Abstract
The impact of cigarette smoking can persist for extended periods following smoking cessation and may involve epigenetic reprogramming. Changes in DNA methylation associated with smoking may help to identify molecular pathways that contribute to the latency between exposure and disease onset. Cross-sectional cohort data from subjects in the International COPD Genetics Network (n = 1085) and the Boston Early-Onset COPD study (n = 369) were analyzed as the discovery and replication cohorts, respectively. Genome-wide methylation data on 27 578 CpG sites in 14 475 genes were obtained on DNA from peripheral blood leukocytes using the Illumina HumanMethylation27K Beadchip in both cohorts. We identified 15 sites significantly associated with current smoking, 2 sites associated with cumulative smoke exposure, and, within the subset of former smokers, 3 sites associated with time since quitting cigarettes. Two loci, factor II receptor-like 3 (F2RL3) and G-protein-coupled receptor 15 (GPR15), were significantly associated in all three analyses and were validated by pyrosequencing. These findings (i) identify a novel locus (GPR15) associated with cigarette smoking and (ii) suggest the existence of dynamic, site-specific methylation changes in response to smoking which may contribute to the extended risks associated with cigarette smoking that persist after cessation.
INTRODUCTION
Tobacco smoking is a leading cause of death and disability worldwide. Smoking attributable deaths are projected to increase over the next two decades and will account for 10% of all deaths globally by 2030 (1). The impact of smoking on cardiovascular (2–4), respiratory (5–9) and malignant (10–12) diseases can persist for extended periods after smoking cessation and may involve epigenetic reprogramming. Identifying molecular pathways that contribute to the delay between exposure and disease may offer opportunities for targeted diagnostic and therapeutic interventions.
In mammals, DNA methylation occurs at the 5′ carbon of cytosine residues in the context of CpG dinucleotides (13) (e.g. a ‘CpG site’) and is a highly plastic, tissue-specific phenomenon which can reflect exposure to exogenous stimuli such as smoking (14). Surprisingly, the number of studies examining the effects of smoking on DNA methylation in blood has been limited. Although relative global hypomethylation has been associated with prenatal exposure to tobacco smoke (15–17), the majority of studies have not demonstrated a significant impact of adult smoking on global DNA methylation content in blood (18–22) (whether assessed directly or through surrogates such as LINE-1 or Alu measurements). Limited numbers of studies have investigated differential methylation in response to cigarette smoking in candidate gene promoters in peripheral blood leukocytes; these studies have been largely restricted to malignancy-related genes and have enrolled small numbers of subjects (23–26). In contrast, Breitling et al. (14) recently reported the results of a genome-wide analysis in peripheral blood leukocytes using a discovery cohort of 177 subjects followed by replication in a second cohort (n = 316); they identified a previously unsuspected CpG site located within the coagulation factor II receptor-like 3 (F2RL3) locus as being strongly associated with current smoking.
We hypothesized that a number of smoking metrics will influence DNA methylation patterns. To test this hypothesis, we sought to identify CpG sites that demonstrate differential methylation with respect to the current smoking status, smoking exposure (pack-years) and time since quitting in a large, well-characterized cohort of former and current smoking subjects from the International COPD Genetics Network (ICGN) (27,28) (n = 1085) followed by replication in the Boston Severe Early-Onset COPD study (EOCOPD) (n = 369) (29). In addition to independently validating the previously reported association of the F2RL3 locus with current smoking, we identify a novel association at the G-protein-coupled receptor 15 (GPR15) locus. We extend our analyses to investigate associations with cumulative smoking exposure (pack-years) and present evidence that suggests the existence of dynamic DNA methylation changes in response to smoking.
RESULTS
The characteristics of the subjects analyzed in the discovery (ICGN) and replication (EOCOPD) cohorts are summarized in Table 1. By design, COPD subjects in the EOCOPD cohort had a lower mean age and FEV1% predicted than COPD subjects in the ICGN cohort. The proportion of male subjects was greater in the ICGN cohort.
Table 1.
Cohort characteristics
| ICGN |
EOCOPD |
|||
|---|---|---|---|---|
| n | 1085 | 369 | ||
| Age | 57.3 (8.1) | 47.5 (7.1) | ||
| Gender | ||||
| Female | 495 (45.6%) | 237 (64.2%) | ||
| Male | 590 (54.4%) | 132 (35.8%) | ||
| Pack-years | 41.7 (26.2) | 28.7 (23.6) | ||
| Age started smokinga | 16.3 (4.1) | 16 (3.1) | ||
| Never smokers | 0 | 68 (18.4%) | ||
| Current smokers | 396 (36.5%) | 103 (27.9%) | ||
| Former smokers | 689 (63.5%) | 198 (53.7%) | ||
| Time since quitting (years)a | 12.3 (10.4) | 7.7 (7.5) | ||
| Obstructive lung diseaseb | Yes = 657 | No = 428 | Yes = 233 | No = 136 |
| FEV1% predicted | 45.4 (17.7) | 104.6 (15.8) | 38.0 (27.6) | 89.7 (16.6) |
ICGN, International COPD Genetics Network; EOCOPD, Early-Onset COPD; FEV1% predicted, forced expiratory volume in the first second expressed as % predicted of normal based on population specific prediction equations.
Data are expressed as mean (SD) or n (%).
aVariable in current or former smokers only (In ICGN, n= 685, four subjects missing data).
bDefined as FEV1/FVC <0.7.
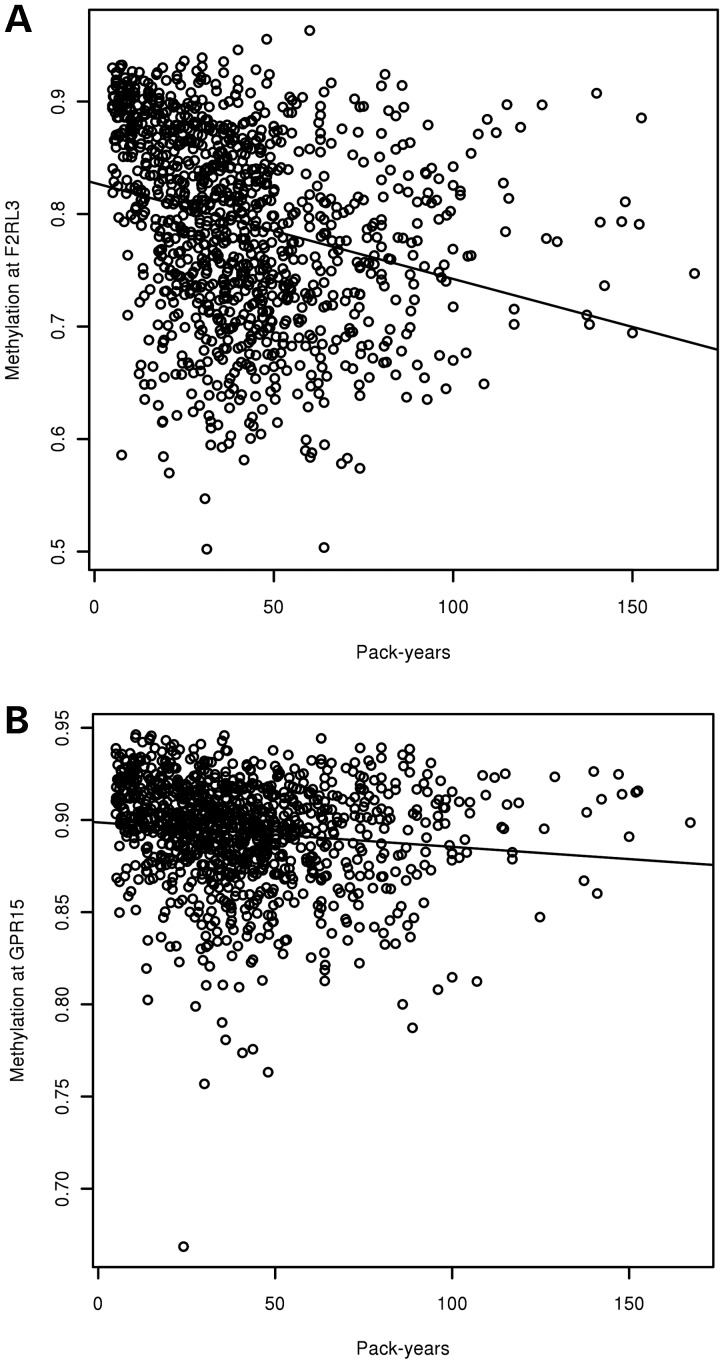
Fifteen CpG sites were significantly associated with current smoking in our cohorts (Table 2). The difference in the mean methylation between current smokers and former and never smokers is illustrated for each cohort. The median and inter-quartile ranges for each of the 15 sites are listed in Supplementary Material, Table S1. Because our discovery cohort compared current versus former smokers, we repeated our analysis after excluding 68 never smokers from our replication cohort (EOCOPD). Nine sites were significantly associated in this subgroup analysis, eight of which overlap with the primary analysis (Supplementary Material, Table S2). For the two most highly associated sites in both analyses, F2RL3 and GPR15, relative hypomethylation was observed with current smoking. Notably, these two CpG sites were also significantly associated with cumulative exposure to cigarette smoke (Table 3). Increasing cumulative exposure to cigarette smoke is associated with relative hypomethylation at both significant loci (Fig. 1).
