Abstract
Thymine glycols are formed in DNA by exposure to ionizing radiation or oxidative stress. Although these lesions are repaired by the base excision repair pathway, they have been shown also to be subject to transcription-coupled repair. A current model for transcription-coupled repair proposes that RNA polymerase II arrested at a DNA lesion provides a signal for recruitment of the repair enzymes to the lesion site. Here we report the effect of thymine glycol on transcription elongation by T7 RNA polymerase and RNA polymerase II from rat liver. DNA substrates containing a single thymine glycol located either in the transcribed or nontranscribed strand were used to carry out in vitro transcription. We found that thymine glycol in the transcribed strand blocked transcription elongation by T7 RNA polymerase ~50% of the time but did not block RNA polymerase II. Thymine glycol in the nontranscribed strand did not affect transcription by either polymerase. These results suggest that arrest of RNA polymerase elongation by thymine glycol is not necessary for transcription-coupled repair of this lesion. Additional factors that recognize and bind thymine glycol in DNA may be required to ensure RNA polymerase arrest and the initiation of transcription-coupled repair in vivo.
Transcription-coupled repair (TCR)1 is a pathway of DNA excision repair that is targeted to removal of DNA lesions present in transcribed strands of expressed genes (1). TCR has been demonstrated in mammalian cells (2), Escherichia coli (3), and Saccharomyces cerevisiae (4–6). TCR was originally observed for lesions repaired by nucleotide excision repair (2). An active RNA polymerase elongation complex is necessary for preferential repair of the transcribed strand (7). The arrest of transcription at DNA lesions has been proposed to serve as a specific signal to direct repair enzymes to the transcribed strand of an active gene to initiate a repair event (8). It is likely that this repair pathway evolved for the dedicated purpose of resolving the impasse of an RNA polymerase arrested at a lesion. The polymerase must be displaced both to permit verification that a lesion caused the arrest and to allow the repair enzymes to operate on the lesion (1).
The role of RNA polymerases in TCR has been examined more directly by comparing the extent of RNA polymerase arrest in vitro by a lesion with TCR of that lesion in vivo. These studies have shown that various types of DNA damage in the template strand of DNA can act as blocks to transcription catalyzed by different RNA polymerases. In several cases, a correlation between the extent of polymerase arrest and the extent of TCR has been found (7). However, most of these studies have been carried out using viral and bacterial RNA polymerases. In an attempt to understand the role of mammalian RNA polymerase II (RNAP II) in TCR, we have developed an in vitro transcription system with templates containing a site-specific lesion positioned downstream of the adenovirus major late promoter (AdMLP) and purified proteins and initiation factors. Using this in vitro transcription system, we have previously shown that a cyclobutane pyrimidine dimer in the transcribed strand in different sequence contexts is a strong block to RNAP II (9, 10). The arrested polymerase is stable and competent to resume transcription. Furthermore, the arrested complex can be displaced from the lesion site during transcript cleavage mediated by elongation factor SII to allow access of a small repair enzyme, photolyase, to the lesion site for repair (11). These results support the original TCR model in which RNAP II must be displaced from the site of the lesion to provide access for repair enzymes to the lesion site (8, 12).
In this study, we have analyzed the effect of thymine glycol (Tg) on transcription elongation. Tgs are formed by oxidation of the 5,6 double bond of thymine on exposure to ionizing radiation or oxidative stress. Tg is a strong blocking lesion for prokaryotic DNA polymerases in vitro (13–16) and can be lethal in vivo (17–20). Tg is normally repaired by the base excision repair system (for review, see Ref. 21). Interestingly, it has been recently shown that Tg in human cells is also subject to TCR (22–24). This represents the first example of an evident coupling of base excision repair to transcription, raising the question of how the RNAP II may react to a thymine glycol compared with a cyclobutane pyrimidine dimer in the transcribed strand.
To study the effect of Tg on transcription, we have prepared DNA substrates containing a single Tg positioned downstream of the T7 promoter or the AdMLP in the transcribed or nontranscribed strand and an in vitro transcription system with purified RNA polymerase and initiation factors. We have found that a Tg located on the transcribed strand is a significant block to T7 RNAP, resulting in polymerase arrest 50% of the time. Surprisingly, and in contrast, a Tg in the transcribed strand has no detectable effect on transcription by RNAP II, suggesting that additional factors that bind to this lesion in the DNA may be involved in RNAP II arrest and initiation of TCR in vivo.
EXPERIMENTAL PROCEDURES
Proteins and Reagents
T7 RNAP was purchased from New England Biolabs. RNAP II and transcription initiation factors were purified from rat liver or recombinant sources as described previously (25, 26). T4 polynucleotide kinase and T4 DNA ligase were from Life Technologies, Inc. E. coli strain MV1184 was a gift from Dr. Joachim Messing (Rutgers University, Piscataway, NJ). D44 IgG anti-RNA antibodies (27) were purified from ascites fluid as described previously (26). Highly purified NTPs and radiolabeled nucleotides were purchased from Amersham Pharmacia Biotech. Formalin-fixed Staphylococcus aureus was obtained from Calbiochem.
