Abstract
T helper (TH) cells integrate biochemical cues present within the tissue microenvironment to orchestrate immunity via production of cytokines. Prior discoveries reveal a qualitative understanding of how TH cells process this biochemical information, as frequently depicted as signaling cartoons. However, the lack of methods for quantifying how well these signaling cartoons apply to a particular cell type presents a major hurdle for translating our knowledge of immunity across systems. In this study, we use model-based inference methods, in conjunction with quantitative cytometry-based methods, to reason about the relative contributions of different putative branches within a signaling network. A cellular model of mouse TH1 cells was used to quantify the functional response to Interleukin-12 (IL-12), a key cytokine that links innate to adaptive immunity. Our results demonstrate that the response of TH cells to IL-12 exhibits a hysteresis and engages a positive feedback mechanism via direct activation of signal transducer and activator of transcription 1. The hysteresis in the dose-response curve to IL-12 creates a transient “memory” by sustaining cytokine secretion following the withdrawal of the stimuli. In summary, this combined experimental and computational approach illustrates how model-based inference can be used to obtain greater fidelity in understanding how cells process and act upon biochemical cues present within the tissue microenvironment.
Introduction
Cell-mediated immunity is tailored to the perceived threat to the host largely through the action of CD4+ T helper (TH) cells [1]. When presented in an appropriate context, TH cells recognize particular antigens associated with the threat and produce a distinct set of biochemical cues in response (e.g., cytokines). The repertoire of receptors that recognize specific antigens is selected within the thymus [2] while the pattern of cytokines produced by TH cells is determined within the periphery [3]. The role that TH cells play within the periphery is defined based upon the pattern of cytokines produced by and master regulatory transcription factors expressed within distinct subsets of TH cells [4]. The particular profile of cytokines produced by TH cells in response to a particular molecular pattern has a strong influence on the outcome of the immune response [1], where alternatives include tolerance, resolution, or autoimmunity. However, understanding how biochemical cues present within the periphery regulate the longevity and phenotype of TH cells (i.e., TH cell fate) remains a challenge in translating basic knowledge of cellular signaling pathways into practical application of this knowledge, including stem cell engineering, regenerative medicine, and immunotherapy.
The fate of TH cells is a quantitative cellular decision making process where biochemical cues present within the periphery act upon the TH cells by modifying intracellular proteins that in turn regulate cell response [5]. Our knowledge of this cellular decision making process is informed by a wealth of experimental data. Generally, the flow of information within a cell can be considered as a network of biochemical reactions that is governed by a propensity for productive interaction, local concentration, and conservation principles [6, 7]. The connectivity among interacting proteins (i.e., the topology) is commonly depicted as graphical signaling pathways. However, the existence of crosstalk among signaling pathways [8, 9], differences in the relative importance of alternative branches within signaling pathways among similar cellular systems [10], and cellular alterations that re-wire signaling pathways [11, 12] complicate interpreting observed data using signaling pathways as a conceptual framework. Moreover, biological systems exhibit intrinsic uncertainty. The uncertainty in characterizing a biological state can be attributed to multiple sources, including the underlying signal-to-noise characteristics of a biological assay [13], the skill of the experimentalist [14], ethical limitations, or cell-to-cell variability that may become important when sampling a finite number of states (i.e., stochasticity) [15, 16]. Given the body of knowledge currently assembled and the intrinsic uncertainty in the data, how do we infer the relative importance of a particular subset of these pathways within a particular system of interest?
Interleukin-12 (IL-12) is an important cytokine that is produced by innate immune cells and acts upon Natural Killer cells, CD8+ Cytotoxic T cells, and TH cells [24, 25]. IL-12 acts via a member of the Janus kinase (JAK) and signal transducer and activator of transcription (STAT) family of signaling pathways [26]. The Janus kinases, JAK2 and TYK2, associate with their corresponding IL-12 receptor subunits, IL-12Rβ1 and IL-12Rβ2, to create a signaling complex that phosphorylates STAT4. The post-translational modification of the STAT family plays a role in both inducing master transcriptional regulators of TH effector function and cytokine production [3]. Sufficient and sustained IL-12 signaling [27] leads to polarization of naïve CD4+ T cells into a TH1 phenotype [28]. Phosphorylated STAT4 translocates to the nucleus to promote the transcription of the cytokines, Interferon-γ (IFN-γ) and IL-10 [29], and the receptor IL-12Rβ2 [30]. IFN-γ is viewed traditionally as a TH1 cytokine while IL-10, produced by TH1 cells, is thought to function as a negative feedback loop to regulate pathogenic TH1 responses [31]. In addition to the direct feedback loop via STAT4, IFN-γ regulates IL-12Rβ2 expression [32], although the sensitivity to IFNγ-may depend on the differentiation state of the TH cell [33, 34]. Recently, Good et al. reported that pSTAT4 promotes the expression of TNF-α in TH1 cells [35]. In addition, an autocrine positive feedback loop regulates the expression of TNF-α [36, 37]. However, the dynamic role of these alternative signaling pathways in shaping the TH1 cell response to IL-12 and in maintaining a TH1 phenotype remains unclear.
Creating new knowledge as to how information flows within signaling networks from a statistical analysis of experimental observations – a process called statistical inference - relies on a spectrum of computational tools [17]. The particular computational approaches vary in how much previous knowledge is used - from none [18] to detailed knowledge regarding the relationships between proteins involved in a signaling pathway [19]. In addition, the particular approach used also influences the quality of inference that can be made about the signaling network - from binary statements regarding the inclusion of possible branches within a signaling network (e.g., Bayesian networks [20]) to quantitative statements about the relative contribution of a specific branch within a defined signaling network (e.g., ODE models [21]). Frequently, the inference problem is a mixture of these two extremes: some prior knowledge of the signaling network thought to play a role in a system exists but there are conflicts in the literature regarding specific details. The lack of quantitative estimates for parameter values also presents an obstacle against the use of quantitative models of signaling networks [22]. Although a single set of parameter values provides a single set of model predictions, inferring new signaling knowledge from data focuses on whether the collection of branches in the signaling network (i.e., topology) is sufficient to explain the observed data for any plausible set of parameter values. This implies two things: this problem of statistical inference must consider (i) the specific data under consideration and (ii) the uncertainty associated with the network predictions, given the associated uncertainty in the parameter estimates and the proposed topology of the network model. Here, we present the use of empirical Bayesian methods in conjunction with quantitative cell signaling models as a solution to this statistical inference problem [23]. It is within this context that we used an empirical Bayesian approach for model-based inference to evaluate competing hypotheses regarding how effector TH1 cells interpret IL-12.
Results
Cell fate varies with time and culture conditions
To explore these signaling questions in the context of TH cell biology, we constructed a quantitative cue-signal-response data set to infer the relative contributions of alternative signaling pathways within our specific system: the mouse 2D6 cell line as a model system for TH1 cells. In total, the quantitative cue-signal-response data set contained 924 data points that included measures of cell fate and key proteins associated with the IL-12 signaling pathway. These measures were obtained at seven time points, under four experimental conditions, and in technical triplicate. In short, cellular response to a biochemical cue is influenced by preexisting biochemical signals within a cell, external biochemical cues, and paracrine feedback mechanisms. A 2×2 factorial experimental design was constructed to parse the cellular response due to the direct effect of IL-12 stimulation from the indirect influence of paracrine feedback mechanisms. The preexisting biochemical signals within a cell is also influenced by dilution within an expanding cell population. We used flow cytometry, as a form of high content assay, to parse the influence of an expanding cell population from the signals elicited within individual cells by a biochemical cue.
