Abstract
The abnormal accumulation of the microtubule-binding protein tau is associated with a number of neurodegenerative conditions, and correlates with cognitive decline in Alzheimer's disease. The ubiquitin ligase carboxy terminus of Hsp70-interacting protein (CHIP) and the molecular chaperone Hsp90 are implicated in protein triage decisions involving tau, and have consequently been targeted for therapeutic approaches aimed at decreasing tau burden. Here, we present evidence that CHIP binds, ubiquitinates and regulates expression of histone deacetylase 6 (HDAC6). As the deacetylase for Hsp90, HDAC6 modulates Hsp90 function and determines the favorability of refolding versus degradation of Hsp90 client proteins. Moreover, we demonstrate that HDAC6 levels positively correlate with tau burden, while a decrease in HDAC6 activity or expression promotes tau clearance. Consistent with previous research on Hsp90 clients in cancer, we provide evidence that a loss of HDAC6 activity augments the efficacy of an Hsp90 inhibitor and drives client degradation, in this case tau. Therefore, our current findings not only identify HDAC6 as a critical factor for the regulation of tau levels, but also indicate that a multi-faceted treatment approach could more effectively arrest tau accumulation in disease.
INTRODUCTION
The microtubule-binding protein tau is central to the regulation of axonal outgrowth and cellular morphology, as well as neuronal transport (1–3). In a number of neurodegenerative diseases classified as tauopathies, which include frontotemporal dementia with parkinsonism associated with chromosome 17, progressive supranuclear palsy, corticobasal degeneration, and Alzheimer's disease (AD), tau becomes hyperphosphorylated and aggregates into filaments, thus losing the ability to bind and stabilize microtubules (4,5). These tau filaments continue to aggregate and form increasingly insoluble deposits referred to as neurofibrillary tangles. Although AD is the most common tauopathy and most frequent cause of dementia, the available treatment options only treat the symptoms of AD, and do nothing to alleviate the underlying pathology. Therefore, understanding the mechanism by which hyperphosphorylated tau is cleared by neurons, and developing therapeutics to eliminate these toxic species are of considerable interest.
Previously, the ubiquitin ligase carboxy terminus of Hsp70-interacting protein (CHIP) and the molecular chaperone Hsp90 have been shown to play pivotal roles in protein triage decisions involving tau (6–9). CHIP has a unique binding affinity for abnormally phosphorylated tau and is required for tau ubiquitination and targeting to proteasomes for degradation (6–8). For its part, Hsp90, a ubiquitous, constitutively expressed protein that constitutes 1–2% of total cellular protein in eukaryotic cells (10,11), functions to maintain its client proteins in a properly folded state and thereby suppresses their aggregation (10). During conditions of stress, this dual function of Hsp90 helps to repair the pool of damaged client proteins, thus serving to reestablish a state of cellular equilibrium (12). Over 100 proteins have been reported to be clients of Hsp90 (12,13), including protein kinases, transcriptional regulators, and steroid receptors (12). Of particular relevance to the current report, tau is also an Hsp90 client (9,14).
Following binding of Hsp90, client proteins either enter a refolding pathway, leading to a functional, properly folded client protein or they are targeted for degradation by the ubiquitin-proteasome system (15). The specific components of the Hsp90 complex ultimately determine whether client refolding or degradation occurs (16). Nucleotide binding to Hsp90 is proposed to alter its conformation and define the subset of chaperones with which it interacts (16). In the ADP-bound conformation, Hsp90 associates with client-bound Hsp70/Hsp40 complexes. At this point, the complex may recruit ubiquitin ligases, such as CHIP, to direct the client to proteasomes for degradation. The replacement of ADP with ATP alters Hsp90 conformation, releasing Hsp70/Hsp40 and allowing the recruitment of other cochaperones, including p23. This complex folds and stabilizes the client now bound by Hsp90. Notably, the acetylation state of Hsp90 modulates Hsp90 function (17–20); specifically, Hsp90 hyperacetylation decreases the affinity of Hsp90 for ATP and oncogenic client proteins, and causes the dissociation of p23 from the Hsp90 complex, leading to an impairment in chaperone function and promoting client degradation (18,21).
Of importance, inhibition or depletion of histone deacetylase 6 (HDAC6) promotes the hyperacetylation of Hsp90, thus augmenting the polyubiquitination and subsequent degradation of Hsp90 client proteins (17–20). Hyperacetylation of Hsp90 due to HDAC6 depletion also leads to an increased binding affinity of Hsp90 for Hsp90 inhibitors (21,22). Hsp90 inhibitors disrupt Hsp90 chaperone function such that client proteins are instead degraded (reviewed in 23). That cotreatment of leukemia cells with HDAC6 and Hsp90 inhibitors synergistically promotes the degradation of Hsp90 client proteins suggests that the hyperacetylation of Hsp90 by HDAC6 augments the pro-degradation effects of Hsp90 inhibitors (21,24). Therefore, it is possible that alterations in Hsp90 acetylation, either through differences in expression or activity of the deacetylase HDAC6, determine the sensitivity of the Hsp90 chaperone complex to chemical modulation, ultimately deciding the fate of Hsp90 client proteins, such as tau. Here, we provide evidence that CHIP interacts with and regulates the half-life of HDAC6 in cells and in mice. In addition, we demonstrate that increased levels of HDAC6 lead to an accumulation of tau, while decreased HDAC6 levels or activity promotes tau clearance and increases the efficacy of Hsp90 inhibitors. Therefore, we hypothesize that CHIP, by regulating HDAC6 levels, influences protein triage decisions by modulating the refolding and degradation activities of Hsp90.
RESULTS
CHIP interacts with and ubiquitinates HDAC6
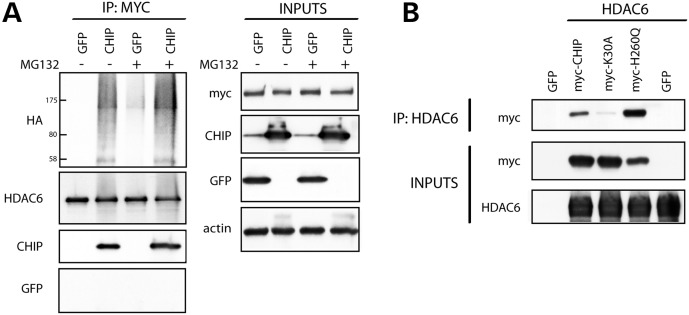
To investigate whether HDAC6 is a substrate for CHIP, co-immunoprecipitation experiments were conducted in the presence and absence of proteasome inhibition (MG132) to slow the degradation of ubiquitinated proteins. HEK293T cells were transfected to express HA-tagged ubiquitin (HA-ubiquitin) and myc-HDAC6, along with either CHIP or GFP, and HDAC6 was then immunoprecipitated from cell lysates using an anti-myc antibody. As shown in Figure 1A, CHIP co-immunoprecipitated with HDAC6, and multiple HDAC6- and HA-ubiquitin-positive high molecular weight species were observed in CHIP-overexpressing cells, likely representing ubiquitinated HDAC6.
Figure 1.
