Abstract
Lipoprotein lipase (LPL) is a 448-amino-acid head-to-tail dimeric enzyme that hydrolyzes triglycerides within capillaries. LPL is secreted by parenchymal cells into the interstitial spaces; it then binds to GPIHBP1 (glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1) on the basolateral face of endothelial cells and is transported to the capillary lumen. A pair of amino acid substitutions, C418Y and E421K, abolish LPL binding to GPIHBP1, suggesting that the C-terminal portion of LPL is important for GPIHBP1 binding. However, a role for LPL's N terminus has not been excluded, and published evidence has suggested that only full-length homodimers are capable of binding GPIHBP1. Here, we show that LPL's C-terminal domain is sufficient for GPIHBP1 binding. We found, serendipitously, that two LPL missense mutations, G409R and E410V, render LPL susceptible to cleavage at residue 297 (a known furin cleavage site). The C terminus of these mutants (residues 298–448), bound to GPIHBP1 avidly, independent of the N-terminal fragment. We also generated an LPL construct with an in-frame deletion of the N-terminal catalytic domain (residues 50–289); this mutant was secreted but also was cleaved at residue 297. Once again, the C-terminal domain (residues 298–448) bound GPIHBP1 avidly. The binding of the C-terminal fragment to GPIHBP1 was eliminated by C418Y or E421K mutations. After exposure to denaturing conditions, the C-terminal fragment of LPL refolds and binds GPIHBP1 avidly. Thus, the binding of LPL to GPIHBP1 requires only the C-terminal portion of LPL and does not depend on full-length LPL homodimers.
INTRODUCTION
GPIHBP1 (glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1) binds lipoprotein lipase (LPL) in the subendothelial spaces and transports it to the capillary lumen, where it hydrolyzes triglycerides within the plasma lipoproteins (1,2). GPIHBP1 has two noteworthy domains—an N-terminal acidic domain enriched in aspartates and glutamates and a cysteine-rich lymphocyte antigen 6 domain (3,4). Mutagenesis studies have established that both of these domains are crucial for GPIHBP1's ability to bind to LPL (5–7). LPL is secreted as a homodimeric enzyme, with each partner monomer consisting of an N-terminal catalytic domain (amino acids 1–312) and a C-terminal region (amino acids 313–448) that plays a role in binding triglyceride substrates (8–11). LPL monomers are catalytically inactive (12,13). Evidence from human genetics suggests that LPL's C terminus plays a role in LPL–GPIHBP1 interactions: a pair of missense mutations in LPL's C-terminal domain, C418Y and E421K, abolishes LPL's capacity to bind to GPIHBP1 (14).
Although some of the sequences necessary for GPIHBP1–LPL interactions are beginning to come into focus (5–7), many questions remain. Certain amino acids within the C-terminal domain of LPL appear important for GPIHBP1 binding, but a role for LPL's N terminus has not been excluded. Also unclear is whether LPL dimers are required for LPL–GPIHBP1 interactions. LPL dimerizes in a head-to-tail fashion; the best evidence for this arrangement is that a single molecule containing two consecutive LPL open reading frames, separated by a 6-amino-acid spacer, is enzymatically active (15). Those experiments implied that the N- and C-terminal ends of LPL are in close proximity, and experiments have implied that the C-terminal triglyceride-binding sequences from one monomer capture triglyceride substrates for cleavage by the N-terminal catalytic domain of the partner monomer (11). At this point, no one knows whether the N- and C-terminal domains both contribute to LPL's GPIHBP1-binding site.
Two considerations led us to suspect that GPIHBP1 might bind only LPL dimers. The first was purely teleological—we were skeptical that nature would devise a system that would allow catalytically inactive LPL monomers to be bound by GPIHBP1 and transported to the capillary lumen. The second consideration was experimental: subjecting LPL dimers to denaturing conditions (which promotes monomer formation) abolishes LPL binding to GPIHBP1 (16). Neither consideration is conclusive; additional experiments are needed to explore this concept.
In the current study, we asked whether the C-terminal portion of LPL, independent of the N-terminal domain, might be capable of binding to GPIHBP1. Addressing this question was facilitated, serendipitously, by the discovery that certain LPL missense mutations render LPL susceptible to an endoproteolytic cleavage event that separates LPL's N- and C-terminal domains.
RESULTS
Amino acid substitutions at LPL residues 409 and 410 render LPL susceptible to endoproteolytic cleavage
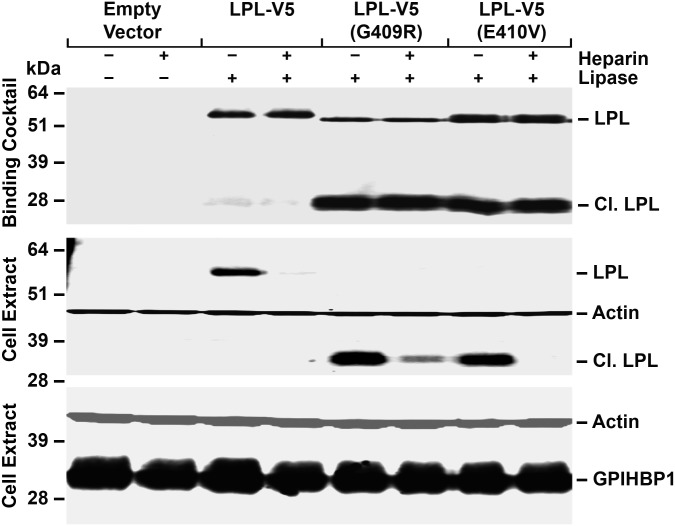
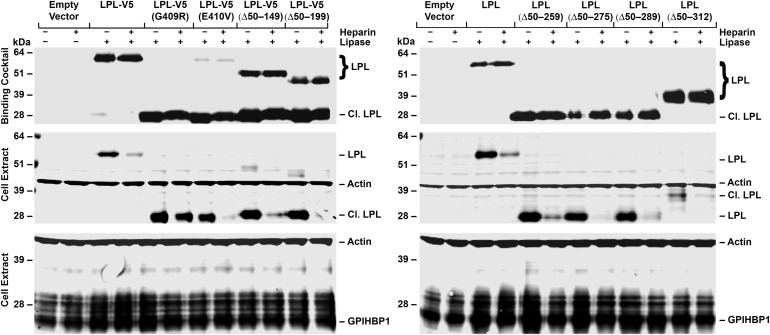
In an earlier study (14), we reported that LPL, C418Y and E421K, mutations abolish LPL's ability to bind to GPIHBP1 and do so without affecting catalytic activity. Two other C-terminal mutations, G409R and E410V, have also been identified in the setting of chylomicronemia (17,18). To define mechanisms underlying the G409R and E410V mutations, we introduced those mutations into the LPL-V5 construct. After expressing the mutant LPL-V5 constructs in CHO-K1 cells, we found that both G409R and E410V mutations rendered LPL highly susceptible to an endoproteolytic cleavage event that releases a 28-kDa C-terminal fragment (containing the V5 tag). This C-terminal fragment bound to GPIHBP1-expressing CHO-K1 cells avidly, as did full-length wild-type LPL-V5 (Fig. 1). The binding of both the full-length LPL-V5 and the 28-kDa C-terminal fragment to GPIHBP1 was inhibited by heparin. [The extent of inhibition of binding by heparin was generally less with the G409R mutation, possibly because that mutation introduces a positively charged residue near LPL's principal heparin-binding domain (residues 403, 405 and 407) (19).] Similar results were obtained in an independent experiment (Supplementary Material, Fig. S1).