Table 2.
Significanta differentially methylated CpG sites by current smoking
| CpG | Gene | ICGN |
EOCOPD |
||
|---|---|---|---|---|---|
| Change in mean methylationb | FDR, P-value | Change in mean methylationb | P-valuec | ||
| cg03636183 | F2RL3 | −0.08 | 2.36 × 10−46 | −0.06 | 6.87 × 10−11 |
| cg19859270 | GPR15 | −0.02 | 1.92 × 10−22 | −0.02 | 3.18 × 10−7 |
| cg09837977 | LRRN3 | −0.02 | 3.88 × 10−11 | −0.02 | 5.45 × 10−5 |
| cg01500140 | LIM2 | 0.01 | 1.88 × 10−10 | 0.01 | 3.83 × 10−5 |
| cg13247990 | MYLK | 0.01 | 3.24 × 10−7 | 0.01 | 2.42 × 10−4 |
| cg01988129 | ADHFE1 | 0.01 | 4.16 × 10−6 | 0.01 | 4.12 × 10−5 |
| cg16254309 | CNTNAP2 | −3.36 × 10−3 | 1.74 × 10−5 | −0.01 | 3.48 × 10−4 |
| cg18881723 | SLAMF1 | −0.01 | 1.69 × 10−4 | −0.01 | 3.56 × 10−4 |
| cg12044210 | APBA2 | 1.74 × 10−3 | 3.47 × 10−4 | 0.01 | 5.51 × 10−4 |
| cg21917349 | APBA2 | 1.66 × 10−3 | 5.49 × 10−4 | 0.01 | 5.73 × 10−5 |
| cg15691199 | CEBPE | −3.87 × 10−3 | 2.89 × 10−3 | −3.22 × 10−3 | 2.66 × 10−4 |
| cg24262469 | TIPARP | −1.96 × 10−3d | 0.01 | 0.01 | 7.27 × 10−5 |
| cg05445326 | TM4SF19 | 0.01 | 0.01 | 0.01 | 7.00 × 10−4 |
| cg15258980 | ARHGAP25 | −3.11 × 10−3 | 0.02 | −4.49 × 10−3 | 4.14 × 10−4 |
| cg10161121 | FASLG | −0.01 | 0.02 | −2.81 × 10−3 | 7.93 × 10−4 |
ICGN, International COPD Genetics Network; EOCOPD, Early-Onset COPD; FDR, false-discovery rate.
aDefined as sites with an FDR adjusted P-value <0.05 in the discovery cohort (ICGN) and a one-sided P-value <10−3 in the replication cohort (EOCOPD).
bChange in mean methylation = (mean methylation in current smokers) – (mean methylation in former and never smokers). Methylation values may range from 0 (no methylation) to 1 (fully methylated).
cOne-sided P-value.
dIn model adjusted for covariates direction of effect is positive.
Table 3.
Significanta differentially methylated CpG sites associated with cumulative smoking exposure (pack-years)
| CpG | Gene | ICGN |
EOCOPD |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean methylation in highest pack-years quartile | Mean methylation in lowest pack-years quartile | Difference | FDR, P-value | Mean methylation in highest pack-years quartile | Mean methylation in lowest pack-years quartile | Difference | P-valueb | ||
| cg03636183 | F2RL3 | 0.76 | 0.84 | −0.07 | 1.91 × 10−8 | 0.79 | 0.85 | −0.07 | 2.15 × 10−8 |
| cg19859270 | GPR15 | 0.89 | 0.90 | −0.01 | 6.35 × 10−6 | 0.85 | 0.89 | −0.04 | 2.09 × 10−8 |
ICGN, International COPD Genetics Network; EOCOPD, Early-Onset COPD; FDR, false-discovery rate.
Methylation values may range from 0 (no methylation) to 1 (fully methylated).
Mean value of pack-years by quartile in ICGN: Quartile 1 = 14.4, Quartile 2 = 30.3, Quartile 3 = 44.4, Quartile 4 = 77.9.
Mean value of pack-years by quartile in EOCOPD: Quartile 1 = 1.3, Quartile 2 = 20.0, Quartile 3 = 34.4, Quartile 4 = 60.2.
aDefined as sites with an FDR adjusted P-value <0.05 in the discovery cohort (ICGN) and a one-sided P-value <10−3 in the replication cohort (EOCOPD).
bOne-sided P-value.
Figure 1.
Scatter plots of methylation by pack-years of smoking in the ICGN cohort. X-axis represents cumulative smoke exposure (pack-years), y-axis is the methylation at (A) cg03636183 (F2RL3) and (B) cg19859270 (GPR15). Methylation values may range from 0 (no methylation) to 1 (complete methylation).