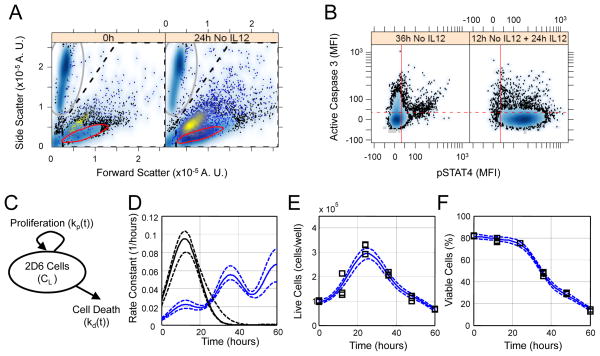
First, we quantified dynamic changes in the number and viability of cells within our system (Fig. 1). We used flow cytometry to assess the viability of cells (which are detected as “events”), using cleavage of caspase 3 as a marker for apoptosis (Fig. 1, A and B). We then used a mathematical cell fate model (Fig. 1C) to infer the time-dependent rate constant for cell proliferation and time-dependent rate constant associated with cell death through apoptosis. The total number of live cells and the percentage of the total number of cells that was viable (that is, the ratio of events contained within red oval in Fig. 1A relative to events contained within the dashed gate) were used to calibrate the cell fate model. We used an empirical Bayesian approach to estimate distributions in the rate constants (Fig. 1D and figs. S1 to S3) and associated model predictions (Fig. 1, E and F, and fig. S4) that are consistent with the experimental observations of the total number of live cells per well and of cell viability (Fig. 1, E and F, and fig. S4). Because these distributions were calculated on the basis of the available data, they are referred to as posterior distributions. The posterior distributions in the time-dependent rate constant for cell proliferation were independent of both cell density and IL-12 (fig. S5A), whereas the time-dependent rate constant for cell death varied with the culture conditions and IL-12 (fig. S5B). Initially, the rate constant for cell death was negligible relative to cell proliferation but it increased over time. The rate constant for cell death increased above the rate constant for cell proliferation after 30 hours in culture and corresponded to the decline in the number of live cells. An increase in the initial number of cells per well shifted the time at which the rate constant for cell death was equal to the rate constant for cell proliferation from 30 hours at the low cell density conditions to less than 25 hours at the high cell density. Although IL-12 suppressed cell death up to 45 hours when applied to cultures of low cell density; the effect of IL-12 was diminished at higher cell densities. These observed dynamics of cell fate within our system led us to develop a quantitative description that integrated the cell population dynamics with an intracellular cue-signal-response model for how IL-12 induces the production of cytokines in a TH1 cell model.
Fig. 1.
Calibration of the cell fate model for a Th1 clone cell line. (A) Forward and side scatter gating strategy for 2D6 cells shown for two time points - 0 hours (left panel) and 24 hours in cRPMI alone (right panel). Calibration bead events (gray oval) were separated from cellular events (dashed pentagon). Live cells (i.e., cleaved caspase 3− cells) were identified using a data-driven gate (red oval). The remaining events contained within the dashed pentagon were considered dead or dying cells, as indicated by back-gating on cleavage of caspase 3 (yellow shading). (B) Data-driven threshold for positive association with cleavage of caspase 3 (red dotted line) was obtained by comparing 2D6 cells cultured for 36 hours in cRPMI alone (left panel) and 2D6 cells cultured in cRPMI alone for 12 hours followed by 24 hours supplemented with IL-12 (right panel). (C) A schematic diagram of the cell fate model that has time-dependent rate constants for proliferation (kp(t)) and cell death (kd(t)). (D) The posterior distribution in the cell fate rate constants (kp(t) black, kd(t) blue, solid line indicates median, dotted lines enclose 95% of the posterior distribution). Posterior distributions in the cell fate model predictions (lines) compared against the observed (squares) change in the cell density (E) and the percentage of flow cytometry events interpreted as live cells (F). In panel B, the solid vertical red line indicates the data-driven threshold for background pSTAT4 (i.e., 95% of unstained cells exhibited a MFI below the threshold) while the dotted horizontal red line indicates the data-driven threshold for background fluorescence associated with cleaved caspase 3 staining (i.e., 95% of the 2D6 cells cultured without IL-12 for 12 hours followed by 24 hours with IL-12 exhibited a MFI below the threshold). Results representative of three technical replicates obtained for each of two biological replicates.
The cue-signal-response model incorporates cell population dynamics
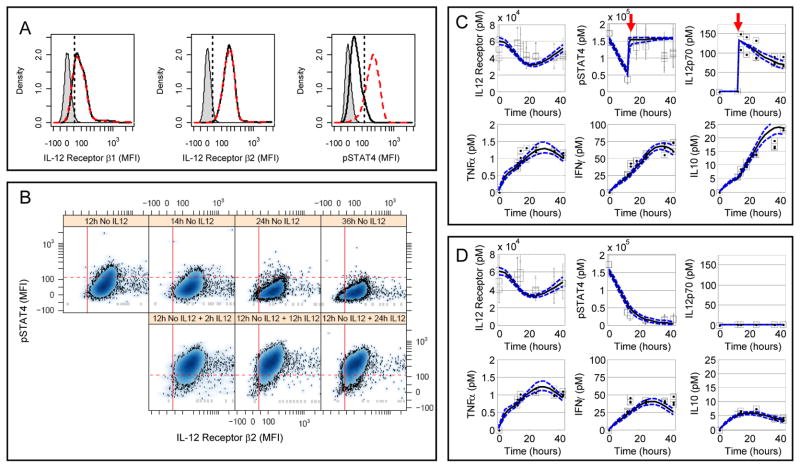
In addition to the cell population dynamics, the quantitative cue-signal-response data set also included key proteins associated with the IL-12 signaling pathway (expression of IL-12Rβ1 and IL-12Rβ2 and phosphorylation of STAT4), integrative measures of cytokine production (IFN-γ, IL-10, TNF-α, IL-6, and MCP-1), and an independent measurement of stimulation by IL-12p70. We used quantitative flow cytometry to observe the nature and dynamics of the response of live 2D6 cells to IL-12. Specifically, we analyzed the IL-12R subunits IL-12Rβ1 and IL-12Rβ2 and the extent of phosphorylation of STAT4 at Tyr693 (Y693) at a series of time points (Fig. 2A). Because the 2D6 cell line is IL-12 dependent, we cultured the 2D6 cells in complete RPMI (cRPMI) medium alone to reduce basal activation of the cells. Upon prolonged culture in cRPMI alone, the mean fluorescence intensity (MFI) of pSTAT4 decreased as a function of time in live IL-12β2-positive cells (Fig. 2B). As expected, STAT4 became phosphorylated, in response to IL-12 in IL-12Rβ2-positive cells, in a graded, dose-dependent manner (fig. S6). Furthermore, the probability distribution functions (PDFs) for IL-12Rβ1, IL-12Rβ2, and pSTAT4 suggested that this cell line could be considered as a single population; thus, median values could be used as appropriate summary statistics (Fig. 2A). The MFI associated with IL-12Rβ1, IL-12Rβ2, and pSTAT4 were used to estimate the protein copy numbers with calibration beads (fig. S7). The median protein copy numbers for IL-12Rβ1 and IL-12Rβ2 were similar at 5500 and 5700 copies per cell, respectively. The median protein copy number for pSTAT4 was 12,100 copies per cell. Because the amount of IL-12 used to stimulate the 2D6 cells was at a saturating concentration, we assumed that 100% of the total STAT4 was phosphorylated. The dynamic response of IL-12Rβ2 was qualitatively similar among the treatment conditions (Fig. 2, C and D, and fig. S8). The abundance of IL-12Rβ1 exhibited dynamics similar to that of IL-12Rβ2 (fig. S9). Because the functional IL-12R consists of both IL-12Rβ1 and IL-12Rβ2 components, we assumed that the observed dynamics of IL-12Rβ2 corresponded to the functional IL-12R.
Fig. 2.