Figure 1.
Western blot assay of LPL binding to GPIHBP1-expressing CHO-K1 cells. We examined the binding of LPL with a C-terminal V5 tag (LPL-V5) and variants of the same construct with G409R or E410V mutations. The G409R and E410V mutations render LPL susceptible to an endoproteolytic cleavage event that releases a ∼28-kDa C-terminal fragment (Cl. LPL, which includes the V5 tag). The medium from LPL-transfected cells (or medium alone) was incubated with CHO-K1 cells expressing S-protein-tagged GPIHBP1 at 4°C in the presence or the absence of heparin (500 U/ml). After 2 h, cell extracts were prepared and western blots were performed. GPIHBP1 was detected with a rabbit antibody against the S-protein tag; LPL was detected with a mouse antibody against the V5 tag. Antibodies against actin were used as a loading control. The relative amounts of LPL added to the cells (the binding cocktail) were assessed with an antibody against the V5 tag (top panel). The intensities of band signals were quantified with a Li-Cor Odessey scanner. The binding of the C-terminal fragment of LPL-V5 (G409R) to GPIHBP1 in the absence of heparin was 3.3 times greater (relative to GPIHBP1) than the binding of full-length LPL-V5 to GPIHBP1. The binding of the C-terminal fragment of LPL-V5 (E410V) to GPIHBP1 in the absence of heparin was 3.1 times greater (relative to the GPIHBP1 signal) than the binding of LPL-V5 to GPIHBP1.
We suspected that the 28-kDa fragment from LPL-V5 (G409R) and LPL-V5 (E410V) resulted from an increased susceptibility to a known furin cleavage site at LPL residue 297 (20,21). To test this possibility, CHO-K1 cells were transfected with LPL-V5, LPL-V5 (G409R) and LPL-V5 (E410V) constructs in the presence or the absence of a furin inhibitor CMK. CMK had no effect on the secretion of LPL-V5 from cells but abolished the secretion of the 28-kDa fragment from LPL-V5 (G409R) and LPL-V5 (E410V) (Supplementary Material, Fig. S2). Introducing an R297A mutation into the constructs [a mutation that blocks the furin cleavage event (20)] had no effect on the secretion of LPL-V5 but abolished the secretion of the 28-kDa fragment from LPL-V5 (G409R) and LPL-V5 (E410V) transfected cells (Supplementary Material, Fig. S2).
The C-terminal cleavage fragment of LPL binds to GPIHBP1
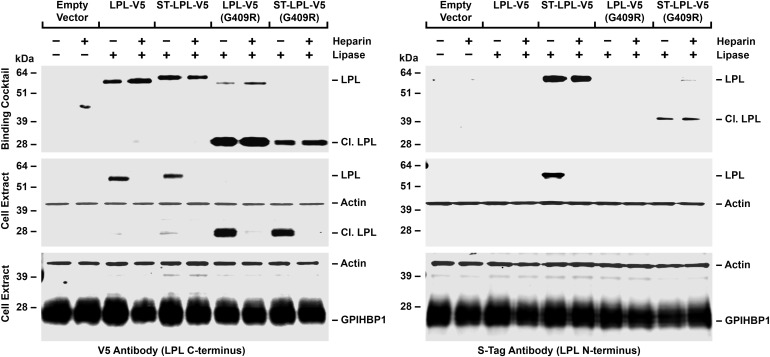
To determine if the other half of LPL-V5 (G409R) (i.e. the N-terminal fragment) is also secreted from cells, and if so, whether it binds to GPIHBP1, we created an LPL-V5 construct containing an N-terminal S-protein tag (ST-LPL-V5). The full-length ST-LPL-V5 was capable of binding to GPIHBP1 as it was detected in cell extracts by both the V5 and S-protein antibodies (Fig. 2). LPL-V5 (G409R) and ST-LPL-V5 (G409R) were susceptible to furin cleavage, releasing the same 28-kDa C-terminal fragment (detected with the V5 antibody) and a 39-kDa N-terminal fragment (detected with the S-protein antibody) (Fig. 2). The C-terminal fragment, but not the N-terminal fragment, bound to GPIHBP1 (Fig. 2), indicating that the two segments of mutant LPL did not remain attached to each other.
Figure 2.
Western blot assays of LPL binding to GPIHBP1-expressing CHO-K1 cells. Cells were incubated at 4°C for 2 h with medium alone or medium containing LPL-V5, a modified LPL-V5 construct with an N-terminal S-protein tag (ST-LPL-V5), LPL-V5 with a G409R mutation or ST-LPL-V5 with a G409R mutation, in the presence or the absence of heparin (500 U/ml). Western blots were performed on cell extracts. GPIHBP1 in the cell extracts was detected with a GPIHBP1-specific rat antibody; LPL was detected with a mouse antibody against the V5 tag or a rabbit antibody against the S-protein tag. Antibodies against actin were used as a loading control. The relative amounts of LPL added to the cells (the binding cocktail) were assessed with antibodies against the V5 tag and S-protein tag (top panel). The intensities of band signals with the V5-tag antibody were quantified with a Li-Cor Odessey scanner. The binding of the C-terminal fragment of LPL-V5 (G409R) to GPIHBP1 in the absence of heparin was 3.4 times greater (relative to the GPIHBP1 signal) than the binding of LPL-V5 to GPIHBP1.
