Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease affecting motor neurons. Mutations in related RNA-binding proteins TDP-43, FUS/TLS and TAF15 have been connected to ALS. These three proteins share several features, including the presence of a bioinformatics-predicted prion domain, aggregation–prone nature in vitro and in vivo and toxic effects when expressed in multiple model systems. Given these commonalities, we hypothesized that a related protein, EWSR1 (Ewing sarcoma breakpoint region 1), might also exhibit similar properties and therefore could contribute to disease. Here, we report an analysis of EWSR1 in multiple functional assays, including mutational screening in ALS patients and controls. We identified three missense variants in EWSR1 in ALS patients, which were absent in a large number of healthy control individuals. We show that disease-specific variants affect EWSR1 localization in motor neurons. We also provide multiple independent lines of in vitro and in vivo evidence that EWSR1 has similar properties as TDP-43, FUS and TAF15, including aggregation–prone behavior in vitro and ability to confer neurodegeneration in Drosophila. Postmortem analysis of sporadic ALS cases also revealed cytoplasmic mislocalization of EWSR1. Together, our studies highlight a potential role for EWSR1 in ALS, provide a collection of functional assays to be used to assess roles of additional RNA-binding proteins in disease and support an emerging concept that a class of aggregation–prone RNA-binding proteins might contribute broadly to ALS and related neurodegenerative diseases.
INTRODUCTION
Amyotrophic lateral sclerosis (ALS, also known as Lou Gehrig's disease or Charcot's disease) is a devastating adult-onset neurodegenerative disease that attacks upper and lower motor neurons (1). A progressive and fatal muscle paralysis usually causes death within 2–5 years of disease onset. ALS is mostly a sporadic disease, but ∼10% of ALS cases are inherited. Because sporadic and inherited forms of ALS are clinically indistinguishable, it is hoped that elucidating the genetic contributors to inherited ALS will also provide insight into the more common sporadic forms of the disease. Pathogenic mutations in several genes have been linked to ALS, including SOD1, TARDBP, FUS/TLS, VAPB, OPTN, VCP, UBQLN2 and C9ORF72 (2–8).
Two of these genes, TARDBP (which encodes TDP-43) and FUS/TLS (FUS), are notable because they code for related RNA-binding proteins (9,10). Moreover, both of these proteins have been identified as components of pathological aggregates in neurons of ALS patients (11–13). An emerging concept suggested by the association of FUS and TDP-43 with ALS is that defects in RNA metabolism might contribute to disease pathogenesis (14,15). Are TDP-43 and FUS lone RNA-binding proteins in ALS or could other proteins with properties like those of TDP-43 and FUS also contribute to disease pathogenesis in similar ways?
To explore this possibility, we recently performed an unbiased functional screen in yeast to identify additional TDP-43- and FUS-like genes. FUS and TDP-43 both aggregate in the cytoplasm and confer cytotoxicity when expressed in yeast (16–22). We surveyed 133 additional human RNA-binding proteins in yeast and identified several others that also aggregated in the cytoplasm and were toxic (23). We further refined this list by identifying a prion-like domain in a subset of these proteins (10,24) and then proceeded to sequence one of these, TAF15, in ALS patients and controls. We identified several missense variants in ALS patients that were absent in over 1500 healthy controls and provided several lines of evidence that these variants are potentially damaging by testing their effects in vitro, in primary motor neurons, and in Drosophila (23). An independent study by Ticozzi et al. (25) also identified TAF15 variants in ALS patients and a role for TAF15 in a related neurodegenerative disorder, frontotemporal lobar degeneration, has been recently demonstrated (26). Together, these findings highlight a potential role for TAF15 in ALS and underscore the utility of the simple yeast model system for predicting new candidate ALS disease genes for further evaluation.
Here, we evaluate the role of EWSR1 (Ewing sarcoma breakpoint region 1) in ALS. We identified EWSR1 as a candidate RNA-binding protein in our yeast functional screen (23) and it is very closely related to TAF15 and FUS. Indeed, all three proteins belong to the same protein family, the FET family (for FUS, EWSR1, TAF15). We analyzed the properties of EWSR1 using diverse approaches, encompassing: in vitro aggregation assays, ability to confer neurodegeneration in Drosophila, sequence analysis of the EWSR1 gene in ALS patients and controls and EWSR1 localization in transfected primary neurons as well as postmortem tissue. These analyses enhance our understanding of the role of FET proteins in ALS and provide a battery of in vitro and in vivo functional assays that can be deployed to test additional ALS candidate genes.
RESULTS
Yeast functional screen identifies additional TDP-43 and FUS-like human RNA-binding proteins
In our initial screen for human RNA-binding proteins with properties similar to TDP-43 and FUS (cytoplasmic aggregation and toxicity and a predicted prion-like domain), we examined 135 out of 213 human RRM-containing proteins (23). We have recently expanded this analysis to include 40 additional RRM genes. Of these 40, 15 aggregate in the cytoplasm and are toxic. Of these, four (CELF4, HNRNPH2, HNRNPH3, TIAL1) also contained a predicted prion-like domain (Supplementary Material, Table S1). Future studies will be required to examine the remaining human RRM genes and this list should also expand to include non-RRM domain-containing RNA-binding proteins, such as KH domain proteins. These data expand the list of human RNA-binding proteins with properties similar to TDP-43 and FUS, providing a resource for further evaluation of some of these proteins in ALS and related neurodegenerative diseases.
Yeast expression of EWSR1 reveals similar properties to TDP-43 and FUS
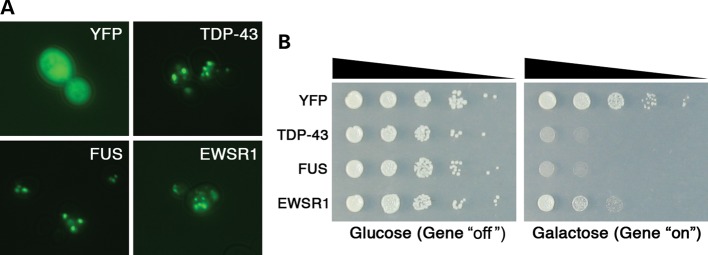
Similar to TDP-43, FUS and TAF15, expression of EWSR1 in yeast resulted in cytoplasmic aggregation and toxicity and was identified as a hit in our original yeast functional screen (23). We expressed YFP alone, or YFP-tagged TDP-43, FUS, and EWSR1 in yeast cells from a high copy 2 µ plasmid under the control of a strong galactose-inducible promoter (Fig. 1). Expressing YFP alone resulted in diffuse localization throughout the cytoplasm and nucleus (Fig. 1A), whereas YFP-tagged TDP-43, FUS or EWSR1 formed multiple cytoplasmic foci (Fig. 1A). EWSR1 was also toxic when expressed in yeast, albeit not as toxic as FUS and TDP-43 (Fig. 1B). Thus, EWSR1 shares key features with ALS proteins TDP-43 and FUS (aggregation–prone and toxic).
Figure 1.
When expressed in yeast, the human RRM RNA-binding protein EWSR1 shows similar properties to FUS and TDP-43. (A) Localization patterns in yeast cells of human TDP-43–YFP, FUS–YFP and EWSR1–YFP fusion proteins, forming multiple cytoplasmic foci. When YFP alone is expressed, it is localized diffusely throughout the cytoplasm. (B) Spotting assays to assess the toxicity of human EWSR1–YFP protein. Transformants were grown on the synthetic media containing either glucose (control, protein expression ‘off’) or galactose (protein expression ‘on’). TDP-43–YFP and FUS–YFP proteins overexpression were very toxic, while EWSR1–YFP was moderately toxic and YFP alone is not toxic (control).
