Abstract
Purpose. Pigment epithelium-derived factor (PEDF) is a multifunctional serpin. The purpose of this study is to identify PEDF protein forms and investigate their biological activities on tumor cell lines. Methods. Recombinant human PEDF proteins were purified by cation- and anion-exchange column chromatography. They were subjected to SDS-PAGE, IEF, deglycosylation, heparin affinity chromatography, and limited proteolysis. Cell viability, real-time electrical impedance of cells, and wound healing assays were performed using bladder and breast cancer cell lines, rat retinal R28, and human ARPE-19 cells. Results. Two PEDF protein peaks were identified after anion-exchange column chromatography: PEDF-1 eluting with lower ionic strength than PEDF-2. PEDF-1 had higher pI value and lower apparent molecular weight than PEDF-2. Both PEDF forms were glycosylated, bound to heparin, and had identical patterns by limited proteolysis. However, PEDF-2 emerged as being highly potent in lowering cell viability in all tumor cell lines tested, and in inhibiting tumor and ARPE-19 cell migration. In contrast, PEDF-1 minimally affected tumor cell viability and cell migration but protected R28 cells against death caused by serum starvation. Conclusion. Two distinct biochemical forms of PEDF varying in overall charge have distinct biological effects on tumor cell viability and migration. The existence of PEDF forms may explain the multifunctional modality of PEDF.
1. Introduction
Pigment epithelium-derived factor (PEDF) is a 50 kDa secreted glycoprotein and a member of the serpin superfamily with no demonstrable protease inhibitory activity [1]. PEDF is associated with several biological processes due to its antiangiogenic, anti-inflammatory, anti-oxidative, neurotrophic, and neuroprotective properties [2]. Moreover, it has been implicated in another interesting role, as an antitumor and antimetastatic agent with applications in multiple malignancies such as retinoblastoma, lung, breast, prostate, ovarian and pancreatic carcinomas, uveal melanoma, glioma, and osteosarcoma [3–8]. PEDF levels are decreased in tumor cells relative to normal cells, and PEDF addition inhibits tumor formation and metastasis, blocks angiogenesis, and induces apoptosis in tumor and endothelial cells. In contrast, PEDF promotes retinal cell survival and neuronal differentiation, protects retinal pigment epithelial cells against oxidative stress [2, 3, 9] and plays a role in expansion of neural stem cells [10]. The mechanisms that mediate the multimodal functionality to PEDF are not clear.
Previous structure-function studies revealed that two peptide regions toward the amino end of the PEDF polypeptide have distinct biological activities [2, 11, 12]. The peptide region termed 34-mer (amino acid residues Asp44-Asn77 of the human PEDF sequence) forms alpha helix A of the 3D structure of human PEDF [13] and confers antiangiogenic and antitumorigenic properties to the PEDF polypeptide [12]. The 44-mer peptide (Val78-Thr121) derived from the region that forms alpha helices B, C, and part of D exhibits neurite-outgrowth activity and protects spinal cord motor neurons against chronic toxicity [11, 14]. A smaller peptide derived from amino acid positions 82–121 exhibits effective neuroprotective properties in retinal ischemia [15].
Interestingly, the native PEDF has several isoforms differing in isoelectric point (pI) values [16, 17] implying differences in posttranslational modifications of the polypeptide backbone. Duh et al. reported that the secreted human recombinant PEDF from human embryonic kidney (HEK) cells has at least two species varying in their carbohydrate composition of the N-glycosylation site and efficacy of suppressing vascular endothelial growth factor-induced proliferation and migration of retinal microvascular endothelial cells [18]. Maik-Rachline and Seger showed that the human plasma PEDF is a phosphoprotein and that extracellular phosphorylation converts the recombinant protein from a neurotrophic to an antiangiogenic factor [19]. Konson et al. demonstrated that a triple phosphomimetic-altered PEDF is more efficient than wild-type PEDF in inhibiting neovascularization and tumor growth in vivo and suppresses cultured endothelial cell proliferation and cell migration much better than the wild-type PEDF [20].
Given the above, the aim of this study was to identify and characterize PEDF isoforms that could contribute to the complexity of PEDF action and its multifunctional modality.
2. Methods
2.1. Cell Culture
Human bladder carcinoma T24 cells (Cell Line collection, Lombardi Comprehensive Cancer Center, passage 33), human breast cancer 231-BR cells [21] (passage 13), mouse breast cancer 4T1-BR5 cells [21] (passage 11), human breast tumor MDA-MB-231 cells (Cell Line Collection, Lombardi Comprehensive Cancer Center, passage 6), mouse breast cancer 4T1 cells (Cell Line Collection, Lombardi Comprehensive Cancer Center, passage 9), and rat retinal R28 (generous gift of Gail Seigel, passages 47–55), were cultured in DMEM medium. Human retinal pigment epithelial ARPE-19 cells (American Type Culture Collection, passages 27–32) were cultured in DMEM-12 medium. Media were supplemented with 10% of fetal bovine serum (FBS) and 1% penicillin/streptomycin and cultures were incubated at 37°C with 5% CO2.
2.2. Protein Purification
Recombinant human PEDF was purified from the culture media of BHK cells harboring an expression plasmid containing full-length PEDF cDNA [22]. The culture media was concentrated by ammonium sulfate precipitation and subjected sequentially to cation- and anion-exchange column chromatography as described before with the following modifications [13]. Cation-exchange column chromatography was performed using a POROS S resin connected to a BioCAD 700E perfusion chromatography system, with buffer S (20 mM Na phosphate, pH 6.5, 50 mM NaCl) and elutions were with a linear gradient of 50–500 mM NaCl in Buffer S. PEDF-containing fractions were pooled, dialyzed against buffer Q (50 mM Tris-HCl, pH 8) and subjected to POROS Q column chromatography in buffer Q and elutions were done with a linear gradient of 100–300 mM NaCl in buffer Q. The PEDF-containing fractions were pooled, concentrated, and the buffer exchanged to PBS using ultrafiltration devices (Centricon-30 or Amicon-30, Millipore). Storage of the final samples was at −80°C.
2.3. Protein Analyses
Proteins were analyzed by SDS-PAGE using 10–20% polyacrylamide in SDS-Tricine gels (Invitrogen) or NuPAGE 10% polyacrylamide gel in Bis-Tris buffer with NuPAGE MOPS-SDS as running buffer (Invitrogen) under reducing conditions, and isoelectric focusing (IEF). Protein detection was accomplished with Coomassie Blue stain. PEDF protein identity was confirmed by Western Blot. After separation by SDS-PAGE electrophoresis, proteins were transferred to a nitrocellulose membrane, blocked for 1 h at room temperature, and incubated with polyclonal antibody to PEDF (Bioproducts MD, Inc.) in blocking solution at 1 : 5,000 or 1 : 10,000 dilution, followed by secondary antibody anti-rabbit IgG (H + L) in a 1 : 1000 dilution and the Vectastatin ABC Kit (Vector Laboratories) with colorimetric detection reagent 4-chloro-1-naphthol (BioRad Laboratories) as substrate. Alternatively, the secondary antibody was affinity-purified peroxidase-labeled goat anti-rabbit IgG (H + L) in a 1 : 200,000 dilution, and chemiluminescence detection with Super Signal West Dura Extended Duration Substrate (Thermo Scientific) on X-ray films.