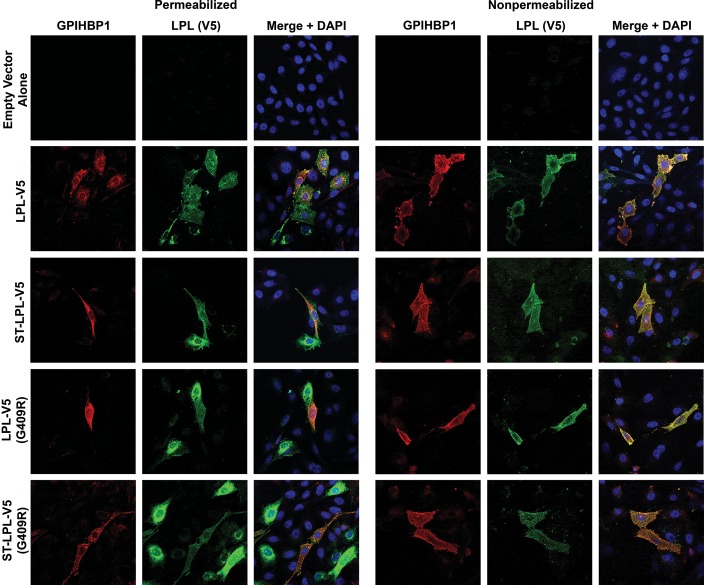
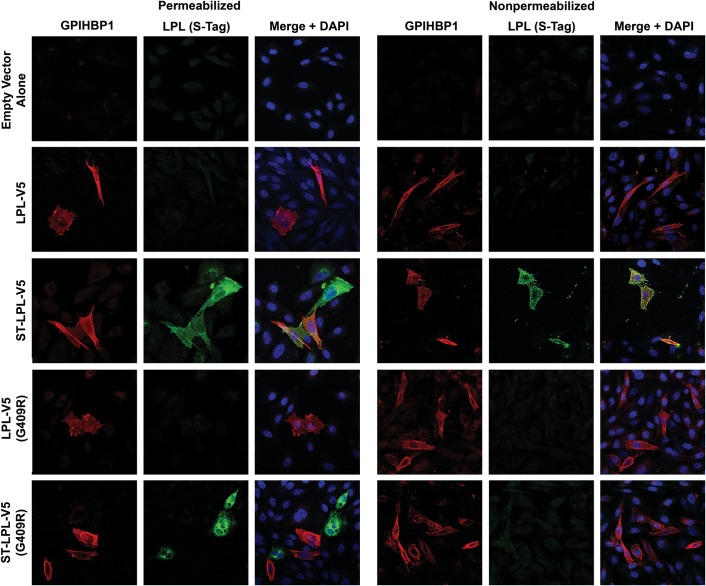
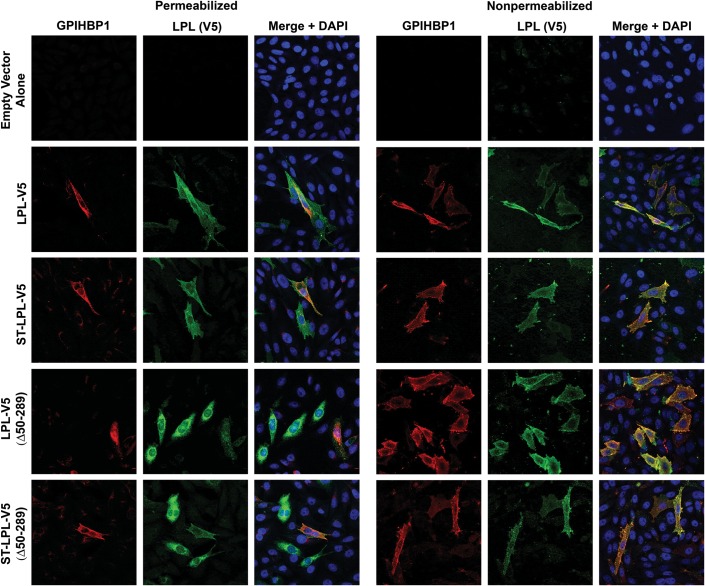
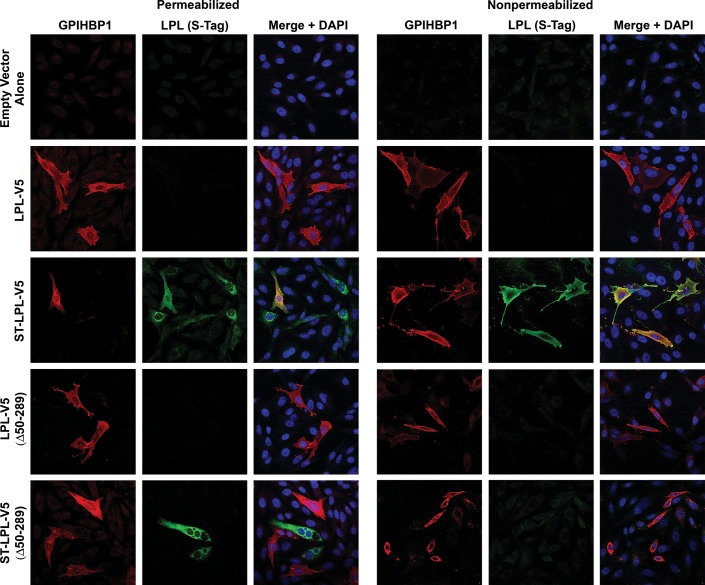
We also assessed the binding of LPL and LPL cleavage fragments to GPIHBP1-expressing cells by immunofluorescence microscopy (Figs 3 and 4). In these studies, we mixed cells that had been transfected with GPIHBP1 with other cells that had been transfected with an LPL construct. The next day, we prepared permeabilized and non-permeabilized cells and used immunocytochemistry to determine if newly secreted LPL (from LPL-transfected cells) bound to GPIHBP1-transfected cells. The binding of LPL-V5 or ST-LPL-V5 (green) to GPIHBP1 (red) was evident by yellow-colored cells on the merged image. In the case of ST-LPL-V5, the binding of LPL to GPIHBP1-transfected cells was evident with both V5 and S-protein antibodies (Figs 3 and 4). With ST-LPL-V5 (G409R), which is cleaved into N- and C-terminal fragments, binding to GPIHBP1 was detectable with the V5 antibody but not the S-protein antibody (Figs 3 and 4), indicating that only LPL's C terminus binds to GPIHBP1 [and that the N- and C-terminal domains of ST-LPL-V5 (G409R) do not remain attached to each other]. Control experiments established the specificity of antibody binding to GPIHBP1 and LPL (Supplementary Material, Fig. S3).
Figure 3.
Immunofluorescence microscopy assay of LPL binding to GPIHBP1-expressing CHO-K1 cells. CHO-K1 cells that had been transfected with GPIHBP1 were mixed with cells that had been transfected with LPL-V5, ST-LPL-V5, LPL-V5 (G409R) or ST-LPL-V5 (G409R). The next day, permeabilized and non-permeabilized cells were analyzed by immunocytochemistry. GPIHBP1 (red) was detected with a rabbit antibody against GPIHBP1; LPL (green) was detected with a mouse antibody against the V5 tag. Nuclei (blue) were stained with DAPI.
Figure 4.
Immunofluorescence microscopy assay of LPL binding to GPIHBP1-expressing CHO-K1 cells. CHO-K1 cells that had been transfected with GPIHBP1 were mixed with cells that had been transfected with LPL-V5, ST-LPL-V5, LPL-V5 (G409R) or ST-LPL-V5 (G409R). The next day, permeabilized and non-permeabilized cells were analyzed by immunocytochemistry. GPIHBP1 (red) was detected with a rat antibody against GPIHBP1; LPL (green) was detected with a rabbit antibody against the S-protein tag. Nuclei (blue) were stained with DAPI.