EWSR1 shares similar domain architecture to TDP-43, FUS and TAF15 and harbors a prion-like domain
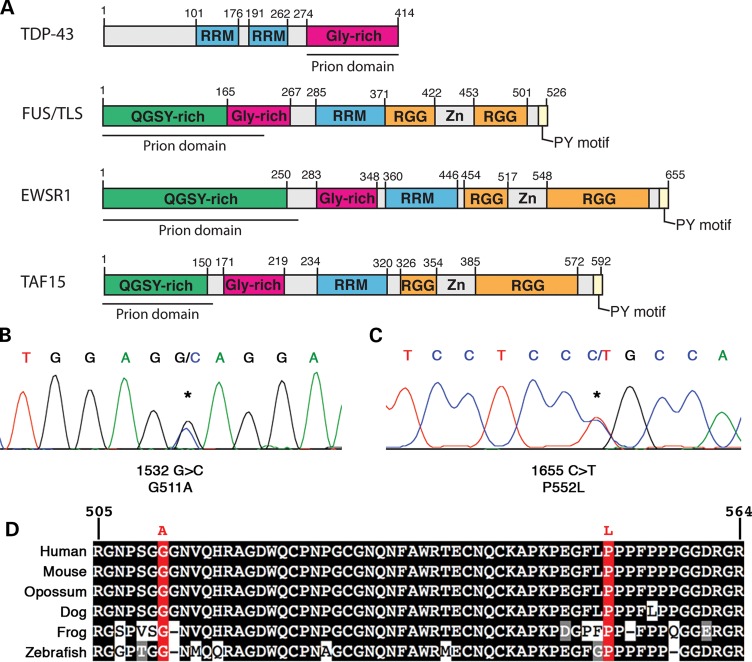
Although the biological functions of EWSR1, TAF15 and FUS are not completely understood and there may be key differences between each protein, EWSR1 contains notably similar domain architecture to TDP-43, and especially to FUS and TAF15. Like FUS and TAF15, EWSR1 contains an RRM, a glycine-rich domain, and a RGG- and PY-motif-containing C-terminal domain, which is important for nuclear localization (27–29). Furthermore, such as TDP-43, FUS and TAF15, EWSR1 harbors a bioinformatics-predicted prion-like domain (amino acids 1–280; Fig. 2A). The prion-like domain is critical for aggregation of TDP-43 and FUS (10,16,17,19,24). Remarkably, out of all 213 human RRM proteins, FUS, TAF15 and TDP-43, respectively, ranked 1st, 2nd and 10th for the prion score [Supplementary Material, Table S1 and (23)]. Using the same prion-domain prediction algorithm, EWSR1's prion score ranked 3rd out of 213 RRM proteins and 25th out of 21 873 human proteins. Moreover, the putative prion domain of EWSR1 also satisfies the criteria of an alternative algorithm for predicted prion formation based on ‘prion propensities’ (derived from random mutagenesis experiments in yeast) (30); namely, they contain a region with predicted disorder and with ‘prion propensity’ >0.05. The core EWSR1 prion domain detected by this algorithm (amino acids 209–249) scores 0.057 [using the collapse–consecutive–prolines option; (30)]. Given these striking commonalities in domain architecture, predicted prion domain and propensity to form cytoplasmic inclusions in yeast, we next sought to analyze the EWSR1 gene for mutations in ALS patients.
Figure 2.
Missense mutations in EWSR1 in ALS patients. (A) Comparison of FUS, EWSR1 and TAF15 demonstrates similar domain architecture. All three proteins contain a single RRM, a glycine-rich domain, a predicted prion domain, RGG domains and a C-terminal PY-motif, which can function as an NLS. Mutations in FUS and TAF15 are examples of those similar to variants found in EWSR1. (B and C) DNA sequence analysis of EWSR1 in North American Caucasian ALS patients identified two missense mutations (shown are electropherograms highlighting the sequence variants). (B) A single base substitution (asterisk) changing the WT guanine at 1532 to cytosine (c.1532 G>C) predicted to lead to an alanine substituting for glycine (p.G511A). (C) Another EWSR1 variant in an ALS case: c.1655 C>T, predicted to lead to a leucine substituted for proline (p.P552L). (D) Sequence alignment of amino acids 505–564 of EWSR1 from diverse vertebrate species indicates that the mutated residues in EWSR1 are highly conserved. Identical amino acids have a black background, similar amino acids are gray and mutation sites are red.
Identification of EWSR1 variants in ALS patients
Since almost all known pathogenic mutations in FUS and TDP-43 are located in the C-terminal domains of the proteins (9), we focused on the last four exons of the EWSR1 gene (exons 15–18; NM_001163285). These exons comprise the RGG- and PY-motif-containing C-terminal domain, which are important for nuclear localization of FUS and EWSR1 (27,28,31,32). Complete sequencing of these exons was performed in 817 individuals diagnosed with ALS and in 1082 geographically matched healthy population control individuals (see Materials and Methods for patient and control demographic information). We followed up this analysis with Taqman single nucleotide polymorphism (SNP) genotyping of any patient-specific mutations in 4608 healthy individuals (Supplementary Material, Table S2). This approach identified two patient-specific missense variants in EWSR1 in two unrelated ALS patients with sporadic disease (Fig. 2B and C, Supplementary Material, Table S2). Missense variants were identified in exon 16 (c.1532G>C, p.G511A) in the first RGG domain and in exon 17 (c.1655C>T, p.P552L) in the second RGG domain (Fig. 2A). These individuals had disease onset of 50 years and 36 years, respectively. Neither of these variants were present in 1082 sequenced controls nor in the 4608-targeted SNP-genotyped controls, strongly supporting clinical significance of these variants. Overall, these specific genetic variants in EWSR1 were detected in 2 out of 817 ALS cases and 0 out of 5690 controls (P = 0.015). Furthermore, none of these variants was present in public SNP databases (e.g. dbSNP), the eight HapMAP individuals sequenced (33), or the 1000 Genomes Project (http://browser.1000genomes.org/index.html). Notably, the two variants are located in highly conserved regions of the first and second RGG domains of EWSR1 (Fig. 2E). Since the EWSR1 variants were identified in sporadic ALS cases, familial evidence for segregation with disease was not possible; however, TARDBP and FUS mutations have also been confirmed in apparent sporadic ALS cases (34). In addition, the parents of the affected individuals were not available to determine whether the mutations occurred de novo or were inherited.
In the process of sequencing these genes in ALS cases and controls, we also identified several synonomous and non-coding variants as summarized in Supplementary Material, Table S2. In addition, we identified one missense variant that was present in both patients and controls (EWSR1 c. 1750G>A, p.G584S) as well as one variant present only in a single control (EWSR1 c.1903C>T, p.R635C). Thus, taken together, we identified two missense variants in EWSR1 that were not present in ALS cases but not in a very large number of healthy controls. However, the presence of some EWSR1 variants in control individuals indicates that additional functional studies are required to evaluate the effects of each variant on EWSR1, in order to assess its potential for pathogenicity.
Functional studies to evaluate the effects of EWSR1 sequence variants
To provide evidence that the EWSR1 variants found in ALS cases might be pathogenic, we next asked if and how they affected the protein. We reasoned that some EWSR1 variants would likely be benign (e.g. polymorphism), whereas others would be deleterious (e.g. pathogenic mutation). We therefore designed an unbiased functional assay to discriminate potentially deleterious variants from benign variants. We had previously found that some ALS-linked TDP-43 mutations increase aggregation and toxicity in vitro and in yeast cells (17) and enhance neurodegeneration in Drosophila (18). On the other hand, in recent experiments with FUS, we found that ALS-linked FUS mutants do not aggregate more rapidly than the wild-type (WT) in vitro and in yeast, and are not more toxic than WT in yeast (19). As with FUS (19) and TAF15 (23), we found that the ALS-linked variants in EWSR1 did not increase aggregation or toxicity in yeast (data not shown). However, ALS-linked mutations in TDP-43 and FUS have been shown to disrupt protein localization, leading to enhanced cytoplasmic accumulation of ALS-linked variants (11,12,28,35–37). Given this common feature, we assessed the effects of the EWSR1 variants on subcellular localization. We tested WT EWSR1 as well as two ALS-specific variants (G511A and P552L), one variant found in both ALS and controls (G584S) and one variant found only in controls (R635C).
We transfected WT and mutant EWSR1 into primary motor neurons cultured from rat embryos. WT EWSR1 primarily localized to the nucleus, whereas both ALS-specific variants resulted in increased cytoplasmic and neuritic accumulation (Fig. 3A and B). We also tested EWSR1 variant G584S, which was found in both ALS patients and controls (1/817 ALS patients and 1/5690 controls) and R635C, which was found in controls (1/1082 controls). Importantly, in contrast to the patient-specific variants, G584S and R635C did not enhance cytoplasmic or neuritic accumulation in this assay and behaved similar to the WT (Fig. 3A and B), providing additional evidence that these variants are not likely to be pathogenic. Additionally, we confirmed that the ALS case with the G584S EWSR1 variant also harbored the recently reported C9ORF72 hexanucleotide expansion (3,8), further evidence that this variant is probably not pathogenic, consistent with the results from our functional assays (Fig. 3).