Protein concentrations were determined using the Protein Assay Kit (BioRad) and Beckman DU 640 Spectrophotometer. Protein concentration was calculated from absorbance values using the formula:
| (1) |
Isoelectric focusing was performed using pH 3–7 or pH 3–10 IEF gels (Invitrogen), following manufacturer's instructions.
2.4. Enzymatic Deglycosylation
Two micrograms of protein was treated with N-glycosidase F (New England Biolabs, Ipswich, MA) following a previously described method [17]. Briefly, the protein sample was denatured by boiling in a solution containing 0.5% SDS, 40 mM DTT. To avoid inactivation of the enzyme by SDS, a total of 1% NP-40 in 50 mM sodium phosphate, pH 7.5 was added to the denatured sample before adding 1000 Units of N-glycosidase F in 20 μL final reaction volume. Enzymatic reactions were incubated at 37°C for 1 h followed by addition of SDS-PAGE sample buffer and samples were boiled for 10 min.
2.5. Limited Proteolysis
PEDF was cleaved with limiting amounts of chymotrypsin. Reaction mixtures contained chymotrypsin (Worthington, Lakewood, NJ) and PEDF substrate: at a w/w ratio of 1 : 10 in 80 mM Tris-HCl, pH 7.5 and 100 mM CaCl2. Protein amounts were 1 μg and incubation temperature was 25°C. Incubation times were as indicated. The reactions were stopped by freezing in dry ice and the addition of SDS-PAGE sample buffer. Reaction products were analyzed by SDS-PAGE analysis of products.
2.6. Glycosaminoglycan-Binding Assays
Heparin-binding assays were performed using heparin affinity column chromatography as described previously [23]. Hyaluronan-binding assays were performed using hyaluronan affinity column chromatography as described before [24].
2.7. RT-CES Assay
The cells were plated on a microplate biosensor platform and real-time electrical impedance of cells was followed with RT-CES system from ACEA Biosciences (San Diego, CA) (http://www.aceabio.com), a system described previously [25, 26]. T24, 231-BR, and 4T1-BR5 cells were seeded at 6,000 cells per well (a 96-well format) and allowed to attach for 18 h. Then the cells were serum-starved for 8 h followed by addition of serum-free media containing PEDF. R28 cells were seeded at a density of 3000 cells/well on 16-well strips of 96-well format with microelectrodes. Background impedance was measured with media alone before adding the cells. The cells were allowed to attach for 8 h with media containing 5% FBS. This was followed by changing to media with or without PEDF at the desired concentration. Real-time electrical cell impendence was monitored every hour in each well for several hours. Data from two replicates was averaged.
2.8. Cell Viability Assays
At the end point of the RT-CES assay, cell viability was measured by determining the relative levels of intracellular ATP as a biomarker for live cells using a CellTiter-Glo kit (Promega, Madison, WI) and following instructions by manufacturer. After 30 min of incubation at room temperature, the incubation solution in each well was transferred into wells in a 96-well microtiter plate. The luminescence intensity was measured using an automated plate reader (Envision, Perkin Elmer, MA). In parallel experiments, cells cultured in 24-well plates in triplicates were imaged and at the end point, cell viability was measured by determining the relative levels of mitochondrial dehydrogenase activity as live cell biomarker using the Cell Counting Kit-8 (Dojindo) following instructions by manufacturer. Cells in each well were incubated with 50 μL of CCK-8 solution diluted 1 : 25 and incubated for 4 h at 37°C. Absorbance of each well was measured at 450 nm using an automated plate reader (Envision). In all the cases, the absorbance reading for background was subtracted from the readings of samples. Data from replicates were averaged and statistical analysis was performed by a t-test. A P value of <0.05 was taken as significant.
2.9. In Vitro Migration Assay
Cell migration was assessed using wound-healing assays. Confluent cultures of ARPE-19 cells on 24-well culture plates and of MDA-MB-231 cells in 6-well plates were scratched with 10-μL pipette tips to create fixed-width linear “wounds” in the cell monolayers, followed by incubation with medium containing indicated concentrations of PEDF. Cell migration was monitored by capturing the images of linear wound closure using a 2X brightfield objective in an Olympus 1 × 70 inverted microscope coupled with a Sony camera. Images were taken immediately after wounding and later at defined time intervals until complete closure in the control cultures.
2.10. Mass Spectrometry
Matrix assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry was performed to determine the average molecular mass of the two PEDF protein forms. Purified samples of PEDF-1 (0.8 μg/μL) and PEDF-2 (2.5 or 1 μg/μL) in PBS were mixed 1 : 3 (v/v) with a saturated solution of sinapinic acid in 50% acetonitrile, 0.3% trifluoroacetic acid, and 0.5 μL of each sample was spotted onto a stainless steel MALDI plate. Mass spectra were manually acquired with 100 laser shots/spectrum using a MALDI Voyager DE-STR (AB Sciex) in positive ion, linear mode. The spectra were calibrated using a mixture of BSA and IgG calibration standards (AB Sciex). Additionally, some samples were mixed 1 : 1 (v/v) with the standard and calibrated internally, postacquisition. Molecular masses of both forms of PEDF were determined by averaging the means of 5 spectra collected from each of three sample spots.
3. Results
3.1. Identification of Two Forms of the PEDF Protein
Human recombinant PEDF was purified from culture media by a two-step procedure involving sequential cation- and anion-exchange column chromatography. The chromatogram from the cation-exchange column showed one broad peak of PEDF eluting with 200–250 mM NaCl, which was pooled and prepared for anion-exchange column chromatography. The anion-exchange column chromatogram showed two main protein peaks (Figure 1). Using conductivity values for the fractions (18.3 mS and 21.96 mS) we determined that protein in the first and second peaks eluted with about 200 mM NaCl and 253 mM NaCl, respectively. Isoelectric focusing of the proteins revealed a higher pI value for the protein in the first peak (peak 1) (pI = 7) than in the second one (peak 2) (pI = 6.1). Proteins in fractions from the two peaks contained PEDF-immunoreactive bands migrating as ~50,000-Mr by SDS-PAGE (see Figure 3). In addition to the IEF gel, Coomassie blue stained SDS-PAGE gels demonstrated that the PEDF forms were highly pure, and the proteins from the first peak migrated slightly faster than those in the second peak (see Figure 3). The protein eluting with lower ionic strength was termed PEDF-1, and the version eluting with higher ionic strength was labeled PEDF-2. As determined by MALDI-TOF mass spectrometry, the average molecular mass (mean ± standard error) of PEDF-1 was 46,063 ± 13 Da and the molecular mass of PEDF-2 was 47,176 ± 87 Da, in agreement with the differences in migration by SDS-PAGE.