A solid-phase immunoassay provided further evidence that the N- and C-terminal domains of ST-LPL-V5 (G409R) are not attached to each other (Supplementary Material, Fig. S4). In this experiment, we tested the ability of LPL-V5, ST-LPL-V5, LPL-V5 (G409R) and ST-LPL-V5 (G409R) to bind to 96-well plates that had been coated with the LPL-specific monoclonal antibody 5D2 (epitope, LPL amino acids 380–410) (22). When ST-LPL-V5 was added to 5D2-coated wells, binding was readily detected with both V5 and S-protein antibodies. However, in the case of the ST-LPL-V5 (G409R) preparation, binding was detectable only with the V5 antibody and not with the S-protein antibody, indicating that LPL's C- and N-terminal fragments do not remain associated with each other after furin cleavage.
LPL proteins with in-frame deletions of the N-terminal catalytic domain are susceptible to endoproteolytic cleavage
To further examine the notion that the C terminus of LPL (detached from the N terminus) binds to GPIHBP1, we expressed mutant LPLs with in-frame deletions of large segments of the N-terminal domain. LPLs with several in-frame deletions [e.g. LPL-V5 (Δ50-199) and LPL-V5 (Δ50-289)] were susceptible to furin cleavage, and the familiar 28-kDa C-terminal fragment was released into the medium (see ‘binding cocktail’ in Fig. 5).
Figure 5.
Western blot assays of LPL binding to CHO-K1 cells expressing S-protein-tagged GPIHBP1. GPIHBP1-transfected cells were incubated at 4°C with cell culture medium alone, LPL-V5 or LPL-V5 proteins with the indicated substitutions or in-frame deletions, in the presence or the absence of heparin (500 U/ml). After 2 h, cell extracts were prepared, and GPIHBP1 was detected with a rabbit antibody against the S-protein tag; LPL was detected with a mouse antibody against the V5 tag. Antibodies against actin were used as a loading control. The relative amounts of LPL added to the cells (the binding cocktail) were assessed with an antibody against the V5 tag (top panel).
When cells expressing LPL-V5 (Δ50-289) were treated with a furin inhibitor, the cleavage event was abolished and the full-length protein was secreted into the medium (Supplementary Material, Fig. S5). Full-length LPL-V5 (Δ50-289) was also secreted into the medium after introducing an R297A mutation (Supplementary Material, Fig. S5).
Binding of LPLs with in-frame deletions to GPIHBP1
As expected, the 28-kDa C-terminal fragment from LPL-V5 (Δ50-289) bound to GPIHBP1-expressing cells avidly, as judged by the western blot assay (Fig. 5 and Supplementary Material, Fig. S6). LPL-V5 (Δ50-289;R297A), which is not cleaved, also bound to GPIHBP1 (Supplementary Material, Fig. S6).
Binding of the C-terminal domain from LPL-V5 (Δ50-289) and ST-LPL-V5 (Δ50-289) to GPIHBP1-expressing cells was also evident from immunofluorescence microscopy assays. The binding of ST-LPL-V5 (green) to GPIHBP1-transfected cells (red) was easily detectable, yielding yellow-colored cells on the merged image [regardless of whether LPL was detected with the V5 antibody (Fig. 6) or with the S-protein antibody (Fig. 7)]. Binding of LPL-V5 (Δ50-289) and ST-LPL-V5 (Δ50-289) (both of which are cleaved at residue 297) could be detected with the V5 antibody (Fig. 6) but not the S-protein antibody (Fig. 7), indicating that the N terminus of ST-LPL-V5 (Δ50-289) does not stay attached to the C terminus.
Figure 6.
Immunofluorescence microscopy assay of LPL binding to GPIHBP1-expressing CHO-K1 cells. CHO-K1 cells that had been transfected with GPIHBP1 were mixed with cells that had been transfected with LPL-V5, ST-LPL-V5, LPL-V5 (Δ50-289) or ST-LPL-V5 (Δ50-289). The next day, permeabilized and non-permeabilized cells were analyzed by immunocytochemistry. GPIHBP1 (red) was detected with a rabbit antibody against GPIHBP1; LPL (green) was detected with a mouse antibody against the V5 tag. Nuclei (blue) were stained with DAPI.
Figure 7.
Immunofluorescence microscopy assay of LPL binding to GPIHBP1-expressing CHO-K1 cells. CHO-K1 cells that had been transfected with GPIHBP1 were mixed with cells that had been transfected with LPL-V5, ST-LPL-V5, LPL-V5 (Δ50-289) or ST-LPL-V5 (Δ50-289). The next day, permeabilized and non-permeabilized cells were analyzed by immunocytochemistry. GPIHBP1 (red) was detected with a rat antibody against GPIHBP1; LPL (green) was detected with a rabbit antibody against the S-protein tag. Nuclei (blue) were stained with DAPI.
Assessing binding to GPIHBP1 of C-terminal LPL fragments harboring C418Y or E421K mutations
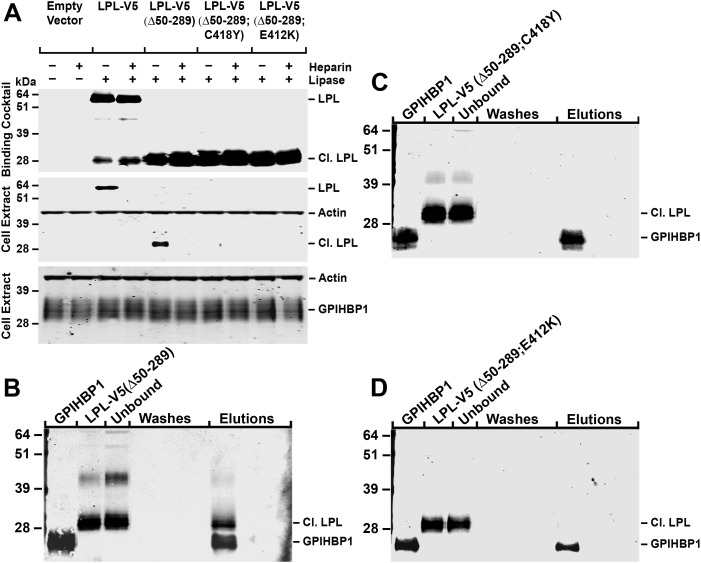
To determine if the binding of LPL's C terminus to GPIHBP1 is specific, we generated LPL-V5 (Δ50-289) expression vectors with a C418Y or an E421K mutation (these mutations block the ability of full-length LPL to bind to GPIHBP1) (14). Like LPL-V5 (Δ50-289), LPL-V5 (Δ50-289;C418Y) is susceptible to furin cleavage, yielding a 28-kDa C-terminal fragment. However, unlike the C-terminus from LPL-V5 (Δ50-289), the C terminus from LPL-V5 (Δ50-289;C418Y) does not bind to GPIHBP1, as judged by the western blot assay (Fig. 8A). Similarly, following furin cleavage, LPL-V5 (Δ50-289;E421K) yielded a 28-kDa fragment that did not bind to GPIHBP1 (Fig. 8A).
