addition of glucose to growing cultures. RNA was collected at various times after shut off and its rate of disappearance was analyzed by Northern blotting (Fig. 4A, quantitated in triplicate in Fig. 4B). This experiment shows that the GAL1 transcript is present for a longer period of time after glucose shutoff in elongation-defective yeast (both single and double mutants) than in wild-type yeast. The transcript disappeared the least rapidly in the rpb2–10 dst1 double mutant, somewhat faster in the rpb2–10 mutant, and the closest to wild-type for the dst1 disruptant (Fig. 4B). This is not consistent with the model in which the reduced level of mRNA seen upon galactose induction in these mutant strains results from an increase in the rate of transcript degradation (Fig. 2). On the contrary, mRNA levels disappeared more slowly in the mutant strains with the double mutant showing the slowest rate of mRNA loss.
Fig. 4. Glucose shutoff of GAL1 in wild-type and mutant yeast.
A, Northern blot analysis was performed on RNA harvested at the indicated times after glucose addition to cultures of DY103 (WILD TYPE), DY106 (dst1), DY105 (rpb2–10), and DY108 (rpb2–10 dst1) cells grown for 2 h in galactose. RNA was also prepared from control cells that were not induced with galactose (−). The blot was probed for GAL1 mRNA, and results were quantitated using a PhosphorImager for three separate experiments. GAL1 transcript levels were corrected for background signal in the same area for each lane. Transcript levels for each strain were calculated relative to its zero time value, and the means and standard deviations (error bars) were plotted (B).
6-Azauracil Treatment Decreases the Rate of GAL1 Transcript Disappearance
Because 6AU treatment affected the induction of GAL1 in a manner similar to that of the mutants, we explored the possibility that 6AU treatment also had an effect similar to that of the mutations on the rate of GAL1 transcript disappearance. Galactose-induced cultures were treated with 75 µg/ml 6AU for 30 min before glucose was added. RNA was prepared for Northern analysis at times thereafter (Fig. 5). 6AU slowed the rate of transcript disappearance in wild-type cells to a rate similar to that of untreated double mutant yeast (rpb2–10 dst1). The rate of disappearance of the GAL1 transcript in the double mutant yeast (rpb2–10 dst1) was also slowed further when treated with 6AU. These data are again consistent with the idea that 6AU phenocopies mutations that perturb transcription elongation and does not operate by increasing the rate of transcript degradation, in further support of the model that delayed GAL1 induction in the rpb2–10 dst1 mutant yeast (Fig. 2) is not due to an increase in GAL1 transcript degradation. The fact that 6AU can make more extreme the defects seen in the rpb2–10 dst1 strain suggests that the process slowed by the drug is additive with the effect of mutations in the transcription elongation pathway.
Fig. 5. The effect of 6AU upon glucose shutoff of GAL1 in wild-type and double mutant (rpb2–10 dst1) yeast.
A, Northern blot analysis was performed on RNA from DY103 (WILD TYPE) and DY108 (rpb2–10 dst1) cells grown for 2 h following addition of galactose. 6AU (75 µg/ml) was added for 30 min as indicated, followed by glucose. RNA was prepared at the indicated times as well as before and after induction by galactose (− gal and + gal lanes). The blot was probed for GAL1 mRNA, and results were quantitated using a PhosphorImager for three separate experiments. GAL1 transcript levels were corrected for background signal in the same area for each lane. The GAL1 transcript levels were expressed as the fraction of starting GAL1 transcript remaining for each culture, and the means and standard deviations (error bars) were plotted (B).