Preparation of a 13-mer DNA Oligonucleotide Containing a Single Thymine Glycol
A 13-mer oligonucleotide of sequence 5′-CCCCGCGTACCGG-3′, containing a single Tg, was prepared by oxidation with osmium tetroxide (OsO4) in the presence of pyridine (16). The reaction mixture contained 200 µg of the oligonucleotide, 3% OsO4, and 5% pyridine in a final volume of a 100 µl. Oxidation was carried out at room temperature for 1 h, after which excess OsO4 was extracted with ether. The 13-mer oligonucleotide containing a single Tg was purified by reverse-phase high pressure liquid chromatography (HPLC) using a Beckman System Gold Nouveau apparatus consisting of an autosampler, a dual pump module, a UV absorbance detector (set to read at 254 nm), and a Hamilton PRP-1 reverse-phase column. Gradient control and data processing were achieved using Beckman System Gold Nouveau software. Optimal separation of the oxidized product was obtained using 0.1 m ammonium acetate, pH 5.8, with a linear gradient of acetonitrile (0–30% over 30 min) at a flow rate of 1 ml/min. (Ref. 28 and see Fig. 2). The presence of a Tg was confirmed by piperidine cleavage of the phosphodiester bond adjacent to the modified base (16) and by enzymatic digestion of the OsO4-treated 13-mer oligonucleotide to deoxynucleosides followed by HPLC (29). The latter treatment revealed the anticipated amounts of dA, dG, and dC, but no detectable dT. As expected, dTg was undetectable because of its weak UV absorbance.
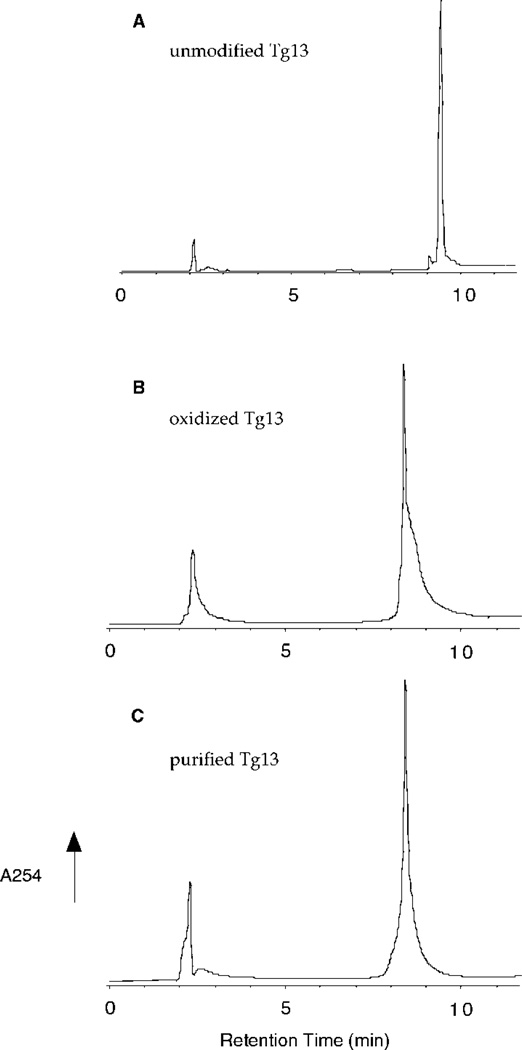
Fig. 2. HPLC profiles of OsO4-treated Tg13 oligonucleotide.
The Tg13 oligonucleotide was eluted on a PRP-1 reverse-phase column using 10 mm ammonium acetate, pH 5.8, with a linear gradient of acetonitrile (0–30% over 30 min) at a flow rate of 1 ml/min. Detection was by UV absorbance at a wavelength of 254 nm. A, unmodified Tg13 oligonucleotide; B, Tg13 oligonucleotide after OsO4 oxidation; C, Tg13 oligonucleotide after further fractionation of B.
DNA Templates for Transcription
DNA templates used for transcription reactions with T7 RNAP consisted of 154-base pair (bp) DNA fragments containing a single Tg in the transcribed (TgTST7) or nontranscribed (TgNTST7) strand downstream of the T7 promoter and of a C-less cassette (Fig. 1A). The C-less cassette resulted in a short stretch of RNA devoid of C. These substrates were constructed from eight oligonucleotides, five in the damage-containing strand and three in the opposite strand (Ref. 30 and Fig. 1A). 50 pmol of each oligonucleotide were phosphorylated with T4 polynucleotide kinase. The mixture was heated 3 min at 95 °C and slowly cooled to room temperature. ATP to a final concentration of 1 mm and 10 units of T4 DNA ligase were added to the mixture containing 50 mm Tris-HCl, pH 7.6, 10 mm MgCl2, 1 mm dithiothreitol, and 5 mm polyethylene glycol-8000. The DNA was ligated overnight at 16 °C. The ligation products were treated with proteinase K and further purified by ethanol precipitation. The DNA samples were resuspended in formamide dye, and the single-stranded 154-bp DNA fragments were purified from an 8% denaturing polyacrylamide gel. To ensure that all samples were double-stranded after reannealing of the complementary strands, the DNA was digested with appropriate restriction enzymes and further purified on a nondenaturing gel if necessary.
Fig. 1. DNA substrates used in this study.