Figure 8.
Binding of LPL-V5, LPL-V5 (Δ50-289) and LPL-V5 (Δ50-289;C418Y) to GPIHBP1. (A) Western blot assay of the binding of LPL preparations to CHO-K1 cells expressing S-protein-tagged GPIHBP1. GPIHBP1-transfected cells were incubated at 4°C for 2 h with medium alone or medium containing LPL-V5, LPL-V5 (Δ50-289), LPL-V5 (Δ50-289;C418Y) or LPL-V5 (Δ50-289;E421K). GPIHBP1 was detected with a rabbit antibody against the S-protein tag, and LPL was detected with a mouse antibody against the V5 tag. Antibodies against actin were used as a loading control. (B–D) Cell-free assays of LPL binding to GPIHBP1. We assessed the binding of LPL-V5 (Δ50-289) (B), LPL-V5 (Δ50-289;C418Y) (C) and LPL-V5 (Δ50-289;E421K) (D) to soluble GPIHBP1 that had been captured on agarose beads coated with a GPIHBP1-specific monoclonal antibody. After 1 h, GPIHBP1 (and any bound LPL) was eluted from the antibody-coated beads with 0.1 M glycine, pH 2.5. The starting material, the material that did not bind to the beads (unbound fraction), the wash fractions and the glycine buffer elution fractions were assessed by western blotting with a rabbit antibody against GPIHBP1 and a mouse antibody against the V5 tag.
Similar results were obtained with a cell-free LPL–GPIHBP1-binding assay (Fig. 8B–D). In this assay, LPL-V5 (Δ50-289), LPL-V5 (Δ50-289;C418Y) and LPL-V5 (Δ50-289;E421K) preparations were incubated with soluble GPIHBP1 that had been captured on agarose beads coated with a GPIHBP1-specific monoclonal antibody. After 1 h, the soluble GPIHBP1 (and any LPL fragments bound to GPIHBP1) were eluted from the antibody-coated beads with 0.1 m glycine, pH 2.5. The C terminus of LPL-V5 (Δ50-289) bound to GPIHBP1 and eluted with GPIHBP1 (Fig. 8B). However, the corresponding C-terminal fragments from LPL-V5 (Δ50-289;C418Y) and LPL-V5 (Δ50-289;E421K) did not bind to GPIHBP1; consequently, all of the LPL was detected in the flow through fraction (unbound fraction) and none could be eluted from the beads with the glycine elution buffer (Fig. 8C–D).
We also used the immunofluorescence microscopy assay to assess the ability of C-terminal LPL fragments with the C418Y or E421K substitutions to bind to GPIHBP1 (Supplementary Material, Fig. S7). Neither LPL-V5 (Δ50-289;C418Y) nor LPL-V5 (Δ50-289;E421K) was capable of binding to GPIHBP1; hence, there was no colocalization of LPL and GPIHBP1 signals on the merged images. Simultaneously, we examined the effect of the R297A mutation on GPIHBP1 binding. Both LPL-V5 (R297A) and LPL-V5 (Δ50-289;R297A) bound normally to GPIHBP1, as judged by the presence of yellow-colored cells on the merged images (Supplementary Material, Fig. S7).
Binding of LPL's C-terminal fragment to GPIHBP1 after exposure to denaturing conditions
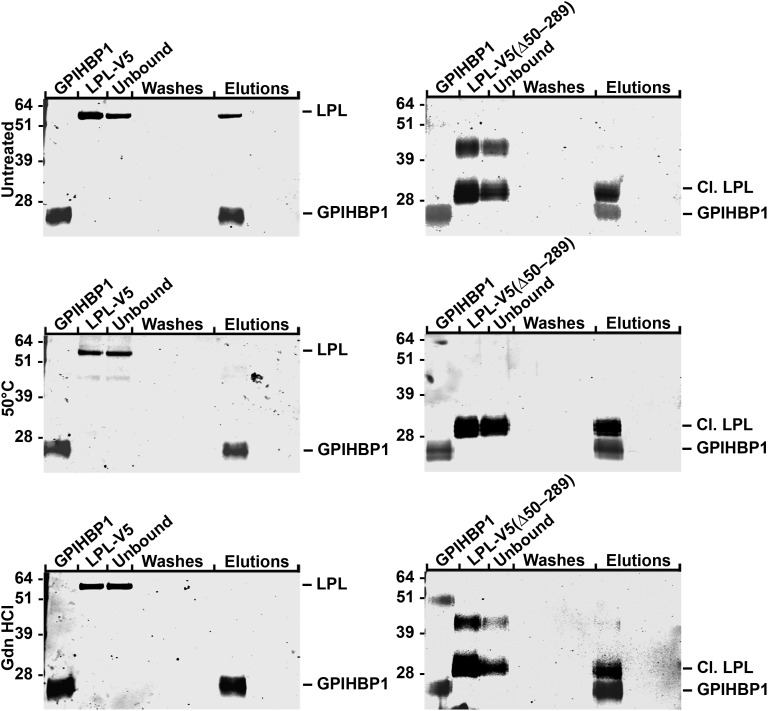
We investigated the effects of denaturing conditions on the ability of LPL-V5 and LPL-V5 (Δ50-289) to bind to GPIHBP1. As reported earlier (16), exposing LPL-V5 to denaturing conditions abolishes its ability to bind to GPIHBP1 (Fig. 9). However, when LPL-V5 (Δ50-289) was exposed to the same denaturing conditions, it was able to bind to GPIHBP1, implying that the C-terminal domain of LPL (when free of the N-terminal domain) is capable of refolding and assuming the proper conformation for GPIHBP1 binding.
Figure 9.
Binding of LPL-V5 and LPL-V5 (Δ50-289) to soluble GPIHBP1 captured by agarose beads coated with a GPIHBP1-specific antibody. Native LPL-V5 and LPL-V5 (Δ50-289), along with the same LPL preparations that had been exposed to denaturing conditions (heating to 50°C or treatment with 3 M guanidine hydrochloride followed by dialysis against PBS), were incubated with soluble GPIHBP1 on antibody-coated beads. After 1 h, GPIHBP1 (and any bound LPL) was eluted from the antibody-coated beads with 0.1 M glycine, pH 2.5. The starting material, the unbound fraction, the wash fractions and the glycine buffer elution fractions were assessed by western blots with a rabbit antibody against GPIHBP1 and a mouse antibody against the V5 tag.