Figure 1.
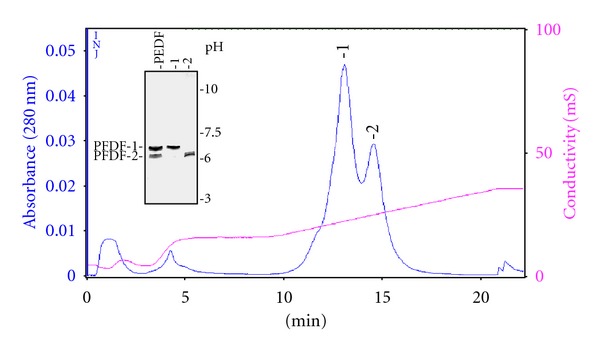
Anion-exchange Column Chromatography of PEDF. Chromatogram of PEDF with a POROS HQ column attached to BioCad700, running buffer of 50 mM Tris-HCl, pH 8, and a flow rate of 3 mL/min. Elution was with 25 column volumes of a linear gradient from 100 to 300 mM NaCl. On the chromatogram, blue indicates the O.D. of the sample read at 280 nm, magenta indicates the conductivity. Resolution of load and peak fractions by IEF is shown in the inset.
Figure 3.
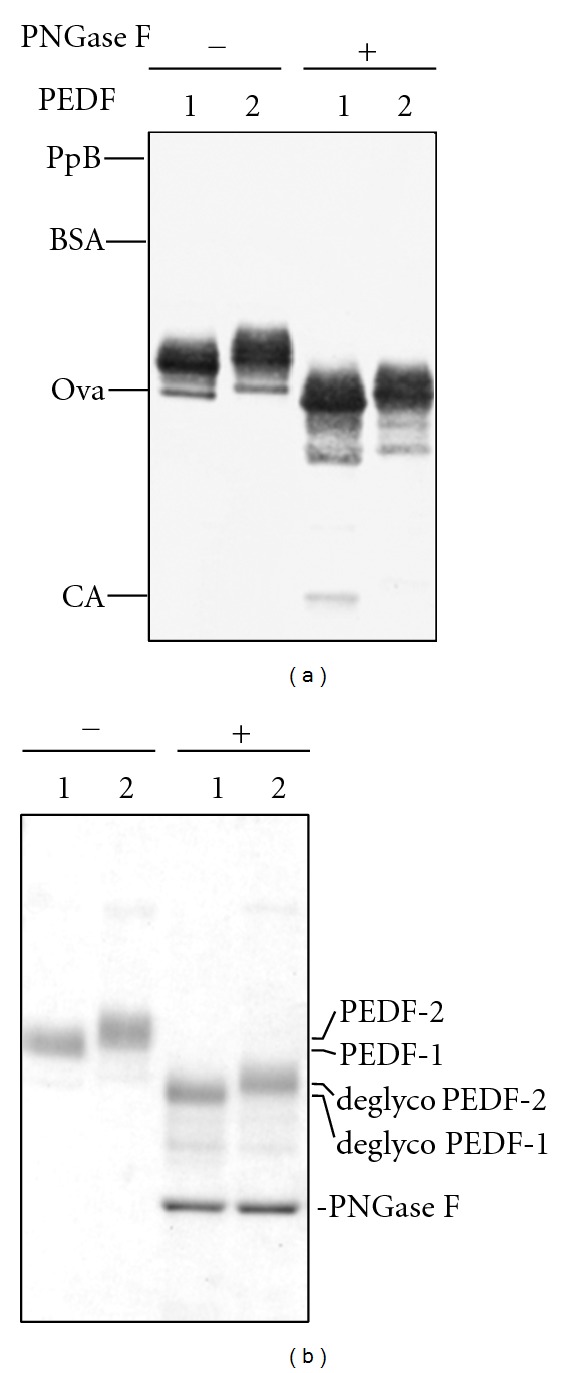
Deglycosylation of PEDF-1 and PEDF-2: deglycosylation of PEDF proteins was performed with PNGase F under denaturing conditions. Proteins were resolved by SDS-PAGE. (a) Western blotting of untreated and treated PEDF proteins (0.15 μg per lane) with anti-PEDF. (b) Coomassie blue staining of untreated and treated PEDF proteins (1.5 μg per lane). Migration position of PNGaseF is indicated on the right side, and molecular weight markers are on the left.
To find out how surface exposed charges affect fractionation of PEDF, alterations at positively charged residues K146/K147/R149 and K189/K191/R194/K197 of PEDF to neutral side chain residue alanine were prepared. Altered proteins were as described before [24]. Fractionation by anion-exchange column chromatography showed that in both cases two peaks of PEDF were eluted with conductivity values similar to those of wild-type PEDF (Figure 2). Recombinant PEDF fused with two tandem repeats of FLAG peptide at the carboxy-end produced by HEK cells [21] was also subjected to anion-exchange column chromatography under identical conditions as above. FLAG peptide (DYKDDDDK) with several acidic amino acid residues would increase the overall negative charge of the PEDF protein. We found that FLAG-PEDF eluted in two peaks with higher ionic strength than the untagged PEDF (with 316 mM NaCl and 367 mM NaCl, resp.) (Figure 2). These results indicate that recombinant human PEDF contains at least two isoforms varying in charge and in apparent molecular weight, and that this characteristic is general.
Figure 2.