mRNA Production in Vivo Is Not Influenced by a Strong in Vitro Arrest Site
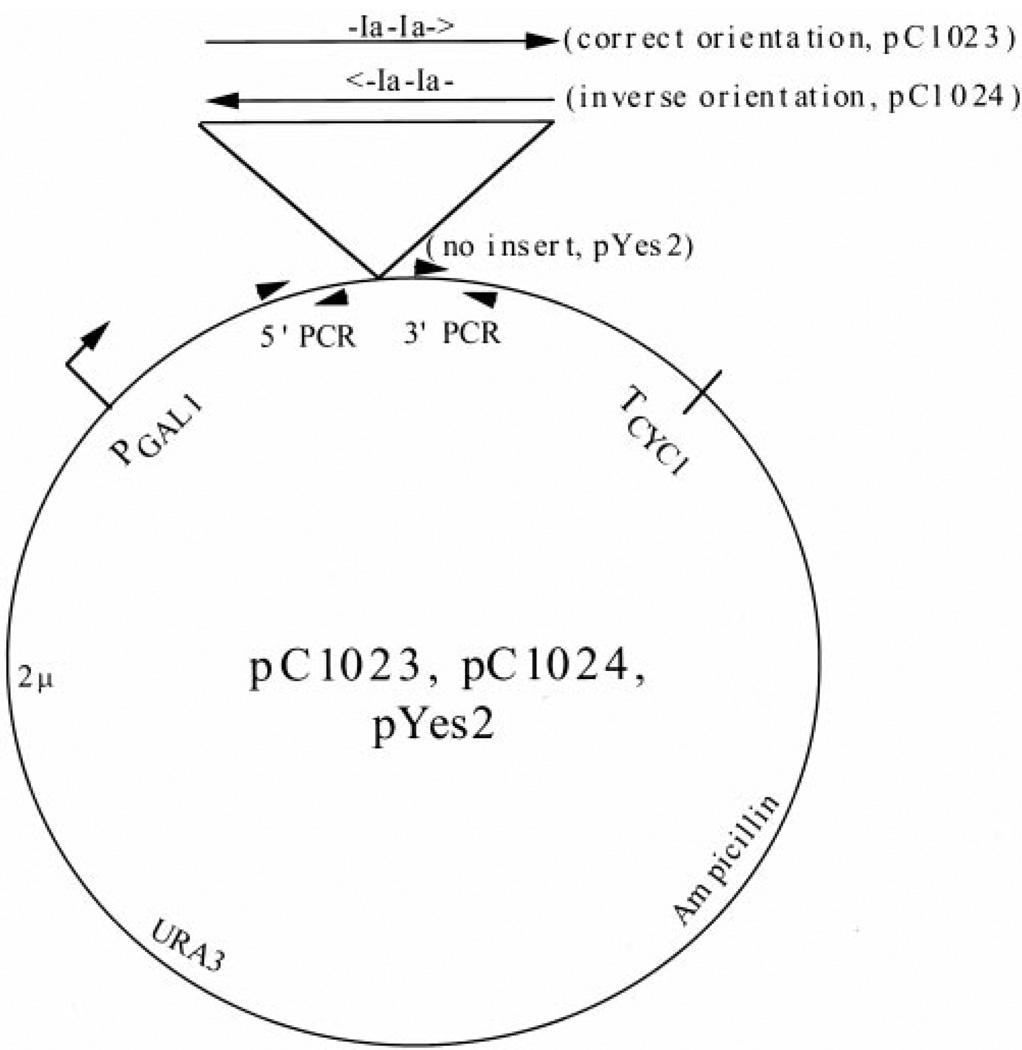
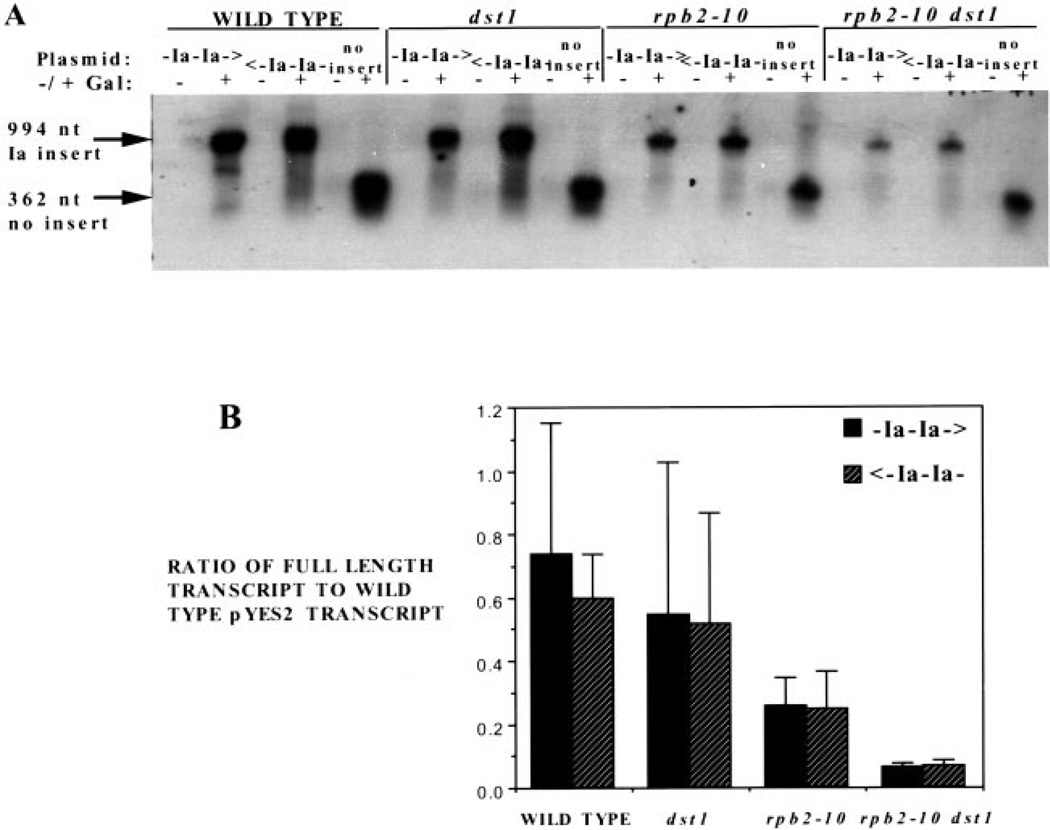
We initially presumed that these elongation mutations and 6AU treatment operated by depleting nucleotides, slowing elongation, enhancing arrest, and reducing the ability of pol II to bypass blocks to elongation, all of which should exacerbate arrest site read through in vivo. Presumably, a gene’s transcription level in this case would be related to the number or location of pause or arrest site sequences within it. Until recently, no arrest sites have been described in yeast gene sequences in vitro nor shown to operate in vivo (22). Hence, we set out to analyze in vivo a well-characterized arrest site (called site Ia; originally derived from a human histone gene), which is known to efficiently arrest transcription by yeast and mammalian pol II as well as phage and bacterial RNA polymerases in vitro (4, 30–35). High copy yeast plasmids were constructed containing a tandem arrangement of two Ia arrest sites inserted downstream of the GAL1 promoter and upstream of the CYC1 polyadenylation/termination signals (pC1023; Fig. 6). Two consecutive Ia sites function additively in stopping 75% of the polymerase enzymes that encounter them in vitro (36). Arrest of pol II at the first or second site would generate 355- and 577-nucleotide transcripts, respectively. As a control, the tandem Ia sites were inserted in the inverse orientation, because its arrest function is strictly orientation-dependent in vitro (pC1024; Fig. 6) (36). Plasmids pC1023 and pC1024, as well as the empty vector (pYES2), were individually