A, DNA template TgTST7 for T7 RNAP transcription. B, DNA template TgTSAd for RNAP II transcription. TgTST7 DNA and TgTSAd DNA, each containing a single Tg in the transcribed strand downstream of T7 or AdMLP promoter, respectively, were constructed as described under “Experimental Procedures.” The oligonucleotides from which the 154-mer TgT7 substrate was synthesized are listed. The DNA sequence of the C-less cassette contained in the 57-mer is underlined. Numbers in parentheses indicate nucleotide positions on the DNA sequence. Runoff RNA (RO) and RNA resulting from transcription arrest at a Tg (Tg) are marked with dashed lines together with their expected sizes. The transcription start site (+1) is represented by a bent arrow. T7, T7 promoter.
DNA templates used for transcription reactions with RNAP II consisted of HindIII-linearized plasmid DNA containing a single Tg downstream of the AdMLP and of a G-less cassette (Fig. 1B). The G-less cassette resulted in a short stretch of RNA devoid of G. To construct plasmids to receive Tg-adducted oligonucleotides, an 18-mer oligonucleotide with the sequence 5′-GATCCCCCGGTACGCGGG-3′ was annealed to the complementary strand and ligated into the BamHI site of pUCH2 (11) in one orientation to yield pUCTg-TS and in the opposite orientation to yield pUCTg-NTS. pUCTg-TS and pUCTg-NTS were transformed into the F′ E. coli strain MV1184 to produce single-stranded DNA for primer extension as described (11).
Covalently closed circular DNA containing a single Tg on the transcribed or nontranscribed strand was generated by priming 10 µg of plus strand of pUCTg-TS or pUCTg-NTS with a 5-fold molar excess of Tg-containing oligonucleotide phosphorylated at the 5′ end in a 300-µl reaction mixture containing 10 mm Tris-HCl, pH 7.9, 50 mm NaCl, 10 mm MgCl2, 1 mm dithiothreitol, 600 µm dATP, dCTP, dTTP, and dGTP, 1 mm ATP, 30 units of T4 DNA polymerase, and 5 units of T4 DNA ligase. Covalently closed circular molecules were purified from an agarose gel containing 0.3 µg/ml ethidium bromide. Under these conditions, covalently closed circular DNA migrates as supercoiled DNA and can be resolved from single-stranded closed circular and nicked double-stranded plasmids. Closed circular DNA molecules were resistant to RsaI, indicating that 100% contained a Tg (data not shown).
T7 RNAP Transcription Reactions
DNA templates were incubated for 5 min at 37 °C in a 10-µl mixture containing 40mm Tris-HCl, pH 7.9, 6 mm MgCl2, 2 mm spermidine, 10 mm dithiothreitol, 10 µCi of [α-32P]GTP, 100 µm A, 212 units of RNAsin, and 50 units of T7 RNAP. Elongation proceeds until T7 RNAP reaches the end of the C-less cassette (nucleotide 7), at which the first UTP is required for incorporation. Heparin was added to prevent further initiation, and then 100 µm ATP, CTP, UTP, and GTP was added to allow elongation to continue, typically for 30 min. Reactions were stopped with SDS and proteinase K, and nucleic acids were precipitated with ethanol. Samples were resuspended in formamide loading dye, heat-denatured, and electrophoresed through an 8% polyacrylamide gel in Tris borate-EDTA (89 mm Tris, 89 mm boric acid, and 1 mm EDTA, pH 8.0) containing 8.3 m urea. Gels were dried and autoradiographed using intensifying screens. Transcripts were quantified by PhosphorImager analysis. All transcripts were labeled up to nucleotide 7 (C-less cassette; Fig. 1), making quantitation independent of their length and G content.
RNAP II Transcription Reactions
DNA templates were incubated for 30 min at 28 °C with rat liver protein fraction D (2 µg, containing TFIID and TFIIH) and rat liver RNAP II (0.5 µg) in a 20-µl mixture containing 20 mm HEPES-NaOH, pH 7.9, 20 mm Tris-HCl, pH 7.9, 2.2 mm polyvinylalcohol, 212 units of RNasin, 0.5 mg/ml acetylated bovine serum albumin, 150 mm KCl, 2 mm dithiothreitol, and 3% glycerol. After incubation, 33 µl of a solution containing fraction B′ (1 µg, containing TFIIF and TFIIE) and recombinant rat TFIIB (3 ng) in the same buffer without KCl were added, and incubation continued for 20 min at 28 °C to form preinitiation complexes. 7 mm MgCl2, 20 µm ATP, 20 µm UTP, and 40 µCi of [α-32P]CTP were added, and incubation continued for 20 min. Elongation proceeded until RNAP II reached the end of the C-less cassette (nucleotide 15), at which the first GTP was required for incorporation. Heparin was added to prevent further initiation, and then 800 µm ATP, CTP, UTP, and GTP was added to allow elongation to continue, typically for 15 min. Elongation complexes were immunoprecipitated with D44 anti-RNA antibodies and formalin-fixed S. aureus and then washed three times in reaction buffer containing 20 mm Tris-HCl, pH 7.9, 3 mm HEPES-NaOH, pH 7.9, 60 mm KCl, 0.5 mm EDTA, 2 mm dithiothreitol, 0.2 mg/ml acetylated bovine serum albumin, and 2.2% (w/v) polyvinyl alcohol. Washed complexes were resuspended in 60 µl of reaction buffer for further treatment. Reactions were stopped with SDS and proteinase K, and nucleic acids were precipitated with ethanol. Samples were resuspended in formamide loading dye, heat-denatured, and electrophoresed through a 6% polyacrylamide gel in Tris borate-EDTA with 8.3 m urea. Gels were dried and autoradiographed using intensifying screens.