CHIP interacts with and ubiquitinates HDAC6 in vitro. (A) HEK293T cells were transfected with either GFP or CHIP in the presence of mycHDAC6 and HA-ubiquitin, and treated with the proteasome inhibitor, MG132, for 24 h. An anti-myc antibody was used to immunoprecipitate mycHDAC6 from cell lysates, and samples were analyzed by western blot. (B) HEK293T cells were transfected with untagged HDAC6 or vector, and either GFP, myc-CHIP, mycCHIP (K30A mutant) or mycCHIP (H260Q mutant). An HDAC6 antibody was used for immunoprecipitation, and cell lysates and immunoprecipitates were probed for anti-myc.
To further characterize the interaction between CHIP and HDAC6, we sought to determine whether CHIP binds HDAC6 directly or whether chaperones Hsp70/Hsp90 serve to bridge the two proteins. Thus, HeLa cells were co-transfected to overexpress HDAC6 and either wild-type (WT) myc-CHIP, myc-CHIP(K30A) or myc-CHIP(H260Q). The K30A mutation prevents CHIP from docking with chaperones Hsp70/Hsp90, while the H260Q mutation, present within the U-box of CHIP, obliterates ubiquitin ligase activity (25). Upon immunoprecipitation for HDAC6, both WT myc-CHIP and myc-CHIP(H260Q) were found to bind HDAC6 (Fig. 1B). In contrast, little myc-CHIP(K30A) co-immunoprecipitated with HDAC6, suggesting that either Hsp70 or Hsp90 is required for the interaction between CHIP and HDAC6.
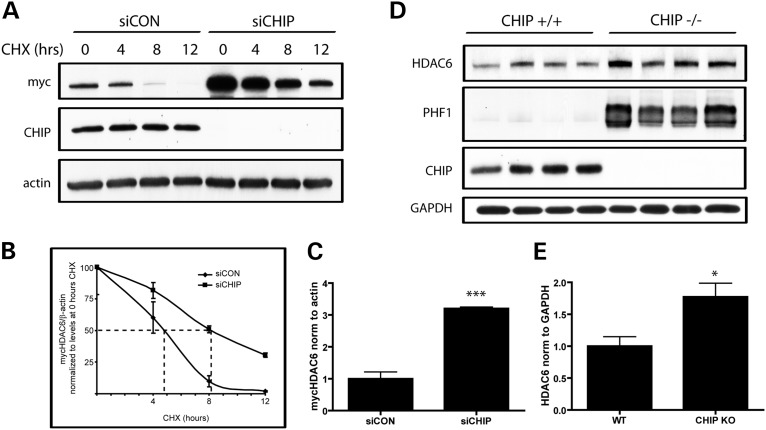
Since CHIP plays a role in promoting the ubiquitination and proteasomal degradation of several proteins, it was next evaluated whether loss of CHIP influences HDAC6 levels. To this end, HeLa cells were transfected with either siCON or siCHIP for 48 h, and subsequently transfected with mycHDAC6 for an additional 24 h. Cells were then treated with cycloheximide (CHX) to inhibit new protein translation, and cell lysates were collected at various time points thereafter. Interestingly, loss of CHIP increased steady-state levels of mycHDAC6 by 2.2-fold in cultured cells, and also prolonged its half-life (Fig. 2A–C).
Figure 2.
Loss of CHIP prolongs the half-life of HDAC6 in vitro and increases HDAC6 levels in vivo. (A) HeLa cells transfected with mycHDAC6 were treated with non-targeting siRNA (siCON) or siCHIP to knock-down endogenous CHIP for 48 h. Cells were then exposed to cycloheximide (CHX; 0.1 mg/ml) for 4, 8 or 12 h to inhibit the continued translation of mycHDAC6, and mycHDAC6 levels were examined by western blot using an anti-myc antibody. (B) Steady-state levels (0 h CHX) of mycHDAC6 were significantly higher in cultures treated with siCHIP compared with siCON-treated cultures (P< 0.005, t-test, n= 3). (C) The rate of degradation of mycHDAC6 post-CHX treatment in siCON- or siCHIP-treated cultures was compared by normalizing values at each time point to their respective levels at 0 h CHX treatment. There was a significant difference in the rate of degradation between siCON- and siCHIP-treated samples as assessed by two-way ANOVA (effect of siCHIP F = 388.3; P< 0.0001; effect of CHX treatment F = 72.39; P< 0.0001; interaction F = 10.71; P = 0.0004). Estimated half-life was the following: siCON, 4.7 h; siCHIP, 7.9 h. (D) HDAC6 and phosphorylated tau levels were examined in brain homogenates from CHIP+/+ and CHIP−/− mice using anti-HDAC6 and PHF1 antibody, respectively. Loss of CHIP leads to increased HDAC6 levels and anti-PHF1 immunoreactive phospho-tau in mice. (E) HDAC6 levels were significantly higher in CHIP−/− compared with CHIP+/+ brain homogenates (t= 3; P< 0.05, n= 4). All data are presented as mean ± SEM. ***P< 0.001 *P< 0.05.
To investigate the impact of decreased CHIP levels in vivo, brain homogenates from CHIP−/− and CHIP+/+ mice were prepared. In agreement with previous reports, loss of CHIP led to an increase in PHF1-immunopositive tau levels (Fig. 2D) (8). In addition, a significant increase in HDAC6 levels was observed in CHIP−/− mice (Fig. 2D and E). Collectively, these results indicate that HDAC6 is a substrate of CHIP, and that the loss of CHIP impacts HDAC6 turnover and leads to increased levels of HDAC6 in vitro and in vivo.
Overexpression of HDAC6 promotes tau accumulation
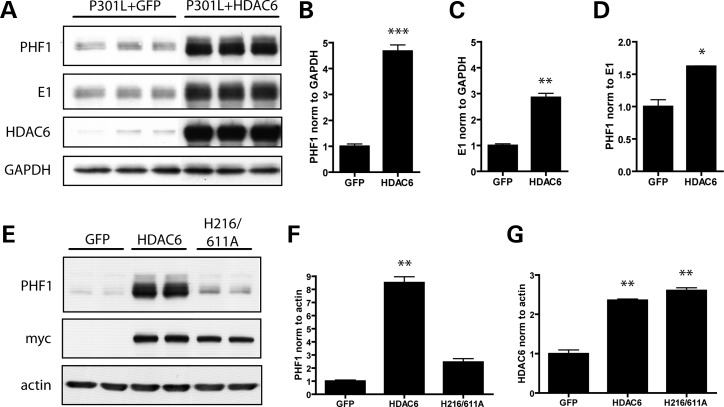
Given that the loss of CHIP is associated with an increase in both HDAC6 and tau levels (Fig. 2), and that HDAC6 levels are increased in AD (26), the effect of HDAC6 overexpression on tau accumulation was evaluated by transfecting HeLa cells to co-express tau with the pathogenic P301L mutation with either GFP or HDAC6. Surprisingly, the co-expression of P301L-tau with HDAC6 resulted in a dramatic upregulation of tau levels, as detected using both PHF1 (pS396/S404) and E1 (human tau specific) antibodies (Fig. 3A–D). As the overexpression of HDAC6 led to a more significant increase in PHF1-tau than total human E1 tau (Fig. 3A–D), HDAC6 overexpression may preferentially increase the accumulation of pathologically phosphorylated tau species. To verify whether HDAC6 overexpression specifically affects tau levels, cells were transfected to coexpress HDAC6 and either GFP or α-synuclein. No changes in GFP or α-synuclein expression were observed in the presence of HDAC6 (Supplementary Material, Fig. S1).