Observed 2D6 response to IL-12 as a context for defining and calibrating the cue-signal-response model. (A) Changes in IL-12Rβ1 (left sub-panel), IL-12Rβ2 (center sub-panel), and phosphorylated STAT4 (right sub-panel) were used to quantify intracellular changes in response to IL-12 stimulation in 2D6 cells. The results from three 2D6 cell populations are shown: unstained cells (gray shaded), cells cultured without IL-12 for 14 hours (black solid line), and cells cultured without IL-12 for 12 hours followed by 2 hours with IL-12 (red dashed line). The straight lines indicate the data-driven threshold for the upper limit of protein expression or activity for 95% of the unstained (IL-12Rβ1 and IL-12Rβ2) or unstimulated (pSTAT4) 2D6 cells. (B) Scatter plots for phosphorylated STAT4 (y-axis) versus IL-12Rβ2 (x-axis). Each sub-panel corresponds to a different time following IL-12 stimulation. (C + D) 2D6 cells exhibited dynamic changes in response to culture conditions and IL-12 stimulation (Panel C - high density + IL-12; Panel D - high density without IL-12). The changes in cellular IL-12Rβ2 and pSTAT4 and IL-12p70, TNFα, IFNγ, and IL-10 in 2D6-conditioned media were represented by median values (squares) and compared against posterior predictions for the calibrated cue-signal-response model (lines). Error bars for IL-12Rβ2 and pSTAT4 enclose 67% of the population. Red arrows indicate the addition of IL-12p70. Results representative of three technical replicates obtained for each of two biological replicates.
The median abundances of IL-12Rβ1 and IL-12Rβ2 and activity of STAT4 for the population of live 2D6 cells were also coupled with changes in biochemical cues enriched in the cell culture media. We assayed IFN-γ, TNF-α, IL-10, IL-6, and MCP-1 at the same time points with cytometric bead arrays. In addition, we assayed IL-12p70 to monitor the biochemical cue that provided an input signal to the system over time. The amounts of IFN-γ, TNF-α and IL-10 secreted by the cells depended on the time and the experimental conditions (Fig. 2, C and D, and fig. S8). Even in the absence of IL-12, we observed a transient increase in these cytokines in the conditioned media. IFN-γ was the most abundant cytokine produced, which was followed by moderate production of IL-10 and a low production of TNF-α, which has not been previously reported. This was not unexpected because the assay used to measure TNF-α in previous studies may have been unable to detect such a small amount of cytokine. Because we observed IL-6 and MCP-1 at the lower end of the dynamic range of the assay and found that they did not vary with experimental conditions, we excluded these data points from subsequent analysis (fig. S9).
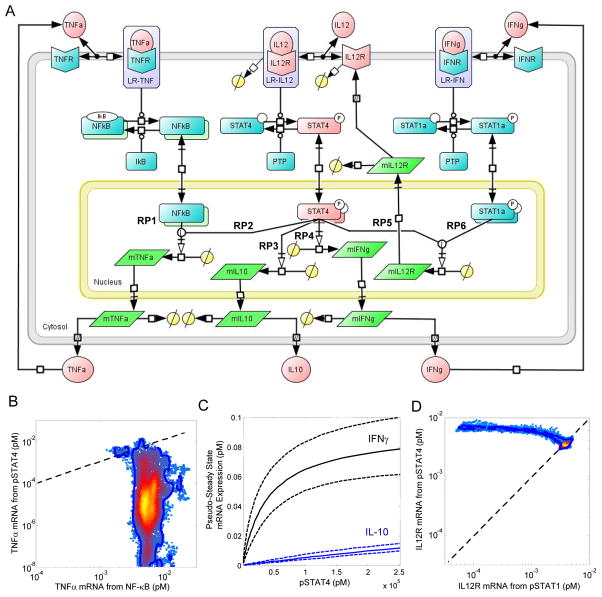
To describe how the 2D6 cells orchestrated a cellular response to IL-12, we developed a deterministic mathematical model to interpret this quantitative cue-signal-response data set. Because this is a closed system, secreted proteins that have a paracrine effect on cellular response may accumulate within the system, and intracellular signals may be diluted because of a cell population that is expanding in numbers. The topology of the model was shaped by our previous study of IL-12 signaling in naïve CD4+ T cells [21, 38], the observed cue-signal-response data set, and putative regulatory pathways that have been reported in the literature (Fig. 3A). In addition to expressing IL-12R, 2D6 cells also have IFN-γ receptor 1 (CD119) and TNF receptor (TNFR) type II (CD120b) (fig. S10). Because the model predictions should be thought of as probabilistic distributions rather than single point estimates [39], we again used an empirical Bayesian approach to estimate the uncertainty in the model predictions, given the available cue-signal-response data (figs. S11 to S13). The posterior distribution in the cue-signal-response model predictions were largely consistent with the quantitative data set (Fig. 2, C and D, and fig. S8). Under prolonged culture without IL-12, the mathematical model predicted an exponential decline in the amount of pSTAT4 and a corresponding reduction in the production of IFN-γ and IL-10. The predicted responses of IFN-γ and IL-10 were consistent with the experimental data. The experimentally determined amounts of pSTAT4 did not approach zero, but decrease to an extent that corresponded to background fluorescence. In our simulation of the response to IL-12, our predictions of the concentrations of pSTAT4, IFN-γ, and IL-10 were consistent with the experimental observations at the low cell density. At the higher cell density, the experimental values for both pSTAT4 and IL-10 were slightly below those predicted by the model at 36 and 42 hours, which was suggestive of a potential paracrine feedback pathway that inhibited cytokine production. In summary, the cue-signal-response model accurately captured the underlying quantitative data set. Systematic differences could be explained by technical limitations of the assays used or were suggestive of paracrine feedback mechanisms that may have influenced the cellular response at a higher cell density. In the following sections, we examine in more detail our interpretation of the data set with the cue-signal-response model.
Fig. 3.
The cue-signal-response model developed to describe the cellular response to IL-12 in 2D6 cells. (A) A schematic diagram of the model includes IFN-γ, IL-12p70, and TNF-α as biochemical cues; intracellular signaling events associated with the canonical IL-12, IFN-γ, and TNF-α signaling pathways; regulation of the IL-12 receptor, and the synthesis of IFN-γ, IL-10, and TNF-α as the cellular response to IL-12. Elements of the model that were calibrated to experimental measurements are highlighted in red. Specific branches within the signaling network that regulate gene expression are annotated with the labels RP1 through RP6. Posterior distributions in the simulated pathway flux associated with NF-κB (RP1) versus STAT4 (RP2) regulation of TNFα mRNA expression (panel B), simulated IL-10 (RP3 - blue lines) and IFN-γ (RP4 - black lines) mRNA expression as a function of STAT4 activation (panel C), and the simulated pathway flux associated with STAT4 (RP5) versus STAT1 (RP6) regulation of IL12Rβ2 mRNA (panel B). Systems Biology Graphical Notation was used to represent the biochemical events encoded within the cue-signal-response model.
The production of TNF-α by 2D6 cells is regulated via an autocrine positive feedback loop
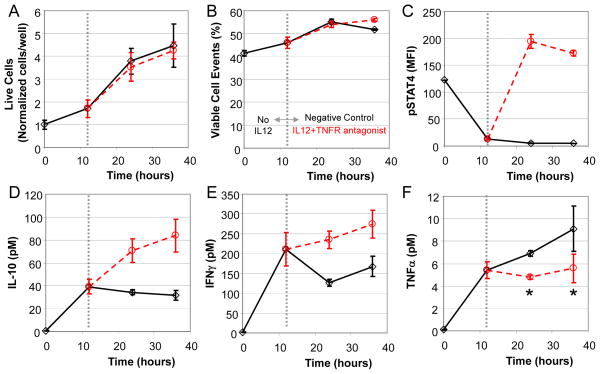
TNF-α is a pleiotropic cytokine whose synthesis by TH1 cells is regulated by STAT4 and an autocrine mechanism that involves members of the nuclear factor κB (NF-κB) family of transcription factors. To gain greater insight into the relative contributions of these parallel pathways in regulating TNF-α production, we obtained a posterior distribution in the simulated pathway flux of the NFκB branch (Fig. 3A, RP1) versus the pSTAT4 branch (Fig. 3A, RP2) in regulating TNF-α synthesis (Fig. 3B). Given the posterior distribution our model predictions, the conditional probability that RP1 was greater than RP2 was 97.7% with kernel density estimation, and the conditional probability that RP1 was a factor of 10 greater than RP2 was greater than 88.3%. To validate this prediction, we used a TNFR antagonist to break the predicted autocrine positive feedback loop without interfering with detection of TNF-α. In combination with the TNFR antagonist (αTNFR), we also stimulated the cells with IL-12 to turn on the competing pathway through the pSTAT4 branch. 2D6 cells exposed to a combination of αTNFR and IL-12 exhibited a cell fate trajectory similar to that of control cells (Fig 4, A and B). In contrast to the control cells, the combined stimulation increased the extent of STAT4 phosphorylation (Fig. 4C), which resulted in increased IL-10 (Fig. 4D) and IFN-γ (Fig. 4E) production. The addition of αTNFR at the 12 hour time point blocked subsequent TNF-α production (Fig. 4F). In summary, the observed cessation of TNF-α secretion and simultaneous doubling of the cell numbers suggested an autocrine positive feedback loop for the production of TNF-α and argued against a role for STAT4 in regulating TNF-α production.