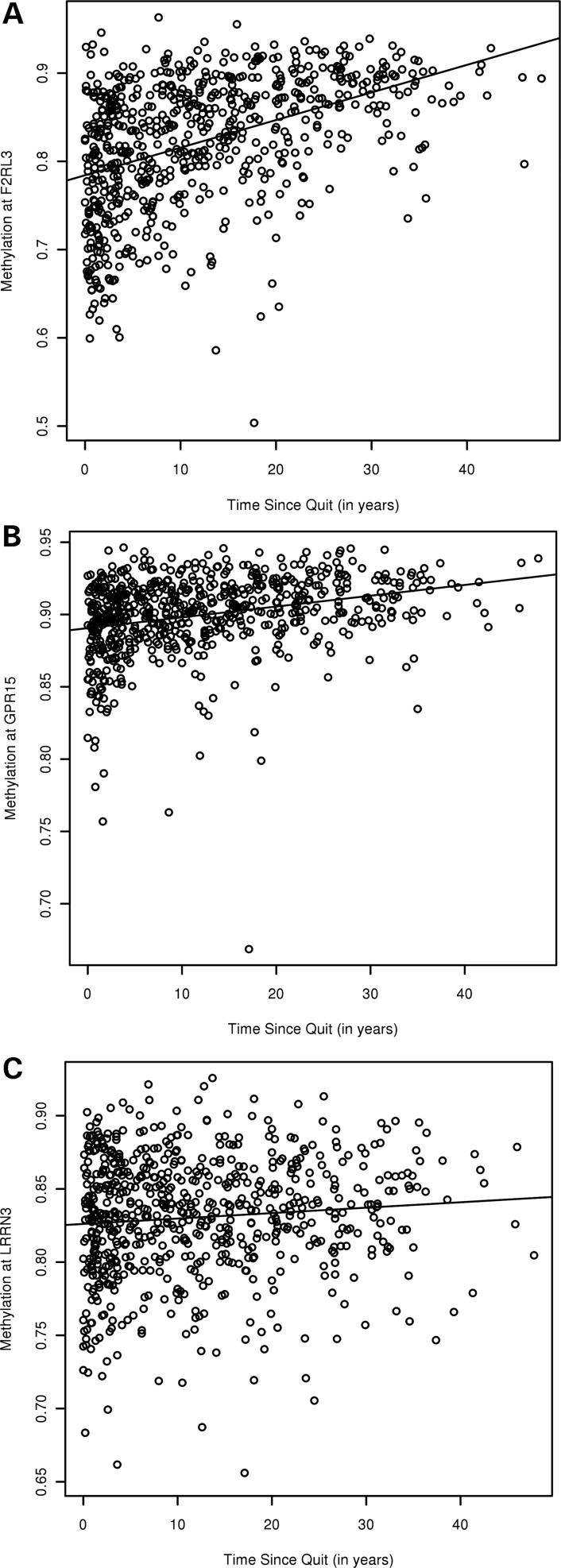
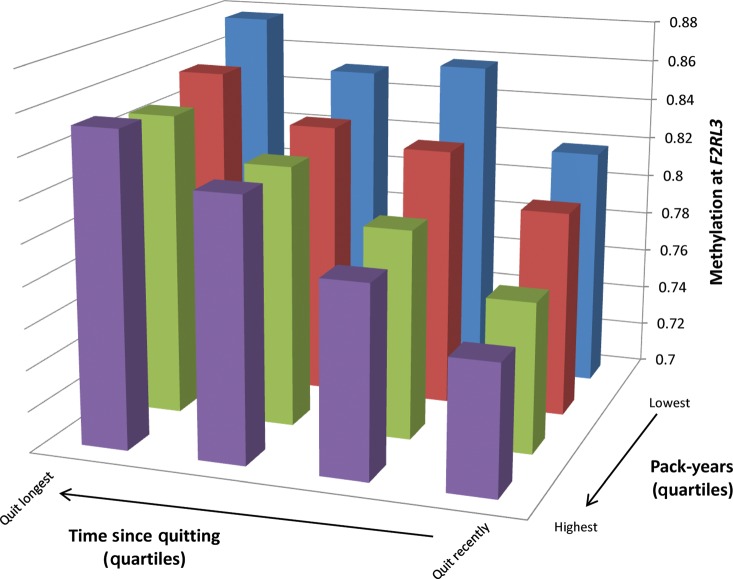
Although data in our cohort are cross-sectional, we analyzed the time since quitting cigarettes in former smokers as a surrogate to evaluate the longitudinal effects of smoking on DNA methylation. The significant differentially methylated sites are listed in Table 4 along with the difference in the mean methylation between the highest and lowest quartiles of time since quitting. The three significant loci in the time since quitting analysis had been previously identified in the current smoking analysis; notably, for each site, current smoking was associated with decreased methylation, whereas time since smoking cessation was associated with increased methylation (Fig. 2). Among former smokers in the ICGN cohort, exploratory analyses revealed a significant interaction between cumulative smoke exposure and the time since quitting under a multiplicative model for F2RL3 and GPR15 (Table 5). This is illustrated qualitatively for the F2RL3 locus in Figure 3; subjects with the highest cumulative smoke exposure and shortest duration of smoking cessation had the lowest mean methylation, whereas subjects with the lowest cumulative smoke exposure and longest duration of cessation had the highest mean methylation.
Table 4.
Significanta differentially methylated sites by time since quitting
| CpG | Gene | ICGN |
EOCOPD |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean methylation in highest time quit quartile | Mean methylation in lowest time quit quartile | Difference | FDR, P-value | Mean methylation in highest time quit quartile | Mean methylation in lowest time quit quartile | Difference | P-valueb | ||
| cg03636183 | F2RL3 | 0.87 | 0.78 | 0.09 | 7.56 × 10−36 | 0.85 | 0.76 | 0.09 | 1.60 × 10−6 |
| cg19859270 | GPR15 | 0.91 | 0.89 | 0.02 | 6.84 × 10−16 | 0.89 | 0.87 | 0.03 | 4.24 × 10−4 |
| cg09837977 | LRRN3 | 0.84 | 0.82 | 0.01 | 3.36 × 10−5 | 0.82 | 0.82 | 5 × 10−3 | 6.69 × 10−4 |
ICGN, International COPD Genetics Network; EOCOPD, Early-Onset COPD; FDR, false-discovery rate
Methylation values amy range from 0 (no methylation) to 1 (fully methylated).
Mean value of time since quit (in years) by quartiles in ICGN: Quartile 1 = 1.5, Quartile 2 = 6.3, Quartile 3 = 14.2, Quartile 4 = 27.4.
Mean value of time since quit (in years) by quartiles in EOCOPD: Quartile 1 = 1.3, Quartile 2 = 3.5, Quartile 3 = 7.4, Quartile 4 = 18.7.
aDefined as sites with an FDR adjusted P-value <0.05 in the discovery cohort (ICGN) and a one-sided P-value <10−3 in the replication cohort (EOCOPD).
bOne-sided P-value.
Figure 2.
Scatter plots of methylation by time since quitting in the ICGN cohort. X-axis represents time since smoking cessation (in years), y-axis is the methylation at (A) cg03636183 (F2RL3), (B) cg19859270 (GPR15) and (C) cg09837977 (LRRN3). Methylation values may range from 0 (no methylation) to 1 (complete methylation).
Table 5.
P-values for multiplicative interaction term between pack-years and time since quitting in ICGN
| CpG | Gene P-value (unadjusted model) | Interaction term | Interaction term P-value (adjusted model)a |
|---|---|---|---|
| cg03636183 | F2RL3 | 7.17 × 10−3 | 4.56 × 10−5 |
| cg19859720 | GPR15 | 0.03 | 0.01 |
aModel adjusted for age, sex and surrogate variables.
Figure 3.
Qualitative illustration of interaction between cumulative smoke exposure and time since smoking cessation at the F2RL3 locus in the ICGN cohort. Quartiles of cumulative smoke exposure (pack-years) are plotted on the x-axis, quartiles of time since smoking cessation (in years) are plotted on the z-axis, while the mean methylation of each quartile is plotted on the y-axis. Methylation values may range from 0 (no methylation) to 1 (complete methylation). The mean methylation for each cell is as follows (from left to right): Blue columns: 0.88 (lowest pack-years, longest duration since cessation), 0.85, 0.86, 0.82. Red columns: 0.86, 0.83, 0.83, 0.80. Green columns: 0.85, 0.83, 0.80, 0.77. Purple columns: 0.85, 0.83, 0.79, 0.76 (highest pack-years, shortest duration since cessation).