Figure 3.
Overexpression of HDAC6 promotes the accumulation of tau in vitro. (A) HeLa cells were transfected with GFP, P301L-V5 or HDAC6 expression constructs for 48 h. Levels of phosphorylated and total tau present in cell lysates were then examined by immunoblotting with PHF1 and E1 antibodies, respectively. HDAC6 overexpression was confirmed by probing for HDAC6. (B) Analysis of PHF1-positive tau levels by student's t-test verified that there was a significant increase in cells overexpressing HDAC6 (t= 14.5; P< 0.0001). (C) Quantification of E1 immunoreactivity normalized to GAPDH also revealed a significant increase in cells overexpressing HDAC6 (t= 11.04; P= 0.0004). (D) The ratio of PHF1-positive tau to E1-positive tau was significantly increased upon overexpression of HDAC6 (t= 5.9; P= 0.004), indicating that PHF1-tau is specifically increased in cells overexpressing HDAC6. (E) HeLa cells were cotransfected with P301L-V5 and either GFP, mycHDAC6 or a catalytically inactive mycHDAC6 construct (H216/611A mutant). Cell lysates were immunoblotted for PHF1-tau, myc or actin to control for protein loading. (F) PHF1-tau levels were quantified and normalized to actin, and analyzed by one-way ANOVA. Overexpression of mycHDAC6 caused a significant increase in PHF1-tau levels (P< 0.001), but there was no effect of the catalytically inactive mycHDAC6 mutant on PHF1-tau (P> 0.05). (G) Anti-myc levels were assessed and normalized to actin, verifying that both mycHDAC6 and the catalytically inactive mutant were expressed at similar levels. All data are presented as mean ± SEM (n= 3). ***P< 0.0001, **P< 0.001, *P< 0.01.
HDAC6-mediated tau accumulation may depend on the deacetylase activity of HDAC6. Alternatively, given that HDAC6 contains a ubiquitin-binding domain and has been shown to play a role in sequestering ubiquitinated proteins and targeting them to the aggresome, it is possible that HDAC6 directly binds and sequesters tau, thereby preventing tau degradation. Thus, to determine whether the deacetylase activity of HDAC6 is required to promote tau accumulation, a catalytically inactive mutant (H216A/H611A) of HDAC6 was generated and cells were transfected to express either this mutant or WT HDAC6. As shown in Figure 3E–G, loss of deacetylase activity prevented HDAC6 from promoting tau accumulation, indicating that the catalytic activity of HDAC6 is required for this effect. These results, in combination with previous reports demonstrating that HDAC6 regulates the acetylation state of Hsp90 (18,20–22), suggest that HDAC6 overexpression increases the affinity of the Hsp90 chaperone complex for pathological tau species by decreasing Hsp90 acetylation, thereby blocking tau degradation.
HDAC6 deficiency promotes tau degradation
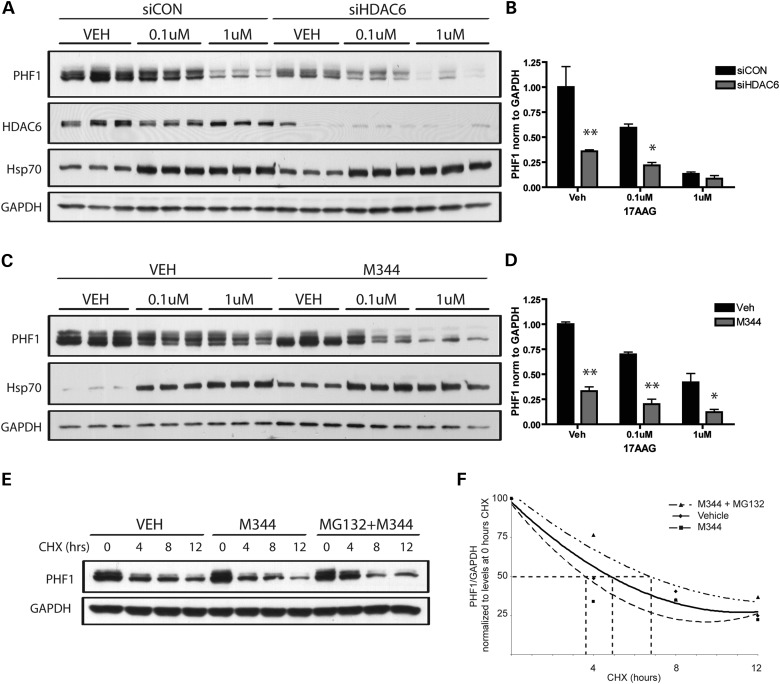
We and others have shown that inhibitors of Hsp90 enhance the degradation of mutated or hyperphosphorylated tau in vitro and in vivo, an effect that is dependent upon the presence of CHIP (9,14,27). More specifically, our work reveals that Hsp90 inhibition does not simply facilitate the proteasomal degradation of tau, but selectively targets aberrantly phosphorylated tau, indicating that the surveillance mechanism of the chaperone network is a highly sensitive and controlled system (9,14,27,28). Since loss of HDAC6 activity has been shown to increase the affinity of Hsp90 for Hsp90 inhibitors, such as 17AAG, as well as enhance 17AAG-mediated degradation of Hsp90 client proteins (21,22), we sought to evaluate whether HDAC6 depletion would likewise enhance the degradation of tau induced by Hsp90 inhibition. HeLa cells were treated with siCON or siHDAC6, and then exposed to 17AAG. To confirm efficient Hsp90 inhibition, Hsp70 induction, a consequence of Hsp90 inhibition (15,29), was also monitored (Fig. 4A and C). In contrast to HDAC6 overexpression, HDAC6 knockdown resulted in a decrease in P301L tau levels compared with tau levels in siCON-treated cells, as assessed using the PHF1 antibody (Fig. 4A and B, compare vehicle-treated samples). In addition, treatment with 17AAG led to a dose-dependent decrease in tau levels, which was enhanced in cells treated with siHDAC6 (Fig. 4A and B). In a similar fashion, enhanced tau clearance was observed upon HDAC6 inhibition. Specifically, treatment with M344, an HDAC6 inhibitor (30), led to a significant decrease in tau levels, and also augmented the ability of 17AAG to promote tau degradation (Fig. 4C and D). As expected, the half-life of tau is decreased in cells treated with M344, an effect that is reversed in the presence of a proteasome inhibitor (Fig. 4E and F). Taken together, these results suggest that HDAC6 deficiency enhances tau degradation induced by Hsp90 inhibitors, which is consistent with previous reports showing that HDAC6 deficiency leads to an increased acetylation of Hsp90, thereby increasing the sensitivity of Hsp90 for Hsp90 inhibitors (21,22).
Figure 4.