Figure 3.
ALS-linked EWSR1 mutations promote cytoplasmic localization in motor neurons with mislocalization to the neurites of primary neurons cultured from mouse spinal cord. Primary mouse neuron cultures were transfected with WT or mutant EWSR1, stained with a-EWSR1 (red) and a-doublecortin (DCX, green). G511A and P552L variants were only found in ALS patients, while R635C was only found in controls and G584S was found in both (and this ALS case also harbored a C9ORF72 hexanucleotide expansion). (A) Endogenous EWSR1 is almost exclusively localized within the nucleus of neurons. Overexpression of WT EWSR1, or variants found in controls, shows localization within the nucleus or cytoplasm of neurons, with rare neurites containing EWSR1. Only the ALS-linked mutant forms of EWSR1 showed increased mislocalization into the neurites, including dendrites and axons. Scale bar is 10 µm. (B) Quantitation of mislocalization of endogenous transfected, WT or mutant, EWSR1 into neuronal processes. *P = 0.0056 for comparing G511A to WT, and P = 0.0008 comparing P552L to WT (two-tailed unequal variance t-test). (C) ES cell-derived neurons were transduced with doxycycline (Dox) inducible lentiviruses expressing WT or ALS-linked mutants of EWSR1, each carrying FLAG and myc epitope tags in their amino- and carboxy-termini, respectively. Five days after induction of expression by Dox, the localization of the proteins was visualized by immunofluorescence microscopy with anti-FLAG antibody (red); nuclei were visualized by 4′,6-diamidino-2-phenylindole (DAPI) staining (blue). The localization of endogenous EWSR1 was performed with anti-EWSR1-specific antibodies (red) in non-transduced neurons. Induced expression of EWSR1 led to accumulation of proteins in the cytoplasm and neuronal processes of transduced cells and this effect was enhanced by the ALS-linked patient mutations. Expression levels and solubility of transduced proteins were determined by immunoblots with anti-FLAG antibodies of radioimmuno precipitation assay buffer (RIPA) and UREA fractions of cell lysates from EWSR1 WT and mutants 5 days after Dox induction. The expression levels of transduced proteins were comparable between WT and mutants with no apparent accumulation of proteins in the UREA fraction.
We observed similar effects on EWSR1 localization when we expressed these proteins in cell cultures of embryonic stem (ES) cell-derived neurons and primary motor neurons isolated from mouse embryos. Transduction of WT EWSR1 in the ES-derived neuronal cultures resulted in a mostly nuclear pattern, with occasional localization to the cytoplasm (Fig. 3C). Strikingly, the two patient-specific EWSR1 variants analyzed (G511A and P552L) resulted in a significant increase in cytoplasmic accumulation, as well as a pattern of coarse neuritic staining that was less prevalent with the WT proteins (Fig. 3C). Immunoblotting confirmed that the transduced WT and variant proteins were expressed at similar levels (Fig. 3C). Thus, in two independent assays in disease-relevant cell types, ALS-specific EWSR1 variants promoted cytoplasmic mislocalization, indicating that these variants are potentially deleterious and could be involved in pathogenesis.
As additional variants in EWSR1 (and related genes) are identified, these functional assays will be a powerful tool for assessing their potential pathogenicity. Thus, like ALS-linked TDP-43 and FUS mutations, the ALS-linked variants of EWSR1 can also promote cytoplasmic accumulation of the protein in motor neurons, a disease-relevant cell type, providing further evidence in support of the pathogenicity of these variants. Interestingly, in addition to the PY-motif, which can function as a NLS (32), sequences in the last RGG domain of EWSR1 have also been shown to be required for proper nuclear localization (31). Notably, one of the two EWSR1 variants we identified is located in this domain (Fig. 2A), suggesting that perhaps these mutations perturb the function of this RGG domain in a way that decreases its ability to interact efficiently with the nuclear localization machinery, resulting in the enhanced cytoplasmic localization observed in the cell cultures (Fig. 3). While the other EWSR1 variant, G511A, is located in a RGG domain that has been suggested to not be required for nuclear localization (31), in other contexts this domain has been shown to contribute to nuclear localization (27). Moreover, the equivalent domain plays a key role in FUS misfolding and aggregation (19). Thus, mutations in this region could exacerbate an intrinsic propensity to misfold (see below). Future work will be required to define the specific requirements of RGG domains in EWSR1 for nuclear localization and to evaluate the effects of disease-linked mutations.
The preceding genetic and functional studies highlight a potential role for EWSR1 in ALS pathogenesis. We next sought additional functional evidence that EWSR1 has properties similar to TDP-43, FUS and TAF15. To evaluate EWSR1 further, we proposed several criteria. First, does EWSR1 spontaneously aggregate in vitro as for TDP-43, FUS and TAF15 (17,19,23)? Secondly, does EWSR1 confer neurodegeneration when expressed in the nervous system, as for TDP-43, FUS and TAF15 (18,38–45)? Thirdly, can EWSR1 mislocalize in ALS patient neurons, as does TDP-43 (13), FUS (11,12) and TAF15 (23)?
EWSR1 is intrinsically aggregation prone
We purified bacterially expressed glutathione S-transferase (GST)-tagged EWSR1 as soluble protein under native conditions, as previously done for TDP-43, FUS and TAF15 (17,19,23). Upon addition of Tobacco Etch Virus (TEV) protease to specifically remove the N-terminal GST tag (Fig. 4A), EWSR1 rapidly aggregated at 25°C with gentle agitation (Fig. 4B). If TEV protease was omitted, then little aggregation occurred (Fig. 4B). EWSR1 aggregated with kinetics similar to FUS and slightly more rapidly than TDP-43, as determined by turbidity (Fig. 4B) and by the amount that entered the pellet fraction after centrifugation (Fig. 4C). Thus, the relative aggregation kinetics of FUS, EWSR1 and TDP-43 were foreshadowed by the prion-domain algorithm (24,46), which ranks FUS above EWSR1 and EWSR1 above TDP-43.
Figure 4.
EWSR1 is intrinsically aggregation–prone and ALS-linked variants accelerate aggregation. (A) Following TEV protease cleavage to remove the N-terminal GST tag, FUS, EWSR1 and TDP-43 proteins were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and Coomassie stained to confirm purity and expected molecular weight. (B) GST-TDP-43, GST-FUS or GST-EWSR1 (3 µm) were incubated in the absence or presence of TEV protease at 25°C for 0–90 min with agitation. At the indicated times, the extent of aggregation was determined by measuring turbidity (absorbance at 395 nm). Note that very little aggregation occurs in the absence of TEV protease. Values represent means ± SEM (n = 3). A human RRM protein that lacks a prion-like domain, DND1, did not aggregate in this assay. (C) GST-TDP-43, GST-FUS or GST-EWSR1 (3 µm) was incubated in the presence of TEV protease at 25°C for 0–60 min with agitation. At the indicated times, reactions were processed for sedimentation analysis. Pellet and supernatant fractions were resolved by SDS–PAGE and stained with Coomassie Brilliant Blue. The amount of protein in the pellet fraction was determined by densitometry in comparison to known quantities of the appropriate protein. Values represent means ± SEM (n = 3). A human RRM protein that lacks a prion-like domain, DND1, did not aggregate in this assay. (D) GST-EWSR1 (3 µm) was incubated in the presence of TEV protease at 25°C for 0–60 min with agitation. At various times, reactions were processed for EM. Small arrows denote pore-shaped oligomers and large arrows denote linear polymers. Bar, 0.5 µm. (E) Gallery of EWSR1 oligomers formed after 10 min of aggregation. Bar, 50 nm. (F) Following TEV protease cleavage to remove the N-terminal GST tag, EWSR1 WT, G511A and P552L proteins were resolved by SDS–PAGE and Coomassie stained to confirm purity and expected molecular weight. (G) GST-EWSR1, GST-EWSR1 (G511A) or GST-EWSR1 (P552L) (3 µm) was incubated in the presence of TEV protease at 25°C for 0–75 min without agitation. At the indicated times, the extent of aggregation was determined by measuring turbidity (absorbance at 395 nm). Note that the ALS-linked EWSR1 variants, G511A and P552L, aggregated with accelerated kinetics. Values represent means ± SEM (n = 3).