Because our cohorts were ascertained on the presence or absence of COPD, we repeated our analyses with the addition of a covariate to adjust for obstructive lung disease. Using the same criteria for significance employed in our original analysis, 13 of the 15 sites remained significantly associated with current smoking. Both F2RL3 and GPR15 remained significantly associated with cumulative smoke exposure as measured in pack-years. F2RL3 remained significant in the time since quitting analysis after adjustment for obstructive lung disease.
Pyrosequencing was successfully completed in the ICGN cohort for the F2RL3 and GPR15 loci; completion rates were 99.6 and 96.9%, respectively. The distributions of the pyrosequencing values with respect to the current smoking status are illustrated in Supplementary Material, Figure S1. Student's t-test comparing DNA methylation by the current smoking status remained highly statistically significant for both F2RL3 (P-value 2.2 × 10−16) and GPR15 (P-value 1.5 × 10−12) and again demonstrated relative hypomethylation associated with current cigarette smoking for each locus. Consistent with the array-based data, a significant inverse correlation between DNA methylation and pack-years of smoking (PF2RL3 = 8.54 × 10−9 and PGPR15 = 7.68 × 10−3) and a significant positive correlation between time since quitting (PF2RL3 = 1.35 × 10−11 and PGPR15 = 7.58 × 10−5) and DNA methylation were also confirmed.
DISCUSSION
Complex chronic exposures such as cigarette smoking can pose challenges in the analysis and interpretation of DNA methylation data. However, the careful analysis of large data sets with detailed phenotype information, along with rigorous statistical thresholds for significance, can reveal novel loci and pathways which may be involved in mediating the effects of smoking in the development of disease. Significantly associated sites from our analyses have been annotated to genes involved in a wide range of functions including cell signaling/apoptosis (FASLG), neurodevelopment (LRRN3, CNTNAP2) and alcohol metabolism (ADHFE1). Myosin light chain kinase (MYLK) has been ascribed putative roles in endothelial (30) and epithelial (31–34) barrier functions; variants in this gene have been associated with acute lung injury (35,36), asthma (37–39) and familial aortic dissections (40). Interestingly, cg09837977 (LRRN3), cg12044210 and cg21917349 (APBA2), and cg10161121 (FASLG) map to loci that have previously been included in a smoking ‘quit-success’ genotype score (41).
F2RL3 and GPR15, the two loci with the strongest associations with current smoking, have also been annotated to seemingly disparate functions. F2RL3, also known as protease-activated receptor 4, has established roles in platelet activation (42,43) and cell signaling (44); differential methylation at this site with respect to current smoking was first reported by Breitling et al. (14). GPR15 is a protein which shares sequence homology with the angiotensin II AT1 and AT2 receptors (45) and has been established as a co-receptor for the human immunodeficiency virus (46–48). Although there have been no previous statistically significant associations of this locus with smoking, cg19859270 was notably ranked second in the analysis conducted by Breitling et al. (14) and demonstrated a similar magnitude and direction of the effect.
We acknowledge that, despite robust statistical significance, the effect estimates of many of the loci reported in this manuscript are modest. Even the effect estimates for F2RL3 and GPR15 do not meet the 15% change often cited as a criterion for significance. However, considerable evidence supports the accuracy of our estimates. The differences in the mean methylation between current smokers and non-smokers for F2RL3 and GPR15 do not appear to be artifacts of the array-based platform as pyrosequencing produced similar differences in the mean methylation for each site. In addition, the differences in the mean methylation are comparable with those reported in the independent populations studied by Breitling et al. (14). We contend, therefore, that large-scale epigenetic studies are a nascent field and that the threshold of change in DNA methylation which correlates with functional significance on the population level has not been firmly established.
To examine the possible functional impact of differential methylation at loci reported in this manuscript, we conducted exploratory analyses using publicly available data sets to assess for changes in gene expression by the current smoking status in blood (49–53), histologically normal lung tissue (54) and bronchial epithelium (55) (Supplementary Material, Table S3). Using Student's t-test to compare the processed, log-transformed expression values in current smokers versus former/never smokers, several loci demonstrated evidence of nominal association (unadjusted P-value < 0.05). Increased gene expression in current smokers was noted for GPR15 in one of two peripheral blood data sets with transcripts available, whereas FASLG, the sole overlapping locus where differential gene expression with respect to the smoking status has previously been reported in blood (50), was found to have decreased expression in smokers in three data sets. Interestingly, increased gene expression in current smokers at LRRN3, a locus which has not been previously reported in either methylation or expression studies, was noted in four of five peripheral blood data sets (with the fifth data set potentially limited by small sample size). FASLG and LRRN3 were not significantly associated in either lung or bronchial epithelial tissue while gene expression at MYLK, which was reported to demonstrate rapidly reversible changes in bronchial epithelium (55), did not vary by the current smoking status in blood. These differences may be attributable to the tissue specificity of gene expression and DNA methylation patterns.
Although our data are cross-sectional in nature, our analyses suggest that dynamic changes in DNA methylation in response to smoking may exist for F2RL3 and GPR15. The inverse correlation between increasing pack-years and methylation at F2RL3 and GPR15 is consistent with progressive demethylation in the setting of higher cumulative smoke exposure. Likewise, the positive correlation between time since quitting and methylation at these loci may be due to remethylation following smoking cessation.
The time course over which DNA methylation changes occur is not known and may vary by CpG site. The concept of differential responses to and recovery from exposure to cigarette smoke was well articulated in a gene expression study conducted in primary bronchial epithelial cells (55); while the majority of gene expression changes due to smoking were found to be rapidly reversible, a subset of loci demonstrated slow or irreversible changes. Studies performed in respiratory epithelial cell cultures exposed to cigarette smoke extract support the concept that distinct loci may be involved in short-term versus chronic exposure to cigarette smoke. In this study, changes in DNA methylation were observable after only 9 months of cigarette smoke exposure (56); none of the differentially methylated sites overlapped with our loci.
To explore whether differential methylation at the loci reported in our study might be involved in rapidly reversible versus slowly reversible-type changes, we conducted additional subgroup analyses in a subset of the public gene expression data sets used in Supplementary Material, Table S3 which had data on former smokers. Although data on time since quitting was not available for former smokers in these cohorts, we hypothesized that genes which were differentially expressed between current and former smokers might represent relatively ‘rapidly reversible’ loci (Supplementary Material, Table S4). Conversely, we speculated that differential expression between former and never smokers might represent ‘slowly reversible’ or irreversible changes (Supplementary Material, Table S5). MYLK, a locus reported to have ‘rapidly reversible’ changes in gene expression in bronchial epithelial cells (55), conforms to our hypotheses well; differential expression is seen between current and former smokers, but not between former and never smokers (in bronchial epithelial cells). LRRN3, relatively hypomethylated in current smokers (Table 2) and relatively hypermethylated following smoking cessation (Table 4), appears to fit this speculative model of a ‘rapidly reversible’ locus with differential (increased) expression evident between current and former smokers in peripheral blood, but no differences between former and never smokers. In contrast, differential expression in peripheral blood at GPR15 and FASLG was observed in both the current/former and former/never smoker analyses and suggests that these loci may demonstrate ‘slowly reversible’ changes. Although intriguing, we acknowledge that only very limited conclusions can be drawn from these unadjusted exploratory analyses conducted in non-contemporaneous data sets and that additional studies are warranted.