HDAC6 deficiency or inhibition decreases tau levels, and functions synergistically with Hsp90 inhibition to promote the degradation of tau. (A) HeLa cells were transfected with siCON or siHDAC6 for 24 h, and subsequently transfected with P301L-V5 for another 24 h. Cells were then treated with 0.1 or 1 μm of 17AAG for 24 h, and cell lysates immunoblotted for PHF1-tau, HDAC6, Hsp70 and GAPDH to control for protein loading. (B) Analysis of PHF1-tau levels by two-way ANOVA verified a significant treatment effect of 17AAG (F = 21.1, P= 0.0001) and siHDAC6 (F = 24.5, P= 0.0003), as well as a significant interaction between 17AAG and siHDAC6 (F= 5.75, P= 0.02). (C) HeLa cells were transfected to overexpress P301L-V5 for 24 h, pretreated with the HDAC6 inhibitor M344 for 1 h and subsequently treated with 0.1–1 μm 17-AAG for an additional 24 h. Levels of PHF1-tau and Hsp70 present in cell lysates were examined by immunoblotting. (D) Analysis of PHF1-tau levels by two-way ANOVA confirmed a significant treatment effect of both 17AAG (F = 33.73, P< 0.0001) and M344 (F= 153.3, P< 0.0001). All data are presented as mean ± SEM (n= 3). (E) HeLa cells were transfected to overexpress P301L-V5 for 24 h, pretreated with 1 μm MG132 for 1 h and subsequently treated with 1 μm M344 and CHX for 4, 8 or 12 h to inhibit the continued translation of tau. Levels of PHF1-tau and GAPDH present in cell lysates were examined by immunoblotting. (F) The rate of degradation of PHF1-tau post-CHX treatment in vehicle, M344 or MG132 + M344-treated cells was compared by normalizing values at each time point to their respective levels at 0 h CHX treatment. There was a significant difference in the rate of degradation between vehicle, M344 and MG132 + M344-treated samples as assessed by two-way ANOVA (effect of M344 or MG132 + M344 treatment F= 6.4; P= 0.01; effect of CHX treatment F= 95.3; P< 0.0001; interaction F= 3.4; P= 0.04). Estimated half-life was the following: vehicle, 5 h; M344, 3.8 h; MG132 + M344, 6.4 h). Data from this experiment are presented as mean ± SEM (n= 2). **P< 0.001, *P< 0.05.
HDAC6 deficiency modulates the expression of the cochaperone p23
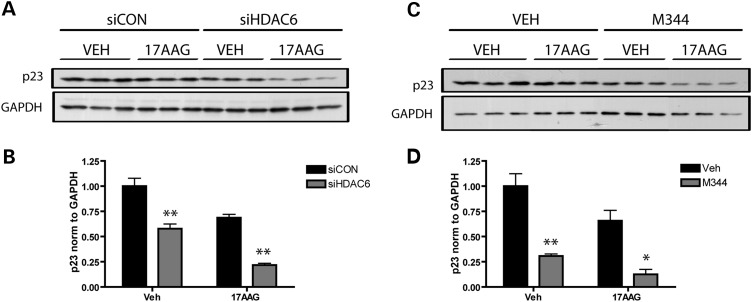
The cochaperone p23 locks Hsp90 in a conformational state that has a high affinity for client proteins (31). Hsp90 complexes that contain p23 promote the folding and stabilization of clients, rather than their degradation. Rao et al. demonstrated that HDAC6 deficiency leads to a decreased interaction between Hsp90 and p23 (21). In addition, we have reported that the loss of p23 expression significantly decreases tau levels (9). Intriguingly, a significant decrease in p23 was detected upon both HDAC6 knockdown (Fig. 5A and B) and HDAC6 inhibition with M344 (Fig. 5C and D), demonstrating that the loss of HDAC6 function is associated with decreased levels of p23. Given that decreased HDAC6 activity has been shown to decrease the interaction between Hsp90 and p23 (21), the current results suggest that HDAC6 deficiency promotes client degradation through the increased formation of Hsp90 chaperone complexes that lack p23, which are characterized by a very low affinity for client proteins, such as tau.
Figure 5.
HDAC6 knockdown and inhibition decrease cochaperone p23 levels. (A) HeLa cells were transfected with siCON or siHDAC6 for 48 h, and then treated with vehicle (DMSO) or 1 μm of 17-AAG for an additional 24 h. Cell lysates were immunoblotted for p23 and GAPDH, the latter as a control for protein loading. (B) Levels of p23 were quantified and normalized to GAPDH, and analysis by two-way ANOVA revealed a significant effect of siHDAC6 on p23 (F= 82.02; P< 0.0001), as well as a significant treatment effect of 17AAG (F= 47.07; P= 0.0001), but no interaction between siHDAC6 and 17AAG (F= 0.25; P= 0.63). (C) HeLa cells were pretreated with the HDAC6 inhibitor M344 for 1 h, and subsequently treated with vehicle (DMSO) or 1 μm 17AAG for an additional 24 h. Levels of p23 and GAPDH in cell lysates were examined by immunoblotting. (D) Levels of p23 were quantified and normalized to GAPDH, and analysis by two-way ANOVA revealed a significant effect of M344 leading to a decrease in p23 (F= 52.6; P< 0.0001), as well as a significant treatment effect of 17AAG (F= 9.6; P= 0.01). All data are presented as mean ± SEM (n= 3). **P< 0.001, *P< 0.01.
Inhibition of HDAC6 decreases tau levels in primary neuronal cultures
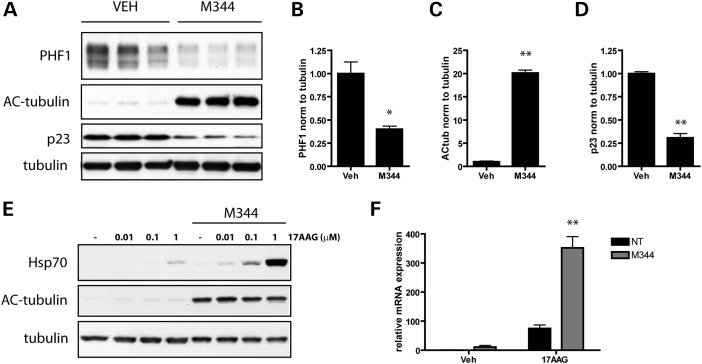
To evaluate the significance of these findings to neurodegenerative diseases characterized by the accumulation of tau in neurons of the brain, the impact of HDAC6 inhibition on tau levels was examined in primary neuronal cultures. Consistent with the results above, a significant decrease in PHF1-positive tau levels was observed in neurons treated with M344 (Fig. 6A and B). Treatment with M344 also led to an increase in the acetylation of tubulin, confirming that M344 did in fact inhibit the deacetylase activity of HDAC6 (Fig. 6C). A significant decrease in p23 levels was also seen upon exposure to M344 (Fig. 6D), suggesting that M344 drives the formation of Hsp90 chaperone complexes favoring degradation, which is sufficient to promote tau clearance in neurons. In addition, consistent with reports demonstrating that the loss of HDAC6 activity increases the affinity of Hsp90 for Hsp90 inhibitors (21,22), we observed an enhanced induction of the inducible Hsp70 isoform in response to 17AAG in the presence of M344 (Fig. 6E and F). Given that the increased induction of inducible Hsp70 is observed at both the RNA and protein levels, this indicates the upregulation is due to increased HSF1-mediated gene transcription, and not due to an altered half-life of Hsp70.
Figure 6.