transformed into wild-type cells and strains with the dst1 and/or rpb2–10 mutations (Table I). Cells were induced with galactose for 1 h, and RNA was collected for Northern analysis using a probe that hybridizes downstream of the Ia sequences (Fig. 6). This probe would detect differences in the total amount of full-length transcript produced. The mutant strains show lower overall levels of transcript than wild-type cells, as expected from the induction defect of the mutants described above (Fig. 2). To facilitate comparison between replicate Northern blot experiments, we measured the amount of full-length transcript that contains the arrest sites in the arresting orientation (pC1023) to that derived from the plasmid with the Ia sites in the nonarresting (pC1024) orientation. Each of these values were divided by the amount of transcript produced by the wild-type strain harboring the vector lacking Ia sites. No significant difference was seen in the relative ability of any strain to transcribe through the Ia site in the correct (arresting) versus the inverse (nonarresting) orientation (Fig. 7A; two-tailed t test, p > 0.025). In other words, within a strain, the level of full-length transcript containing the Ia insert in the sense orientation was similar to that generated from the plasmid with the Ia array in the inverse orientation (Fig. 7B). Using a probe complementary to the transcript upstream of the inserted Ia sequences, we were unable to detect shortened transcripts of the size expected if pol II became arrested at the Ia sites (data not shown). This indicates that an in vitro arrest site does not have a strong effect upon transcription in vivo.