RESULTS
Purification of the 13-mer Oligonucleotide Containing a Single Tg
Oxidation of a 13-mer oligonucleotide with OsO4 in the presence of pyridine (16) yielded one major peak (retention time, 8.37 min), as detected by reverse-phase HPLC (Ref. 28 and Fig. 2B), that was completely shifted from the position of the undamaged Tg13 oligonucleotide (retention time, 9.34 min; Fig. 2A). Fractions were collected at 20-s intervals and further analyzed by HPLC; those containing only the oxidized product were pooled to generate ~20 µg of pure Tg-containing oligonucleotide (Fig. 2C). The presence of a TG in the purified oligonucleotide was confirmed by HPLC analysis of enzymatic hydrolysates (29) and by piperidine cleavage of the TG-containing oligonucleotide (Ref. 16 and data not shown).
Construction of DNA Templates Containing a Single Tg in the Transcribed or Nontranscribed Strand
The Tg-containing oligonucleotide was used to construct DNA substrates for T7 and RNAP II transcription. The 154-mer DNA substrate used for T7 RNAP transcription was obtained by hybridization and ligation of eight oligonucleotides, five in the damage-containing strand and three in the complementary strand (Fig. 1A; see “Experimental Procedures”). A single Tg was positioned in the transcribed or nontranscribed strand 52 bp downstream of the T7 promoter. The substrate used for RNAP II transcription consisted of an HindIII-linearized plasmid obtained by primer extension, using the Tg-containing oligonucleotide as primer (see “Experimental Procedures”). A single Tg in the transcribed or nontranscribed strand was positioned 167 bp downstream of the AdMLP (Fig. 1B) to ensure that RNAP II complexes had completed any transitions out of the initiation state before encountering a Tg (31). The presence of a single Tg in the T7 and RNAP II DNA templates was confirmed by resistance to cleavage by RsaI restriction enzyme and by E. coli endonuclease III digestion (data not shown).
Effect of a Single Thymine Glycol in the Transcribed or Nontranscribed Strand of Template DNA on Transcription Elongation by T7 RNAP and Mammalian RNAP II
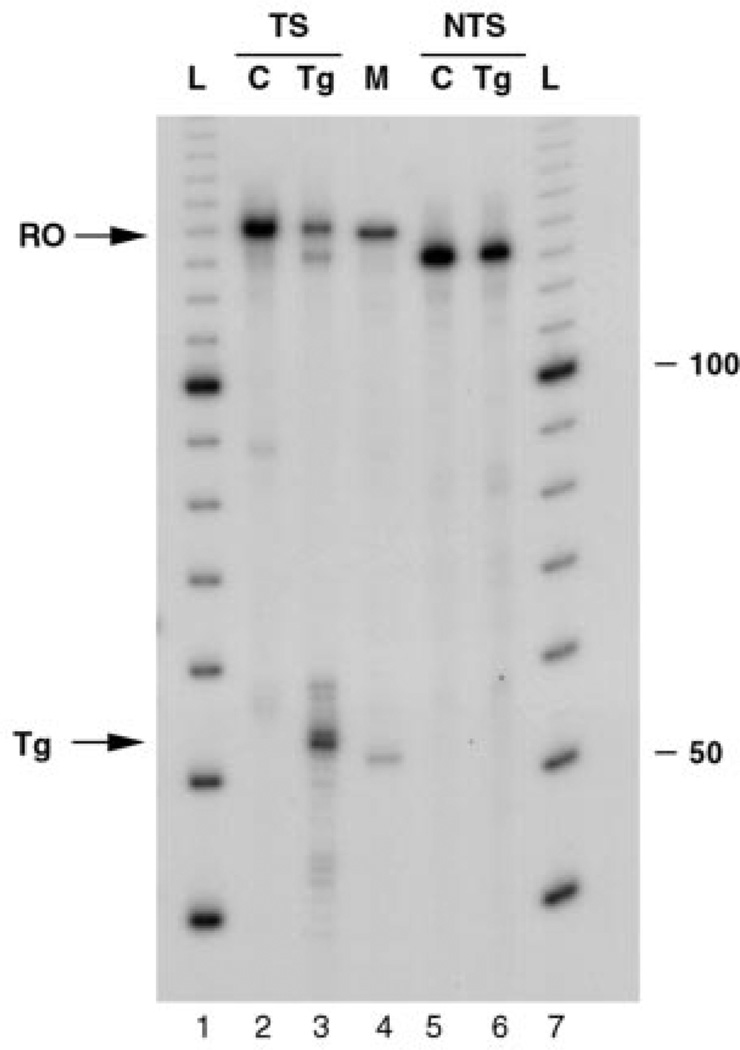
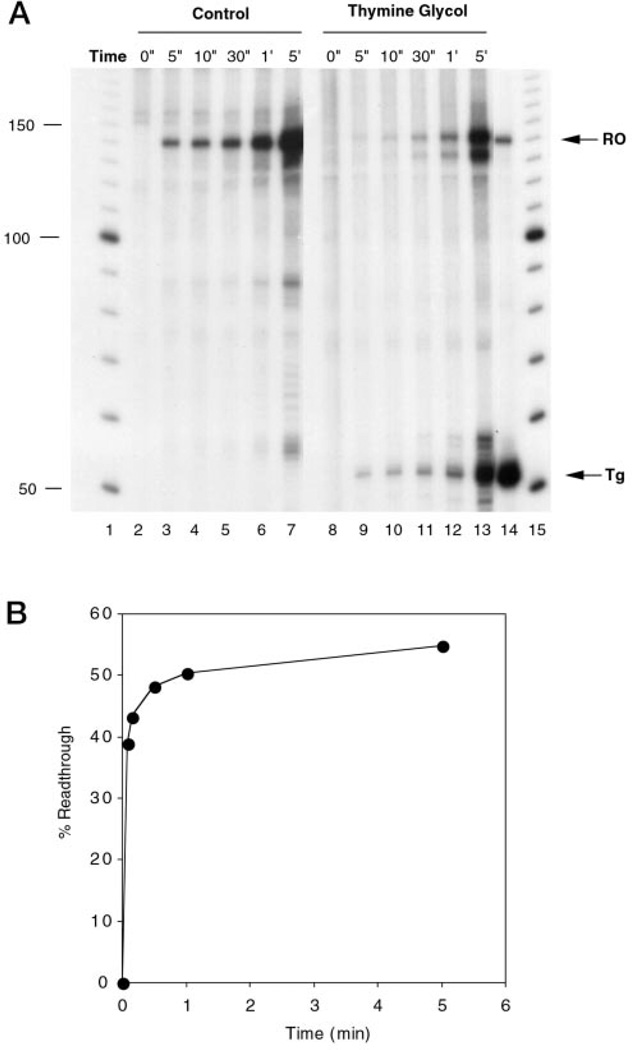
To analyze the behavior of T7 RNAP and RNAP II when they encounter a Tg, single round transcription experiments were carried out. In these experiments, each template molecule is transcribed only once by a single molecule of RNA polymerase so that the transcription products represent a single promoter-dependent elongation event (32). T7 RNAP and RNAP II were stalled downstream of the T7 promoter or AdMLP, respectively, after synthesis of a short 32P-labeled RNA, followed by addition of heparin to prevent further initiation. Then NTPs were added to