Electron microscopy (EM) revealed that EWSR1 rapidly accessed oligomeric forms (Fig. 4D), which would frequently adopt a pore-like conformation (Fig. 4D, small arrows; Fig. 4E), similar to those formed by TDP-43, FUS and TAF15 (17,19,23). Furthermore, EWSR1 also assembled into linear polymers with a cross-sectional diameter of ∼15–20 nm (Fig. 4D, large arrows) that increased in length over time and would often become tangled into large masses by 60 min (Fig. 4D). In general, the morphology of EWSR1 aggregates was more similar to FUS and TAF15 than to TDP-43, which over this time frame formed shorter polymers that would clump together to form large masses (17).
We also purified bacterially expressed GST-tagged EWSR1 ALS-associated variants: G511A and P552L, as soluble protein under native conditions, as previously done for the WT protein. The N-terminal GST tag was specifically removed by TEV treatment (Fig. 4F). To distinguish subtle differences in aggregation kinetics, we tracked EWSR1 misfolding in reactions without agitation at 25°C. The ALS-associated EWSR1 variants aggregated more rapidly than the WT protein, as determined by turbidity (Fig. 4G). Thus, similar to TDP-43, FUS and TAF15, and concordant with the yeast data, EWSR1 is inherently aggregation–prone and the patient-specific EWSR1 variants, G511A and P552L, exhibit enhanced aggregation propensity.
EWSR1 causes neurodegeneration when expressed in Drosophila
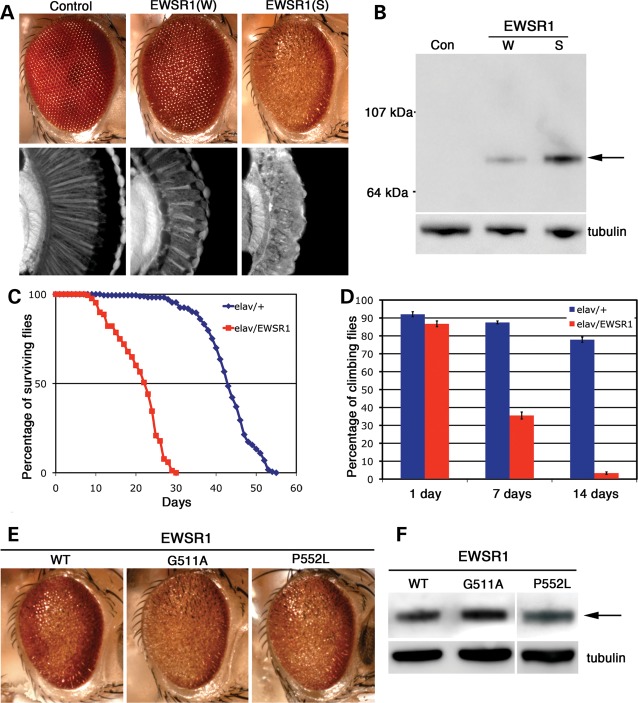
To analyze the effects of EWSR1 in the nervous system, we used Drosophila. We and others have previously shown that directing TDP-43, FUS or TAF15 expression to the fly nervous system causes neurodegeneration (18,23,38–45). We generated a series of transgenic lines that expressed human WT and mutant EWSR1. Directing expression of EWSR1 to the eye of the fly caused degeneration of the structure, with higher levels of expression resulting in a stronger effect (Fig. 5A and B). Directing EWSR1 expression throughout the nervous system led to a shortened lifespan (Fig. 5C) and progressive loss of motility (Fig. 5D). Expressing the ALS-associated variants (G511A and P552L) had similar effects as WT on the fly eye (Fig. 5E and F) and lifespan (data not shown). That the EWSR1 mutants did not confer a stronger phenotype than WT EWSR1 could argue against pathogenicity. Alternatively, it could represent a limitation of the model system. Importantly, EWSR1 possesses activity sufficient to confer neurodegeneration in the nervous system, in a manner similar to that of TDP-43, FUS and TAF15. Furthermore, these experiments do not indicate per se that EWSR1 causes disease. Rather, they provide additional evidence that EWSR1 has similar properties as the known ALS disease-causing RNA-binding proteins TDP-43 and FUS.
Figure 5.
WT and disease-related mutants of EWSR1 lead to neural degeneration and dysfunction in Drosophila. (A) Expression of EWSR1 caused a dose-dependent disruption of retinal structure. (W) and (S) refer to weak and strong EWSR1 expression levels. (B) Western immunoblot showing the level of expression of EWSR1. β-Tubulin levels were used as the loading control. (A and B) Genotypes: control (Con) is driver line alone, gmr-GAL4(YH3)/+. EWSR1(W) is UAS-EWSR1(W)/gmr-GAL4(YH3). EWSR1(S) is UAS-EWSR1(S)/+; gmr-GAL4(YH3)/+. (C) Expression of EWSR1 in the nervous system reduces lifespan (red, compared with normal in blue). (D) EWSR1 caused progressive loss of climbing behavior when expressed in the nervous system. (C and D) Genotypes: elav is elav3A-GAL4/+. elav/EWSR1 is UAS-EWSR1(S)/+; elav3A-GAL4/+. (E) External eyes from 1 day adult flies expressing WT and mutant EWSR1.G511A under control of the gmr-GAL4 driver. (F) Immunoblot of head homogenates showing EWSR1 WT and mutant expression levels in transgenic flies. β-Tubulin served as loading control. (E and F) Genotypes: WT is UAS-EWSR1/+; gmr-GAL4(YH3)/+. G511A is UAS-EWSR1(G511A)/+; gmr-GAL4(YH3)/+. P552L is UAS-EWSR1(P552L)/gmr-GAL4(YH3).
EWSR1 mislocalization in ALS patient spinal cord
We next performed immunohistochemistry on human postmortem spinal cord tissue to determine whether EWSR1 was present in motor neurons and whether its localization was affected in ALS, as for TDP-43 and FUS. Interestingly, EWSR1 mislocalization was recently reported in some frontotemporal lobar degeneration cases [FTLD-FUS cases; (26)]. EWSR1 was robustly expressed in spinal cord motor neurons and localized to the nucleus, as for TDP-43, FUS and TAF15 (Fig. 6A and B). We examined EWSR1 localization in autopsy tissue from six sporadic ALS cases. Immunostaining of formalin-fixed spinal cord sections from ALS patients revealed a predominantly nuclear localization for EWSR1 within motor neurons. However, in the ALS patient sections, we examined there was also significant cytoplasmic staining in either a punctate granular (Fig. 6C–E) or diffuse (Fig. 6F–H) pattern. The strong punctate granular localization for EWSR1 was not seen in any of the control cases that we examined (n = 3). Thus, EWSR1 is expressed in a disease-relevant cell type and can be mislocalized to the cytoplasm in ALS, further supporting the notion that EWSR1 might contribute to the disease.
Figure 6.
Immunostaining to visualize localization of EWSR1 in spinal cord of control or ALS patients. (A and B), In control spinal cord neurons, EWSR1 is localized predominantly to the nucleus while in ALS spinal cord neurons, EWSR1 was present in cytoplasmic punctate granular structures (E, arrows) or in a diffuse pattern throughout the cytoplasm (C–H). Scale bar is 25 µm.
DISCUSSION
There has recently been tremendous progress in defining the genetic landscape of ALS (2). The promise of the genomics revolution is being realized as state-of-the-art sequencing technologies are being combined with tried and true genetics approaches to elucidate new disease genes (47). To add to these efforts, we recently designed a simple yeast functional screen to identify additional RNA-binding proteins with basic properties shared by the known ALS disease genes FUS and TDP-43, and to evaluate their potential role in ALS (23). We focused on one gene from this screen, TAF15, which possesses a prominent prion-like domain, and provided evidence for a connection to ALS. Independent studies by others also support a potential role of TAF15 in ALS (25) and FTLD (26).