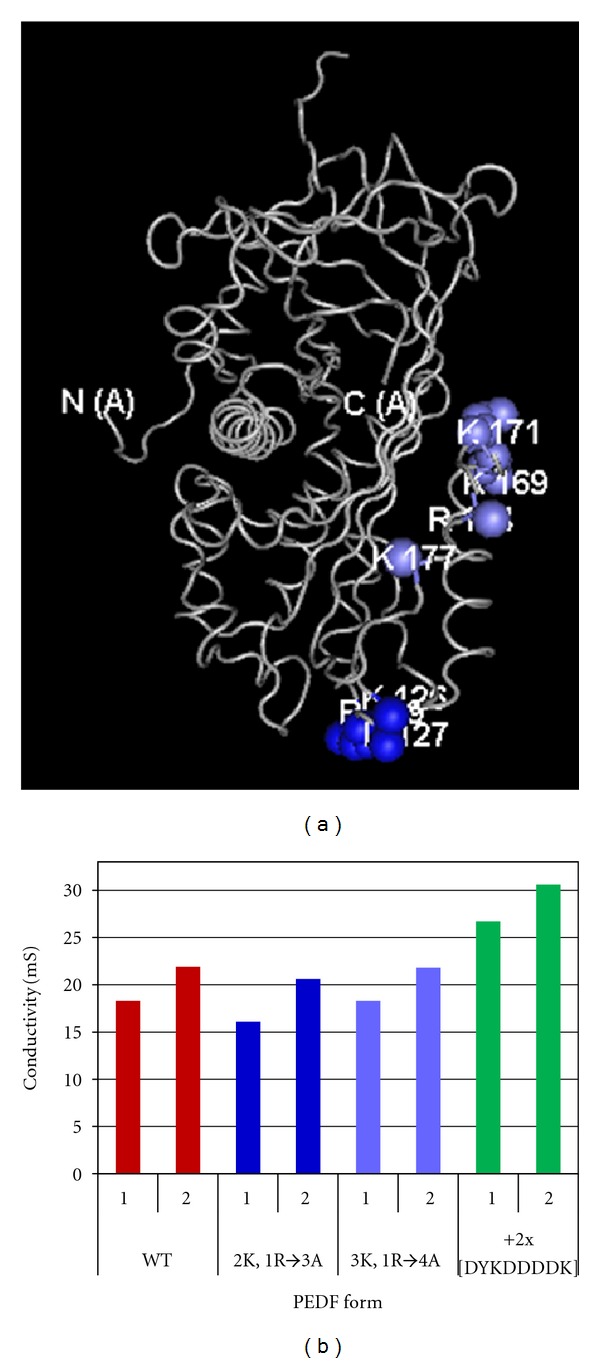
Recombinant altered PEDF proteins secreted to the culturing media were analyzed by anion-exchange column chromatography. Altered PEDF proteins were K146/K147/R149 (2 K,1R→3A) and K189/K191/R194/K197 (3 K,1R→4A), and FLAG-PEDF. (a) the backbone of the 3D structure of PEDF (tube in grey) from PDB 1IMV with highlighted location for amino acids K146/K147/R149 (blue) and K189/K191/R194/K197 (light blue). FLAG tagged PEDF had two tandem repeats of DYKDDDDK on the carboxy-end (C(A)). (b) Conductivity of PEDF fractions at peaks by anion-exchange column chromatography was determined and plotted.
3.2. Deglycosylation of PEDF-1 and PEDF-2
To determine whether the glycosylation attachments on the PEDF-1 and PEDF-2 polypeptide backbones are involved in their differences, the proteins were treated with N-glycosidase F to release Asn-linked oligosaccharides from glycoproteins. Figure 3 shows that N-glycosidase F digested both PEDF-1 and PEDF-2 causing them to migrate faster by SDS-PAGE. It indicates that the enzymatic treatment decreased their apparent molecular weight by identical values of ~8000. Both PEDF forms were glycosylated, and the deglycosylated PEDF-1 still migrated slightly faster than PEDF-2 by SDS-PAGE. These observations demonstrate that each PEDF-1 and PEDF-2 molecule had similar carbohydrate content of about 15% attached to an internal asparagine residue and that it does not account for the differences between the two forms PEDF-1 and PEDF-2.
3.3. Limited Proteolysis and Heparin Binding
The overall protein confirmation of both PEDF forms was analyzed by limited proteolysis. Treatment with limiting amounts of chymotrypsin revealed that both PEDF forms had identical patterns after digestion. By incubation with chymotrypsin at a protease : substrate ratio of 1 : 10 (w/w), both PEDF forms of about 50 kDa were digested in a one-step fashion into a product of about 46 kDa, that is, a decrease of ~4 kDa. Complete substrate utilization was achieved by 60 min (Figure 4(a)). A band corresponding to the released low-molecular-weight peptide was not readily identified by Coomassie Blue staining of the gel. Given that PEDF has affinity for heparin, we examined binding to heparin by both forms. Heparin affinity column chromatography showed that both forms bound to heparin-conjugated beads with similar affinity (Figure 4(b)). Hyaluronan binding assays were performed with PEDF-1 and PEDF-2 (data not shown) and showed similar results. Altogether, the results indicate that there are no differences in the overall protein conformation and in binding affinity to glycosaminoglycans between the two PEDF forms.
Figure 4.
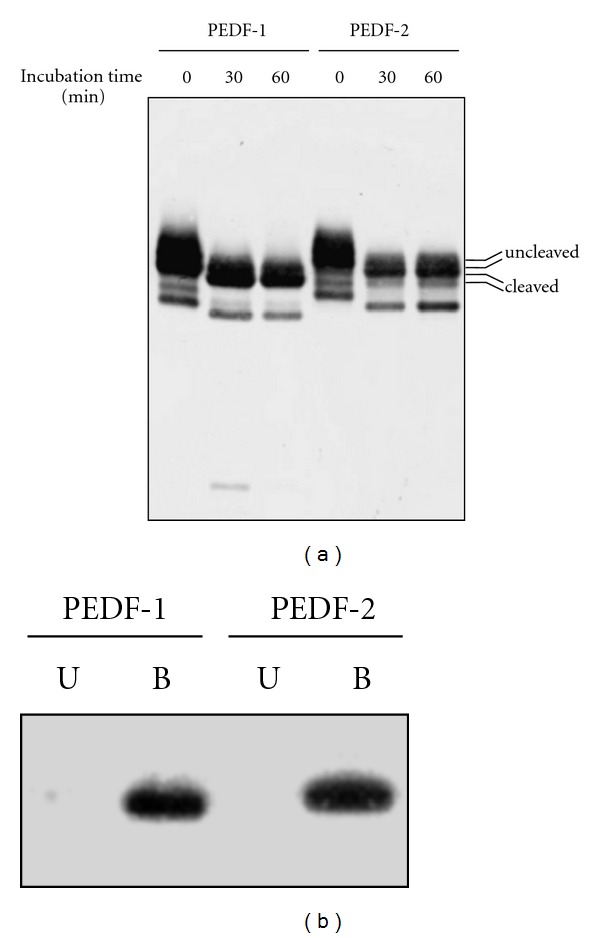
(a) Chymotrypsin Limited Proteolysis: reactions for the indicated times were performed with mixtures containing α-chymotrypsin and PEDF substrate at a ratio of 1 : 10 (w/w). Reactions were resolved by SDS-PAGE (0.1 μg per lane) followed by western blotting with anti-PEDF. Cleaved and the uncleaved forms are shown to the right. (b) Binding to Heparin. PEDF-1 and PEDF-2 were subjected to heparin-affinity column chromatography. Unbound (U) and bound (B) material was analyzed by SDS-PAGE and immunostained with anti-PEDF.