allow elongation to continue. When a Tg was located in the transcribed strand, T7 RNAP transcription produced a significant proportion of transcripts shorter then the full-length RNA present in the control (Fig. 3, lanes 2 and 3). Comparison of the size of these transcripts with those obtained from an RsaI-digested undamaged substrate indicated that these RNAs were extended up to and one nucleotide past the site of the Tg (Fig. 3, lanes 3 and 4). A small fraction of transcripts arrested around the lesion was also observed. These transcripts were likely the product of nucleotide addition or loss by T7 RNAP (33). A Tg located in the nontranscribed strand was not a block to T7 RNAP, as shown by the generation of full-length runoff transcripts (Fig. 3, lane 6). To determine whether the rate of synthesis of the full-length and truncated transcripts would vary with time, we monitored the amounts of transcripts generated from 0 to 5 min after the start of the elongation phase. The undamaged control templates produced a full-length runoff product as expected (Fig. 4A, lanes 2–7), which increased as a function of time. On the other hand, transcription of the Tg-containing template (Fig. 4A, lanes 8–13) resulted in the synthesis of transcripts stalled at the lesion and full-length runoff transcripts, with a read-through frequency up to 50% (Fig. 4B).
Fig. 3. Effect of a single Tg on transcription by T7 RNAP.
DNA templates were transcribed in vitro such that transcripts were labeled with 32P as described under “Experimental Procedures.” Elongation was allowed to proceed for 30 min after addition of NTPs to the reaction mixture. RNA was isolated and electrophoresed through a 8% denaturing polyacrylamide gel. Lanes 1 and 7, 10-bp DNA ladder (L); lanes 2 and 5, unadducted templates (C); lanes 3 and 6, templates containing a specific Tg in the transcribed and nontranscribed strand, respectively (Tg); lane 4, unadducted template partially digested with RsaI. RO, full-length runoff transcript; Tg, transcripts arrested at a Tg. The position and length in bases of the 10-bp molecular mass marker is given to the right. TS, transcribed strand; NTS, nontranscribed strand.
Fig. 4. Time course of T7 RNAP transcription of templates containing a single Tg.
A, Templates containing either an unadducted T (lanes 2–7) or a Tg in the transcribed strand (lanes 8–13) were transcribed in vitro. Samples were removed from each reaction mixture at the indicated times, and transcripts were analyzed as described in the legend to Fig. 3. Lane 14, unadducted template partially digested with RsaI; lanes 1 and 15, 10-bp DNA ladder. RO, full-length runoff transcript; Tg, transcript arrested at a Tg. B, quantitation of the read-through frequency past a Tg.