HDAC6 inhibition decreases tau levels in primary neuronal cultures. (A) Primary neurons from non-transgenic mice (129SVE × FVB) were cultured for 9 days, treated with vehicle (50% ethanol) or M344 (1 μm) for 24 h and harvested on the 10th day in vitro (DIV). Cell lysates were immunoblotted for PHF1-tau, acetylated tubulin, p23, and total tubulin. (B) Treatment with 1 μm M344 for 24h led to a significant decrease in PHF1-tau levels in primary neurons (t= 4.7; P= 0.01). (C) Assessment of acetylated-tubulin levels verified that M344 does inhibit the deacetylase activity of HDAC6 and promotes increased acetylation of tubulin (t= 29.7; P< 0.0001). (D) Levels of p23 were also quantified and normalized to tubulin, and statistical analysis revealed a significant effect of M344 leading to a decrease in p23 (t= 13.74; P= 0.0002). (E) On DIV9, primary neurons were treated with vehicle (50% ethanol) or M344 (1 μm) for 1 h, and subsequently treated with vehicle (DMSO) or the indicated concentration of 17AAG for an additional 23 h. Cell lysates were immunoblotted for Hsp70, acetylated tubulin and total tubulin. (F) HSPA1A mRNA expression was evaluated in primary neuronal cultures pretreated with vehicle (50% ethanol) or M344 (1 μm) for 1 h and subsequently treated with vehicle (DMSO) or 17AAG (1 μm) for an additional 23 h. GAPDH was used as an endogenous loading control for each sample (n= 3 per condition), and each sample was analyzed in quadruplicate. Results were analyzed by two-way ANOVA, revealing a significant effect of 17AAG (F= 62.31; P< 0.0001), as well as a significant effect of M344 (F= 29.82; P< 0.0001) on Hsp70 mRNA levels. All data are presented as mean ± SEM (n= 3). **P≤ 0.0002, *P= 0.01.
DISCUSSION
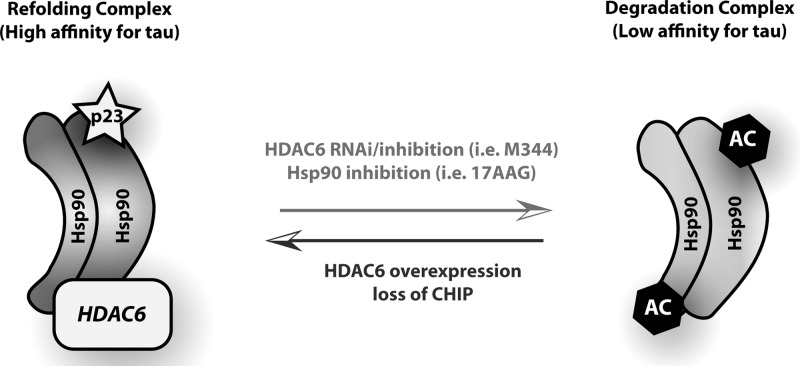
CHIP is intimately involved in the mechanisms of Hsp-mediated client degradation, and seems to be an integral participant in the removal of hyperphosphorylated tau. It has a unique binding affinity for tau phosphorylated at disease-specific phospho-epitopes, and is required for the ubiquitination of these tau species, leading to their proteasome-mediated degradation (6–8). In addition to directly promoting tau clearance by ubiquitination, CHIP may also facilitate tau degradation by abrogating the protein folding function of Hsp90. Indeed, CHIP has been shown to elicit the release of p23 from Hsp90 upon binding (32), thereby causing a shift in the function of Hsp90 from refolding client proteins to promoting their degradation. We have now identified an additional, novel mechanism by which CHIP regulates protein triage decisions involving Hsp90. Specifically, our data suggest that CHIP facilitates tau clearance by decreasing the steady-state levels of HDAC6, thereby increasing the sensitivity of Hsp90 to 17-AAG through acetylation and promoting client degradation (Fig. 7).
Figure 7.
CHIP and HDAC6 levels modulate Hsp90 chaperone complex. Schematic diagram depicting how CHIP and HDAC6 levels influence formation of an Hsp90 chaperone complex either favoring refolding or degradation of client proteins, and the resulting impact on tau accumulation. Specifically, decreased HDAC6 activity favors formation of a ‘degradation’ complex, driving tau clearance. Conversely, upregulation of HDAC6 or loss of CHIP promotes formation of a ‘refolding’ complex, which leads to tau accumulation.
In this report, we present evidence demonstrating that CHIP interacts with and ubiquitinates HDAC6 (Fig. 1). HDAC6 likely undergoes CHIP-mediated proteasomal degradation, given that knocking-down CHIP in cultured cells and knocking-out CHIP in mice both result in elevated HDAC6 levels (Fig. 2). Increases in HDAC6, in turn, have detrimental consequences on tau levels. Indeed, we show that HDAC6 overexpression promotes the accumulation of tau (Fig. 3); conversely, HDAC6 deficiency decreases tau (Figs 4 and 6). We propose that changes in HDAC6 indirectly influence whether or not tau is degraded by modulating the acetylation state of Hsp90. As mentioned, Hsp90 hyperacetylation promotes client degradation (18,21). Since Hsp90 is a substrate for the deacetylase activity of HDAC6, loss of HDAC6 expression would promote Hsp90 acetylation, thereby favoring tau degradation. In contrast, an increase in HDAC6 would decrease Hsp90 acetylation, and therefore cause a decrease in tau degradation. Given that HDAC6 can form an Hsp90/HDAC6/HSF1 complex (33,34), it is possible that alterations in HDAC6 expression disrupt Hsp90 chaperone function due to changes in complex formation, and not due to HDAC6-mediated deacetylation of Hsp90. However, because the catalytically inactive HDAC6 mutant (H216A/H611A) fails to promote tau accumulation (Fig. 3D), it is most likely that HDAC6-mediated tau accumulation is dependent on the deacetylase function of HDAC6.
Not only do HDAC6 depletion and inhibition enhance tau clearance, they also augment Hsp90 inhibitor-mediated tau degradation (Figs 4 and 6). This is consistent with findings that hyperacetylation of Hsp90 due to HDAC6 depletion leads to an increased binding affinity of Hsp90 for Hsp90 inhibitors (21,22). Indeed, we show that Hsp70 induction, a consequence of effective Hsp90 inhibition by 17AAG, is enhanced in primary neurons treated with the HDAC6 inhibitor, M344 (Fig. 6E). That a similar increase in 17AAG-mediated induction of Hsp70 following HDAC6 inhibition in HeLa cells was not observed (Fig. 4) is most likely due to the difference in basal levels of inducible Hsp70 between HeLa cells (in which it is high) and primary neurons (in which it is low).