Fig. 6. Plasmid constructs used for in vivo transcription of the Ia arrest site.
pC1023 and pC1024 contain two tandem Ia sites in the sense orientation (-Ia-Ia→) and the inverse orientation (←Ia-Ia-), respectively. pYES2 is the vector used for construction of pC1023 and pC1024 and was used as a no-insert control in these experiments. Primers used for PCR of 5′- and 3′-fragments used as probes are indicated with the arrowheads.
Fig. 7. Transcription of the Ia arrest site in wild-type and mutant yeast.
A, Northern blot analysis was carried out on RNA prepared from DY500, DY510, DY520 (WILD TYPE); DY501, DY511, DY521 (dst1); DY502, DY512, DY522 (rpb2–10); and DY503, DY513, DY523 (rpb2–10 dst1) yeast after 60 min of galactose (+) or no (−) induction. The blot was probed with the PCR product 3′ of the Ia site, shown in Fig. 6. Arrows indicate the full-length transcripts produced from the different plasmids. B, the mean (n = 3) transcript level for galactose-induced cultures was determined by PhosphorImager, corrected for background levels, and normalized to transcript levels in wild-type cells containing pYES2. The means and standard deviations (error bars) were plotted.
6AU Does Not Cause pol II to Arrest in Wild-type or Elongation-defective Yeast
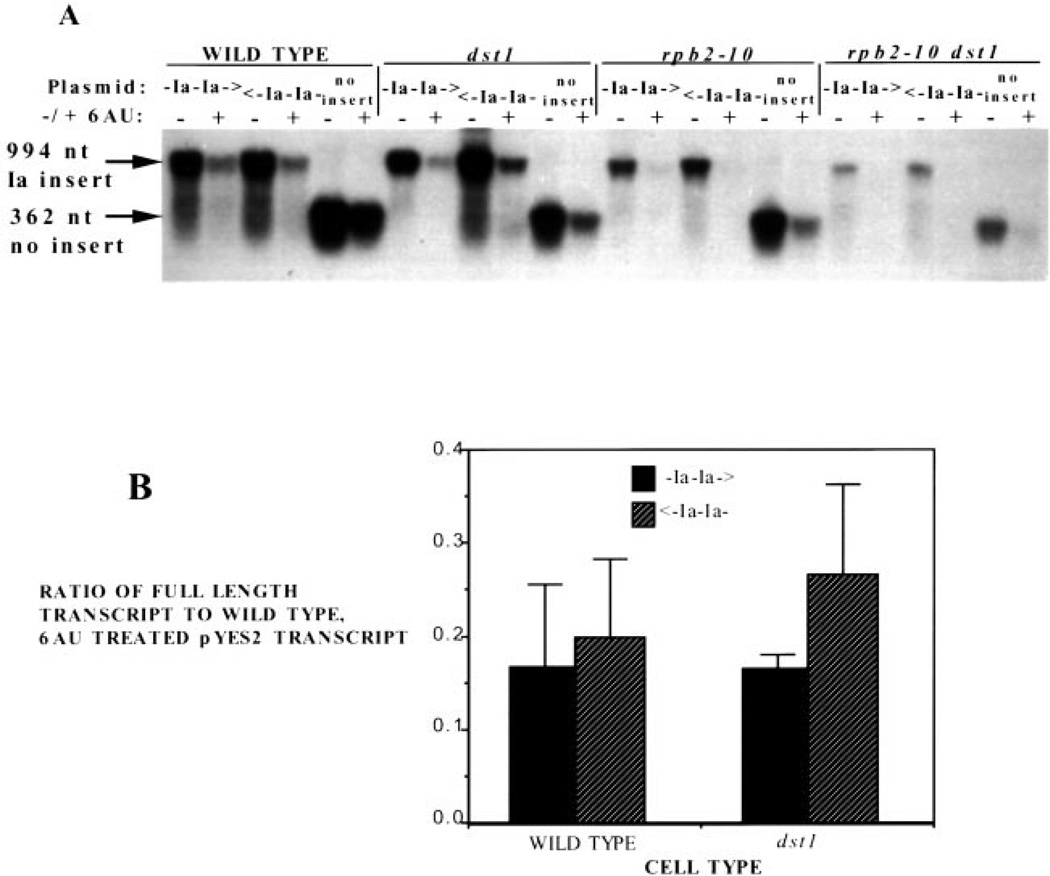
If a reduction in intracellular NTP pools increases arrest in vivo as it does in in vitro, then we reasoned that 6AU treatment of these strains might reveal an attenuation in transcription at these tandem arrest sites, particularly in the mutants. Drug-treated cultures were pretreated with 6AU for 30 min. Following drug treatment, cells were induced with galactose for 1 h, and samples were collected for Northern analysis. As observed above for galactose induction of the chromosomal GAL1 gene (Fig. 3), the addition of 6AU decreased the overall yield of transcript from the plasmids (Fig. 8A). The treatment, however, did not make detectable an effect of the arrest site on attenuating transcription when the total amount of full-length transcript produced from these two plasmids was compared (Fig. 8B; two-tailed t test, p > 0.025). Similar levels of full-length transcript were produced when the Ia sites were transcribed in the arresting orientation versus the