DISCUSSION
In an earlier study, we showed that the exposure of LPL-V5 to denaturing conditions abolished its capacity to bind to GPIHBP1 (16). That finding suggested that LPL dimers might be important for GPIHBP1 binding (i.e. that LPL's GPIHBP1-binding domain might involve sequences from the N-terminal domain of one monomer and the C-terminal domain of the partner monomer). However, our current studies show that this is not the case. We showed that LPL-V5 (G409R), LPL-V5 (G410V) and multiple LPL mutants with in-frame deletions of LPL's catalytic domain are highly susceptible to furin cleavage and that the C-terminal domain released by this cleavage event (amino acids 298–448) binds GPIHBP1 avidly (and independently of the N-terminal fragment). Binding of LPL's C terminus to GPIHBP1 was observed in two cell-based assays (the western blot and immunofluorescence microscopy assays) along with a cell-free assay of LPL binding to soluble GPIHBP1. Like full-length LPL (2,14), the binding of LPL's C-terminal fragment to GPIHBP1 was inhibited by heparin and was abolished by C418Y or E421K mutations. Given that the C terminus is sufficient for GPIHBP1 binding, our LPL denaturation studies need to be interpreted differently. When full-length LPL is subjected to denaturing conditions, it would appear its C-terminal sequences are unable to refold properly and therefore cannot bind to GPIHBP1.
No one has yet determined the structure of LPL, likely because LPL dimers are notoriously unstable (12). However, our current experiments suggest that it might be possible to move forward with the C-terminal fragments of LPL. First, it is encouraging that LPL's C terminus, free of the N terminus, is secreted from cells quite well. Previous studies have shown that missense mutations in LPL's N terminus can interfere with LPL secretion (23,24), but we showed that C-terminal LPL fragments are secreted from cells efficiently. Second, it is encouraging that, unlike full-length LPL, the C-terminal domain of LPL refolds and binds GPIHBP1 after exposure to denaturing conditions. Structural studies of the C terminus should make it possible to understand both LPL's GPIHBP1-binding sequences and its triglyceride-binding sequences. Also, these studies should reveal how specific amino acid substitutions in LPL's C terminus (e.g. C418Y and E421K) interfere with GPIHBP1 binding.
The current studies define an entirely new mechanism by which LPL mutations can cause disease—rendering the enzyme susceptible to endoproteolytic cleavage. The E410V mutation was originally identified in a 3-year-old boy with chylomicronemia (17), and a G412R mutation was identified in a mutant line of domestic cats with chylomicronemia (Gly-412 in the cat LPL corresponds to Gly-409 in human LPL) (18). Both mutations are associated with very low levels of LPL mass and activity in the post-heparin plasma (17,18). However, the mechanism for those findings was not clear, and no one had examined the mutant LPLs with western blots. Our studies demonstrated that these mutations render LPL susceptible to furin cleavage at residue 297. Wild-type LPL is also cleaved by furin (Figs 1, 2, 5 and 8 and Supplementary Material, Figs S1, S2 and S6) but only to a minimal degree. The E410V and G409R substitutions apparently alter conformation so as to expose LPL's furin cleavage site. Interestingly, there is evidence that LPL's furin cleavage site is physiologically important. ANGPTL3 inactivates LPL by rendering it susceptible to furin cleavage, and it is noteworthy that a similar LPL fragment can be detected in human post-heparin plasma (21). While the furin cleavage of LPL can occur in the secretory pathway, it also occurs at the surface of cells after secretion (21).
In the case of wild-type LPL-V5, the inhibition of furin cleavage with CMK or an R297A mutation had little effect on LPL secretion from cells. However, in the case of G409R and E410V mutations, CMK (or the R297A mutation) abolished LPL secretion. It would appear that, in the setting of the G409R or E410V mutations, furin cleavage actually allows the LPL fragments to be secreted. Presumably, when furin cleavage is inhibited, these LPL mutants are more susceptible to being degraded within the secretory pathway. From a therapeutic standpoint, this finding was somewhat disappointing. Any attempt to treat a chylomicronemia patient harboring Gly-409 or Glu-410 mutations with a furin inhibitor would likely be fruitless. While a potent inhibitor might well prevent the cleavage of LPL, it would probably also abolish LPL secretion from cells.
The fact that the G409 and E410 increased susceptibility to furin cleavage will give clinical geneticists more to think about when considering mechanisms by which LPL mutations (as well as commonly occurring LPL polymorphisms) affect plasma triglyceride levels. We would not be surprised if future studies uncovered LPL mutations with subtle effects on susceptibility to furin cleavage, or perhaps mutations that render LPL more susceptible to inactivation by ANGPTL3 or ANGPTL4.
In summary, we have shown that the C terminus of LPL, detached from the N terminus, is sufficient for specific binding to GPIHBP1. We also demonstrate a new mechanism for LPL mutations—enhanced susceptibility to a furin cleavage.
MATERIALS AND METHODS
GPIHBP1 and LPL expression vectors and cell transfections
Expression vectors for human and mouse GPIHBP1 (an untagged version and a version with an N-terminal S-protein tag) have been described previously (2,5–7,16). An expression construct for human LPL with a C-terminal V5 tag (LPL-V5) (2) was obtained from Dr Mark Doolittle (University of California, Los Angeles). LPL-V5 was modified by adding an S-protein tag to its N terminus (generating ST-LPL-V5). Site-directed mutagenesis (QuikChange kit, Agilent Technologies) was used to introduce point mutations into LPL constructs. All constructs were validated by DNA sequencing.
To express GPIHBP1 or LPL in CHO-K1 cells, 5 × 106 cells were electroporated with 5 μg of plasmid DNA with the Nucleofector II apparatus (Lonza). Following the electroporation, cells were grown for 24 h in serum-free medium containing protease inhibitors. LPL from the conditioned medium was concentrated 10-fold with an Amicon filter [Amicon Ultra 10 000 nominal molecular weight limit (NMWL) filter] and used for GPIHBP1-binding assays. The LPL preparations were detected by western blotting with a mouse monoclonal antibody against the V5 tag (1:200, Invitrogen) or a rabbit polyclonal antibody against the S-protein tag (1:1000, Abcam).
Binding of LPL to GPIHBP1-transfected CHO cells
CHO-K1 cells were transfected with an untagged or an S-protein-tagged GPIHBP1 expression vector and grown in 24-well tissue culture plates; the cells were then incubated for 2 h at 4°C with LPL-V5 (400 μl/well) or various mutant LPLs. In some wells, heparin (500 U/ml) was added to the medium. Cells were then washed six times with ice-cold PBS containing 1 mm MgCl2 and 1 mm CaCl2 (PBS/Mg/Ca), and cell extracts were collected in RIPA buffer (PBS containing 1% Triton X-100, 0.5% deoxycholate and 0.02% SDS) with complete mini EDTA-free protease inhibitors (Roche). The LPL preparations used in the binding assays, the amount of LPL bound to GPIHBP1-transfected cells (i.e. the amount of LPL in the cell extracts) and the amount of GPIHBP1 in the cell extracts were assessed by western blotting (5,6,16). Proteins in cell extracts were size-fractionated on 12 or 4–12% Bis–Tris SDS-polyacrylamide gels; the separated proteins were then transferred to nitrocellulose for western blotting. The antibody dilutions for western blots were 1:1000 for a rabbit polyclonal antibody against the S-protein tag (Abcam); 1:200 for a mouse monoclonal antibody against the V5 tag (Invitrogen); 1:500 for a rabbit polyclonal antibody against β-actin (Abcam); 1:500 for a mouse monoclonal antibody against β-actin (Abcam); 1:500 for a rat monoclonal antibody against GPIHBP1 (6,16); 1:500 for an IRdye800-conjugated donkey anti-mouse IgG; 1:1000 for an IRdye800-conjugated donkey anti-rat IgG; 1:1000 for an IRdye800-conjugated donkey anti-rabbit IgG; 1:2000 for an IRdye680-conjugated donkey anti-mouse IgG; and 1:2000 for an IRdye680-conjugated donkey anti-rabbit IgG (all from Li-Cor). Antibody binding was visualized with an Odyssey infrared scanner (Li-Cor).