We recognize the limitation of association studies as providing only correlative data; we cannot establish whether the changes in methylation reported herein are primary (i.e. established directly by smoking exposure) or secondary to more generalized processes (i.e. established as a consequence of perturbations in other cellular processes). Gene expression (55) and DNA methylation (56) changes may both persist in non-malignant tissues for extended periods following smoking cessation; the mechanism by which these changes are established and maintained is a topic of active research (57). Changes in methylation marks at specific loci may arise from a change in DNA methyltransferase expression, abundance or activity. Indeed, changes in DNA methyltransferase expression and levels have been reported in neurons (58) and human bronchial epithelial cell cultures (56) exposed to nicotine or cigarette smoke extract. However, we found no compelling evidence for differential expression in the DNA methyltransferases by current smoking status in blood (49–53,55), histologically normal lung tissue (54) or bronchial epithelium (55) based on our exploration of public data sets (Supplementary Material, Table S6). Alternative mechanisms, beyond changes in methyltransferase gene expression, may contribute to the establishment of locus-specific differential methylation observed in our study; additional in vitro and in vivo studies are needed to explore this question.
Potential limitations of this study include the inability to quantify the degree to which cell type heterogeneity in blood contributes to our results and the reliance upon self-reported smoking metrics. Epidemiological studies have found self-reported smoking to be reliable except in cases where non-smoking is perceived as socially desirable (59–61). Because misclassification due to under-reporting of current smoking would bias our results towards the null, we believe our results are robust even in the absence of biochemical confirmation of current smoking status (i.e. serum cotinine measurements).
Lastly, the frequency and impact of genetic and structural variants on the array-based platforms used to assess DNA methylation in our study is unknown and is an area of active research (62). We performed sequence alignment of the probes of significantly associated sites against a consensus human genome (NCBI37/hg19) and noted a number of single nucleotide polymorphisms, copy number variants and repetitive elements overlapping with our probes (Supplementary Material, Table S7). To assess the impact of these variants on our methylation data, we plotted the distribution of the raw methylation values of current and non-smokers (Supplementary Material, Fig. S2) and the distribution of the raw methylation values against the continuous variable of pack-years of smoking (Supplementary Material, Fig. S3). We saw no evidence of distinct subpopulations indicative of genetic or structural variants among significantly associated sites. Additionally, despite the report of structural variants involving the GPR15 locus, the association of this locus remained robust when assessed through a second technology (pyrosequencing).
Given the profound physiological impact of smoking as well as considerable epidemiological evidence linking smoking to a number of systemic diseases beyond directly exposed organs, the effects of smoking on peripheral blood DNA methylation appear to impact a limited number of loci in our cohorts. In addition, since gene expression changes were not noted in our exploratory analyses at the majority of loci reported in our study, the impact of differential methylation at these loci remains to be established. Thus, the integration of epigenetic data with genetic and genomic lines of investigation will likely yield additional biological insights into clinical challenges such as smoking cessation and smoking-related complex diseases.
MATERIALS AND METHODS
Subjects/cohorts
The discovery cohort consisted of subjects participating in the ICGN (27,28), a family-based study which recruited probands in the age range of 45–65, with ≥5 pack-years of cigarette smoking and airflow limitation (defined as an FEV1 <60% predicted and an FEV1/VC <90% predicted). Subjects with alpha-1 antitrypsin genotypes PiZZ, Z-Null, Null–Null or SZ were excluded from the study. In addition, all probands were required to have at least one eligible sibling with ≥5 pack-years of cigarette smoking. All subjects completed spirometry, questionnaire data and had whole blood drawn for DNA analysis. Subjects included in the current analysis were of self-reported Caucasian ancestry. The protocol was approved by the local Internal Review Board at each of the recruitment sites and all subjects provided written informed consent. Details regarding the number of samples excluded at each step during data cleaning have been previously described (63) and are summarized in Supplementary Material, Methods; the number of subjects analyzed from this cohort was 1085.
The replication cohort consisted of subjects recruited from the Boston Severe EOCOPD (29), a family-based study which recruited probands who had an FEV1 <40% predicted, by age <53 in the absence of severe alpha-1 antitrypsin deficiency. Although extended pedigrees were recruited, DNA methylation data were obtained only on probands and their siblings. Subjects completed spirometry, questionnaire data and provided blood for DNA analysis. The protocol was approved by the Brigham and Women's Hospital Human Research Committee; all subjects provided written informed consent. Sample processing and quality control measures were identical to those employed for the ICGN cohort as outlined above; the number of subjects from the EOCOPD cohort included in our analysis was 369.
Variable definitions
Current smoking was defined as an affirmative response to the question: ‘Do you currently smoke cigarettes (as of 1 month ago)?’ The average number of cigarettes smoked per day was the self-reported number of cigarettes smoked per day during the period when the subject smoked. Pack-years was calculated according to the number of years smoked multiplied by the average number of cigarettes smoked per day divided by 20. In former smokers, the time since quitting was calculated as the subject's age minus the age when they stopped smoking (in years). Obstructive lung disease was defined as an FEV1/FVC ratio <0.7.
DNA extraction and bisulfite treatment
For both cohorts, DNA extraction from white blood cells was performed using the Gentra Puregene blood kit (Qiagen, Inc., Valencia, CA, USA). Bisulfite modification of 1 mg of DNA was performed using the 96-well EZ DNA Methylation Gold Deep-well kit (Zymo Research, Orange, CA, USA). The protocol is outlined in Supplementary Material, Methods. Less than 2% of DNA samples failed bisulfite conversion.
DNA methylation array
Both cohorts were assessed using the Illumina (San Diego, CA, USA) Infinium HumanMethylation27K BeadChip, which assays 27 578 CpG sites in 14 475 consensus coding sequences in the NCBI Database (Genome Build 36). Bisulfite converted DNA was amplified, fragmented and hybridized to locus-specific 50-mer probes on the arrays, followed by single base extension using biotin (C and G) or dinitrophenyl (A and T) labeled nucleotides, per routine. All arrays were imaged using an Illumina BeadArray Reader. Beadstudio software (methylation module version 3.2) was used to assemble the workspace. Each locus results in an intensity value for methylation (M) and unmethylated (U) alleles. The methylation status was expressed as a beta value (β), which represents a ratio of the M to U fluorescent signals such that β = Max (M,0)/[Max(M,0) + Max(U,0) + 100]. Using this metric, DNA methylation is represented by a variable between 0 (no methylation) to 1 (complete methylation). Raw average beta values were exported and analyzed without normalization. CpG sites will be discussed using the industry assigned identifier (‘cg’) as outlined in the Illumina HumanMethylation27K manifest. Quality control measures for array data are described in Supplementary Material, Methods.
Pyrosequencing
We performed pyrosequencing using the Pyromark96MD (Qiagen, Inc.) for technical validation of cg03636183 (F2RL3) and cg19859270 (GPR15) in the ICGN cohort. Details are provided in Supplementary Material. CpG methylation percentages were calculated using the height of the T and C peaks at the methylation site and applying the formula C/(C + T) × 100 as implemented in the Pyromark CpG software (version 1.0.11).