3.4. Effects of PEDF Versions on Tumor Cell Viability
To compare the antitumorigenic effects of PEDF-1 and PEDF-2, T24, 231-BR, and 4T1-BR5 cell cultures were incubated with serum-free medium containing increasing concentrations of each PEDF form, and monitored, in parallel, in real time, for cell impedance using a microelectronic system and under the microscope. Cells treated without PEDF increased the electrical impedance represented as cell index value with time up to 72–96 h. However, the cell index decreased in those treated with PEDF-2 in a dose-dependent fashion, while this was not observed with PEDF-1 (Figure 5), suggesting a differential effect on lowering the cell number. Examination under the microscope revealed a decline in cell number with increasing PEDF-2 additions and not with PEDF-1 with the three types of cells (Figure 6). The numbers of cells with 100 nM PEDF-2 in each case were significantly decreased in each field relative to those without or with PEDF-1. Quantification of relative cell numbers using two different biomarkers for live cells, intracellular ATP content, and mitochondrial dehydrogenase activity at end point corroborated the observation that PEDF-2 decreased the viability of each cell type, while PEDF-1 did not have a significant effect (Figure 7). Altogether, treatment with PEDF-2 resulted in a dramatic direct negative effect in tumor cell viability in contrast to PEDF-1.
Figure 5.
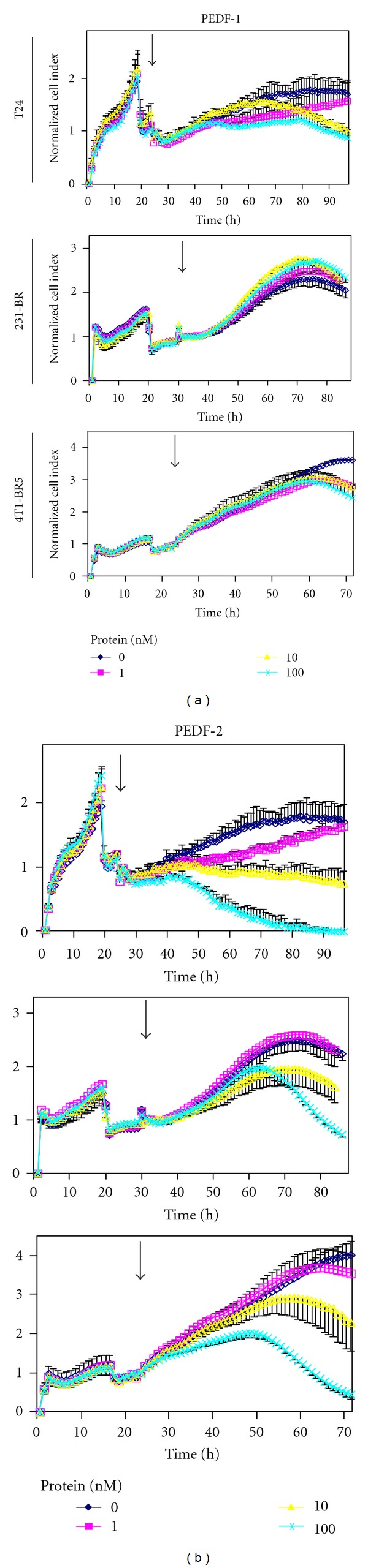
Effects of PEDF proteins on real-time electrical cell impedance of T24, 231-BR, and 4T1-BR5 cells in culture: cells were cultured in complete media to subconfluency, followed by serum starvation for 8 h, and then media was replaced with fresh media without serum containing indicated concentrations of recombinant human PEDF-1 or PEDF-2 proteins. Real-time cell impedance was monitored in cells treated with indicated concentrations of PEDF. Cell index was normalized relative to the cell numbers at the time of PEDF addition (shown by arrow). Each point is the average of triplicate assays. Error bars indicate average ± SD.
Figure 6.
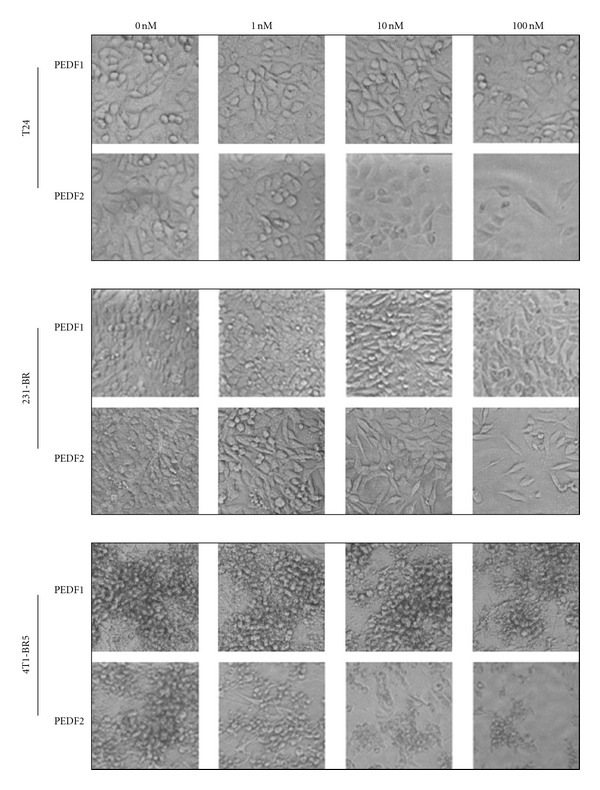
Imaging of T24, 231-BR, and 4T1-BR5 cells treated with PEDF-1 and PEDF-2 proteins: bright field images were taken at the end point of cells treated with PEDF proteins at concentrations indicated on the top.
Figure 7.
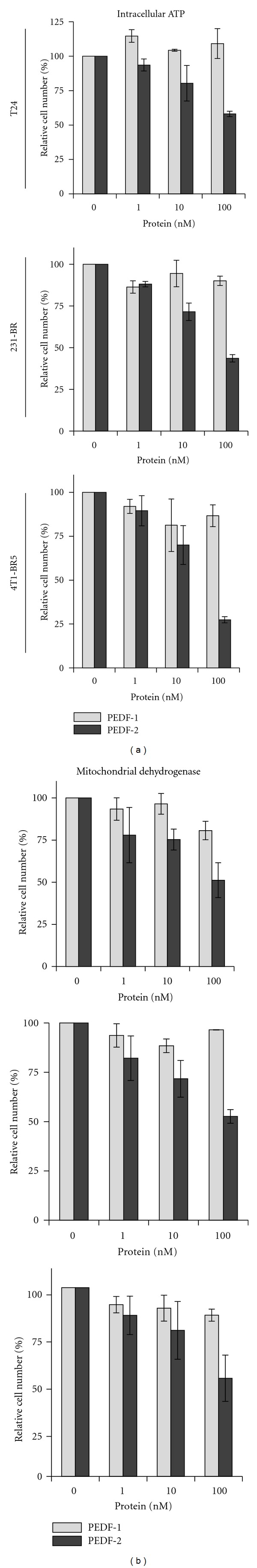
Effects of PEDF proteins on viability of T24, 231-BR, and 4T1-BR5 cells in culture: effect of PEDF on cell viability at the treatment end point. Plot of relative cell numbers quantified based on intracellular ATP content (shown on the left) and mitochondrial dehydrogenase activity (shown on the right) as biomarkers of live cells. Cell viability was expressed as percentage of luminescence values relative to untreated controls, and percentage of absorbance at 490 nm relative to untreated controls, respectively. Each point is the average of triplicate assays. Error bars indicate average ± SD.