Immunofluorescence microscopy
CHO-K1 cells (1 × 106) were electroporated with 2 μg of either a mouse GPIHBP1 expression vector or an LPL expression vector. After the transfections, the two groups of cells were mixed together in roughly equal numbers and plated on coverslips in 24-well plates. Twenty-four hours later, the cells were washed extensively and fixed in 3% paraformaldehyde. After a 1-h incubation in blocking buffer (PBS/Mg/Ca containing 10% donkey serum), the cells were incubated with a rabbit polyclonal antibody against the S-protein tag (1:500, Abcam), a rat monoclonal against GPIHBP1 (1:1000), a rabbit polyclonal against GPIHBP1 (1:500) and/or a mouse monoclonal antibody against the V5 tag (1:100, Invitrogen) for 1 h. After washing, the cells were incubated for 30 min with an Alexa 568-conjugated donkey anti-rabbit IgG (1:800, Invitrogen), a DyLight 550-conjugated donkey anti-rat IgG (1:800, Abcam), an Alexa 488-conjugated donkey anti-mouse IgG (1:800, Invitrogen) and/or an Alexa 488-conjugated donkey anti-rabbit IgG (1:800, Invitrogen). After a final washing step, the cells were stained with DAPI to visualize DNA. In some experiments, cells were permeabilized with 0.2% Triton X-100.
Images were recorded with an Axiovert 200M microscope (equipped with an LSM 700 confocal scanning module) and processed with the Zen 2010 software (all from Zeiss). The exposure conditions for each image within an experiment were fixed and identical.
Binding of LPL preparations to 96-well plates coated with an LPL-specific monoclonal antibody
A 96-well plate was coated overnight with the LPL-specific antibody 5D2, which binds to the C terminal region of LPL (amino acids 380–410) (22). After washing the plates and blocking any remaining binding sites with Starting Block (Pierce), the plates were incubated with LPL preparations for 2 h at 4°C. Wells were then washed with PBS/Mg/Ca containing 0.1% Tween-20, and LPL binding to antibody 5D2 was detected with a horseradish peroxidase (HRP)-labeled rabbit polyclonal antibody against the V5 tag (0.5 μg/ml, Abcam) or an HRP-labeled rabbit polyclonal antibody against the S-protein tag (0.5 μg/ml, Abcam) followed by an incubation with TMB 1-Component HRP Microwell Substrate (SUB1, ImmunoChemistry Technologies).
Effect of furin inhibition on secretion of LPL from transfected cells
CHO-K1 cells were electroporated with different LPL expression vectors and were then grown in a medium in the presence or the absence of 50 μm decanoyl-RVKR-chloromethylketone (CMK, Calbiochem) for 24 h. Cell extracts and samples of conditioned medium were then analyzed by western blotting. Simultaneously, we assessed the effect of an LPL missense mutation, R297A, on the secretion of LPL. The R297A mutation blocks the furin cleavage of LPL (20).
Binding of LPL to soluble GPIHBP1 captured on agarose beads coated with a GPIHBP1-specific monoclonal antibody
CHO-K1 cells were electroporated with a vector encoding a soluble version of mouse GPIHBP1 (truncated after Q197 and therefore not GPI-anchored) (6). After 48 h, conditioned medium was collected and concentrated 10-fold with an Amicon Ultra 10 000 filter. Binding of LPL proteins to soluble GPIHBP1 was assessed with a cell-free binding assay (6). Medium containing soluble mouse GPIHBP1 was mixed with LPL preparations and agarose beads coated with the GPIHBP1-specific monoclonal antibody 11A12 in PBS containing 0.2% NP-40 (6). After 2 h, beads were washed, and the soluble GPIHBP1, together with any bound LPL, was eluted from the beads with 0.1 m glycine, pH 2.5. The amounts of GPIHBP1 and LPL in the starting material, the unbound fraction and each wash and elution fraction were assessed by western blotting. This same assay was used to test the GPIHBP1-binding capacity of LPL preparations that had been exposed to denaturing conditions (heating to 50°C for 16 h or incubation with 3 m guanidine hydrochloride for 16 h followed by dialysis against PBS).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by a Scientist Development Award from the American Heart Association, National Office (to A.P.B.); a Postdoctoral Fellowship Award from the American Heart Association, Western States Affiliate (to C.N.G.); National Institutes of Health (R01 HL094732 to A.P.B., RO1 HL089781 to L.G.F., P01 HL090553 to S.G.Y. and R01 HL087228 to S.G.Y.).