Statistical analysis
Surrogate variable analysis (64,65), which identifies and estimates the effects of known and unknown sources of variation in high-throughput experiments, was performed to account for batch effects. To account for familial correlations in the data, generalized linear mixed models (66–69) as implemented in R (release 2.12) were used to analyze the methylation data. We used an identity link function in our models and the penalized quasi-likelihood method (66) to estimate the model parameters. The Illumina ‘beta’ value was modeled as the dependent variable, whereas the smoking metric (current smoking, pack-years and time since quitting cigarettes) was included as the independent variable. All models were adjusted for age, gender and significant surrogate variables. CpG sites with a false-discovery rate (FDR) adjusted P-value <0.05 (70) in the discovery cohort (ICGN) were examined using the same models in the replication cohort (EOCOPD). CpG sites demonstrating a P-value <10−3 in the replication cohort with the same direction of effect were considered significant. To assess the impact of COPD on our results, we repeated our analysis with the addition of a covariate to account for obstructive lung disease.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. E.S.W., H.B. W.Q., V.J.C., A.B. and D.L.D. have no conflicts of interest to report. D.A.L. has received grant funding, consultancy fees and honoraria from GlaxoSmithKline. W.A. was an employee of GlaxoSmithKline at the time of the analyses. S.R. has consulted or participated in advisory boards for: ABIM, Adelphi Research, Almirall/Forest, APT, AstraZeneca, Boehringer Ingelheim/Pfizer, BoomCom, Capital Research, CommonHealth, Decision Resources, Easton Associates, Equinox, Forest (Cory Paeth), Fulcrum, Gerson Lehman, GSK Eclipse, Guidepoint, Informed, Insyght, MedImmune, Novartis, Oriel, Pearl, Pfizer (Varenicline), PharmaVentures, Prescott, Propagate Pulmonary Reviews, Roche, Schering Plough, Smith Research, Talecris, UBC. S.R. has given lectures for: American College of Osteopathic Physicians, Asan Medical Center, American Thoracic Society, COPD Foundation, Creative Education Concepts, Duke, Information TV, Nycomed, Otsuka, University of Washington, University of Alabama-Birmingham. S.R. has received industry-sponsored grants from: AstraZeneca, Otsuka, Pfizer, Nabi, Merck. A.A. a board member of GlaxoSmithKline, Almirall, Boheringer-Ingelheim, Astra-Zeneca, Esteve, Novartis, Nycomed and Roche. A.A. has grants (current or pending) from GlaxoSmithKline, Almirall and Nycomed. A.A. has received payment for lectures and educational presentations from GlaxoSmithKline, Almirall, Boheringer-Ingelheim, Astra-Zeneca, Esteve and Nycomed.
FUNDING
This work was supported by the National Institutes of Health (R01 HL089438, R01 HL075478, T32 HL007427, K12 HL089990, P01 HL105339); the Doris Duke Foundation Clinician Scientist Development Award (to D.L.D.); and the National Institute of Environmental Health Sciences-Harvard School of Public Health New Investigator Fund (ES00002 to A.B.). None of the funding sources played a role in the management, analysis or interpretation of the data, nor did they influence the preparation, review or decision to submit for publication.
Supplementary Material
REFERENCES
- 1.Mathers C.D., Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3:e442. doi: 10.1371/journal.pmed.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conen D., Everett B.M., Kurth T., Creager M.A., Buring J.E., Ridker P.M., Pradhan A.D. Smoking, smoking status, and risk for symptomatic peripheral artery disease in women: a cohort study. Ann. Intern. Med. 2011;154:719–726. doi: 10.1059/0003-4819-154-11-201106070-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawachi I., Colditz G.A., Stampfer M.J., Willett W.C., Manson J.E., Rosner B., Speizer F.E., Hennekens C.H. Smoking cessation and decreased risk of stroke in women. JAMA. 1993;269:232–236. [PubMed] [Google Scholar]
- 4.Rea T.D., Heckbert S.R., Kaplan R.C., Smith N.L., Lemaitre R.N., Psaty B.M. Smoking status and risk for recurrent coronary events after myocardial infarction. Ann. Intern. Med. 2002;137:494–500. doi: 10.7326/0003-4819-137-6-200209170-00009. [DOI] [PubMed] [Google Scholar]
- 5.Bouloukaki I., Tsiligianni I.G., Tsoumakidou M., Mitrouska I., Prokopakis E.P., Mavroudi I., Siafakas N.M., Tzanakis N. Sputum and nasal lavage lung-specific biomarkers before and after smoking cessation. BMC Pulm. Med. 2011;11:35. doi: 10.1186/1471-2466-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braber S., Henricks P.A., Nijkamp F.P., Kraneveld A.D., Folkerts G. Inflammatory changes in the airways of mice caused by cigarette smoke exposure are only partially reversed after smoking cessation. Respir. Res. 2010;11:99. doi: 10.1186/1465-9921-11-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Louhelainen N., Stark H., Mazur W., Rytila P., Djukanovic R., Kinnula V.L. Elevation of sputum matrix metalloproteinase-9 persists up to 6 months after smoking cessation: a research study. BMC Pulm. Med. 2010;10:13. doi: 10.1186/1471-2466-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vernooy J.H., Kucukaycan M., Jacobs J.A., Chavannes N.H., Buurman W.A., Dentener M.A., Wouters E.F. Local and systemic inflammation in patients with chronic obstructive pulmonary disease: soluble tumor necrosis factor receptors are increased in sputum. Am. J. Respir. Crit. Care Med. 2002;166:1218–1224. doi: 10.1164/rccm.2202023. [DOI] [PubMed] [Google Scholar]
- 9.Willemse B.W., ten Hacken N.H., Rutgers B., Lesman-Leegte I.G., Postma D.S., Timens W. Effect of 1-year smoking cessation on airway inflammation in COPD and asymptomatic smokers. Eur. Respir. J. 2005;26:835–845. doi: 10.1183/09031936.05.00108904. [DOI] [PubMed] [Google Scholar]
- 10.Luo J., Margolis K.L., Wactawski-Wende J., Horn K., Messina C., Stefanick M.L., Tindle H.A., Tong E., Rohan T.E. Association of active and passive smoking with risk of breast cancer among postmenopausal women: a prospective cohort study. BMJ. 2011;342:d1016. doi: 10.1136/bmj.d1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vineis P., Alavanja M., Buffler P., Fontham E., Franceschi S., Gao Y.T., Gupta P.C., Hackshaw A., Matos E., Samet J., et al. Tobacco and cancer: recent epidemiological evidence. J. Natl Cancer Inst. 2004;96:99–106. doi: 10.1093/jnci/djh014. [DOI] [PubMed] [Google Scholar]
- 12.Newcomb P.A., Carbone P.P. The health consequences of smoking. Cancer Med. Clin. North Am. 1992;76:305–331. doi: 10.1016/s0025-7125(16)30355-8. [DOI] [PubMed] [Google Scholar]