Currently, modulation of the Hsp90 chaperone complex with chemical inhibitors to promote the degradation of Hsp90 client proteins is being tested in clinical trials as a therapeutic approach for the treatment of cancer (reviewed in 35). In cancer, cell death pathways are often blocked by the overexpression of prosurvival Hsp90 client proteins that promote uncontrolled cell proliferation (16,36,37). Hsp90 inhibition results in the ubiquitination and proteasomal degradation of these deleterious client proteins. It is noteworthy that Hsp90 chaperone complexes from tumor cells exhibit a high affinity for Hsp90 inhibitors when compared with Hsp90 chaperone complexes from normal cells (38,39), demonstrating that treatment with Hsp90 inhibitors can be used to selectively target diseased cells. These findings form the basis for the use of Hsp90 inhibitors in the treatment of cancer. Of importance, there is a significant amount of evidence indicating that Hsp90 inhibitors may also hold clinical relevance for the treatment of neurodegenerative diseases characterized by abnormal tau accumulation. For instance, the increase in phosphorylated tau associated with disease, as well as the ability of Hsp90 inhibition to promote the preferential degradation of phosphorylated tau species in pathogenic models, conclusively identifies tau as an Hsp90 client protein, and further implicates Hsp90 dysfunction in the pathogenesis of disease (9,14,27,28). Moreover, the demonstration that the Hsp90 chaperone complex exhibits a high binding affinity for Hsp90 inhibitors only in affected brain regions of AD patients, and not in unaffected regions (9), suggests that similarly to cancer, Hsp90 inhibitors can be used to selectively target Hsp90 chaperone complexes in diseased areas of the brain. Certainly, with the potential clinical applications of Hsp90 inhibitors for tauopathies, deciphering the mechanisms that cause Hsp90 to adopt a high affinity state for Hsp90 inhibitors is of considerable interest. Hsp90 has a higher binding affinity for Hsp90 inhibitors when acetylated (21,22), and we hypothesize that CHIP modulates the affinity of Hsp90 for Hsp90 inhibitors by regulating levels of the deacetylase, HDAC6. In addition to ubiquitinating clients for targeting to the proteasome (6–8), CHIP may regulate protein triage decisions by modulating the folding and degradation functions of Hsp90 via HDAC6.
Given that a treatment paradigm combining HDAC6 and Hsp90 inhibition has been shown to synergistically and selectively augment Hsp90 client degradation in leukemia cells but not normal cells (21), it is possible that a similar approach could be utilized in neurodegenerative disease to selectively target Hsp90 chaperone complexes present in cells expressing pathological client proteins, such as phosphorylated tau. While exploring the therapeutic benefit of modulating HDAC6 function, the relationship among tau pathology, HDAC6 activity and Hsp90 acetylation should be more fully characterized. It has been demonstrated that tau overexpression inhibits HDAC6 in a cell culture model (40). Additionally, the overexpression of tau leads to an increase in tubulin acetylation (41), which is indicative of a loss of HDAC6 function. By inhibiting HDAC6, tau may indirectly regulate the acetylation of Hsp90. Given that increased acetylation of Hsp90 increases the binding affinity of the Hsp90 chaperone complex for Hsp90 inhibitors, it is possible that the increased binding affinity of the Hsp90 chaperone complex observed in affected regions of AD brain (9) could be due to hyperacetylation of Hsp90 as a result of pathological tau-mediated inhibition of HDAC6. However, the level of Hsp90 acetylation in affected areas of the AD brain remains to be determined. Regardless, given the importance of protein quality control in neurodegenerative diseases, the current findings will not only benefit the tau field, but are also expected to have broad implications for all diseases characterized by the accumulation and aggregation of abnormal Hsp90 client proteins.
MATERIALS AND METHODS
Antibodies, chemicals and reagents
PHF1 (1:500; anti-S396/S404 p-tau) was provided by P. Davies (Albert Einstein College of Medicine, New York, NY, USA). Anti-myc (1:1000) and anti-HA (1:1000) were obtained from Roche Applied Science. Anti-HDAC6 (1:1000) was purchased from Millipore. Anti-GFP (1:2000) and anti-V5 (1:1000) were obtained from Life Technologies. Anti-actin (1:10 000) was obtained from Sigma Aldrich. Anti-hsp70 (1:1000) was obtained from Enzo Life Sciences. Anti-GAPDH (1:10 000) was obtained from Meridian Life Science, Inc. Anti-p23 (1:1000) was obtained from Fisher Scientific. Anti-CHIP (1:1000) and E1 (1:1000; human specific tau antibody) were generated by our group. 17AAG was purchased from A.G. Scientific, M344 and cycloheximide were obtained from Sigma-Aldrich and MG132 was purchased from Cayman Chemical.
All siRNAs were obtained from Qiagen, with the proprietary sequence of the non-silencing control not disclosed by the company. The target sequence for siCHIP (STUB1) is TGCCGCCACTATCTGTGTAAT, and the target sequence for siHDAC6 is CACCGTCAACGTGGCATGGAA. siRNA efficiency for protein knockdown was validated by western blot.
Expression plasmids
GFP, untagged and myc-tagged CHIP, P301L-V5 tau and HA-ubiquitin were generated by our lab. A human HDAC6 clone in a pCMV SPORT6 vector was purchased from Life Technologies. The QuikChange Mutagenesis kit (Stratagene) was utilized to generate a catalytically inactive HDAC6 mutant (H216A/H611A) following the manufacturer's protocol. The QuikChange Mutagenesis kit was also utilized to generate K30A and H260Q mutations in myc-CHIP. The integrity of all constructs was confirmed by automated sequencing.
Cell culture and transient transfections
HeLa and HEK-293T were maintained in Opti-Mem (Life Technologies) supplemented with 10% heat-inactivated FBS (Life Technologies) and passaged every 3–4 days based on 90% confluency.
siRNA experiments were carried out in six-well plates using human gene-specific validated and genome-wide siRNAs. Final siRNA concentration per well was 20 nm in Opti-Mem, with 4 μl of siLentFect transfection reagent (Bio-Rad) used per well. This mixture was incubated in a final volume of 500 μl for 20 min and then added to 40–50% confluent HeLa cells in six-well plates for a final in-well volume of 2 ml. Forty-eight hours after transfection, the complete medium was removed and replaced with serum-free Opti-Mem for subsequent plasmid transfection.
For plasmid transfections, 1–2 μg plasmid DNA was combined with Lipofectamine 2000 reagent for 15 min in 500 μl Opti-Mem, and this mixture was added to either untreated or siRNA-transfected cells for 4h. The transfection mixture was then replaced with fresh complete media.
Immunoprecipitation
For coimmunoprecipitation studies, 90% of confluent cells in 10 cm dishes were transfected using Lipofectamine 2000 with the combinations of HA-ubiquitin, HDAC6, myc-HDAC6, GFP, CHIP and myc-CHIP (WT, K30A and H260Q mutants) for 4 h and then maintained in complete media for 48 h. Cells were collected in coimmunoprecipitation buffer (50 mm Tris, 274 mm NaCl, 5 mm KCl, 5 mm EDTA, 1% Triton X-100, 1 mm PMSF and a protease and phosphatase inhibitor cocktail), supernatants were precleared with protein G and subsequently incubated with anti-myc antibody or anti-HDAC6 antibody and 20 μl protein G overnight at 4°C. Protein G beads were washed three times in coimmunoprecipitation buffer, resuspended in 2× tris-glycine SDS sample buffer with 5% beta-mercaptoethanol, heated for 5min at 95°C and subjected to western blot analyses following SDS–PAGE electrophoresis.
Mice
CHIP–/– mice were generated as previously described (42). CHIP–/– and CHIP+/+ mice were humanely euthanized at 30 days of age, and their brains were quickly removed for dissection and frozen on dry ice for subsequent biochemical analyses. Brain tissue was homogenized in buffer containing 50 mm Tris, 274 mm NaCl, 5 mm KCl, 5 mm EDTA, 1% SDS, 1% Triton X-100, 1 mm PMSF and a protease and phosphatase inhibitor cocktail (pH 7.4). Homogenates were centrifuged at 15 400g for 15 min, and supernatants were collected and subjected to western blot analyses.