Supplementary Material
REFERENCES
- 1.Davies B.S.J., Beigneux A.P., Barnes R.H., II, Tu Y., Gin P., Weinstein M.M., Nobumori C., Nyrén R., Goldberg I.J., Olivecrona G., et al. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 2010;12:42–52. doi: 10.1016/j.cmet.2010.04.016. doi:10.1016/j.cmet.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beigneux A.P., Davies B., Gin P., Weinstein M.M., Farber E., Qiao X., Peale P., Bunting S., Walzem R.L., Wong J.S., et al. Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 2007;5:279–291. doi: 10.1016/j.cmet.2007.02.002. doi:10.1016/j.cmet.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ioka R.X., Kang M.-J., Kamiyama S., Kim D.-H., Magoori K., Kamataki A., Ito Y., Takei Y.A., Sasaki M., Suzuki T., et al. Expression cloning and characterization of a novel glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein, GPI-HBP1. J. Biol. Chem. 2003;278:7344–7349. doi: 10.1074/jbc.M211932200. doi:10.1074/jbc.M211932200. [DOI] [PubMed] [Google Scholar]
- 4.Young S.G., Davies B.S., Voss C.V., Gin P., Weinstein M.M., Tontonoz P., Reue K., Bensadoun A., Fong L.G., Beigneux A.P. GPIHBP1, an endothelial cell transporter for lipoprotein lipase. J. Lipid Res. 2011;52:1869–1884. doi: 10.1194/jlr.R018689. doi:10.1194/jlr.R018689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beigneux A.P., Davies B.S., Tat S., Chen J., Gin P., Voss C.V., Weinstein M.M., Bensadoun A., Pullinger C.R., Fong L.G., et al. Assessing the role of the glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) three-finger domain in binding lipoprotein lipase. J. Biol. Chem. 2011;286:19735–19743. doi: 10.1074/jbc.M111.242024. doi:10.1074/jbc.M111.242024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beigneux A.P., Gin P., Davies B.S.J., Weinstein M.M., Bensadoun A., Fong L.G., Young S.G. Highly conserved cysteines within the Ly6 domain of GPIHBP1 are crucial for the binding of lipoprotein lipase. J. Biol. Chem. 2009;284:30240–30247. doi: 10.1074/jbc.M109.046391. doi:10.1074/jbc.M109.046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gin P., Yin L., Davies B.S., Weinstein M.M., Ryan R.O., Bensadoun A., Fong L.G., Young S.G., Beigneux A.P. The acidic domain of GPIHBP1 is important for the binding of lipoprotein lipase and chylomicrons. J. Biol. Chem. 2008;284:29554–29562. doi: 10.1074/jbc.M802579200. doi:10.1074/jbc.M802579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong H., Davis R.C., Thuren T., Goers J.W., Nikazy J., Waite M., Schotz M.C. Lipoprotein lipase domain function. J. Biol. Chem. 1994;269:10319–10323. [PubMed] [Google Scholar]
- 9.Wong H., Schotz M.C. The lipase gene family. J. Lipid Res. 2002;43:993–999. doi: 10.1194/jlr.r200007-jlr200. doi:10.1194/jlr.R200007-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Lookene A., Groot N.B., Kastelein J.J., Olivecrona G., Bruin T. Mutation of tryptophan residues in lipoprotein lipase. Effects on stability, immunoreactivity, and catalytic properties. J. Biol. Chem. 1997;272:766–772. doi: 10.1074/jbc.272.2.766. doi:10.1074/jbc.272.2.766. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi Y., Nakajima T., Inoue I. Molecular modeling of the dimeric structure of human lipoprotein lipase and functional studies of the carboxyl-terminal domain. Eur. J. Biochem. 2002;269:4701–4710. doi: 10.1046/j.1432-1033.2002.03179.x. doi:10.1046/j.1432-1033.2002.03179.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L., Lookene A., Wu G., Olivecrona G. Calcium triggers folding of lipoprotein lipase into active dimers. J. Biol. Chem. 2005;280:42580–42591. doi: 10.1074/jbc.M507252200. doi:10.1074/jbc.M507252200. [DOI] [PubMed] [Google Scholar]
- 13.Sukonina V., Lookene A., Olivecrona T., Olivecrona G. Angiopoietin-like protein 4 converts lipoprotein lipase to inactive monomers and modulates lipase activity in adipose tissue. Proc. Natl Acad. Sci. USA. 2006;103:17450–17455. doi: 10.1073/pnas.0604026103. doi:10.1073/pnas.0604026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voss C.V., Davies B.S., Tat S., Gin P., Fong L.G., Pelletier C., Mottler C.D., Bensadoun A., Beigneux A.P., Young S.G. Mutations in lipoprotein lipase that block binding to the endothelial cell transporter GPIHBP1. Proc. Natl Acad. Sci. USA. 2011;108:7980–7984. doi: 10.1073/pnas.1100992108. doi:10.1073/pnas.1100992108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong H., Yang D., Hill J.S., Davis R.C., Nikazy J., Schotz M.C. A molecular biology-based approach to resolve the subunit orientation of lipoprotein lipase. Proc. Natl Acad. Sci. USA. 1997;94:5594–5598. doi: 10.1073/pnas.94.11.5594. doi:10.1073/pnas.94.11.5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gin P., Beigneux A.P., Voss C., Davies B.S., Beckstead J.A., Ryan R.O., Bensadoun A., Fong L.G., Young S.G. Binding preferences for GPIHBP1, a glycosylphosphatidylinositol-anchored protein of capillary endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2011;31:176–182. doi: 10.1161/ATVBAHA.110.214718. doi:10.1161/ATVBAHA.110.214718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Previato L., Guardamagna O., Dugi K.A., Ronan R., Talley G.D., Santamarina-Fojo S., Brewer H.B., Jr A novel missense mutation in the C-terminal domain of lipoprotein lipase (Glu410–>Val) leads to enzyme inactivation and familial chylomicronemia. J. Lipid Res. 1994;35:1552–1560. [PubMed] [Google Scholar]
- 18.Ginzinger D.G., Lewis M.E., Ma Y., Jones B.R., Liu G., Jones S.D. A mutation in the lipoprotein lipase gene is the molecular basis of chylomicronemia in a colony of domestic cats. J. Clin. Invest. 1996;97:1257–1266. doi: 10.1172/JCI118541. doi:10.1172/JCI118541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sendak R.A., Melford K., Kao A., Bensadoun A. Identification of the epitope of a monoclonal antibody that inhibits heparin binding of lipoprotein lipase: new evidence for a carboxyl-terminal heparin-binding domain. J. Lipid Res. 1998;39:633–646. [PubMed] [Google Scholar]
- 20.Jin W., Fuki I.V., Seidah N.G., Benjannet S., Glick J.M., Rader D.J. Proprotein convertases [corrected] are responsible for proteolysis and inactivation of endothelial lipase. J. Biol. Chem. 2005;280:36551–36559. doi: 10.1074/jbc.M502264200. doi:10.1074/jbc.M502264200. [DOI] [PubMed] [Google Scholar]
- 21.Liu J., Afroza H., Rader D.J., Jin W. Angiopoietin-like protein 3 inhibits lipoprotein lipase activity through enhancing its cleavage by proprotein convertases. J. Biol. Chem. 2010;285:27561–27570. doi: 10.1074/jbc.M110.144279. doi:10.1074/jbc.M110.144279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang S.-F., Reich B., Brunzell J.D., Will H. Detailed characterization of the binding site of the lipoprotein lipase-specific monoclonal antibody 5D2. J. Lipid Res. 1998;39:2350–2359. [PubMed] [Google Scholar]
- 23.Kobayashi J., Inadera H., Fujita Y., Talley G., Morisaki N., Yoshida S., Saito Y., Fojo S.S., Brewer H.B., Jr A naturally occurring mutation at the second base of codon asparagine 43 in the proposed N-linked glycosylation site of human lipoprotein lipase: in vivo evidence that asparagine 43 is essential for catalysis and secretion. Biochem. Biophys. Res. Commun. 1994;205:506–515. doi: 10.1006/bbrc.1994.2694. doi:10.1006/bbrc.1994.2694. [DOI] [PubMed] [Google Scholar]
- 24.Hayden M.R., Ma Y., Brunzell J., Henderson H.E. Genetic variants affecting human lipoprotein and hepatic lipases. Curr. Opin. Lipidol. 1991;2:104–109. doi:10.1097/00041433-199104000-00008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.