3.5. Effect of PEDF-1 and PEDF-2 on Cancer and RPE Cell Motility
It has been reported that PEDF inhibits the migration of breast tumor MDA-MB-231 cells and retinal pigment epithelial (RPE) cells [20, 27]. Thus, the effect of the PEDF-1 and PEDF-2 proteins on cell migration was examined in MDA-MB-231 cells and ARPE-19 cells. For this wound healing assay, a fixed-width scratch in a cell monolayer was created and the advancement of the migrating front was followed in the presence of either PEDF-1 or PEDF-2. PEDF-2 affected negatively the migration of the MDA-MB-231 cells, while PEDF-1 was not significant (Figure 8(a)). Wound healing assays performed with ARPE-19 cells showed similar results as with the cancer cells (Figure 8(b)). These observations indicate that PEDF-2 was more efficient than PEDF-1 in inhibiting cancer and RPE cell migration.
Figure 8.
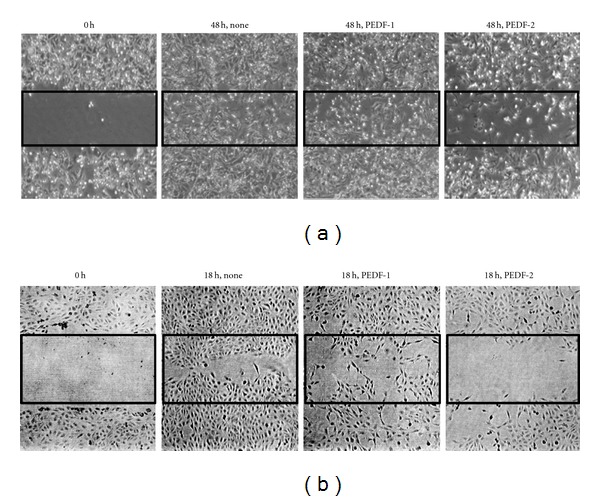
Effect of PEDF proteins on wound healing assay: MDA-MB-231 (a) and ARPE-19 (b) cell monolayers were scratched with a 10-μL pipette tip to make a wound. Cells were then incubated with media alone or PEDF proteins (20 ng/mL) for 48 h in (a) and 18 h in (b). Images were taken at the end points and compared to 0 h to measure wound healing.
3.6. PEDF-1 Is Active on Retina R28 Cells against Serum-Starvation-Induced Death
The above results suggest that PEDF-2 is more efficient than PEDF-1, implying that PEDF-1 may have lost its activity. It has been reported that PEDF also protects retina cells against trophic factor depletion [28, 29]. Thus the activity of PEDF-1 was also examined using a method in which PEDF protects retina cells against death induced by serum starvation. Figure 9 shows cell viability of retina R28 cells treated with increasing amounts of PEDF in serum-free media for 48 hours. Figure 9(a) shows that real-time cell index of R28 cells decreased almost linearly between the 20th hour and the 48th hour after plating. However treatments with increasing concentrations of PEDF-1 prevented the cell index decrease, which with the highest dose of PEDF (100 nM) even increased from the 20th to the 35th hour after plating. Quantification of relative cell numbers at end point using intracellular ATP content as biomarker of live cells corroborated the observation that PEDF-1 increased viability of R28 cells in a dose-dependent fashion (Figure 9(b)). The effects on the retina survival activity of PEDF-2 were similar to those of PEDF-1 (Figures 9(c) and 9(d)). Altogether, the results show that both PEDF forms PEDF-1 and PEDF-2 behaved as active survival factor for retina cells in culture.
Figure 9.
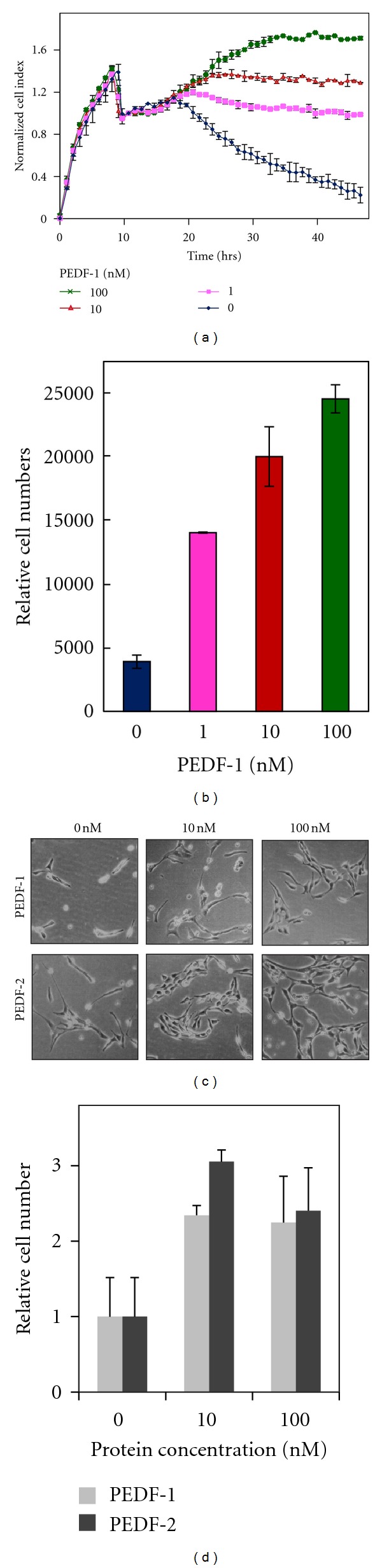
Retina R28 Cell Survival Assays: (a) real-time cell impedance monitoring of PEDF cell survival activity in serum-starved R28 cells in the presence of increasing concentrations of PEDF. The cell index was normalized relative to the cell numbers at the time of PEDF addition. (b) Plot of relative cell numbers quantified based on intracellular ATP content performed at end point (see (a)). (c) Bright field images of R28 cells were taken at end point (48 h) of cells treated with PEDF proteins at concentrations indicated on the top. (d) Plot of relative cell numbers quantified based on intracellular ATP content performed at 48 h. Each point is the average of duplicate wells. Error bars indicate average ± SD.