Sample preparation and immunoblotting procedure
Cell samples for immunoblotting were suspended in 100–200 µl of homogenate buffer [50 mm Tris–HCl, pH 7.4, 300 mm NaCl, 5 mm EDTA, 1% Triton X-100, 1% SDS, 1 mm PMSF, protease inhibitor cocktail, phosphatase inhibitors I and II (Fisher Scientific)]. Mice brains were weighed and homogenized in 10× volume of homogenate buffer. All samples were sonicated, and following sonication, samples were centrifuged at 16 000g for 15 min, and a BCA protein assay (Fisher Scientific) performed on the supernatant. Thirty microgram of protein from each sample was diluted in dH2O, 2× tris-glycine SDS sample buffer (Life Technologies), 5% beta-mercaptoethanol (Sigma Aldrich) and heat-denatured for 5 min at 95°C. Samples were run on 4–20% tris-glycine gels (Life Technologies), and transferred to PVDF membrane (Millipore). Membranes were blocked in 5% milk in TBS/0.1% Triton X-100, and incubated overnight in primary antibody diluted in 5% milk in TBS/0.1% Triton X-100 rocking at 4°C. Membranes were incubated in HRP-conjugated secondary antibodies (1:5000; Jackson Immuno) for 1 h at room temperature, and detected by ECL (PerkinElmer).
Primary neuronal culture
For primary neuronal cultures, hippocampi from postnatal day 2 mouse pups were removed and stored at 4°C in HIBERNATE™ A media without calcium (BrainBits), supplemented with B27 (Life Technologies), 0.5 mm GMAX (Life Technologies) and gentamicin (Life Technologies). Excised hippocampi were digested in papain (1 mg/ml; Fisher Scientific), triturated with a Pasteur pipet (bore size 0.8–1 mm), centrifuged to collect cell pellet, and resuspended in Neurobasal A (Life Technologies), supplemented with B27, GMAX, gentamicin and bFGF (Life Technologies). Following determination of cell number, neurons were plated on poly-D-lysine-coated cover slips within 24-well plates for immunocytochemical studies (seeded at a density of 2.5 × 104 cells/cover slip), or seeded at a density of 3 × 105 cells/well on poly-D-lysine-coated six-well plates for immunoblotting.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by Mayo Clinic Foundation (L.P.), National Institutes of Health/National Institute on Aging [5R01AG026251-04 (L.P.) and AG17216-10JP3 (L.P.)], National Institutes of Health/National Institute of Neurological Disorders and Stroke [R01 NS 063964-01 (L.P.), R01 NS077402 (L.P.)], PSP Foundation (L.P.) and ADRC AG016574 (C.C. and T.F.G.). Funding to pay the Open Access publication charges for this article was provided by Mayo Clinic.
Supplementary Material
ACKNOWLEDGEMENTS
Dr Cam Patterson kindly provided the CHIP knockout mice which were used in our previous studies (4,5). Dr Peter Davies kindly provided us with the PHF-1 antibodies.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Drubin D.G., Feinstein S.C., Shooter E.M., Kirschner M.W. Nerve growth factor-induced neurite outgrowth in PC12 cells involves the coordinate induction of microtubule assembly and assembly-promoting factors. J. Cell Biol. 1985;101:1799–1807. doi: 10.1083/jcb.101.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esmaeli-Azad B., McCarty J.H., Feinstein S.C. Sense and antisense transfection analysis of tau function: tau influences net microtubule assembly, neurite outgrowth and neuritic stability. J. Cell Sci. 1994;107:869–879. doi: 10.1242/jcs.107.4.869. [DOI] [PubMed] [Google Scholar]
- 3.Stamer K., Vogel R., Thies E., Mandelkow E., Mandelkow E.M. Tau blocks traffic of organelles, neurofilaments, and APP vesicles in neurons and enhances oxidative stress. J. Cell Biol. 2002;156:1051–1063. doi: 10.1083/jcb.200108057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buee L., Bussiere T., Buee-Scherrer V., Delacourte A., Hof P.R. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res. Brain Res. Rev. 2000;33:95–130. doi: 10.1016/s0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 5.Dickson D.W. Tau and synuclein and their role in neuropathology. Brain Pathol. 1999;9:657–661. doi: 10.1111/j.1750-3639.1999.tb00548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petrucelli L., Dickson D., Kehoe K., Taylor J., Snyder H., Grover A., De Lucia M., McGowan E., Lewis J., Prihar G., et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum. Mol. Genet. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 7.Shimura H., Schwartz D., Gygi S.P., Kosik K.S. CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J. Biol. Chem. 2004;279:4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- 8.Dickey C.A., Yue M., Lin W.L., Dickson D.W., Dunmore J.H., Lee W.C., Zehr C., West G., Cao S., Clark A.M., et al. Deletion of the ubiquitin ligase CHIP leads to the accumulation, but not the aggregation, of both endogenous phospho- and caspase-3-cleaved tau species. J. Neurosci. 2006;26:6985–6996. doi: 10.1523/JNEUROSCI.0746-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickey C.A., Kamal A., Lundgren K., Klosak N., Bailey R.M., Dunmore J., Ash P., Shoraka S., Zlatkovic J., Eckman C.B., et al. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J. Clin. Invest. 2007;117:648–658. doi: 10.1172/JCI29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchner J. Hsp90 & Co.—a holding for folding. Trends Biochem. Sci. 1999;24:136–141. doi: 10.1016/s0968-0004(99)01373-0. [DOI] [PubMed] [Google Scholar]
- 11.Welch W.J., Feramisco J.R. Purification of the major mammalian heat shock proteins. J. Biol. Chem. 1982;257:14949–14959. [PubMed] [Google Scholar]
- 12.Pratt W.B., Gehring U., Toft D.O. Molecular chaperoning of steroid hormone receptors. EXS. 1996;77:79–95. doi: 10.1007/978-3-0348-9088-5_6. [DOI] [PubMed] [Google Scholar]
- 13.Lattouf J.B., Srinivasan R., Pinto P.A., Linehan W.M., Neckers L. Mechanisms of disease: the role of heat-shock protein 90 in genitourinary malignancy. Nat. Clin. Pract. Urol. 2006;3:590–601. doi: 10.1038/ncpuro0604. [DOI] [PubMed] [Google Scholar]
- 14.Dickey C.A., Dunmore J., Lu B., Wang J.W., Lee W.C., Kamal A., Burrows F., Eckman C., Hutton M., Petrucelli L. HSP induction mediates selective clearance of tau phosphorylated at proline-directed Ser/Thr sites but not KXGS (MARK) sites. FASEB J. 2006;20:753–755. doi: 10.1096/fj.05-5343fje. [DOI] [PubMed] [Google Scholar]