Fig. 4.
An autocrine feedback loop regulates TNFα expression in 2D6 cells that is independent of STAT4 activation. Following a 12 hour pre-conditioning with cRPMI, 2D6 cells were stimulated with IL-12 in combination with a TNF receptor antagonist (circles). Untreated cells were used as a negative control (diamonds). Changes in live cell density (A), viable cellular events (B), and STAT4 activation (C) were quantified as a function of time by flow cytometry. Enrichment of IL-10 (D), IFNγ (E), and TNFα (F) in the 2D6-conditioned media were quantified as a function of time using cytometric bead array. The dotted vertical line indicates the switch from pre-conditioning to stimulation conditions. Mean response (± standard deviation) at each time point and condition were used to create trend lines (solid and dotted lines). A Student’s t test was used to assess statistical significance, where * indicates p<0.05.
The dynamics of STAT4 deactivation creates a hysteresis in the cellular response to IL-12
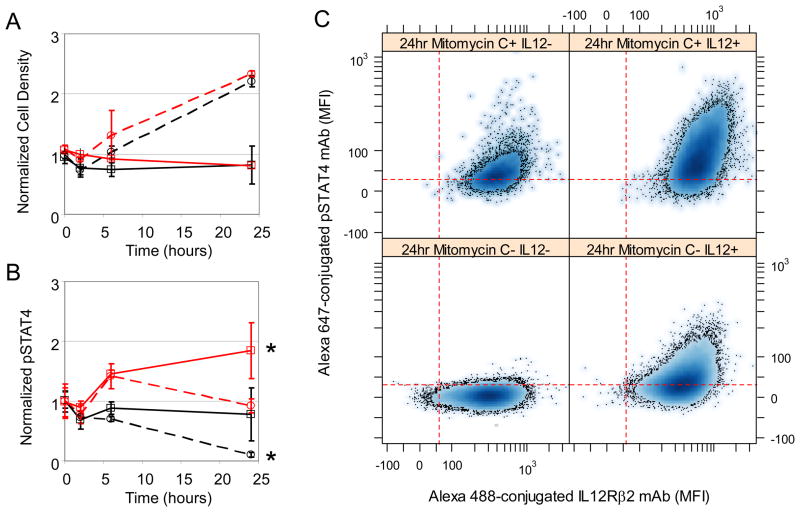
Conventionally, the activity of a STAT protein reflects the kinetic balance between activating events, for example, through the actions of kinases, and deactivating events, for example, through the actions of phosphatases. However, the effective dilution of a post-translationally modified protein within an expanding cell population might also be interpreted as a deactivation event and is a confounding factor. With model-based inference, we wanted to distinguish the effects of population dilution from biochemical events that activate and deactivate STAT4. Experimentally, we observed that the amount of pSTAT4 declined with time upon culture in cRPMI alone (that is, a chase experiment) and was rapidly increased in response to IL-12 (that is, a pulse experiment) (Fig. 2B). Functionally, the extent of production of IFN-γ and IL-10 by 2D6 cells in cRPMI alone correspondingly decreased with time but was restored upon exposure to IL-12 (Fig. 2, C and D). Because the exponential decline in the amount of pSTAT4 in cRPMI alone corresponded to a period of exponential growth of the cells, we used the cue-signal-response model to infer the relative contributions of potential phosphatase action and dilution within the expanding population. From the posterior distributions (fig. S14), we determined that the decline in the amount of pSTAT4 as a result of dilution within the cellular population accounted for 30% to the net rate of decline. To validate this prediction, we pretreated cells with mitomycin C, a potent DNA cross-linker that inhibits cell division. Untreated cells doubled in number by 24 hours, whereas pretreatment with mitomycin C stopped cell proliferation (Fig. 5). When cells were cultured without IL-12 (the chase experiment) and allowed to proliferate, the amount of pSTAT4 decreased as a function of time. In contrast, when the cells were cultured without IL-12 and treated with mitomycin C, the amount of pSTAT4 remained constant. Both IL-12-treated conditions exhibited an increase in the abundance of pSTAT4 at the 6 hour time point. If the cells were allowed to subsequently divide, the amount of pSTAT4 in IL-12 stimulated cells returned to the basal amounts; however, if cell division was inhibited, the cell appeared to have twice the amount of pSTAT4. Collectively, these data suggested that the observed reduction in the amount of pSTAT4 was as a result of cell proliferation-induced dilution of the protein in an expanding cell population rather than as a result of phosphatase activity.
Fig. 5.
Dilution due to cell proliferation influences the decline in STAT4 phosphorylation. Dynamic changes in cell density (Panel A) and phosphorylated STAT4 (Panel B) in 2D6 cells (solid lines) were compared against 2D6 cells cultured with Mitomycin C (dotted lines). IL-12-stimulated cells (red) were also compared against cells cultured in cRPMI alone (black). Representative flow cytometry assays of pSTAT4 versus IL12Rβ2 expression at the 24 hr time point are shown in Panel C. Results reported as mean response (± standard deviation) and are representative of at least three technical replicates and two biological replicates. A finding of statistical significance (p<0.05) is indicated by *.
In the cue-signal-response model, the functional response to pSTAT4 includes the regulation of the expression of il10 (Fig. 3A, RP3) and ifng (RP4 in Fig. 3A, RP4). We used model-based inference of the pulse-chase experiment to estimate a quantitative relationship between STAT4 activation and the expression of these genes (Fig. 3C). Regulation of il10 expression was sensitive to changes in the activation of STAT4 that were observable with flow cytometry. In contrast, regulation of ifng expression became less sensitive to changes in STAT4 activation because the cellular response was saturated. This suggested that the range of STAT4 phosphorylation in which STAT4 activation linearly correlated with gene expression was at the lower limit of detection of our flow cytometry assay. That is, increases in the abundance of pSTAT4 as observed by flow cytometry correlated with STAT4-dependent gene expression, but loss of pSTAT4 did not imply a lack of STAT4-dependent gene expression. In summary, the decay in intracellular signal and a nonlinear dependence of gene expression on STAT4 activity created a hysteresis in the cellular response to IL-12 for the production of cytokines such that the response depended on the current level of stimulation and on past exposure to IL-12. This hysteresis in the cellular response to IL-12 enabled the cells to sustain STAT4-dependent gene expression in the absence of IL-12 and encoded a transient “memory” of past encounters with biochemical cues. Moreover, the duration of this memory was reduced by an increase in the rate of cell proliferation.
IL-12 activates both STAT1 and STAT4
Sustained signaling through the IL-12 pathway is important for driving the polarization of naïve CD4+ T cells toward a TH1 phenotype and for sustained production of TH1-type cytokines. We hypothesized that the cue-signal-response model could be used to infer the relative contribution of different pathways that regulate the abundance of IL-12Rβ2 from the quantitative data set. Two alternative pathways were encoded within the cue-signal-response model. The first pathway encoded the observation that IL-12 exhibited a direct positive feedback mechanism in which pSTAT4 promoted the induction of IL-12Rβ2 (Fig. 3A, RP5) [30]. The second pathway encoded the observation that IFN-γ regulated the abundance of IL-12Rβ2 through an autocrine mechanism that includes the phosphorylation of STAT1 (Fig. 3A, RP6) [33]. To discriminate between these alternative pathways, we used the posterior distribution in the cue-signal-response model predictions to infer the pathway flux associated with the pSTAT4 branch (RP5) from that associated with the pSTAT1 branch (RP6) in regulating the amount of cell surface IL-12Rβ2 (Fig. 3D). The posterior distributions exhibited a non-normal distribution that was centered around an equal contribution from both pathways, and the tail of the distribution was above the diagonal, which implied that the contribution of RP5 is greater than RP6 and that the model predictions are more sensitive to parameters that influenced RP5 than parameters that influenced RP6. In summary, the data set was not sufficient to clearly distinguish between these competing pathways because most of the posterior distribution in fluxes associated with regulating IL-12R abundance was contained on the diagonal.