nonarresting orientation in wild-type and dst1 disruptant yeast (Fig. 8B). Levels of full-length transcript in double mutant or rpb2–10 mutant yeast, were so low they could not be quantitated. Northern blots were also hybridized with a radiolabeled probe complementary to a region upstream of the Ia sequences to determine whether any intermediate-length-arrested transcripts were produced, and again none were detected in any of the yeast strains examined (data not shown). Under our assay conditions, 6AU strongly affects the absolute levels of transcripts produced, but it did not reveal a role of the Ia site in reducing pol II’s ability to elongate in vivo.
Fig. 8. The effect of 6AU upon transcription of the Ia site in wild-type and mutant yeast.
A, Northern blot analysis was carried out on RNA from DY500, DY510, DY520 (WILD TYPE); DY501, DY511, DY521 (dst1); DY502, DY512, DY522 (rpb2–10); and DY503, DY513, DY523 (rpb2–10 dst1) yeast untreated (−) or treated with 75 µg/ml 6AU (+) for 30 min and challenged with galactose for 1 h. The blot was probed with the PCR product 3′ of the Ia site, shown in Fig. 6. Arrows indicate the full-length transcripts produced by the different plasmids. B, the means and standard deviations (n = 3) were calculated, normalized to the level of transcript in untreated wild-type cells containing pYES2, and plotted.
DISCUSSION
The role of eukaryotic elongation factors in facilitating elongation in vivo is uncertain. In this report we have examined inducible transcription systems and a synthetic transcription unit containing a known arrest site, to study elongation in living yeast. We provide evidence that mutations in the elongation machinery negatively impact the ability of yeast to carry out two transcriptional induction processes, consistent with prior reports describing the role of other elongation factors in gene induction and mRNA biosynthesis in yeast (5, 9–12). Curiously, when transcription of a specific arrest site known to block elongation in vitro was examined, we did not find an effect upon RNA production in vivo. This is surprising, because the DNA sequence used is recognized as a particularly strong arrest signal that impedes elongation by a number of RNA polymerases in vitro, including yeast pol II.