- 13.Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M., et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breitling L.P., Yang R., Korn B., Burwinkel B., Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am. J. Hum. Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breton C.V., Byun H.M., Wenten M., Pan F., Yang A., Gilliland F.D. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am. J. Respir. Crit. Care Med. 2009;180:462–467. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerrero-Preston R., Goldman L.R., Brebi-Mieville P., Ili-Gangas C., Lebron C., Witter F.R., Apelberg B.J., Hernandez-Roystacher M., Jaffe A., Halden R.U., et al. Global DNA hypomethylation is associated with in utero exposure to cotinine and perfluorinated alkyl compounds. Epigenetics. 2010;5:539–546. doi: 10.4161/epi.5.6.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terry M.B., Ferris J.S., Pilsner R., Flom J.D., Tehranifar P., Santella R.M., Gamble M.V., Susser E. Genomic DNA methylation among women in a multiethnic New York City birth cohort. Cancer Epidemiol. Biomarkers Prev. 2008;17:2306–2310. doi: 10.1158/1055-9965.EPI-08-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi J.Y., James S.R., Link P.A., McCann S.E., Hong C.C., Davis W., Nesline M.K., Ambrosone C.B., Karpf A.R. Association between global DNA hypomethylation in leukocytes and risk of breast cancer. Carcinogenesis. 2009;30:1889–1897. doi: 10.1093/carcin/bgp143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang F.F., Cardarelli R., Carroll J., Fulda K.G., Kaur M., Gonzalez K., Vishwanatha J.K., Santella R.M., Morabia A. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics. 2011;6:623–629. doi: 10.4161/epi.6.5.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsiung D.T., Marsit C.J., Houseman E.A., Eddy K., Furniss C.S., McClean M.D., Kelsey K.T. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol. Biomarkers Prev. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 21.Moore L.E., Pfeiffer R.M., Poscablo C., Real F.X., Kogevinas M., Silverman D., Garcia-Closas R., Chanock S., Tardon A., Serra C., et al. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case-control study. Lancet Oncol. 2008;9:359–366. doi: 10.1016/S1470-2045(08)70038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Z.Z., Hou L., Bollati V., Tarantini L., Marinelli B., Cantone L., Yang A.S., Vokonas P., Lissowska J., Fustinoni S., et al. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int. J. Epidemiol. 2012;41:126–139. doi: 10.1093/ije/dyq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ally M.S., Al-Ghnaniem R., Pufulete M. The relationship between gene-specific DNA methylation in leukocytes and normal colorectal mucosa in subjects with and without colorectal tumors. Cancer Epidemiol. Biomarkers Prev. 2009;18:922–928. doi: 10.1158/1055-9965.EPI-08-0703. [DOI] [PubMed] [Google Scholar]
- 24.Belinsky S.A., Klinge D.M., Dekker J.D., Smith M.W., Bocklage T.J., Gilliland F.D., Crowell R.E., Karp D.D., Stidley C.A., Picchi M.A. Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin. Cancer Res. 2005;11:6505–6511. doi: 10.1158/1078-0432.CCR-05-0625. [DOI] [PubMed] [Google Scholar]
- 25.Brait M., Ford J.G., Papaiahgari S., Garza M.A., Lee J.I., Loyo M., Maldonado L., Begum S., McCaffrey L., Howerton M., et al. Association between lifestyle factors and CpG island methylation in a cancer-free population. Cancer Epidemiol. Biomarkers Prev. 2009;18:2984–2991. doi: 10.1158/1055-9965.EPI-08-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Umemura S., Fujimoto N., Hiraki A., Gemba K., Takigawa N., Fujiwara K., Fujii M., Umemura H., Satoh M., Tabata M., et al. Aberrant promoter hypermethylation in serum DNA from patients with silicosis. Carcinogenesis. 2008;29:1845–1849. doi: 10.1093/carcin/bgn169. [DOI] [PubMed] [Google Scholar]
- 27.Patel B.D., Coxson H.O., Pillai S.G., Agusti A.G., Calverley P.M., Donner C.F., Make B.J., Muller N.L., Rennard S.I., Vestbo J., et al. Airway wall thickening and emphysema show independent familial aggregation in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2008;178:500–505. doi: 10.1164/rccm.200801-059OC. [DOI] [PubMed] [Google Scholar]
- 28.Zhu G., Warren L., Aponte J., Gulsvik A., Bakke P., Anderson W.H., Lomas D.A., Silverman E.K., Pillai S.G. The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am. J. Respir. Crit. Care Med. 2007;176:167–173. doi: 10.1164/rccm.200611-1723OC. [DOI] [PubMed] [Google Scholar]
- 29.Silverman E.K., Chapman H.A., Drazen J.M., Weiss S.T., Rosner B., Campbell E.J., O'Donnell W.J., Reilly J.J., Ginns L., Mentzer S., et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease. Risk to relatives for airflow obstruction and chronic bronchitis. Am. J. Respir. Crit. Care Med. 1998;157:1770–1778. doi: 10.1164/ajrccm.157.6.9706014. [DOI] [PubMed] [Google Scholar]
- 30.Dudek S.M., Jacobson J.R., Chiang E.T., Birukov K.G., Wang P., Zhan X., Garcia J.G. Pulmonary endothelial cell barrier enhancement by sphingosine 1-phosphate: roles for cortactin and myosin light chain kinase. J. Biol. Chem. 2004;279:24692–24700. doi: 10.1074/jbc.M313969200. [DOI] [PubMed] [Google Scholar]
- 31.Clayburgh D.R., Rosen S., Witkowski E.D., Wang F., Blair S., Dudek S., Garcia J.G., Alverdy J.C., Turner J.R. A differentiation-dependent splice variant of myosin light chain kinase, MLCK1, regulates epithelial tight junction permeability. J. Biol. Chem. 2004;279:55506–55513. doi: 10.1074/jbc.M408822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Connell L.E., Helfman D.M. Myosin light chain kinase plays a role in the regulation of epithelial cell survival. J. Cell Sci. 2006;119:2269–2281. doi: 10.1242/jcs.02926. [DOI] [PubMed] [Google Scholar]
- 33.Russo J.M., Florian P., Shen L., Graham W.V., Tretiakova M.S., Gitter A.H., Mrsny R.J., Turner J.R. Distinct temporal-spatial roles for rho kinase and myosin light chain kinase in epithelial purse-string wound closure. Gastroenterology. 2005;128:987–1001. doi: 10.1053/j.gastro.2005.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang F., Graham W.V., Wang Y., Witkowski E.D., Schwarz B.T., Turner J.R. Interferon-gamma and tumor necrosis factor-alpha synergize to induce intestinal epithelial barrier dysfunction by up-regulating myosin light chain kinase expression. Am. J. Pathol. 2005;166:409–419. doi: 10.1016/s0002-9440(10)62264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao L., Grant A., Halder I., Brower R., Sevransky J., Maloney J.P., Moss M., Shanholtz C., Yates C.R., Meduri G.U., et al. Novel polymorphisms in the myosin light chain kinase gene confer risk for acute lung injury. Am. J. Respir. Cell Mol. Biol. 2006;34:487–495. doi: 10.1165/rcmb.2005-0404OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christie J.D., Ma S.F., Aplenc R., Li M., Lanken P.N., Shah C.V., Fuchs B., Albelda S.M., Flores C., Garcia J.G. Variation in the myosin light chain kinase gene is associated with development of acute lung injury after major trauma. Crit. Care Med. 2008;36:2794–2800. doi: 10.1097/ccm.0b013e318186b843. [DOI] [PubMed] [Google Scholar]
- 37.Flores C., Ma S.F., Maresso K., Ober C., Garcia J.G. A variant of the myosin light chain kinase gene is associated with severe asthma in African Americans. Genet. Epidemiol. 2007;31:296–305. doi: 10.1002/gepi.20210. [DOI] [PubMed] [Google Scholar]
- 38.Lee S.O., Cheong H.S., Park B.L., Bae J.S., Sim W.C., Chun J.Y., Isbat M., Uh S.T., Kim Y.H., Jang A.S., et al. MYLK polymorphism associated with blood eosinophil level among asthmatic patients in a Korean population. Mol. Cells. 2009;27:175–181. doi: 10.1007/s10059-009-0022-2. [DOI] [PubMed] [Google Scholar]