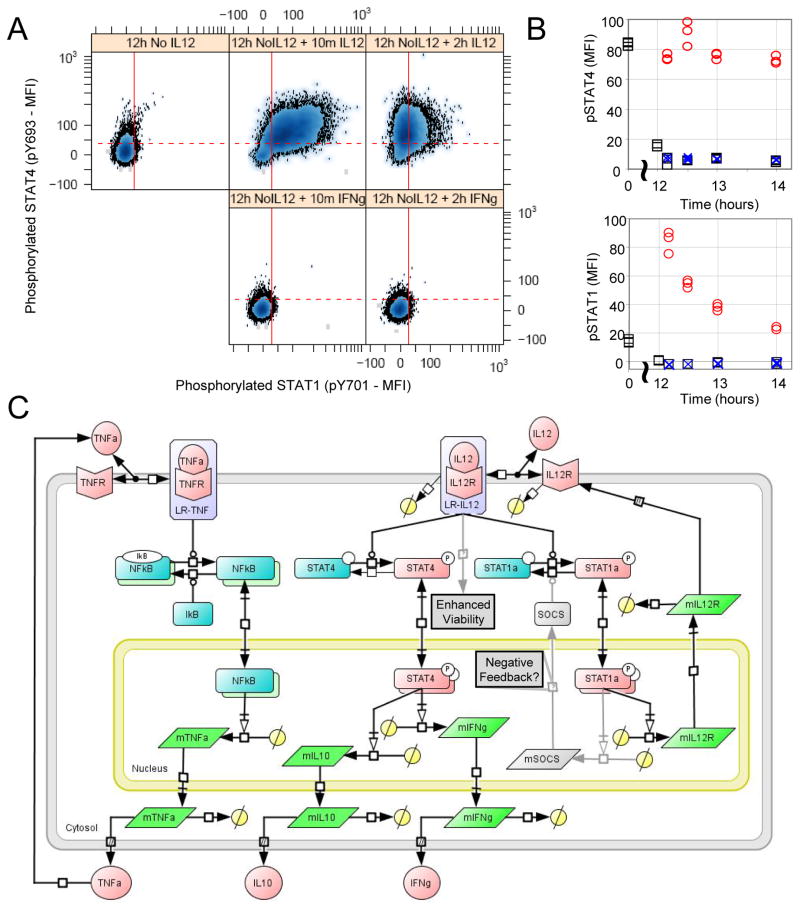
In formulating the cue-signal-response model, we assumed that IFN-γ, which is secreted in response to IL-12, activates STAT1 to form an autocrine or paracrine positive feedback pathway. While naïve CD4+ T cells exhibit phosphorylation of STAT1 in response to IFN-γ, effector TH cells are typically considered to be unresponsive to IFN-γ. It is unclear where the 2D6 cell line falls along this polarization spectrum. Moreover, STAT1 activation is typically reported within minutes after the addition of a stimulus. The perceived nonresponsiveness of TH1 cells to IFN-γ could be a result of differences in the particular time points used to measure pSTAT1 or a result of the constitutive activation of pSTAT1 through an autocrine or paracrine feedback mechanism. To test for the presence of an autocrine feedback loop, we used blocking antibodies against IFN-γ (fig. S15). To test for the presence of a paracrine feedback loop, we hypothesized that the paracrine IFN-γ feedback pathway could be confirmed by analyzing cells at a low density and observing STAT1 phosphorylation within minutes of adding IFN-γ back. We thought that stimulation of the cells with IL-12 would provide an additional negative control for STAT1 activation. Instead, we observed that stimulation with IL-12 activated both STAT4 and STAT1 within 10 min and that IFN-γ did not activate STAT1 (Fig. 6, A and B, and fig. S15). In primary cells, IL-12 also phosphorylated STAT1 (fig. S16). STAT1 and STAT4 activation also exhibited different dynamics, such that STAT4 remained phosphorylated during the duration of the experiment, whereas STAT1 was maximally activated at the first time point, which subsequently declined. Phosphorylation of STAT1 in response to IL-12 persisted for at least 24 hours, whereas the addition of a neutralizing antibody against IFN-γ had no effect on the activation of STAT1 (fig. S15). Given the rapid dynamics of STAT1 activation after stimulation with IL-12, the simplest interpretation of this observation was that IL-12 activated the IL-12R-JAK complex that subsequently phosphorylated both STAT1 and STAT4. IL-12-induced activation of STAT1 created a direct positive feedback loop to regulate the expression of IL-12Rβ2 at the cell surface. Phosphorylation of STAT1 is also controlled by a selective negative feedback loop, a common motif for regulating target gene expression. Collectively these findings suggest a revised cue-signal-response model with altered topology related to TNF-α production, enhanced viability in response to stimulation with IL-12, and regulation of IL-12Rβ2 surface expression in 2D6 cells (Fig. 6C). The mechanistic details associated with the dynamic regulation of STAT1 and enhanced cell viability upon stimulation with IL-12 remain to be resolved.
Fig. 6.
STAT1 and STAT4 are rapidly phosphorylated in 2D6 cells in response to IL-12 stimulation. 2D6 cells were stimulated with either IL-12, IFN-γ or left untreated. (A) Scatter plots for phosphorylated STAT4 (y-axis) versus phosphorylated STAT1 (x-axis). Each sub-panel corresponds to a different time following IL-12 stimulation. The dotted horizontal line indicates the upper limit of pSTAT4 MFI for 95% of the 2D6 cells following culture without IL-12 for 14 hours. The solid vertical line indicates the upper limit of pSTAT1 MFI for 95% of the 2D6 cells following culture without IL-12 for 14 hours. (B) STAT4 (top subpanel) and STAT1 (bottom subpanel) activation exhibited different dynamics following IL-12 stimulation (red circle) compared to IFNγ stimulated (blue x) and unstimulated (black squares) cells. Flow cytometry results are summarized by the median values of the distribution. (C) A revised schematic diagram of the cue-signal-response model that incorporates topological changes associated with the response of 2D6 cells to IL-12. Details associated with enhanced viability and the differential regulation of STAT1 versus STAT4 in response to IL-12 remain to be elucidated.
Discussion
IL-12 is an important cytokine that is produced by innate immune cells and influences adaptive immunity by polarizing naïve and activating a discrete subset of effector CD4+ TH cells. Active TH cells, in turn, orchestrate the adaptive immune response through the production of cytokines. The specific profile of cytokines produced reflects the combined effects of genetic and epigenetic influences on how a specific TH cell interprets the dose and dynamics of biochemical cues. Mathematical models can aid in interpreting observed data by providing a quantitative context for encoding previous knowledge of the cellular system, a process called model-based inference. How well a particular mathematical model describes the cellular decision process, given the particular data at hand and the previous knowledge of the cellular system, corresponds to reasoning within a Bayesian framework. Here, we obtained a quantitative cue-signal-response data set, developed a mathematical model that encoded alternative pathways associated with how TH1 cells orchestrate a cellular response to IL-12, and used an empirical Bayesian approach to reason about the relative contributions of these alternative pathways. The results clarify the biochemical basis for two emerging concepts regarding cellular decision making: feedback control and transient memory.