- 15.Kamal A., Boehm M.F., Burrows F.J. Therapeutic and diagnostic implications of Hsp90 activation. Trends Mol. Med. 2004;10:283–290. doi: 10.1016/j.molmed.2004.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isaacs J.S., Xu W., Neckers L. Heat shock protein 90 as a molecular target for cancer therapeutics. Cancer Cell. 2003;3:213–217. doi: 10.1016/s1535-6108(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 17.Bali P., Pranpat M., Bradner J., Balasis M., Fiskus W., Guo F., Rocha K., Kumaraswamy S., Boyapalle S., Atadja P., et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J. Biol. Chem. 2005;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 18.Kovacs J.J., Murphy P.J., Gaillard S., Zhao X., Wu J.T., Nicchitta C.V., Yoshida M., Toft D.O., Pratt W.B., Yao T.P. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol. Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 19.Murphy P.J., Morishima Y., Kovacs J.J., Yao T.P., Pratt W.B. Regulation of the dynamics of hsp90 action on the glucocorticoid receptor by acetylation/deacetylation of the chaperone. J. Biol. Chem. 2005;280:33792–33799. doi: 10.1074/jbc.M506997200. [DOI] [PubMed] [Google Scholar]
- 20.Scroggins B.T., Robzyk K., Wang D., Marcu M.G., Tsutsumi S., Beebe K., Cotter R.J., Felts S., Toft D., Karnitz L., et al. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol. Cell. 2007;25:151–159. doi: 10.1016/j.molcel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao R., Fiskus W., Yang Y., Lee P., Joshi R., Fernandez P., Mandawat A., Atadja P., Bradner J.E., Bhalla K. HDAC6 inhibition enhances 17-AAG–mediated abrogation of hsp90 chaperone function in human leukemia cells. Blood. 2008;112:1886–1893. doi: 10.1182/blood-2008-03-143644. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y., Rao R., Shen J., Tang Y., Fiskus W., Nechtman J., Atadja P., Bhalla K. Role of acetylation and extracellular location of heat shock protein 90alpha in tumor cell invasion. Cancer Res. 2008;68:4833–4842. doi: 10.1158/0008-5472.CAN-08-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amolins M.W., Blagg B.S. Natural product inhibitors of Hsp90: potential leads for drug discovery. Mini Rev. Med. Chem. 2009;9:140–152. doi: 10.2174/138955709787316056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.George P., Bali P., Annavarapu S., Scuto A., Fiskus W., Guo F., Sigua C., Sondarva G., Moscinski L., Atadja P., et al. Combination of the histone deacetylase inhibitor LBH589 and the hsp90 inhibitor 17-AAG is highly active against human CML-BC cells and AML cells with activating mutation of FLT-3. Blood. 2005;105:1768–1776. doi: 10.1182/blood-2004-09-3413. [DOI] [PubMed] [Google Scholar]
- 25.Xu W., Marcu M., Yuan X., Mimnaugh E., Patterson C., Neckers L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl Acad. Sci. USA. 2002;99:12847–12852. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding H., Dolan P.J., Johnson G.V. Histone deacetylase 6 interacts with the microtubule-associated protein tau. J. Neurochem. 2008;106:2119–2130. doi: 10.1111/j.1471-4159.2008.05564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo W., Dou F., Rodina A., Chip S., Kim J., Zhao Q., Moulick K., Aguirre J., Wu N., Greengard P., et al. Roles of heat-shock protein 90 in maintaining and facilitating the neurodegenerative phenotype in tauopathies. Proc. Natl Acad. Sci. USA. 2007;104:9511–9516. doi: 10.1073/pnas.0701055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dickey C.A., Eriksen J., Kamal A., Burrows F., Kasibhatla S., Eckman C.B., Hutton M., Petrucelli L. Development of a high throughput drug screening assay for the detection of changes in tau levels – proof of concept with HSP90 inhibitors. Curr. Alzheimer Res. 2005;2:231–238. doi: 10.2174/1567205053585927. [DOI] [PubMed] [Google Scholar]
- 29.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 30.Heltweg B., Dequiedt F., Marshall B.L., Brauch C., Yoshida M., Nishino N., Verdin E., Jung M. Subtype selective substrates for histone deacetylases. J. Med. Chem. 2004;47:5235–5243. doi: 10.1021/jm0497592. [DOI] [PubMed] [Google Scholar]
- 31.McLaughlin S.H., Sobott F., Yao Z.P., Zhang W., Nielsen P.R., Grossmann J.G., Laue E.D., Robinson C.V., Jackson S.E. The co-chaperone p23 arrests the Hsp90 ATPase cycle to trap client proteins. J. Mol. Biol. 2006;356:746–758. doi: 10.1016/j.jmb.2005.11.085. [DOI] [PubMed] [Google Scholar]
- 32.Connell P., Ballinger C.A., Jiang J., Wu Y., Thompson L.J., Hohfeld J., Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 33.Boyault C., Zhang Y., Fritah S., Caron C., Gilquin B., Kwon S.H., Garrido C., Yao T.P., Vourc'h C., Matthias P., et al. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 2007;21:2172–2181. doi: 10.1101/gad.436407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westerheide S.D., Morimoto R.I. Heat shock response modulators as therapeutic tools for diseases of protein conformation. J. Biol. Chem. 2005;280:33097–33100. doi: 10.1074/jbc.R500010200. [DOI] [PubMed] [Google Scholar]
- 35.Kim Y.S., Alarcon S.V., Lee S., Lee M.J., Giaccone G., Neckers L., Trepel J.B. Update on Hsp90 inhibitors in clinical trial. Curr. Top. Med. Chem. 2009;9:1479–1492. doi: 10.2174/156802609789895728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu W., Neckers L. Targeting the molecular chaperone heat shock protein 90 provides a multifaceted effect on diverse cell signaling pathways of cancer cells. Clin. Cancer Res. 2007;13:1625–1629. doi: 10.1158/1078-0432.CCR-06-2966. [DOI] [PubMed] [Google Scholar]
- 37.Kamal A., Burrows F.J. Hsp90 inhibitors as selective anticancer drugs. Discov. Med. 2004;4:277–280. [PubMed] [Google Scholar]
- 38.Biamonte M.A., Shi J., Hong K., Hurst D.C., Zhang L., Fan J., Busch D.J., Karjian P.L., Maldonado A.A., Sensintaffar J.L., et al. Orally active purine-based inhibitors of the heat shock protein 90. J. Med. Chem. 2006;49:817–828. doi: 10.1021/jm0503087. [DOI] [PubMed] [Google Scholar]
- 39.Kamal A., Thao L., Sensintaffar J., Zhang L., Boehm M.F., Fritz L.C., Burrows F.J. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 40.Perez M., Santa-Maria I., Gomez de Barreda E., Zhu X., Cuadros R., Cabrero J.R., Sanchez-Madrid F., Dawson H.N., Vitek M.P., Perry G., et al. Tau–an inhibitor of deacetylase HDAC6 function. J. Neurochem. 2009;109:1756–1766. doi: 10.1111/j.1471-4159.2009.06102.x. [DOI] [PubMed] [Google Scholar]
- 41.Takemura R., Okabe S., Umeyama T., Kanai Y., Cowan N.J., Hirokawa N. Increased microtubule stability and alpha tubulin acetylation in cells transfected with microtubule-associated proteins MAP1B, MAP2 or tau. J. Cell. Sci. 1992;103:953–964. doi: 10.1242/jcs.103.4.953. [DOI] [PubMed] [Google Scholar]
- 42.Dai Q., Zhang C., Wu Y., McDonough H., Whaley R.A., Godfrey V., Li H.H., Madamanchi N., Xu W., Neckers L., Cyr D., Patterson C. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003;22:5446–5458. doi: 10.1093/emboj/cdg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.