- 39.Leguillette R., Laviolette M., Bergeron C., Zitouni N., Kogut P., Solway J., Kachmar L., Hamid Q., Lauzon A.M. Myosin, transgelin, and myosin light chain kinase: expression and function in asthma. Am. J. Respir. Crit. Care Med. 2009;179:194–204. doi: 10.1164/rccm.200609-1367OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L., Guo D.C., Cao J., Gong L., Kamm K.E., Regalado E., Li L., Shete S., He W.Q., Zhu M.S., et al. Mutations in myosin light chain kinase cause familial aortic dissections. Am. J. Hum. Genet. 2010;87:701–707. doi: 10.1016/j.ajhg.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rose J.E., Behm F.M., Drgon T., Johnson C., Uhl G.R. Personalized smoking cessation: interactions between nicotine dose, dependence and quit-success genotype score. Mol. Med. 2010;16:247–253. doi: 10.2119/molmed.2009.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kahn M.L., Nakanishi-Matsui M., Shapiro M.J., Ishihara H., Coughlin S.R. Protease-activated receptors 1 and 4 mediate activation of human platelets by thrombin. J. Clin. Invest. 1999;103:879–887. doi: 10.1172/JCI6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu W.F., Andersen H., Whitmore T.E., Presnell S.R., Yee D.P., Ching A., Gilbert T., Davie E.W., Foster D.C. Cloning and characterization of human protease-activated receptor 4. Proc. Natl Acad. Sci. USA. 1998;95:6642–6646. doi: 10.1073/pnas.95.12.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen H.T., Tsou H.K., Tsai C.H., Kuo C.C., Chiang Y.K., Chang C.H., Fong Y.C., Tang C.H. Thrombin enhanced migration and MMPs expression of human chondrosarcoma cells involves PAR receptor signaling pathway. J. Cell Physiol. 2010;223:737–745. doi: 10.1002/jcp.22083. [DOI] [PubMed] [Google Scholar]
- 45.Heiber M., Marchese A., Nguyen T., Heng H.H., George S.R., O'Dowd B.F. A novel human gene encoding a G-protein-coupled receptor (GPR15) is located on chromosome 3. Genomics. 1996;32:462–465. doi: 10.1006/geno.1996.0143. [DOI] [PubMed] [Google Scholar]
- 46.Blaak H., Boers P.H., Gruters R.A., Schuitemaker H., van der Ende M.E., Osterhaus A.D. CCR5, GPR15, and CXCR6 are major coreceptors of human immunodeficiency virus type 2 variants isolated from individuals with and without plasma viremia. J. Virol. 2005;79:1686–1700. doi: 10.1128/JVI.79.3.1686-1700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cilliers T., Willey S., Sullivan W.M., Patience T., Pugach P., Coetzer M., Papathanasopoulos M., Moore J.P., Trkola A., Clapham P., et al. Use of alternate coreceptors on primary cells by two HIV-1 isolates. Virology. 2005;339:136–144. doi: 10.1016/j.virol.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 48.Farzan M., Choe H., Martin K., Marcon L., Hofmann W., Karlsson G., Sun Y., Barrett P., Marchand N., Sullivan N., et al. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J. Exp. Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buttner P., Mosig S., Funke H. Gene expression profiles of T lymphocytes are sensitive to the influence of heavy smoking: A pilot study. Immunogenetics. 2007;59:37–43. doi: 10.1007/s00251-006-0177-3. [DOI] [PubMed] [Google Scholar]
- 50.Charlesworth J.C., Curran J.E., Johnson M.P., Goring H.H.H., Dyer T.D., Diego V.P., Kent J.W., Mahaney M.C., Almasy L., MacClue J.W., et al. Transcriptomic epidemiology of smoking: the effect of smoking on gene expression in lymphocytes. BMC Med. Genomics. 2010;3:29. doi: 10.1186/1755-8794-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dumeaux V., Olsen K.S., Nuel G., Paulssen R.H., Borresen-Dale A.L., Lund E. Deciphering normal blood gene expression variation—the NOWAC postgenome study. PLoS Genet. 2010;6:e1000873. doi: 10.1371/journal.pgen.1000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rotunno M., Hu N., Su H., Wang C., Goldstein A.M., Bergen A.W., Consonni D., Pesatori A.C., Bertazzi P.A., Wacholder S., et al. A gene expression signature from peripheral whole blood for stage I lung adenocarcinoma. Cancer Prev. Res. (Phila) 2011;4:1599–1608. doi: 10.1158/1940-6207.CAPR-10-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Showe M.K., Vachani A., Kossenkov A.V., Yousef M., Nichols C., Nikonova E.V., Chang C., Kucharczuk J., Tran B., Wakeam E., et al. Gene expression profiles in peripheral blood mononuclear cells can distinguish patients with non-small cell lung cancer from patients with nonmalignant lung disease. Cancer. Res. 2009;69:9202–9210. doi: 10.1158/0008-5472.CAN-09-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Landi M.T., Dracheva T., Rotunno M., Figueroa J.D., Liu H., Dasgupta A., Mann F.E., Fukuoka J., Hames M., Bergen A.W., et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS ONE. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beane J., Sebastiani P., Liu G., Brody J.S., Lenburg M.E., Spira A. Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biol. 2007;8:R201. doi: 10.1186/gb-2007-8-9-r201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu F., Killian J.K., Yang M., Walker R.L., Hong J.A., Zhang M., Davis S., Zhang Y., Hussain M., Xi S., et al. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene. 2010;29:3650–3664. doi: 10.1038/onc.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schembri F., Sridhar S., Perdomo C., Gustafson A.M., Zhang X., Ergun A., Lu J., Liu G., Bowers J., Vaziri C., et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc. Natl Acad. Sci. USA. 2009;106:2319–2324. doi: 10.1073/pnas.0806383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Satta R., Maloku E., Zhubi A., Pibiri F., Hajos M., Costa E., Guidotti A. Nicotine decreases DNA methyltransferase 1 expression and glutamic acid decarboxylase 67 promoter methylation in GABAergic interneurons. Proc. Natl Acad. Sci. USA. 2008;105:16356–16361. doi: 10.1073/pnas.0808699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murray R.P., Connett J.E., Lauger G.G., Voelker H.T. Error in smoking measures: effects of intervention on relations of cotinine and carbon monoxide to self-reported smoking. The Lung Health Study Research Group. Am. J. Public Health. 1993;83:1251–1257. doi: 10.2105/ajph.83.9.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patrick D.L., Cheadle A., Thompson D.C., Diehr P., Koepsell T., Kinne S. The validity of self-reported smoking: a review and meta-analysis. Am. J. Public Health. 1994;84:1086–1093. doi: 10.2105/ajph.84.7.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wagenknecht L.E., Burke G.L., Perkins L.L., Haley N.J., Friedman G.D. Misclassification of smoking status in the CARDIA study: a comparison of self-report with serum cotinine levels. Am. J. Public Health. 1992;82:33–36. doi: 10.2105/ajph.82.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bock C., Tomazou E.M., Brinkman A.B., Muller F., Simmer F., Gu H., Jager N., Gnirke A., Stunnenberg H.G., Meissner A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat. Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qiu W., Baccarelli A., Carey V.J., Boutaoui N., Bacherman H., Klanderman B., Rennard S., Agusti A., Anderson W., Lomas D.A., et al. Variable DNA methylation is associated with COPD and lung function. Am J Respir Crit Care Med. 2012;185:373–381. doi: 10.1164/rccm.201108-1382OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leek J.T., Storey J.D. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leek J.T., Storey J.D. A general framework for multiple testing dependence. Proc. Natl Acad. Sci. USA. 2008;105:18718–18723. doi: 10.1073/pnas.0808709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Breslow N., Clayton D. Approximate inference in generalized linear mixed models. J. Am. Stat. Assoc. 1993;88:9–25. [Google Scholar]
- 67.Scall R. Estimation in generalized linear models with random effects. Biometrika. 1991;78:719–727. [Google Scholar]
- 68.Wolfinger R., O'Connell M. Generalized linear mixed models: a pseudo-likelihood approach. J. Stat. Comp. Simulat. 1993;48:233–243. [Google Scholar]
- 69.Venables W.N., Ripley B.D. Modern Applied Statistics with S-PLUS. 4th edn. Springer, New York, NY; 2002. [Google Scholar]
- 70.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.