Here, we focused on EWSR1, another top candidate from our yeast screen that harbors a prion-like domain (ranks 25th in the entire human genome, and 3rd among RRM-bearing prion candidates), and a member of the same protein family as FUS and TAF15. We identified three missense variants in EWSR1, in unrelated ALS patients and two missense variants in unrelated healthy controls. We provide evidence that the variants found in ALS cases alter EWSR1 localization in motor neurons, whereas the variants found in controls do not. We also provide in vitro and in vivo evidence that EWSR1 has properties similar to those of TDP-43, FUS and TAF15 and can confer neurodegeneration in Drosophila. These genetic and functional data are correlative and further studies will be required to directly test if the variants identified in EWSR1 represent bona fide pathogenic disease mutations for ALS. The functional assays described here will empower the evaluation of additional EWSR1 variants for potential pathogenicity.
It is important to note that our studies alone do not prove that EWSR1 is an ALS disease gene. Instead, they contribute to our understanding of the properties of this FUS-related protein and provide the basis for its further investigation in disease. If mutations in EWSR1 can cause ALS, based on our genetic analyses and those of Ticozzi et al. (25), they will likely be much rarer than FUS and TARDBP mutations. Therefore, the ALS genetic landscape might be comprised of relatively common genetic causes [e.g. C9ORF72, (3,8)] and other very rare causes. Moreover, there may even be extremely rare, perhaps even private mutations that are disease causing, a concept that is emerging for other human diseases (48). Our conceptual model predicts a delicate balance in RNA processing within motor neurons such that slight perturbations from any one of several different aggregation–prone RNA-binding proteins could cause disease. Multiple weak mutations in different RNA-binding proteins might also be required to produce the same effect as a single strong mutation in one RNA-binding protein [e.g. FUS P525L and R495X, which cause extremely aggressive ALS pathogenesis, with disease onset in the teenage years (28,37,49)]. We suspect that these studies may have merely scratched the surface and predict that many other aggregation–prone RRM or other RNA-binding proteins, such as EWSR1, TAF15, FUS and TDP-43, could also contribute to ALS and related neurodegenerative disorders (50). The yeast functional screen, prion-domain algorithm and in vitro and in vivo assays described here and previously (23,50) will hopefully contribute to efforts to unravel the genetic underpinnings of ALS and potentially other neurodegenerative diseases.
MATERIALS AND METHODS
Plasmids, yeast strains and media
The yeast strain used was BY4741 [genotype, Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0]. Strains were manipulated and media prepared using standard techniques (51). The EWSR1 expression construct was generated by Gateway® cloning (Invitrogen), starting with entry clones in pDONR221 and shuttled from entry clones into a modified pGW vector (for motor neuron transfection experiments), created by incorporating the Gateway B cassette into the SmaI site of the pGW vector using the Gateway conversion kit (Invitrogen). ALS patient mutations in EWSR1 were introduced by site-directed mutagenesis using the QuickChange Site Directed Mutagenesis kit (Stratagene). For yeast expression, EWSR1 was shuttled into the 2 μm galactose-inducible yeast expression plasmid pAG426Gal-ccdB-EYFP by Gateway® LR cloning reaction (46) to generate a C-terminally tagged EWSR1–YFP fusion. Restriction digest and DNA sequencing were used to confirm the integrity of each expression construct.
Yeast transformation and spotting assays
The PEG/lithium acetate method was used to transform yeast with each plasmid DNA from the RRM ORF library (52). For spotting assays, yeast cells were grown overnight at 30°C in liquid media containing raffinose (SRaf/-Ura) until log or mid-log phase. Cultures were then normalized for OD600, serially diluted and spotted onto synthetic solid media containing glucose or galactose lacking uracil, and were grown at 30°C for 2–3 days.
Microscopy
For fluorescence microscopy experiments, single colony isolates of the yeast strains were grown to mid-log phase in SRaf/-Ura media at 30°C. Cultures were spun down and resuspended in the same volume of SGal/-Ura to induce expression of the TDP-43 constructs. Cultures were induced with galactose for 4–6 h and processed for microscopy. Images were obtained using an Olympus IX70 inverted microscope and a Photometrics CoolSnap HQ 12-bit CCD camera.
Prion-prediction algorithm
Proteins were parsed into prion-like and non-prion-like regions using a hidden Markov model developed to identify regions that have the unusual amino acid composition characteristic of yeast prions (24,53). Prion-like regions of length ≥60 were given a prion-domain score, defined as the maximum log-likelihood for the prion-like state vs. the non-prion-like state over any 60 consecutive amino acids within the regions (53). Among the 21 873 human genes analyzed (Ensembl GrCh37.59), 246 had prion-like regions of length ≥60, and were ranked by the prion-domain score. For genes with multiple transcripts, the longest one was used, with the one with lowest Ensembl Transcript ID used in case of ties.
Sequencing the EWSR1 gene in ALS patients and controls
Genomic DNA from non-Latino Caucasian individuals with sporadic ALS (n = 552) was obtained from the Coriell Institute for Medical Research (Coriell, Camden, NJ, USA) distributed in 96-well plates NDPT025, NDPT026, NDPT030, NDPT100, NDPT103 and NDPT106. Additional genomic DNA samples from patients verified to meet El Escorial criteria for definite or probable ALS by a neurologist (n = 258) or with neuropathologic findings consistent with ALS (n = 88) were obtained from the University of Pennsylvania (PENN) Center for Neurodegenerative Disease Research. All subjects were collected with the PENN Institutional Review Board approval. The racial background of the PENN subjects was 90% non-Latino Caucasian, 5% black and 5% mixed or other. The PENN subjects were 57% male and had an average age of onset of 57 years (8–89) and an average duration of disease of 4 years (range 1–46). A family history of ALS (FALS) was present in 29 of 250 patients (11.6%) for which family history was available. Mutations in SOD1 and FUS/TLS were excluded in all the familial ALS cases and TARDBP mutations excluded in all PENN cases. All cases with potentially pathogenic variants in EWSR1 were also sequenced for TARDBP, FUS, SOD1 and C9ORF72.
Six hundred seventy-nine neurologically normal control samples from Coriell were distributed in 96-well plates NDPT084, NDPT090, NDPT093, NDPT094, NDPT095, NDPT096, NDPT098 and NDPT099. An additional 90 neurologically normal control samples were obtained from the Children's Hospital of Philadelphia (CHOP). One hundred seventy-nine DNA samples from cognitively normal individuals >60 years of age were obtained from the National Cell Repository for Alzheimer's Disease (NCRAD, Indianapolis, IN, USA).
We sequenced exons 15–18 of EWSR1 which encode the C-terminal domains of EWSR1. EWSR1 was successfully sequenced in 817 ALS cases (n = 514 Coriell and n = 303 PENN) and 1082 controls. Bidirectional sequencing was performed by separately amplifying EWSR1 exons 15–16, 17 and 18 from samples using the polymerase chain reaction (PCR). PCR primers and cycling conditions used for amplification and sequencing are available upon request. Amplicons were purified, processed and sequenced using Big-Dye® Terminator v3.1 sequencing (Applied Biosystems). All variants identified were confirmed by repeat sequencing. Sequence analysis was performed using Sequencher DNA Software.
SNP genotyping
DNA samples from 4811 de-identified healthy control subject of European ancestry who were recruited from the CHOP Health Care Networks (parents of children cared for at CHOP) were screened for mutations in the EWSR1 gene, using a custom-designed TaqMan SNP genotyping assay from Applied Biosystems:
| Marker name | Assay ID | SNP alleles | AA change | Gene name |
|---|---|---|---|---|
| 29694840 | AHLJH6F | G/C | G511A | EWSR1 |
| 29695301 | AHMSGCN | C/T | P552L | EWSR1 |
| 29695663 | AHN1EIV | G/A | G584S | EWSR1 |
A total of 10 ng of DNA was used as a template for the PCR reactions. Samples were run on the 7900HT analyzer from Applied Biosystems, after pooling three samples per run to expedite the screening process. Along with the pooled samples, each 384-well plate contained a positive and non-template control. Subsequent endpoint allelic discrimination was performed, using the SDSv2.4 software from Applied Biosystems. To ensure mutations were captured from the pooled approach, a test plate was run with mixtures of 1 heterozygous (het) positive and 1 homozygous (hom) negative sample; 1 het pos and 2 hom neg; and 1 het pos and 3 hom neg. The positive alleles were detected in all pools and a mixture of 3 unknowns was ultimately chosen for the study.