Our findings extend to the ENA1 and GAL1 induction systems, recent reports describing a role for SII in the transcription of the SSM1 gene, the function of which is not known, and the IMD2 (PUR5) gene (12, 22). The pol II mutation rpb2–10, which changes a conserved proline near the active site to a serine and generates an arrest-prone enzyme that transcribes slowly in vitro, confers a similar “slow-induction” phenotype upon yeast as do the ELP1, ELP3, and DST1 deletions. The original description of this allele included inositol auxotrophy and reduced levels of INO1 transcription (21). Combining these two mutations (rpb2–10 and dst1) consistently resulted in a more severe defect than either alone, in agreement with earlier findings that the double mutant displayed synthetic 6AU hypersensitivity, contained less poly(A)+ mRNA, and was more severely compromised in its ability to induce transcription of IMD2 (PUR5), than either single mutant (5, 12). The inefficiency with which the mutant strains induce transcription of ENA1, GAL1, and IMD2 (PUR5) strengthens the idea that the efficiency of transcription elongation contributes to the rate of mRNA synthesis and that SII accelerates the process in vivo. These data may be the most direct evidence to date showing that SII augments transcription by pol II in vivo and is notable in that we observe an influence of SII and the rpb2–10 mutation on RNA metabolism in the absence of drug treatment.
Our studies on glucose repression of the GAL1 gene were prompted by the possibility that the reduction in GAL1 mRNA seen in elongation mutants could be due to enhanced mRNA degradation. Surprisingly, yeast with both single and double mutations in their elongation machinery (dst1, rpb2–10, and rpb2–10 dst1) showed the opposite effect, the GAL1 transcript took longer to disappear for the double mutant than either single mutant, in which GAL1 mRNA was more long-lived than in wild-type cells. This effect was exacerbated in the presence of 6AU. One interpretation of these findings is that, subsequent to the repression of new initiation events by glucose, there may be a residual contribution to the mRNA pool by slowly transcribing template-engaged pol II enzymes that have yet to reach the end of the transcription unit. Partially completed primary transcripts would not be readily observed in Northern assays due to their heterogeneity.
To determine if the mutations in the elongation machinery affect yeast transcription in a site-specific manner, the ability to transcribe a known in vitro arrest site was studied in vivo. The effect of transcribing tandem Ia arrest sites was determined, as well as the effect of 6AU treatment on the transcription of these Ia sites. Why this arrest site does not attenuate transcription in vivo is unclear. If the in vitro strength of the tandem Ia sites was recapitulated in vivo, we would expect one-fourth of the polymerases to complete synthesis of full-length transcripts in the absence of SII (dst1). The efficiency of arrest would be even higher than that in the rpb2–10 dst1 double mutants due to the “hyper-arresting” phenotype of this polymerase and the cell’s lack of SII (4). 6AU treatment of yeast has been reported to reduce the level of GTP ≈13-fold in yeast (8) which, based upon measurements of intracellular GTP concentration, represents a drop of intracellular GTP from 600–1500 µm to 50–115 µm (37). This would bring the GTP concentration to a level below 200 µm, the Ks of mammalian pol II for this substrate (38).