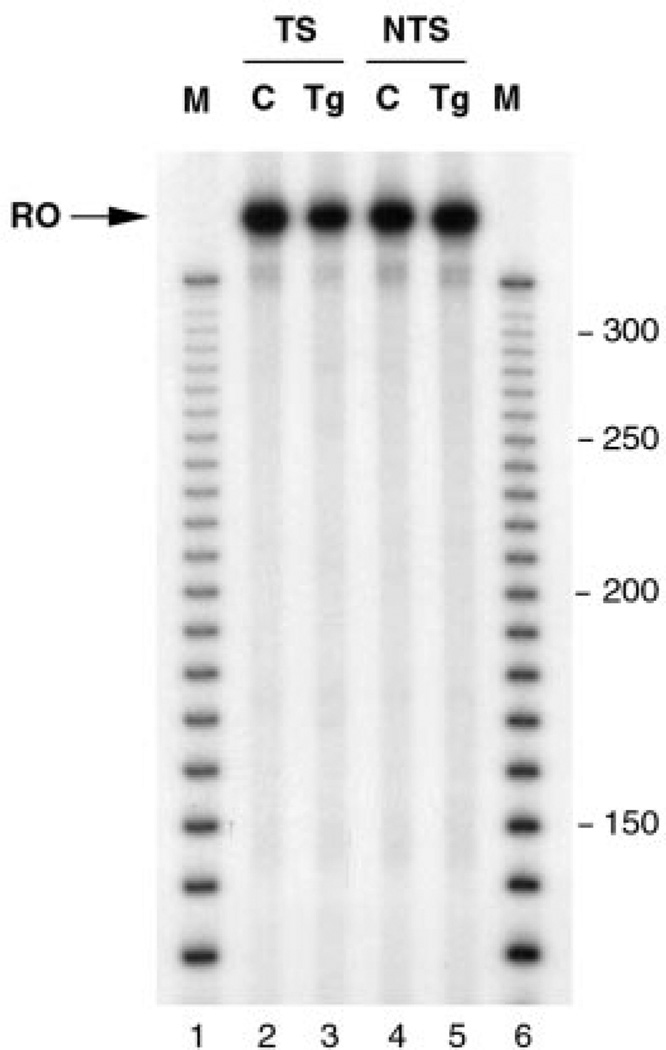
To study the effect of a Tg on transcription by RNAP II, we used an in vitro reconstituted system containing purified RNAP II and initiation factors. In this system, repair of the lesion cannot occur because of the lack of repair proteins. Surprisingly, in contrast with the results obtained with T7 RNAP, the presence of a Tg in the transcribed strand did not affect RNAP II transcription, as indicated by the generation of a full-length runoff product (Fig. 5, lane 3). Tg bypass was also observed when transcription of Tg-containing substrates was carried out for shorter times (data not shown), indicating that a Tg was not even a temporary block to RNAP II elongation. The presence of a Tg in the nontranscribed strand also did not affect transcription by RNAP II (Fig. 5, lane 5).
Fig. 5. Effect of a single Tg on transcription by RNAP II.
Templates were transcribed in vitro such that transcripts were labeled with 32P as described under “Experimental Procedures.” Elongation was allowed for 15 min, after addition of NTPs to the reaction mixture. RNA was then isolated and electrophoresed through a 5% polyacrylamide gel. Lanes 1 and 6, 10-bp DNA ladder; lanes 2 and 4, unadducted templates (C); lanes 3 and 5, templates containing a single Tg in the transcribed or nontranscribed strand, respectively (Tg). RO, full-length runoff transcript.
DISCUSSION
We have studied the effect of Tg on transcription elongation by T7 RNAP and mammalian RNAP II using an in vitro assay with DNA substrates containing a single Tg in the transcribed or nontranscribed strand located downstream of the T7 promoter or the AdMLP and purified proteins to carry out transcription. We have found that a Tg in the transcribed strand of template DNA was a significant block to the progression of T7 RNAP, in general agreement with the results of others (34, 35), resulting in polymerase arrest 50% of the time. Surprisingly, a Tg in the transcribed strand was efficiently bypassed by RNAP II. A Tg in the nontranscribed strand was bypassed by both polymerases.
The ability of a Tg to block transcription by T7 RNAP is similar to the effect of this lesion on in vitro DNA synthesis by prokaryotic DNA polymerases (13–16). These studies have shown that the extent of DNA polymerase blockage at a Tg is modulated by the sequence context of the lesion (15, 20). Lesion bypass by E. coli DNA polymerase I has been observed at 5′-C-Tg-pur-3′ and at 5′-C-Tg-C-3′, suggesting that both nucleotides 5′ and 3′ to the Tg influence the ability of the polymerase to bypass the lesion (15). Interestingly, when a Tg is located in the same sequence context we have analyzed, 5′-G-Tg-T-3′, it can be bypassed, although at low frequency, by E. coli DNA polymerase Klenow fragment lacking proofreading activity (20). It is likely that sequence context effects may also modulate the efficiency of T7 RNAP transcription arrest by Tg, and this could explain the different extent of transcription blockage we observed compared with the 100% blockage reported by Hatahet et al. (34).