Cell-mediated immunity is a tightly controlled process with severe consequences for the host when dysregulated and is initiated after a productive interaction between an antigen-presenting cell (APC) and a TH cell. Because cell-cell contact can be considered as a random process, the probability of a productive interaction between an APC and a TH cell within a given time interval is directly related to the number of interacting cells per unit volume (that is, the precursor frequency). Thus, cell-mediated immunity also includes strong stochastic influences because the frequency of naïve CD4+ T cells specific for a particular antigen varies from 20 to 200 cells per mouse, depending on the antigen [40]. Noisy processes, such as the stochastic steps associated with cell-mediated immunity, can be made more predictable with a paracrine feedback control mechanism [41]. In the case of TH1 cell differentiation, reported differences in IL-12 dependence may be dependent on quantitative differences in the dose of antigen. Paracrine action of IFN-γ can be thought to function as a feedback control mechanism to mitigate stochastic variation in antigen recognition. Strong antigen stimulation promotes the development of TH1 cells that produce IFN-γ through an IL-12 independent mechanism [42, 43]. Because individual T cells recognize multiple presented antigens [44, 45], the paracrine action of IFN-γ signaling on other naive CD4+ subsets may amplify cell-mediated immunity to include a broader repertoire of TH cells. Within the broader repertoire, the paracrine action of IFN-γ enables the sustained IL-12 signaling required for TH1 polarization [46, 27]. Our data suggests that the shift in IFN-γ- to IL-12-dependent activation of STAT1 corresponds to a shift in dependence from an external control mechanism to a cell-autonomous control mechanism for maintaining responsiveness to IL-12. Moreover, the different dynamics associated with STAT1 and STAT4 activation after stimulation with IL-12 suggest an additional intracellular feedback mechanism, such as spatial differences in signaling as a result of receptor trafficking [47] or the increased abundance of inhibitors of STATs (for example, SOCS or PIAS), that differentially regulate STAT activation.
Upon receiving stimulation by antigen, effector TH cells differentiate into TH subsets that are defined on the basis of the transcription factor that they express [4] and the cytokines that they produce. TH cell fate was viewed traditionally as a series of irreversible transitions from one stable discrete state to another. However, another picture is emerging: TH cell fate is plastic and is determined by the combination of biochemical cues and genetic influences on how a cell interprets this information [4]. Implied in these two perspectives is a time dependency associated with the assay of TH cell phenotype. Using a synergistic combination of quantitative experiments and model-based inference methods, we found one mechanism that may contribute to the observed degree of plasticity in the cellular response to IL-12, a hysteresis in the dose-response curve to IL-12. Hysteresis is indicated by the finding that the initial stimulation with IL-12 dose-dependently activated STAT4 and subsequently increased ifng and il10 expression, and that the expression of ifng and il10 remained despite the withdrawal of IL-12. In this TH1 cell model, STAT4 was present in sufficiently large amounts to saturate the threshold for STAT4-dependent gene induction, particularly in the case of ifng. Cessation of STAT4-dependent gene expression did not occur until the concentration of active STAT4 fell below the threshold, which occurred mainly because of dilution through cell division. Thus, the cellular response to IL-12 depends on both the current extend of IL-12 stimulation and the previous exposure to IL-12. Ras activation in T cells has also been suggested to exhibit hysteresis as a mechanism to inhibit spurious T cell activation in response to weak stimulation and to integrate interrupted serial encounters within the lymph node with APCs bearing the cognate antigen [48]. Here, a hysteresis in the dose-response to IL-12 may provide a dynamic robustness to inhibitory signals in the peripheral tissues.
Finally, our analysis also suggests that manipulation of protein copy number and the degree of reversibility in post-translational modification of STAT proteins provides an additional layer to the epigenetic landscape associated with TH cell polarization. Cell division, as a mechanism to facilitate epigenetic imprinting, is also required to enable cytokine production by effector TH cells [49, 50]. STAT family members may play a role in this epigenetic imprinting because they associate with transcriptional coactivators that regulate chromatin structure [51, 52, 53]. Moreover, our integrated in vitro and in silico approach may help to answer fundamental questions regarding the fate and plasticity of primary TH cells as well as provide a framework for integrating our emerging understanding of the epigenetic regulation of T cell differentiation [54] with the dynamics of signal transduction within an expanding cell population.
Materials and Methods
Antibodies and reagents
Cytokines, drugs, and pharmacological inhibitors were from commercial sources and used according to suppliers’ recommendations or as indicated. V450-conjugated rabbit anti-active Caspase-3 (C92-605), phycoerythrin-conjugated mouse anti-mouse CD212 (IL-12Rβ1), purified hamster anti-mouse IL-12 receptor β2, fluorescein isothiocyanate (FITC)-conjugated mouse anti-hamster immunoglobulin G1 (IgG1, G94-56), Alexa Fluor 647-conjugated mouse anti-STAT4 (pY693) and BD Cytometric Bead Array (CBA) Mouse Inflammation Kit were purchased from BD Biosciences (San Diego, CA, U.S.A.). Alexa 488-conjugated mouse anti-mouse IL-12 receptor β2 (clone 305719) was purchased from RnD Systems (Minneapolis, MN). ChromPure mouse IgG (whole molecule) and ChromPure rat IgG (whole molecule) were purchased from Jackson Immuno Research (West Grove, PA, U.S.A.). Armenian hamster anti-mouse CD120a (55R-170 - TNFR1p55) was purchased from eBioscience (San Diego, CA, U.S.A.). All cell cultures were maintained at 37°C in 5% CO2 in RPMI 1640 plus supplements (referred to as complete RPMI or cRPMI) as described in [21]. AccuCount Fluorescent Particles (8.0 – 12.9 μm) were purchased from Spherotech (Lake Forest, IL). Quantum Simply Cellular uniform microspheres conjugated to anti-mouse IgG were purchased from Bangs Laboratories (Fishers, IN).
Cell culture and stimulation
The TH1 cell line, 2D6 [29], was provided by M. Grusby (Harvard University, Cambridge, MA) and grown in 25 cm2 tissue culture flasks with 15ml cRPMI and supplemented with 90 pM rmIL-12p70. Tissue culture media was replaced every 24 hours. To assess the cellular response to IL-12, 2×2 factorial experimental design was used with cell density and IL-12 stimulation as the two factors. We selected a saturation concentration of 150 pM IL-12 for the stimulation conditions. This concentration of IL-12 was greater than the effective concentration necessary to elicit a 50% of maximal response in STAT4 at 2 hours (EC50 = 0.2 pM - see fig. S6). 2D6 cells were washed and plated at two cell densities, 6×104 (Lo) and 1.2×105 (Hi) cells per well, in 96-well U-bottom plates for 12 hours with cRPMI. The volume in each well was 120 μl. After 12 hours, rmIL-12p70 (150 pM) was added to the IL-12 treatment groups and DPBS was added into negative controls. Cells were cultured at 37°C in 5% CO2 for appropriate time period. To validate the model predictions, we plated 1.2×105 cells per well in 96-well U-bottom plates for 12 hours with cRPMI. After 12 hours, cells were stimulated using combinations of DPBS (negative control), rmIL-12p70 (150 pM), or an inhibitor and cultured for the indicated times. The inhibitors included Mitomycin C (50 μg/ml) and neutralizing antibody against TNFR1p55 (50 μg/ml). To characterize the putative IFN-γ autocrine or paracrine feedback loops, 6×104 cells per well were plated in 96-well U-bottom plates for 12 hours with cRPMI. After 12 hours, cells were exposed to one of two experimental designs. In the first experimental design, a 2×2 factorial experimental design was used with the two factors being rmIL-12p70 (150 pM) and two neutralizing antibodies against IFN-γ (15 ng/ml) and IL-18 (53g/ml). In the second experimental design, 2D6 cells were exposed to three treatment conditions - DPBS, rmIL-12p70 (150 pM), or rmIFN-γ (10 ng/ml) - and cultured for the indicated times. At each time point, the plate was centrifuged 1250 RPM for 5 minutes at 4°C. The supernatant from each well was collected and stored at −20°C for subsequent analysis with cytometric beads. At least two biological replicates were performed for each experiment, and two technical replicates were obtained for each biological replicate.