EWSR1 protein purification
EWSR1 was expressed and purified from E. coli as a GST-tagged protein. EWSR1 was cloned into GV13 to yield GST-TEV-EWSR1 and overexpressed in E. coli BL21 Star (Invitrogen). Protein was purified over a glutathione-sepharose column (GE) according to the manufacturer's instructions. GST-EWSR1 was eluted from the glutathione sepharose with 50 mm Tris–HCl, pH 7.4, 100 mm potassium acetate, 200 mm trehalose, 0.5 mm ethylenediaminetetraacetic acid (EDTA) and 20 mm glutathione. After purification, protein was concentrated to 10 µm or greater using Amicon Ultra-4 centrifugal filter units (10 kDa molecular weight cut-off; Millipore). Protein was then filtered through a 0.22 µm filter to remove any aggregated material. After filtration, the protein concentration was determined by the Bradford assay (Bio-Rad) and the proteins were used immediately for aggregation reactions.
EWSR1 in vitro aggregation assays
Filtered, purified GST-EWSR1 protein was used immediately for aggregation assays. Aggregation was initiated by the addition of TEV protease (Invitrogen) to EWSR1 (3 µm) in assembly buffer: 50 mm Tris–HCl, pH 7.4, 100 mm potassium acetate, 200 mm trehalose, 0.5 mm EDTA and 20 mm glutathione. Aggregation reactions were incubated at 25°C for 0–90 min with agitation at 700 rpm in an Eppendorf Thermomixer. No aggregation occurred unless TEV protease was added to separate GST from EWSR1. Turbidity was used to assess aggregation by measuring absorbance at 395 nm. For sedimentation analysis, reactions were centrifuged at 16 100g for 20 min at 25°C. Supernatant and pellet fractions were then resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and stained with Coomassie Brilliant Blue, and the amount in either fraction determined by densitometry in comparison to known quantities of EWSR1. For EM of in vitro aggregation reactions, protein samples (20 µl of a 3 µm solution) were adsorbed onto glow-discharged 300-mesh Formvar/carboncoated copper grid (Electron Microscopy Sciences) and stained with 2% (w/v) aqueous uranyl acetate. Excess liquid was removed, and grids were allowed to air dry. Samples were viewed using a JEOL 1010 transmission electron microscope.
Drosophila experiments
Transgenic flies expressing human EWSR1 were generated by standard techniques using the pUAST vector. To direct transgene expression to the eye, gmr-GAL4 driver was used. To direct expression to motor neurons, D42-GAL4 driver was used. Locomotor activity was assessed using a climbing assay as described in ref. (18).
EWSR1 plasmid and cell culture
EWS WT and mutants were amplified by PCR using the following primers: hEWS-specific primers (forward, hEWSKozMFlagf 5′-TCA CCA TGG ACT ACA AGG ACG ACG ATG ACA AAA TGG CGT CCA CGG ATT ACA G-3′; reverse, hEWSMycNotIr 5′-CAC GCG GCC GCC TAC AGA TCC TCT TCT GAG ATG AGT TTT TGT TCG TAG GGC CGA TCT CTG-3′). PCR-amplified fragments were cloned into the pEN-Tmcs entry vector using T4 DNA ligase (Promega) and recombined by LR-clonase (Invitrogen) into pSLIK-Neo destination vector (Signaling-gateway) to obtain pSLIK-EWS WT and mutants plasmids. Mouse ES cells were maintained in the ES medium [Dulbecco's modified Eagle's medium (DMEM), 15% fetal bovine serum (FBS), 1× penicillin/streptomycin, 1× glutamax, 1× non-essential amino acid, 1× sodium pyruvate, 0.1 mm beta-mercaptoethanol, 1000 μ/ml LIF, 25 μm PD98059] on a gelatinized plate without feeder cells. For differentiation, the protocol of Wichterle et al. (54) was followed. Briefly, 1 × 106 ES cells were cultured with the ADFNK medium (45% advanced DMEM/F12, 45% neurobasal medium, 10% knockout serum replacement, 1× penicillin/streptomycin, 1× L-glutamine, 0.1 mm beta-mercaptoethanol) for 5 days to form embryonic bodies (EBs)-small floating aggregates of ES cells. EBs were utilized for neurons culture on day 6 of differentiation. ES cells differentiated into neuron were dissociated into single cells and plated on a poly-l-lysine/laminin-coated 12-well chamber or Nunc 8-well chambers with the ADFNB + GDNF medium [49% advanced DMEM/F12, 49% neurobasal medium, 2% B27 supplement, 1× penicillin/streptomycin, 1× l-glutamine, 5 ng/ml glial cell line-derived neurotrophic factor (GDNF)].
Lentivirus production and transduction
The pSLIK expression lentivector was transfected along with lentivirus packaging and pseudotyping plasmids into 293T cells using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer's instructions. 293T cells were cultured in DMEM (GIBCO Invitrogen) and 10% fetal plex serum (Gemini). Plasmids were cotransfected by using 6 μg of pSLIK plasmid, 4.5 μg of the packaging plasmid psPAX2 (Addgene) and 3 μg of the vesicular stomatitis virus (VSV) G envelope plasmid pMD2 (Addgene) diluted in Opti-MEM (Gibco Invitrogen). The viral supernatant was collected 48 h after transfection, passed through 0.45 μm pore size filters and concentrated by ultracentrifugation onto a 20% sucrose gradients using SW41 rotor (Beckman) at 20 000 rpm for 2 h at 4°C. Viral pellets were resuspended in the ADFNB medium and stored in aliquots at −80°C. For transduction, cells were mixed with the virus at a low MOI to ensure <30% infection frequency such that the majority of transduced cells contained single viral integrants. Four micrograms of polybrene/ml (Sigma) was included and cells were plated on either a 12-well chamber or Nunc 8-well chambers (56815-1PAK; Fisher). To induce protein expression, 1 μg/ml Doxycycline (DOX; Millipore) was added to the cells 24 h after transduction and for 5 days.
Immunofluorescence analysis
For immunostaining, neurons were washed 3× with 1× phosphate buffered saline (PBS), fixed for 30 min with 3.7% paraformaldehyde, permeabilized using 0.1% Triton X-100 for 10 min, blocked with 3% bovine serum albumin in PBS for 15 min and incubated overnight at 4°C with primary antibodies as follows: anti-EWS (1:4000; Santa Cruz, sc-28327) and anti-FLAG M2 (1:5000; Sigma, F1804-1MG). Cells were washed 3× with 1× PBS and incubated with either Alexa-Fluor 555 goat anti-rabbit IgG (1:1000; Molecular Probes, A21429) or Alexa-Fluor 555 goat anti-mouse IgG (1:1,000; Molecular Probes, A21424). Cells were mounted with prolong plus 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen) and were visualized with an Olympus BX-60 microscope. Images were recorded with a Spot Digital camera.
Cell fractionation and immunoblot analysis
For western blot analysis, 5 × 105 cells of neurons, transduced with pSLIK lentivectors expressing TAF15 or EWS WT and mutants and induced with 1 μg/ml DOX for 5 days, were lysed using radioimmuno precipitation assay buffer (RIPA) buffer (0.1% SDS, 0.5% deoxycholate, 1% NP-40, 150 mm NaCl, 50 mm Tris–HCl, pH 8.0) for 10 min on ice and centrifuged at 13 000 rpm for 10 min to obtain the soluble fractions. Pellets were lysed with urea buffer (7 m urea, 2 m thiourea, 4% CHAPS, 30 mm Tris, pH 8.5) and were sonicated to obtain the insoluble fractions. Cell lysates were separated by Nu-PAGE (4–12% gradient gels; Invitrogen), transferred to nitrocellulose membranes and analyzed by western blotting with ECL plus detection reagents (GE Healthcare). Primary antibodies used were as follows: anti-FLAG M2 (1:10 000; Sigma), anti-GAPDH (1:20 000; Sigma). A secondary anti-mouse IgG HRP antibody was used at a dilution of 1:10 000. Membranes were developed using X-ray film (Kodak).