The structural modification of DNA caused by the presence of a Tg in the double helix may explain how T7 RNAP arrest at this lesion occurs. NMR studies on Tg-containing oligonucleotides indicate that the presence of a Tg in the double helix induces a significant and highly localized alteration in the structure of the DNA, with the Tg and the complementary base being extrahelical (29, 36); these structural perturbations are likely to affect DNA-protein interactions. Molecular modeling studies have revealed that, although Tg stabilizes interactions with the base pair 3′ to the lesion, the stacking interactions between Tg and the base pair 5′ to the lesion are considerably destabilized. This is attributable to an unfavorable steric overlap between the base 5′ to the lesion and the methyl group of Tg, which in turn results in a significant increase of the tilt angle of the base 5′ to the Tg. The structural perturbation caused by Tg in DNA has been proposed to result in a reduced rate of base insertion beyond Tg and, as a consequence, to DNA polymerase blockage by this lesion. This effect is modulated by the sequence context surrounding the lesion (37). Similarly, the presence of a Tg in the DNA may affect the formation and stability of the RNA·DNA hybrid, an essential component of the elongation complex necessary to maintain the RNA 3′ terminus in contact with the active site of RNAP (38, 39). This effect may shift the kinetic competition between incorporation of the next nucleotide and decay into the inactive state toward transcription arrest (40).
Our finding that a Tg does not cause RNAP II arrest but rather allows for efficient bypass raises the question of how the initiation of TCR of Tg occurs. A current TCR model proposes that RNAP II arrest at a lesion represents a signal to recruit the repair enzymes to the lesion site to initiate a repair event. This model predicts that lesions that block RNAP II transcription will be subject to TCR. Consistent with this model, a correlation between transcription arrest by a lesion in vitro and TCR of the lesion in vivo has been found in most cases analyzed (7). Thus, cyclobutane pyrimidine dimers are subject to TCR and are also absolute blocks to elongation by RNAP II (9–10). Furthermore, the ternary complex is stable (9), and it can resume elongation past the thymines after reversal of the cyclobutane pyrimidine dimer by photoreactivation (11). Similarly, acetylaminofluorene is not repaired by TCR, consistent with the instability of the RNAP II ternary complex arrested at this lesion (41). Some clues to this question come from studies carried out on 8-oxoguanine (8-oxoG), another oxidative lesion that has been shown to be repaired in a transcription-coupled manner (24). Similar to our results, 8-oxoG has been shown to partially block transcription by T7 RNAP (33) but to be completely bypassed by E. coli RNAP in vitro (32), suggesting that this lesion may not block RNAP II transcription. However, the presence of a single 8-oxoG in a shuttle vector downstream of the SV40 promoter impedes transcription by RNAP II in human cells (24). It was speculated that 8-oxoG does not block RNAP II directly but rather through binding of a protein to the lesion. In support of this hypothesis, it was found that the activity of purified human 8-oxoguanine-DNA glycosylase on an 8-oxoG-containing oligonucleotide in vitro was inhibited by addition of whole cell extract, suggesting the existence of an 8-oxoG binding protein in human cells (42). It is possible that similar to the putative mechanism of initiation of TCR of 8-oxoG, additional factors that bind to a Tg may be required to ensure RNAP II arrest at Tg and consequent initiation of TCR.
Acknowledgments
We thank Ann K. Ganesan and C. Allen Smith for helpful discussion and critical reading of the manuscript. We are indebted to Joyce Hunt and John Mote, Jr., for expert technical assistance.
Footnotes
This work was supported by Grant CA-77712 from the NCI, National Institutes of Health.
The abbreviations used are: TCR, transcription-coupled repair; Tg, thymine glycol; 8-oxoG, 8-oxoguanine; RNAP”, RNA polymerase”; AdMLP, adenovirus major late promoter; T7 RNAP, T7 RNA polymerase; OsO4, osmium tetroxide; HPLC, high pressure liquid chromatography; bp, base pair.