By a number of criteria, the defects seen in these yeast strains appear fairly general, including reduced levels of total poly(A)+ mRNA, reduced levels of a number of individual mRNAs, and the failure of transcriptional induction in at least three systems. All of these general defects are exacerbated after 6AU treatment (5, 12). The in vitro data made the relative inactivity of arrest sites in vivo somewhat surprising. Nevertheless, the in vitro assay may exaggerate the strength of the arrest site resulting in a more subtle effect in vivo. It is also possible that a compensating activity present in yeast but not in the in vitro reaction allows for efficient elongation through arrest sites. Close homologues of SII are not apparent in the yeast genome and no eukaryotic proteins other than SII are known that activate the nascent RNA cleavage reaction employed for arrest site read through. The most closely related gene products are Rpa12, Rpb9, and Rpc11, subunits of RNA polymerases I, II, and III, respectively. Rpb9 and Rpc11 have been implicated in transcription elongation (7, 33, 39). pol II, lacking the Rpb9 subunit, is relatively read through-proficient (33). Perhaps the conditional dissociation of this subunit from elongation complexes can compensate for the absence of SII in vivo. As well, in the absence of SII, pol II possesses low but detectable amounts of nascent RNA cleavage activity, which may suffice to support read through in vivo. Alternatively, other proteins known to increase the average elongation rate such as TFIIF, Elongator, or elongin may minimize or pre-empt arrest by helping pol II sustain high elongation rates in vivo (9, 17, 40, 41). In vitro transcription of arrest sites by yeast pol II has generally employed promoter-less “tailed” templates. Activated transcription that takes place from the GAL1 promoter in vivo may result in a different, more processive form of pol II that may be immune from arrest in vivo, possibly through modification of pol II at the promoter (42). A synthetic lethal screen has recently revealed activities that may compensate for the absence of SII in vivo and has drawn a connection between SII and chromatin structure (43). Further work will be required to test these possibilities.
Acknowledgments
We thank Dr. Cale Lennon for generating some of the strains and clones used in this study and Drs. Jerry Boss, Sue Jinks-Robertson, Paul Doetsch, Charlie Moran, and Frank Gordon for helpful discussions.
Footnotes
This work was supported by Grant GM46331 from the National Institutes of Health.
The abbreviations used are: pol II, polymerase II; 6AU, 6-azauracil; PCR, polymerase chain reaction.
REFERENCES
- 1.Uptain SM, Kane CM, Chamberlin MJ. Annu. Rev. Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 2.Wind M, Reines D. BioEssays. 2000;22:327–336. doi: 10.1002/(SICI)1521-1878(200004)22:4<327::AID-BIES3>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archambault J, Lacroute F, Ruet A, Friesen JD. Mol. Cell. Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powell W, Reines D. J. Biol. Chem. 1996;271:6866–6873. doi: 10.1074/jbc.271.12.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lennon JC, III, Wind M, Saunders L, Hock MB, Reines D. Mol. Cell. Biol. 1998;18:5771–5779. doi: 10.1128/mcb.18.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishiguro A, Nogi Y, Hisatake K, Muramatsu M, Ishihama A. Mol. Cell. Biol. 2000;20:1263–1270. doi: 10.1128/mcb.20.4.1263-1270.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hemming SA, Jansma DB, Macgregor PF, Goryachev A, Friesen JD, Edwards AM. J. Biol. Chem. 2000;275:35506–355011. doi: 10.1074/jbc.M004721200. [DOI] [PubMed] [Google Scholar]