4. Discussion
We have identified two distinct biochemical forms of recombinant PEDF, which we termed PEDF-1 and PEDF-2. The distinction is based on ionic potential, pI values, and apparent molecular weight and demonstrates that PEDF-2 has higher overall negative charge than PEDF-1. PEDF-2 elution required higher ionic potential from anion-exchange column chromatography; the protein has a lower pI and migrated slower in SDS-PAGE than PEDF-1. This characteristic is shared with PEDF from different sources and even when it is altered or fused to tags. Altered and FLAG-tagged PEDF yielded two peaks similar to the wild-type PEDF (Figure 2), confirming that the existence as two forms is a general feature of PEDF. Because the protein derives from one PEDF cDNA sequence transfected into BHK cells, the sequence of the polypeptide backbone is identical in both forms. Upon maturation and secretion, the PEDF versions may have adopted modifications posttranslationally that confer charge differences, and in turn grant diversity in biological activities to PEDF. Isoforms of PEDF have been described by different laboratories. Recombinant PEDF isoforms were reported to have differences in carbohydrate composition at its glycosylation site [18]. PEDF isoforms have also been reported in peripheral neuropathies, with fucose residues on the oligosaccharide chain found only in the isoforms downregulated in neuropathic patients without pain [30]. Although both PEDF-1 and PEDF-2 are glycosylated, they differ in apparent molecular weight even after deglycosylation, suggesting that this is not the main difference between them. More recently, it was reported that the triple phosphomimetic PEDF, EEE-PEDF, containing three negatively charged glutamic acids instead of serines, possesses significantly increased antiangiogenic, and anticancer activities than wild-type PEDF [20, 31]. However, our PEDF forms were not phosphorylated (data not shown). The significance of these findings is that PEDF isoforms have been identified in ocular, tumor, and nontumor tissues [16, 17, 32], and interestingly their distribution varies between normal and tumor tissues.
Several reports on posttranslational modifications of PEDF offer explanations for the multifunctional modality of PEDF. Similar to our results, Duh et al. [18] purified two forms of PEDF, A and B, using cation-exchange column. Form A eluted at a lower NaCl concentration, that is, less positively charged and had a slightly larger molecular weight than from B; and PEDF A was a more efficient antiangiogenic form than PEDF B. This implies a match of PEDF-1 to PEDF B and PEDF-2 to PEDF A. Petersen et al. [33] reported that a complete map of all post-translational modifications revealed that authentic plasma PEDF carries an N-terminal pyroglutamate blocking group and an N-linked glycan at position Asn266. They proposed that the pyroglutamate residue may regulate the activity of PEDF analogously to the manner in which it regulates thyrotropin-releasing hormone. Maik-Rachline et al. [34] demonstrated that extracellular phosphorylation converts PEDF from a neurotrophic to an antiangiogenic factor. Later they proposed that differential phosphorylation induces variable effects of PEDF, and therefore contributes to the complexity of PEDF action [19]. Although the observations with PEDF phosphomimetics match our results, our PEDF forms were not found to be phosphorylated and therefore, another chemical difference is sought for PEDF-1 and PEDF-2 that is yet to be determined. These observations are in agreement with the idea that the multifunctional modality of PEDF may be explained by differences in posttranslational modifications of the PEDF polypeptide, which may regulate their biological activities. Given that synthetic peptides 34-mer and 44-mer derived from PEDF are biologically active, these regions may require to be properly exposed in the folded protein to interact with cell surface receptors and trigger the necessary signals for activity. Posttranslational modifications may be tools to open up or expose the active regions in the folded PEDF protein. It is envisioned that in PEDF-2 the active region for antitumorigenic activity is more exposed than in PEDF-1. Other factors that enhance efficacy in PEDF-2 may include post-translational modification(s) that confers an increase in negative charge to the protein. Further studies to identify the chemical differences between the PEDF forms will be of great interest for the development of second generation PEDF molecules.
Authors' Contribution
P. Subramanian and M. Deshpande share first coauthorship and they contributed equally to this paper.
Acknowledgments
This work was supported, in part, by National Institutes of Health NEI Intramural Research Program and by National Cancer Institute Grant CA134727 (to V. Notario). The authors thank Natalia Balko for assistance in purification of PEDF protein and Gail Seigel for generously providing R28 cells.
References
- 1.Becerra SP, Sagasti A, Spinella P, Notario V. Pigment epithelium-derived factor behaves like a noninhibitory serpin. neurotrophic activity does not require the serpin reactive loop. Journal of Biological Chemistry. 1995;270(43):25992–25999. doi: 10.1074/jbc.270.43.25992. [DOI] [PubMed] [Google Scholar]
- 2.Patricia Becerra S. Focus on molecules: pigment epithelium-derived factor (PEDF) Experimental Eye Research. 2006;82(5):739–740. doi: 10.1016/j.exer.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 3.Barnstable CJ, Tombran-Tink J. Neuroprotective and antiangiogenic actions of pedf in the eye: molecular targets and therapeutic potential. Progress in Retinal and Eye Research. 2004;23(5):561–577. doi: 10.1016/j.preteyeres.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Broadhead ML, Dass CR, Choong PF. In vitro and in vivo biological activity of pedf against a range of tumors. Expert Opinion on Therapeutic Targets. 2009;13(12):1429–1438. doi: 10.1517/14728220903307475. [DOI] [PubMed] [Google Scholar]
- 5.Doll JA, Stellmach VM, Bouck NP, et al. Pigment epithelium-derived factor regulates the vasculature and mass of the prostate and pancreas. Nature Medicine. 2003;9(6):774–780. doi: 10.1038/nm870. [DOI] [PubMed] [Google Scholar]
- 6.Ek ETH, Dass CR, Contreras KG, Choong PFM. Pigment epithelium-derived factor overexpression inhibits orthotopic osteosarcoma growth, angiogenesis and metastasis. Cancer Gene Therapy. 2007;14(7):616–626. doi: 10.1038/sj.cgt.7701044. [DOI] [PubMed] [Google Scholar]
- 7.Palmieri D, Fitzgerald D, Shreeve SM, et al. Analyses of resected human brain metastases of breast cancer reveal the association between up-regulation of hexokinase 2 and poor prognosis. Molecular Cancer Research. 2009;7(9):1438–1445. doi: 10.1158/1541-7786.MCR-09-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H, Grossniklaus HE. Constitutive overexpression of pigment epithelium-derived factor inhibition of ocular melanoma growth and metastasis. Investigative Ophthalmology and Visual Science. 2010;51(1):28–34. doi: 10.1167/iovs.09-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukherjee PK, Marcheselli VL, Barreiro S, Hu J, Bok D, Bazan NG. Neurotrophins enhance retinal pigment epithelial cell survival through neuroprotectin d1 signaling. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(32):13152–13157. doi: 10.1073/pnas.0705949104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramirez-Castillejo C, Sanchez-Sanchez F, Andreu-Agullo C, et al. Pigment epithelium-derived factor is a niche signal for neural stem cell renewal. Nature Neuroscience. 2006;9(3):331–339. doi: 10.1038/nn1657. [DOI] [PubMed] [Google Scholar]