REFERENCES
- 1.Hanawalt PC. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 2.Mellon I, Spivak G, Hanawalt PC. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 3.Mellon I, Hanawalt PC. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 4.Leadon SA, Lawrence DA. J. Biol. Chem. 1992;267:23175–23182. [PubMed] [Google Scholar]
- 5.Sweder KS, Hanawalt PC. Proc. Natl. Acad. Sci. U.S.A. 1992;89:10696–10700. doi: 10.1073/pnas.89.22.10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smerdon MJ, Thoma F. Cell. 1990;61:675–684. doi: 10.1016/0092-8674(90)90479-x. [DOI] [PubMed] [Google Scholar]
- 7.Tornaletti S, Hanawalt PC. Biochimie. 1999;81:139–146. doi: 10.1016/s0300-9084(99)80046-7. [DOI] [PubMed] [Google Scholar]
- 8.Mellon I, Bohr VA, Smith CA, Hanawalt PC. Proc. Natl. Acad. Sci. U.S.A. 1986;83:8878–8882. doi: 10.1073/pnas.83.23.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donahue BA, Yin S, Taylor J-S, Reines D, Hanawalt PC. Proc. Natl. Acad. Sci. U.S.A. 1994;91:8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tornaletti S, Donahue BA, Reines D, Hanawalt PC. J. Biol. Chem. 1997;272:31719–31724. doi: 10.1074/jbc.272.50.31719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tornaletti S, Reines D, Hanawalt PC. J. Biol. Chem. 1999;274:24124–24130. doi: 10.1074/jbc.274.34.24124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanawalt PC. Proceedings Alfred Benzon Symposium 35 on. DNA Repair Mechanisms. In: Bohr VA, Wasserman K, Kraemer KH, editors. Munksgaard: Copenhagen; 1993. pp. 231–242. [Google Scholar]
- 13.Ide H, Kow YW, Wallace SS. Nucleic Acids Res. 1985;13:8035–8052. doi: 10.1093/nar/13.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rouet P, Essigmann JM. Nucleic Acids Res. 1985;45:6113–6118. [PubMed] [Google Scholar]
- 15.Hayes RC, LeClerc JE. Nucleic Acids Res. 1986;14:1045–1061. doi: 10.1093/nar/14.2.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark JM, Beardsley GP. Biochemistry. 1987;26:5398–5403. doi: 10.1021/bi00391a027. [DOI] [PubMed] [Google Scholar]
- 17.Achey PM, Wright CF. Radiat. Res. 1983;93:609–612. [PubMed] [Google Scholar]
- 18.Moran E, Wallace SS. Mutat. Res. 1985;146:229–241. doi: 10.1016/0167-8817(85)90063-x. [DOI] [PubMed] [Google Scholar]
- 19.Laspia MF, Wallace SS. J. Bacteriol. 1988;170:3359–3366. doi: 10.1128/jb.170.8.3359-3366.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purmal AA, Lampman GW, Bond JP, Hatahet Z, Wallace SS. J. Biol. Chem. 1998;273:10026–10035. doi: 10.1074/jbc.273.16.10026. [DOI] [PubMed] [Google Scholar]
- 21.Lindahl T, Wood RD. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 22.Leadon SA, Barbie SL, Dunn AB. Mutat. Res. 1995;337:169–178. doi: 10.1016/0921-8777(95)00021-b. [DOI] [PubMed] [Google Scholar]
- 23.Cooper P, Nouspikel T, Clarkson SG, Leadon S. Science. 1997;275:990–993. doi: 10.1126/science.275.5302.990. [DOI] [PubMed] [Google Scholar]
- 24.Le Page F, Kwoh EE, Avrutskaya A, Gentil A, Leadon SA, Sarasin A, Cooper PK. Cell. 2000;101:159–170. doi: 10.1016/s0092-8674(00)80827-2. [DOI] [PubMed] [Google Scholar]
- 25.Gu W, Reines D. J. Biol. Chem. 1995;270:11238–11244. doi: 10.1074/jbc.270.19.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reines D. J. Biol. Chem. 1991;266:10510–10517. [PMC free article] [PubMed] [Google Scholar]
- 27.Eilat D, Hochberg M, Fischel R, Laskov R. Proc. Natl. Acad. Sci. U.S.A. 1982;79:3818–3822. doi: 10.1073/pnas.79.12.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basu AK, Loechler EL, Leadon S, Essigmann JM. Proc. Natl. Acad. Sci. U.S.A. 1989;86:7677–7681. doi: 10.1073/pnas.86.20.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao JY, Goljer I, Phan TA, Bolton PH. J. Biol. Chem. 1993;268:17787–17793. [PubMed] [Google Scholar]
- 30.Shi Y-b, Gamper H, Hearst J. J. Biol. Chem. 1988;263:527–534. [PubMed] [Google Scholar]
- 31.Samkurashvili I, Luse D. J. Biol. Chem. 1996;271:23495–23505. doi: 10.1074/jbc.271.38.23495. [DOI] [PubMed] [Google Scholar]
- 32.Viswanathan A, Doetsch PW. J. Biol. Chem. 1998;273:21276–21281. doi: 10.1074/jbc.273.33.21276. [DOI] [PubMed] [Google Scholar]
- 33.Jacques J, Kolakofsky D. Genes Dev. 1991;5:707–713. doi: 10.1101/gad.5.5.707. [DOI] [PubMed] [Google Scholar]
- 34.Hatahet Z, Purmal AA, Wallace SS. Ann. NY. Acad. Sci. 1994;726:346–348. doi: 10.1111/j.1749-6632.1994.tb52847.x. [DOI] [PubMed] [Google Scholar]
- 35.Htun H, Johnston BH. Methods Enzymol. 1992;212:272–294. doi: 10.1016/0076-6879(92)12017-k. [DOI] [PubMed] [Google Scholar]
- 36.Kung HC, Bolton PH. J. Biol. Chem. 1997;272:9227–9236. doi: 10.1074/jbc.272.14.9227. [DOI] [PubMed] [Google Scholar]
- 37.Clark JM, Pattabiraman N, Jarvis W, Beardsley GP. Biochemistry. 1987;26:5404–5409. doi: 10.1021/bi00391a028. [DOI] [PubMed] [Google Scholar]
- 38.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 39.Sidorenkov I, Komissarova N, Kashlev M. Mol. Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 40.von Hippel PH. Science. 1998;281:660–665. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- 41.Donahue BA, Fuchs RP, Reines D, Hanawalt PC. J. Biol. Chem. 1996;271:10588–10594. doi: 10.1074/jbc.271.18.10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hazra TK, Izumi T, Maidt L, Floyd RA, Mitra S. Nucleic Acids Res. 1998;26:5116–5122. doi: 10.1093/nar/26.22.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]