Flow cytometry and Cytometric Bead Array
Fluorescence-activated cell sorting (FACS) was performed as described previously [21]. Three sets of Quantum Simply Cellular calibration beads that contain five Quantum Simply Cellular microsphere populations with different mouse IgG antibody binding capacities were also simultaneously stained with fluorophore-conjugated monoclonal antibodies specific for IL12Rβ1, IL12Rβ2, pSTAT1, or pSTAT4. AccuCount calibration beads (1.7×104 beads in a volume of 50 μL) were added to each of the tubes containing stained 2D6 cells that were subsequently analyzed using a FACSAria flow cytometer and FACSDiva version 6.1.1 software (Becton Dickinson). Cellular events were identified by forward and side scatter characteristics and Spherotech AccuCount beads were used to calibrate the cellular density among replicates and time points. On average, 2×104 events were analyzed at each time point. Cytometric bead array (BD Bioscience) was performed according to the manufacturers instructions and analyzed on a FACSCaliber with CellQuest Pro software. To determine the concentration of cytokines in each sample, BD CBA software was used.
Data Analysis
Flow cytometry data was exported as FCS3.0 files and analyzed using R/Bioconductor [55]. The fluorescence intensity for each parameter was reported as a pulse area using 18-bit resolution. The statistical difference between two treatment conditions was assessed using a two sample t-Test that assumes equal variance, where a p-value of less than 0.05 was considered statistically significant.
Models and modeling
A description of the mathematical models and model-based inference can be found in the Supplementary Materials.
Supplementary Material
Acknowledgments
We thank Andrew Yates, Kathy Brundage and Chris Cuff for comments on the manuscript. The authors thank K. Brundage for assistance with the experimental techniques and J. Barnett for access to laboratory facilities.
Funding: This work was supported by grants from the National Science Foundation (CAREER 1053490 to DJK), the National Cancer Institute (R15CA132124 to DJK), and the National Institute of Allergy and Infectious Diseases (R56AI076221 to DJK), and National Institute of Health (RR016440 and RR020866). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Science Foundation, the National Cancer Institute, the National Institute of Allergy and Infectious Diseases, or the National Institutes of Health.
Footnotes
This Supplemental PDF file includes:
www.sciencesignaling.org/cgi/content/full/5/220/ra32/DC1
Section 1. Models and modeling.
Section 2. Model parameters. References
Table S1. Values for cell fate model parameters and initial conditions.
Table S2. List of cue-signal-response model variables and initial conditions.
Table S3. Reaction rate laws for the cue-signal-response model.
Table S4. Differential equations that define the cue-signal-response model.
Table S5. List of cue-signal-response model parameters and corresponding values.
Fig. S1. Convergence of AMCMC results for cell fate model.
Fig. S2. AMCMC summary plots for each of the cell fate model parameters.
Fig. S3. Pairwise comparison of posterior distribution in cell fate rate parameters.
Fig. S4. Calibration of cell fate model.
Fig. S5. The balance between proliferative and apoptotic cell fates depends on time and culture conditions.
Fig. S6. IL-12 phosphorylates STAT4 in a graded dose-dependent manner in 2D6 cells.
Fig. S7. Copy number estimation for IL-12Rβ1, IL-12Rβ2, STAT1, and STAT4 in 2D6 cells.
Fig. S8. Calibration of the cue-signal-response model.
Fig. S9. IL-12Rβ1 expression exhibited similar dynamics as IL-12Rβ2 while MCP-1 and IL-6 were unchanged across experimental conditions.
Fig. S10. 2D6 cells express IFN-γ and TNF-α receptors.
Fig. S11. Convergence of AMCMC results for the cue-signal-response model.
Fig. S12. AMCMC summary plots for each of the cue-signal-response model parameters.
Fig. S13. Pairwise comparison of posterior distribution in rate parameters associated with cue-signal-response model.
Fig. S14. Posterior distributions in the rate constants associated with the decline in phosphorylated STAT4.
Fig. S15. STAT4 and STAT1 are phosphorylated in response to IL-12 stimulation.
Fig. S16. IL-12 and IFN-γ phosphorylate STAT1 in primary cells.
Fig. S17. IL-12 binding enhanced IL-12 receptor down-regulation.
Author contributions: DJK conceived and coordinated the study; NC and EC performed the experiments; NC and DJK analyzed the data; DJK wrote the paper; and all of the authors discussed the results and edited the manuscript.
Competing interests The authors declare that they have no competing financial interest.
References and Notes
- 1.Sher A, Coffman RL. Regulation of immunity to parasites by T cells and T cell-derived cytokines. Annu Rev Immunol. 1992;10:385–409. doi: 10.1146/annurev.iy.10.040192.002125. [DOI] [PubMed] [Google Scholar]
- 2.Ladi E, Yin X, Chtanova T, Robey EA. Thymic microenvironments for T cell differentiation and selection. Nat Immunol. 2006;7:338–343. doi: 10.1038/ni1323. [DOI] [PubMed] [Google Scholar]
- 3.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Garra A, Gabrysova L, Spits H. Quantitative events determine the differentiation and function of helper T cells. Nat Immunol. 2011;12:288–294. doi: 10.1038/ni.2003. [DOI] [PubMed] [Google Scholar]
- 6.Asthagiri AR, Lauffenburger DA. Bioengineering models of cell signaling. Ann Rev Biomed Eng. 2000;2:31–53. doi: 10.1146/annurev.bioeng.2.1.31. [DOI] [PubMed] [Google Scholar]
- 7.Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maruyama T, Mirando AJ, Deng CX, Hsu W. The balance of WNT and FGF signaling influences mesenchymal stem cell fate during skeletal development. Sci Signal. 2010;3:ra40. doi: 10.1126/scisignal.2000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qualls JE, Neale G, Smith AM, Koo MS, DeFreitas AA, Zhang H, Kaplan G, Watowich SS, Murray PJ. Arginine usage in mycobacteria-infected macrophages depends on autocrine-paracrine cytokine signaling. Sci Signal. 2010;3:ra62. doi: 10.1126/scisignal.2000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexopoulos LG, Saez-Rodriguez J, Cosgrove BD, Lauffenburger DA, Sorger PK. Networks inferred from biochemical data reveal profound differences in Toll-like receptor and inflammatory signaling between normal and transformed hepatocytes. Mol Cell Proteomics. 2010;9:1849–1865. doi: 10.1074/mcp.M110.000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pawson T, Warner N. Oncogenic re-wiring of cellular signaling pathways. Oncogene. 2007;26:1268–1275. doi: 10.1038/sj.onc.1210255. [DOI] [PubMed] [Google Scholar]
- 12.Klinke DJ. Signal transduction networks in cancer: Quantitative parameters influence network topology. Cancer Res. 2010;70:1773–1782. doi: 10.1158/0008-5472.CAN-09-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klinke DJ, Ustyugova IV, Brundage KM, Barnett JB. Modulating temporal control of NF-kappaB activation: Implications for therapeutic and assay selection. Biophys J. 2008;94:4249–4259. doi: 10.1529/biophysj.107.120451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorger PK. A reductionist’s systems biology: opinion. Curr Opin Cell Biol. 2005;17:9–11. doi: 10.1016/j.ceb.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Fiering S, Northrop JP, Nolan GP, Mattila PS, Crabtree GR, Herzenberg LA. Single cell assay of a transcription factor reveals a threshold in transcription activated by signals emanating from the T-cell antigen receptor. Genes Devel. 1990;4:1823–1834. doi: 10.1101/gad.4.10.1823. [DOI] [PubMed] [Google Scholar]
- 16.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ideker T, Lauffenburger D. Building with a scaffold: emerging strategies for high- to low-level cellular modeling. Trends Biotechnol. 2003;21:255–262. doi: 10.1016/S0167-7799(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 18.Lau KS, Juchheim AM, Cavaliere KR, Philips SR, Lauffenburger DA, Haigis KM. In vivo systems analysis identifies spatial and temporal aspects of the modulation of TNF-alpha-induced apoptosis and proliferation by MAPKs. Sci Signal. 2011;4:ra16. doi: 10.1126/scisignal.2001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sturm OE, Orton R, Grindlay J, Birtwistle M, Vyshemirsky V, Gilbert D, Calder M, Pitt A, Kholodenko B, Kolch W. The mammalian MAPK/ERK pathway exhibits properties of a negative feedback amplifier. Sci Signal. 2010;3:153. doi: 10.1126/scisignal.2001212. [DOI] [PubMed] [Google Scholar]