- 8.Exinger F, Lacroute F. Curr. Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- 9.Otero G, Fellows J, Li Y, deBizemont T, Dirac AMG, Gustafsson CM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Mol. Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 10.Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis CD, Tempst P, Svejstrup JQ. Mol. Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- 11.Hartzog GA, Wada T, Handa H, Winston F. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw RJ, Reines D. Mol. Cell. Biol. 2000;20:7427–7437. doi: 10.1128/mcb.20.20.7427-7437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa PJ, Arndt KM. Genetics. 2000;156:535–547. doi: 10.1093/genetics/156.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiest DK, Hawley DK. Mol. Cell. Biol. 1990;10:5782–5792. doi: 10.1128/mcb.10.11.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiest DK, Wang D, Hawley DK. J. Biol. Chem. 1992;267:7733–7744. [PubMed] [Google Scholar]
- 16.Izban MG, Luse DS. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- 17.Gu W, Reines D. J. Biol. Chem. 1995;270:11238–11244. doi: 10.1074/jbc.270.19.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loison G, Losson R, Lacroute F. Curr. Genet. 1980;2:39–44. doi: 10.1007/BF00445692. [DOI] [PubMed] [Google Scholar]
- 19.Padilla PA, Fuge EK, Crawford ME, Errett A, Werner-Washburne M. J. Bacteriol. 1998;180:5718–5726. doi: 10.1128/jb.180.21.5718-5726.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakanishi T, Nakano A, Nomura K, Sekimizu K, Natori S. J. Biol. Chem. 1992;267:13200–13204. [PubMed] [Google Scholar]
- 21.Scafe C, Nonet M, Young R. Mol. Cell. Biol. 1990;10:1010–1016. doi: 10.1128/mcb.10.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimoaraiso M, Nakanishi T, Kubo T, Natori S. J. Biol. Chem. 2000;275:29623–29627. doi: 10.1074/jbc.M910371199. [DOI] [PubMed] [Google Scholar]
- 23.Gietz D, St. Jean A, Woods RA, Schiestl RH. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherman F. Methods Enzymol. 1991;194:3–20. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 25.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York, NY: Appendix 2, Greene Publishing Associates/Wiley-Interscience; 1988. [Google Scholar]
- 26.Haro R, Garciadeblas B, Rodriguez-Navarro A. FEBS Lett. 1991;291:189–191. doi: 10.1016/0014-5793(91)81280-l. [DOI] [PubMed] [Google Scholar]
- 27.Johnston M, Flick JS, Pexton T. Mol. Cell. Biol. 1994;14:3834–3841. doi: 10.1128/mcb.14.6.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuras L, Struhl K. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 29.Li XY, Virbasius A, Zhu X, Green MR. Nature. 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 30.Reines D, Wells D, Chamberlin MJ, Kane CM. J. Mol. Biol. 1987;196:299–312. doi: 10.1016/0022-2836(87)90691-7. [DOI] [PubMed] [Google Scholar]
- 31.Christie KR, Awrey DE, Edwards AM, Kane CM. J. Biol. Chem. 1994;269:936–943. [PubMed] [Google Scholar]
- 32.Reeder TC, Hawley DK. Cell. 1996;87:767–777. doi: 10.1016/s0092-8674(00)81395-1. [DOI] [PubMed] [Google Scholar]
- 33.Awrey DE, Weilbaecher RG, Hemming SA, Orlicky SM, Kane CM, Edwards AM. J. Biol. Chem. 1997;272:14747–14754. doi: 10.1074/jbc.272.23.14747. [DOI] [PubMed] [Google Scholar]
- 34.Mote J, Jr, Reines D. J. Biol. Chem. 1998;27:16843–16852. doi: 10.1074/jbc.273.27.16843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samkurashvili I, Luse DS. J. Biol. Chem. 1996;271:23495–23505. doi: 10.1074/jbc.271.38.23495. [DOI] [PubMed] [Google Scholar]
- 36.Reines D, Ghanouni P, Gu W, Mote J, Jr, Powell W. Cell. Mol. Biol. Res. 1993;39:331–338. [PubMed] [Google Scholar]
- 37.Ditzelmuller G, Wohrer W, Kubicek CP, Rohr M. Arch. Microbiol. 1983;135:63–67. doi: 10.1007/BF00419484. [DOI] [PubMed] [Google Scholar]
- 38.Kadesch TR, Chamberlin MJ. J. Biol. Chem. 1982;257:5286–5295. [PubMed] [Google Scholar]
- 39.Chedin S, Riva M, Schultz P, Sentenac A, Carles C. Genes Dev. 1998;12:3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bengal E, Flores O, Krauskopf A, Reinberg D, Aloni Y. Mol. Cell. Biol. 1991;11:1195–1206. doi: 10.1128/mcb.11.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koth CM, Botuyan MV, Moreland RJ, Jansma DB, Conaway JW, Conaway RC, Chazin WJ, Friesen JD, Arrowsmith CH, Edwards AM. J. Biol. Chem. 2000;275:11174–11180. doi: 10.1074/jbc.275.15.11174. [DOI] [PubMed] [Google Scholar]
- 42.Akhtar A, Faye G, Bentley DL. EMBO J. 1996;15:4654–4664. [PMC free article] [PubMed] [Google Scholar]
- 43.Davie JK, Kane CM. Mol. Cell. Biol. 2000;20:5960–5973. doi: 10.1128/mcb.20.16.5960-5973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]