Mouse primary motor neuron transfection and immunofluorescence
Primary neuron cultures were transfected after 5 days in vitro using Lipofectamine LTX with PLUS reagent (Invitrogen) according to the manufacturer’s protocol in media lacking antibiotics. Media were replaced 12 h following transfection with media containing antibiotics. Cells were harvested for immunoflourescence 96 h after transfection. Briefly, cultures were washed in PBS and fixed in 4% paraformaldehyde 15 min, then washed in 1× PBS 4×. Cells were blocked for 1 h in blocking solution (2% fetal bovine serum, 0.02% Triton X-100, 1× PBS), and then incubated 1 h in primary antibody at RT. Cells were then washed 3× in PBS, and then incubated with secondary antibody 1 h RT. Cells were then washed with blocking solution and mounted in Vectashield mounting media with DAPI (Vector). Antibodies used were: α-EWSR1 mouse antibody (Santa Cruz), 1:1000; α-Doublecortin goat antibody (Santa Cruz) 1:500; Cy-3 conjugated α-mouse IgG (Jackson Immunoresearch), 1:250; Cy-3 conjugated α-rabbit IgG (Jackson Immunoresearch), 1:250; and Cy-2 conjugated α-goat IgG (Jackson Immunoresearch), 1:250. Cells were visualized by light microscopy. Localization of EWSR1 was quantified using blinded analysis of random fields of cells. The number of neurons with EWSR1 staining in processes was divided by the total number of neurons counted to yield the percent of neurons with EWSR1 in neuronal processes. More than 50 neurons were analyzed for each condition. Neurons were identified using morphology and doublecortin staining.
Immunohistochemistry
Formalin-fixed, paraffin-embedded human spinal cord sections were deparaffinized before pre-treatment using heat antigen retrieval with Bull's Eye Decloaker (BioCare Medical). Endogenous peroxidase was then blocked with 3% hydrogen peroxide in PBS for 10 min. After washing with 0.1%, PBST blocking was performed with 10% goat serum, and 0.5% PBST for 30–60 min at 25°C. Sections were incubated with mouse anti-EWSR1 (1:125; Santa Cruz Biotechnology) in 0.1% PBST overnight at 4°C. After washing with 0.1% PBST, sections were incubated with biotinylated goat anti-mouse IgG (1:200; Vector Laboratories) for 1h at 25°C. After washing with 0.1% PBST, sections were then incubated with Vectastain ABC (Vector Laboratories) for 45 min. After washing with 0.1% PBST followed by 0.1 m Tris (pH 7.5) and 0.3 m NaCl. Peroxidase activity was then detected with DAB (Sigma). Detailed immunohistochemistry protocols are available at http://www.med.upenn.edu/mcrc/histology_core/.
Statistical analysis
Two-tailed Fisher's exact tests were used to evaluate genetic association between sequence variants and ALS.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by National Institutes of Health Director's New Innovator Awards 1DP2OD004417 (A.D.G.) and 1DP2OD002177 (J.S.), National Institutes of Health grants 1R01NS065317 (A.D.G.), AG17586 (V.M.V.D., J.Q.T., R.G.), AG10124 (V.M.V.D., J.Q.T.), P01-AG-09215 (N.M.B.), NS056070, NS072561 (Z.M.), T32-AG00255 (to F.I.; V.M.Y.L. program director), the University of Pennsylvania Institute on Aging and Alzheimer Disease Core Center (AG10124) Pilot Grant Program (V.M.V.D.), an Ellison New Scholar in Aging Award (J.S.) and a grant from the Packard Center for ALS Research at Johns Hopkins (A.D.G. and J.S.). A.D.G. is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts and a Rita Allen Foundation Scholar. J.Q.T is the William Maul Measey-Truman G. Schnabel, Jr, Professor of Geriatric Medicine and Gerontology. N.M.B. is an Investigator of the HHMI. In Australia, the work was supported by the National Health and Medical Research Council of Australia (1004670, 511941, 570957) and a Peter Stearne grant from the Motor Neurone Disease Research Institute of Australia. This research was conducted while J.C. was an Ellison Medical Foundation/AFAR Postdoctoral Fellow. Samples from the National Cell Repository for Alzheimer's Disease (NCRAD), which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the National Institute on Aging (NIA), were used in this study.
Supplementary Material
ACKNOWLEDGEMENTS
We are truly grateful for the dedication of the patients and their families and for their invaluable contributions to this research. We thank contributors, including the Alzheimer's Disease Centers who collected samples used in this study.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Cleveland D.W., Rothstein J.D. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat. Rev. Neurosci. 2001;2:806–819. doi: 10.1038/35097565. doi:10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 2.Andersen P.M., Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat. Rev. Neurol. 2011;7:603–615. doi: 10.1038/nrneurol.2011.150. doi:10.1038/nrneurol.2011.150. [DOI] [PubMed] [Google Scholar]
- 3.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J., et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. doi:10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng H.X., Chen W., Hong S.T., Boycott K.M., Gorrie G.H., Siddique N., Yang Y., Fecto F., Shi Y., Zhai H., et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. doi:10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fecto F., Siddique T. Making connections: pathology and genetics link amyotrophic lateral sclerosis with frontotemporal lobe dementia. J. Mol. Neurosci. 2011;45:663–675. doi: 10.1007/s12031-011-9637-9. doi:10.1007/s12031-011-9637-9. [DOI] [PubMed] [Google Scholar]
- 6.Gijselinck I., Van Langenhove T., van der Zee J., Sleegers K., Philtjens S., Kleinberger G., Janssens J., Bettens K., Van Cauwenberghe C., Pereson S., et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11:54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 7.Ince P.G., Highley J.R., Kirby J., Wharton S.B., Takahashi H., Strong M.J., Shaw P.J. Molecular pathology and genetic advances in amyotrophic lateral sclerosis: an emerging molecular pathway and the significance of glial pathology. Acta Neuropathol. 2011;122:657–671. doi: 10.1007/s00401-011-0913-0. doi:10.1007/s00401-011-0913-0. [DOI] [PubMed] [Google Scholar]
- 8.Renton A.E., Majounie E., Waite A., Simon-Sanchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L., et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. doi:10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagier-Tourenne C., Cleveland D.W. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. doi:10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gitler A.D., Shorter J. RNA-binding proteins with prion-like domains in ALS and FTLD-U. Prion. 2011;5:179–197. doi: 10.4161/pri.5.3.17230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwiatkowski T.J., Jr, Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T., et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. doi:10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 12.Vance C., Rogelj B., Hortobagyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. doi:10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. doi:10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 14.Hanson K.A., Kim S.H., Tibbetts R.S. RNA-binding proteins in neurodegenerative disease: TDP-43 and beyond. Wiley Interdiscip. Rev. RNA. 2012;3:265–285. doi: 10.1002/wrna.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee E.B., Lee V.M., Trojanowski J.Q. Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat. Rev. Neurosci. 2011;13:38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson B.S., McCaffery J.M., Lindquist S., Gitler A.D. A yeast TDP-43 proteinopathy model: exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc. Natl Acad. Sci. USA. 2008;105:6439–6444. doi: 10.1073/pnas.0802082105. doi:10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson B.S., Snead D., Lee J.J., McCaffery J.M., Shorter J., Gitler A.D. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. doi:10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elden A.C., Kim H.J., Hart M.P., Chen-Plotkin A.S., Johnson B.S., Fang X., Armakola M., Geser F., Greene R., Lu M.M., et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. doi:10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Z., Diaz Z., Fang X., Hart M.P., Chesi A., Shorter J., Gitler A.D. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. doi:10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ju S., Tardiff D.F., Han H., Divya K., Zhong Q., Bosco D.A., Hayward L.J., Brown R.H., Jr, Lindquist S.L., Ringe D., et al. A Yeast model of FUS/TLS-dependent cytotoxicity. PLoS Biol. 2011;9:e1001052. doi: 10.1371/journal.pbio.1001052. doi:10.1371/journal.pbio.