- 11.Bilak MM, Patricia Becerra S, Vincent AM, Moss BH, Aymerich MS, Kuncl RW. Identification of the neuroprotective molecular region of pigment epithelium-derived factor and its binding sites on motor neurons. Journal of Neuroscience. 2002;22(21):9378–9386. doi: 10.1523/JNEUROSCI.22-21-09378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filleur S, Volz K, Nelius T, et al. Two functional epitopes of pigment epithelial-derived factor block angiogenesis and induce differentiation in prostate cancer. Cancer Research. 2005;65(12):5144–5152. doi: 10.1158/0008-5472.CAN-04-3744. [DOI] [PubMed] [Google Scholar]
- 13.Simonovic M, Gettins PGW, Volz K. Crystal structure of human pedf, a potent anti-angiogenic and neurite growth-promoting factor. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(20):11131–11135. doi: 10.1073/pnas.211268598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alberdi E, Aymerich MS, Becerra SP. Binding of pigment epithelium-derived factor (pedf) to retinoblastoma cells and cerebellar granule neurons. evidence for a pedf receptor. Journal of Biological Chemistry. 1999;274(44):31605–31612. doi: 10.1074/jbc.274.44.31605. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Tran VV, Hu Y, Mark Saltzman W, Barnstable CJ, Tombran-Tink J. A pedf n-terminal peptide protects the retina from ischemic injury when delivered in plga nanospheres. Experimental Eye Research. 2006;83(4):824–833. doi: 10.1016/j.exer.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 16.Tombran-Tink J, Shivaram SM, Chader GJ, Johnson LV, Bok D. Expression, secretion, and age-related downregulation of pigment epithelium-derived factor, a serpin with neurotrophic activity. Journal of Neuroscience. 1995;15(7 I):4992–5003. doi: 10.1523/JNEUROSCI.15-07-04992.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu YQ, Notario V, Chader GJ, Becerra SP. Identification of pigment epithelium-derived factor in the interphotoreceptor matrix of bovine eyes. Protein Expression and Purification. 1995;6(4):447–456. doi: 10.1006/prep.1995.1060. [DOI] [PubMed] [Google Scholar]
- 18.Duh EJ, Yang HS, Suzuma I, et al. Pigment epithelium-derived factor suppresses ischemia-induced retinal neovascularization and vegf-induced migration and growth. Investigative Ophthalmology and Visual Science. 2002;43(3):821–829. [PubMed] [Google Scholar]
- 19.Maik-Rachline G, Seger R. Variable phosphorylation states of pigment-epithelium-derived factor differentially regulate its function. Blood. 2006;107(7):2745–2752. doi: 10.1182/blood-2005-06-2547. [DOI] [PubMed] [Google Scholar]
- 20.Konson A, Pradeep S, D’Acunto CW, Seger R. Pigment epithelium-derived factor and its phosphomimetic mutant induce jnk-dependent apoptosis and p38-mediated migration arrest. Journal of Biological Chemistry. 2011;286(5):3540–3551. doi: 10.1074/jbc.M110.151548. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Fitzgerald DP, Subramanian P, Deshpande M, et al. Opposing effects of pigment epithelium-derived factor on breast cancer cell versus neuronal survival: implication for brain metastasis and metastasis-induced brain damage. Cancer Research. 2012;72(1):144–153. doi: 10.1158/0008-5472.CAN-11-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stratikos E, Alberdi E, Gettins PGW, Becerra SP. Recombinant human pigment epithelium-derived factor (PEDF): characterization of pedf overexpressed and secreted by eukaryotic cells. Protein Science. 1996;5(12):2575–2582. doi: 10.1002/pro.5560051220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alberdi E, Hyde CC, Becerra SP. Pigment epithelium-derived factor (PEDF) binds to glycosaminoglycans: Analysis of the binding site. Biochemistry. 1998;37(30):10643–10652. doi: 10.1021/bi9802317. [DOI] [PubMed] [Google Scholar]
- 24.Becerra SP, Perez-Mediavilla LA, Weldon JE, et al. Pigment epithelium-derived factor binds to hyaluronan: mapping of a hyaluronan binding site. Journal of Biological Chemistry. 2008;283(48):33310–33320. doi: 10.1074/jbc.M801287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abassi YA, Jackson JA, Zhu J, Oconnell J, Wang X, Xu X. Label-free, real-time monitoring of ige-mediated mast cell activation on microelectronic cell sensor arrays. Journal of Immunological Methods. 2004;292(1-2):195–205. doi: 10.1016/j.jim.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 26.Solly K, Wang X, Xu X, Strulovici B, Zheng W. Application of real-time cell electronic sensing (RT-CES) technology to cell-based assays. Assay and Drug Development Technologies. 2004;2(4):363–372. doi: 10.1089/adt.2004.2.363. [DOI] [PubMed] [Google Scholar]
- 27.Ma X, Pan L, Jin X, et al. Microphthalmia-associated transcription factor acts through PEDF to regulate RPE cell migration. Experimental Cell Research. 2012;318(3):251–261. doi: 10.1016/j.yexcr.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Murakami Y, Ikeda Y, Yonemitsu Y, et al. Inhibition of nuclear translocation of apoptosis-inducing factor is an essential mechanism of the neuroprotective activity of pigment epithelium-derived factor in a rat model of retinal degeneration. American Journal of Pathology. 2008;173(5):1326–1338. doi: 10.2353/ajpath.2008.080466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Notari L, Miller A, Martinez A, et al. Pigment epithelium-derived factor is a substrate for matrix metalloproteinase type 2 and type 9: implications for downregulation in hypoxia. Investigative Ophthalmology & Visual Science. 2005;46(8):2736–2747. doi: 10.1167/iovs.04-1489. [DOI] [PubMed] [Google Scholar]
- 30.Conti A, Ricchiuto P, Iannaccone S, et al. Pigment epithelium-derived factor is differentially expressed in peripheral neuropathies. Proteomics. 2005;5(17):4558–4567. doi: 10.1002/pmic.200402088. [DOI] [PubMed] [Google Scholar]
- 31.Konson A, Pradeep S, Seger R. Phosphomimetic mutants of pigment epithelium-derived factor with enhanced antiangiogenic activity as potent anticancer agents. Cancer Research. 2010;70(15):6247–6257. doi: 10.1158/0008-5472.CAN-10-0434. [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez-Piñeiro AM, Blanco-Prieto S, Sánchez-Otero N, Rodríguez-Berrocal FJ, Páez de la Cadena M. On the identification of biomarkers for non-small cell lung cancer in serum and pleural effusion. Journal of Proteomics. 2010;73(8):1511–1522. doi: 10.1016/j.jprot.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 33.Petersen SV, Valnickova Z, Enghild JJ. Pigment-epithelium-derived factor (pedf) occurs at a physiologically relevant concentration in human blood: purification and characterization. Biochemical Journal. 2003;374(1):199–206. doi: 10.1042/BJ20030313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maik-Rachline G, Shaltiel S, Seger R. Extracellular phosphorylation converts pigment epithelium-derived factor from a neurotrophic to an antiangiogenic factor. Blood. 2005;105(2):670–678. doi: 10.1182/blood-2004-04-1569. [DOI] [PubMed] [Google Scholar]


