Nucleostemin is a novel cell reprogramming factor that promotes self-renewal and pluripotency through effects on gene expression and cell cycle progression.
Abstract
Nucleostemin (NS) is a nucleolar GTP-binding protein that was first identified in neural stem cells, the functions of which remain poorly understood. Here, we report that NS is required for mouse embryogenesis to reach blastulation, maintenance of embryonic stem cell (ESC) self-renewal, and mammary epithelial cell (MEC) reprogramming to induced pluripotent stem (iPS) cells. Ectopic NS also cooperates with OCT4 and SOX2 to reprogram MECs and mouse embryonic fibroblasts to iPS cells. NS promotes ESC self-renewal by sustaining rapid transit through the G1 phase of the cell cycle. Depletion of NS in ESCs retards transit through G1 and induces gene expression changes and morphological differentiation through a mechanism that involves the MEK/ERK protein kinases and that is active only during a protracted G1. Suppression of cell cycle inhibitors mitigates these effects. Our results implicate NS in the maintenance of ESC self-renewal, demonstrate the importance of rapid transit through G1 for this process, and expand the known classes of reprogramming factors.
Introduction
The nucleostemin gene (Ns, also known as Gnl3) was discovered during a search for genes that are preferentially expressed in neural stem cells (Tsai and McKay, 2002). Ns encodes a GTP binding protein (NS) that resides principally in the nucleolus (Tsai and McKay, 2002), but can apparently shuttle to and from the nucleoplasm in response to various cues (Meng et al., 2008). The biological function of NS is far from clear. In particular, it is not known whether the protein plays a specific role in stem cells. We sought to determine the role of Ns in the production and maintenance of embryonic stem cells (ESCs). To this end, we performed a detailed examination of early embryogenesis in the absence of Ns, depleted NS from ESCs in vitro, and tested the ability of Ns to participate in the reprogramming of differentiated somatic cells to induced pluripotent stem (iPS) cells. The results implicate Ns in the maintenance of ESC self-renewal, suggest a mechanism by which Ns might sustain self-renewal, demonstrate the importance of rapid transit through G1 to the preservation of self-renewal by ESCs, and expand the known classes of reprogramming factors.
Results
Ns is essential for the transition from morula to blastocyst
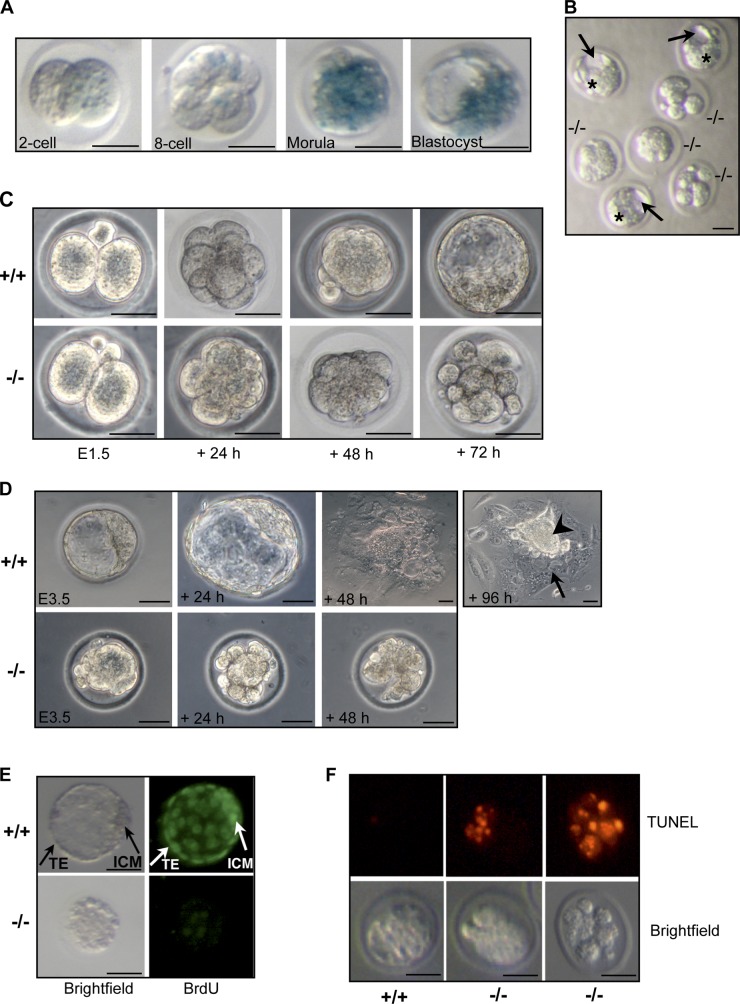
We used a clonal line of ESCs with a well-characterized gene trap insertion to develop a mouse strain that is null for Ns and, instead, expresses β-galactosidase (β-gal) under the control of the Ns regulatory elements (Fig. S1 A). We affirmed the null genotype by analyzing the topography of the gene trap insertion in the Ns locus (Fig. S1 B) and the anticipated effects on expression of NS protein in heterozygous embryos (Fig. S1 C) and Ns RNA in homozygous nulls (Fig. S1 D). By using β-gal activity as a surrogate indicator for zygotic Ns expression, we detected weak expression as early as the two-cell embryonic stage, and vigorous expression in morulae and blastocysts (Fig. 1 A). The expression of Ns at the two-cell stage was also detectable by quantitative real-time PCR (QPCR) analysis (unpublished data). In agreement with previous reports (Beekman et al., 2006; Zhu et al., 2006), the absence of Ns in homozygous null mice (Ns βgeo/βgeo, hereafter Ns−/−) resulted in early embryonic lethality (Fig. S1 E).