- 20.Sachs K, Perez O, Pe’er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 21.Finley SD, Gupta D, Cheng N, Klinke DJ. Inferring relevant control mechanisms for Interleukin-12 signaling within naive CD4+ T cells. Immunol Cell Biol. 2011;89:100–110. doi: 10.1038/icb.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakraborty AK, Das J. Pairing computation with experimentation: a powerful coupling for understanding T cell signalling. Nat Rev Immunol. 2010;10:59–71. doi: 10.1038/nri2688. [DOI] [PubMed] [Google Scholar]
- 23.Klinke DJ. An empirical bayesian approach for model-based inference of cellular signaling networks. BMC Bioinform. 2009;10:371. doi: 10.1186/1471-2105-10-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gately MK, Renzetti LM, Magram J, Stern AS, Adorini L, Gubler U, Presky DH. The Interleukin-12/Interleukin-12-receptor system: role in normal and pathologic immune responses. Annu Rev Immunol. 1998;16:495–521. doi: 10.1146/annurev.immunol.16.1.495. [DOI] [PubMed] [Google Scholar]
- 25.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 26.Murray PJ. The JAK-STAT signaling pathway: Input and output integration. J Immunol. 2007;178:2623–2629. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- 27.Athie-Morales V, Smits HH, Cantrell DA, Hilkens CMU. Sustained IL-12 signaling is required for Th1 development. J Immunol. 2004;172:61–69. doi: 10.4049/jimmunol.172.1.61. [DOI] [PubMed] [Google Scholar]
- 28.Jacobson NG, Szabo SJ, Weber-Nordt RM, Zhong Z, Schreiber RD, Darnell JE, Jr, Murphy KM. Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (STAT)3 and STAT4. J Exp Med. 1995;181:1755–1762. doi: 10.1084/jem.181.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maruo S, Ahn HJ, Yu WG, Tomura M, Wysocka M, Yamamoto N, Kobayashi M, Hamaoka T, Trinchieri G, Fuijiwara H. Establishment of an IL-12-responsive T cell clone: its characterization and utilization in the quantitation of IL-12 activity. J Leukoc Biol. 1997;61:346–352. doi: 10.1002/jlb.61.3.346. [DOI] [PubMed] [Google Scholar]
- 30.Becskei A, Grusby MJ. Contribution of IL-12r mediated feedback loop to Th1 cell differentiation. FEBS Letters. 2007;581:5199–5206. doi: 10.1016/j.febslet.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gabrysova L, Nicolson KS, Streeter HB, Verhagen J, Sabatos-Peyton CA, Morgan DJ, Wraith DC. Negative feedback control of the autoimmune response through antigen-induced differentiation of IL-10-secreting Th1 cells. J Exp Med. 2009;206:1755–1767. doi: 10.1084/jem.20082118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szabo SJ, Dighe AS, Gubler U, Murphy KM. Regulation of the Interleukin (IL)-12r beta 2 subunit expression in developing T helper 1 (Th1) and Th2 cells. J Exp Med. 1997;185:817–824. doi: 10.1084/jem.185.5.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12r expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 34.Schulz EG, Mariani L, Radbruch A, Hofer T. Sequential polarization and imprinting of type 1 T helper lymphocytes by Interferon-gamma and Interleukin-12. Immunity. 2009;30:673–683. doi: 10.1016/j.immuni.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 35.Good SR, Thieu VT, Mathur AN, Yu Q, Stritesky GL, Yeh N, O’Malley JT, Perumal NB, Kaplan MH. Temporal induction pattern of STAT4 target genes defines potential for Th1 lineage-specific programming. J Immunol. 2009;183:3839–3847. doi: 10.4049/jimmunol.0901411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulbe H, Thompson R, Wilson JL, Robinson S, Hagemann T, Fatah R, Gould D, Ayhan A, Balkwill F. The inflammatory cytokine Tumor Necrosis Factor-alpha generates an autocrine tumor-promoting network in epithelial ovarian cancer cells. Cancer Res. 2007;67:585–592. doi: 10.1158/0008-5472.CAN-06-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janes KA, Gaudet S, Albeck JG, Nielsen UB, Lauffenburger DA, Sorger PK. The response of human epithelial cells to TNF involves an inducible autocrine cascade. Cell. 2006;124:1225–1239. doi: 10.1016/j.cell.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 38.Klinke DJ, Finley SD. Timescale analysis of rule-based biochemical reaction networks. Biotech Prog. 2011 doi: 10.1002/btpr.704. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen WW, Niepel M, Sorger PK. Classic and contemporary approaches to modeling biochemical reactions. Genes Dev. 2010;24:1861–1875. doi: 10.1101/gad.1945410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coffman RL, Reiner SL. Instruction, selection, or tampering with the odds? Science. 1999;284:1283–1285. doi: 10.1126/science.284.5418.1283. [DOI] [PubMed] [Google Scholar]
- 42.Constant S, Pfeiffer C, Woodard A, Pasqualini T, Bottomly K. Extent of T cell receptor ligation can determine the functional differentiation of naive CD4+ T cells. J Exp Med. 1995;182:1591–1596. doi: 10.1084/jem.182.5.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hosken NA, Shibuya K, Heath AW, Murphy KM, O’Garra A. The effect of antigen dose on CD4+ T helper cell phenotype development in a T cell receptor-alpha beta-transgenic model. J Exp Med. 1995;182:1579–1584. doi: 10.1084/jem.182.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evavold BD, Sloan-Lancaster J, Wilson KJ, Rothbard JB, Allen PM. Specific T cell recognition of minimally homologous peptides: evidence for multiple endogenous ligands. Immunity. 1995;2:655–663. doi: 10.1016/1074-7613(95)90010-1. [DOI] [PubMed] [Google Scholar]
- 45.Felix NJ, Donermeyer DL, Horvath S, Walters JJ, Gross ML, Suri A, Allen PM. Alloreactive T cells respond specifically to multiple distinct peptide-MHC complexes. Nat Immunol. 2007;8:388–397. doi: 10.1038/ni1446. [DOI] [PubMed] [Google Scholar]
- 46.Sinigaglia F, D’Ambrosio D, Panina-Bordignon P, Rogge L. Regulation of the IL-12/IL-12R axis: a critical step in T-helper cell differentiation and effector function. Immunol Rev. 1999;170:65–72. doi: 10.1111/j.1600-065x.1999.tb01329.x. [DOI] [PubMed] [Google Scholar]
- 47.Lei JT, Martinez-Moczygemba M. Separate endocytic pathways regulate IL-5 receptor internalization and signaling. J Leukoc Biol. 2008;84:1–11. doi: 10.1189/jlb.1207828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Das J, Ho M, Zikherman J, Govern C, Yang M, Weiss A, Chakraborty AK, Roose JP. Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell. 2009;136:337–351. doi: 10.1016/j.cell.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR, Gajewski TF, Wang CR, Reiner SL. Helper t cell differentiation is controlled by the cell cycle. Immunity. 1998;9:229–237. doi: 10.1016/s1074-7613(00)80605-6. [DOI] [PubMed] [Google Scholar]
- 50.Gett AV, Hodgkin PD. Cell division regulates the T cell cytokine repertoire, revealing a mechanism underlying immune class regulation. Proc Natl Acad Sci USA. 1998;95:9488–9493. doi: 10.1073/pnas.95.16.9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horvai AE, Xu L, Korzus E, Brard G, Kalafus D, Mullen TM, Rose DW, Rosenfeld MG, Glass CK. Nuclear integration of JAK/STAT and RAS/AP-1 signaling by CBP and p300. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, Mullen TM, Glass CK, Rosenfeld MG. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 53.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston DM. Cooperation of STAT2 and p300/CBP in signalling induced by Interferon-alpha. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 54.O’Shea JJ, Lahesmaa R, Vahedi G, Laurence A, Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat Rev Immunol. 2011;11:239–250. doi: 10.1038/nri2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klinke DJ, Brundage KM. Scalable analysis of flow cytometry data using R/Bioconductor. Cytometry A. 2009;75:699–706. doi: 10.1002/cyto.a.20746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.