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fushimi K., Long C., Jayaram N., Chen X., Li L., Wu J.Y. Expression of human FUS/TLS in yeast leads to protein aggregation and cytotoxicity, recapitulating key features of FUS proteinopathy. Protein Cell. 2011;2:141–149. doi: 10.1007/s13238-011-1014-5. doi:10.1007/s13238-011-1014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kryndushkin D., Wickner R.B., Shewmaker F. FUS/TLS forms cytoplasmic aggregates, inhibits cell growth and interacts with TDP-43 in a yeast model of amyotrophic lateral sclerosis. Protein Cell. 2011;2:223–236. doi: 10.1007/s13238-011-1525-0. doi:10.1007/s13238-011-1525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Couthouis J., Hart M.P., Shorter J., DeJesus-Hernandez M., Erion R., Oristano R., Liu A.X., Ramos D., Jethava N., Hosangadi D., et al. A yeast functional screen predicts new candidate ALS disease genes. Proc. Natl Acad. Sci. USA. 2011;108:20881–20890. doi: 10.1073/pnas.1109434108. doi:10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cushman M., Johnson B.S., King O.D., Gitler A.D., Shorter J. Prion-like disorders: blurring the divide between transmissibility and infectivity. J. Cell Sci. 2010;123:1191–1201. doi: 10.1242/jcs.051672. doi:10.1242/jcs.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ticozzi N., Vance C., Leclerc A.L., Keagle P., Glass J.D., McKenna-Yasek D., Sapp P.C., Silani V., Bosco D.A., Shaw C.E., et al. Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2011;156B:285–290. doi: 10.1002/ajmg.b.31158. [DOI] [PubMed] [Google Scholar]
- 26.Neumann M., Bentmann E., Dormann D., Jawaid A., DeJesus-Hernandez M., Ansorge O., Roeber S., Kretzschmar H.A., Munoz D.G., Kusaka H., et al. FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain. 2011;134:2595–2609. doi: 10.1093/brain/awr201. doi:10.1093/brain/awr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Araya N., Hirota K., Shimamoto Y., Miyagishi M., Yoshida E., Ishida J., Kaneko S., Kaneko M., Nakajima T., Fukamizu A. Cooperative interaction of EWS with CREB-binding protein selectively activates hepatocyte nuclear factor 4-mediated transcription. J. Biol. Chem. 2003;278:5427–5432. doi: 10.1074/jbc.M210234200. doi:10.1074/jbc.M210234200. [DOI] [PubMed] [Google Scholar]
- 28.Dormann D., Rodde R., Edbauer D., Bentmann E., Fischer I., Hruscha A., Than M.E., Mackenzie I.R., Capell A., Schmid B., et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29:2841–2857. doi: 10.1038/emboj.2010.143. doi:10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee B.J., Cansizoglu A.E., Suel K.E., Louis T.H., Zhang Z., Chook Y.M. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. doi:10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toombs J.A., McCarty B.R., Ross E.D. Compositional determinants of prion formation in yeast. Mol. Cell Biol. 2010;30:319–332. doi: 10.1128/MCB.01140-09. doi:10.1128/MCB.01140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw D.J., Morse R., Todd A.G., Eggleton P., Lorson C.L., Young P.J. Identification of a tripartite import signal in the Ewing Sarcoma protein (EWS) Biochem. Biophys. Res. Commun. 2009;390:1197–1201. doi: 10.1016/j.bbrc.2009.10.120. doi:10.1016/j.bbrc.2009.10.120. [DOI] [PubMed] [Google Scholar]
- 32.Zakaryan R.P., Gehring H. Identification and characterization of the nuclear localization/retention signal in the EWS proto-oncoprotein. J. Mol. Biol. 2006;363:27–38. doi: 10.1016/j.jmb.2006.08.018. doi:10.1016/j.jmb.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 33.Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C., Shaffer T., Wong M., Bhattacharjee A., Eichler E.E., et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. doi:10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagier-Tourenne C., Polymenidou M., Cleveland D.W. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 2010;19:R46–R64. doi: 10.1093/hmg/ddq137. doi:10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barmada S.J., Skibinski G., Korb E., Rao E.J., Wu J.Y., Finkbeiner S. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J. Neurosci. 2010;30:639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. doi:10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabashi E., Lin L., Tradewell M.L., Dion P.A., Bercier V., Bourgouin P., Rochefort D., Bel Hadj S., Durham H.D., Vande Velde C., et al. Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum. Mol. Genet. 2010;19:671–683. doi: 10.1093/hmg/ddp534. doi:10.1093/hmg/ddp534. [DOI] [PubMed] [Google Scholar]
- 37.Bosco D.A., Lemay N., Ko H.K., Zhou H., Burke C., Kwiatkowski T.J., Jr, Sapp P., McKenna-Yasek D., Brown R.H., Jr, Hayward L.J. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum. Mol. Genet. 2010;19:4160–4175. doi: 10.1093/hmg/ddq335. doi:10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanson K.A., Kim S.H., Wassarman D.A., Tibbetts R.S. Ubiquilin modifies TDP-43 toxicity in a Drosophila model of amyotrophic lateral sclerosis (ALS) J. Biol. Chem. 2010;285:11068–11072. doi: 10.1074/jbc.C109.078527. doi:10.1074/jbc.C109.078527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y., Ray P., Rao E.J., Shi C., Guo W., Chen X., Woodruff E.A., 3rd, Fushimi K., Wu J.Y. A Drosophila model for TDP-43 proteinopathy. Proc. Natl Acad. Sci. USA. 2010;107:3169–3174. doi: 10.1073/pnas.0913602107. doi:10.1073/pnas.0913602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu Y., Ferris J., Gao F.B. Frontotemporal dementia and amyotrophic lateral sclerosis-associated disease protein TDP-43 promotes dendritic branching. Mol. Brain. 2009;2:30. doi: 10.1186/1756-6606-2-30. doi:10.1186/1756-6606-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritson G.P., Custer S.K., Freibaum B.D., Guinto J.B., Geffel D., Moore J., Tang W., Winton M.J., Neumann M., Trojanowski J.Q., et al. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J. Neurosci. 2010;30:7729–7739. doi: 10.1523/JNEUROSCI.5894-09.2010. doi:10.1523/JNEUROSCI.5894-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lanson N.A., Jr, Maltare A., King H., Smith R., Kim J.H., Taylor J.P., Lloyd T.E., Pandey U.B. A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum. Mol. Genet. 2011;20:2510–2523. doi: 10.1093/hmg/ddr150. doi:10.1093/hmg/ddr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J.W., Brent J.R., Tomlinson A., Shneider N.A., McCabe B.D. The ALS-associated proteins FUS and TDP-43 function together to affect Drosophila locomotion and life span. J. Clin. Invest. 2011;121:4118–4126. doi: 10.1172/JCI57883. doi:10.1172/JCI57883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y., Yang M., Deng J., Chen X., Ye Y., Zhu L., Liu J., Ye H., Shen Y., Li Y., et al. Expression of human FUS protein in Drosophila leads to progressive neurodegeneration. Protein Cell. 2011;2:477–486. doi: 10.1007/s13238-011-1065-7. doi:10.1007/s13238-011-1065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miguel L., Avequin T., Delarue M., Feuillette S., Frebourg T., Campion D., Lecourtois M. Accumulation of insoluble forms of FUS protein correlates with toxicity in Drosophila. Neurobiol. Aging. 2012;33:1008.e1–1008.e15. doi: 10.1016/j.neurobiolaging.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 46.Alberti S., Gitler A.D., Lindquist S. A suite of Gateway((R)) cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007;24:913–919. doi: 10.1002/yea.1502. doi:10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng S.B., Buckingham K.J., Lee C., Bigham A.W., Tabor H.K., Dent K.M., Huff C.D., Shannon P.T., Jabs E.W., Nickerson D.A., et al. Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 2009;42:30–35. doi: 10.1038/ng.499. doi:10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lupski J.R., Belmont J.W., Boerwinkle E., Gibbs R.A. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. doi:10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang E.J., Zhang J., Geser F., Trojanowski J.Q., Strober J.B., Dickson D.W., Brown R.H., Jr, Shapiro B.E., Lomen-Hoerth C. Extensive FUS-immunoreactive pathology in juvenile amyotrophic lateral sclerosis with basophilic inclusions. Brain Pathol. 2010;20:1069–1076. doi: 10.1111/j.1750-3639.2010.00413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.King O.D., Gitler A.D., Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012 doi: 10.1016/j.brainres.2012.01.016. DOI:10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guthrie C., Fink G.R. Methods in ezymology: guide to yeast genetics and molecular and cell biology. Academic Press. 2002;169:169–182. [Google Scholar]
- 52.Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alberti S., Halfmann R., King O., Kapila A., Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. doi:10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wichterle H., Peljto M., Nedelec S. Xenotransplantation of embryonic stem cell-derived motor neurons into the developing chick spinal cord. Methods Mol. Biol. 2009;482:171–183. doi: 10.1007/978-1-59745-060-7_11. doi:10.1007/978-1-59745-060-7_11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.