Figure 1.
Nucleostemin is essential for preimplantation development. (A) X-gal staining of Ns+/− (Ns+/βgeo) preimplantation embryos derived from intercrosses of Ns+/− males with wild-type females. The thick transparent layer surrounding the embryos is the zona pellucida. For A–F, individual embryos or cultures were genotyped by PCR after photomicroscopy. (B) Morphology of E3.5 embryos derived from Ns+/− intercrosses. Embryos were sorted into morphologically normal and abnormal sets. The first set was a mixture of Ns+/+ and Ns+/− genotypes, the second set was exclusively Ns−/−. Although littermate wild-type or heterozygous embryos formed blastocysts with a discernable blastocoel cavity (arrows), ICM (stars), and TE (the outer cell layer surrounding the embryos) at E3.5, Ns−/− embryos resembled morulae and exhibited various degrees of morphological degeneration. (C) In vitro culture demonstrating failure of Ns−/− embryos to develop from compacted morulae to blastocysts. Embryos were collected at E1.5 from Ns+/− intercrosses and cultured individually. Shown are littermate embryos at matched time points. (D) Ns−/− embryos fail to blastulate and hatch in outgrowth cultures. Shown are E3.5 littermate embryos freshly isolated from Ns+/− intercrosses, and after 1–4 d of culture. The Ns−/− embryo shown here was chosen for genotyping because of its degeneration after 48 h of culture. The arrow and arrowhead indicate trophoblast giant cells and ICM-derived cells, respectively. (E) BrdU incorporation assay showing decreased proliferation in Ns−/− embryos. E3.5 embryos were cultured overnight in M16 medium containing BrdU and analyzed. After overnight culture, wild-type embryos (top) developed into late blastocysts with expanded blastocoel and demonstrated extensive proliferation with strong BrdU staining in both the ICM and TE. In contrast, Ns−/− embryos (bottom) failed to develop into blastocysts and incorporated little BrdU. (F) TUNEL staining of embryos indicating increased apoptosis in Ns−/− embryos. Freshly isolated E3.5 embryos were examined by TUNEL staining. Although wild-type embryos (left) showed few labeled cells, Ns−/− embryos demonstrated intense labeling of disorganized blastomeres in partially degenerating embryos (middle), and in most cells of apparently fragmented embryos (right), indicating apoptotic cell death in these mutants. Bars, 40 µm.
To better understand the events contributing to the lethality in Ns−/− embryos, we assessed preimplantation embryos from Ns+/− intercrosses. E3.5 stage Ns+/+ and Ns+/− blastocysts contained a discernable inner cell mass (ICM), outer trophectoderm (TE) cell layer, and blastocoel cavity (Fig. 1 B). In contrast, the E3.5 Ns−/− embryos appeared developmentally delayed, resembled morulae, and exhibited various degrees of morphological degeneration (Fig. 1 B). We enlarged upon these observations by using in vitro embryo culture. Ns−/− embryos isolated at the 2- to 4-cell stage reached compaction, but then degenerated without having blastulated (Fig. 1 C). Littermate wild-type controls developed appropriately into blastocysts (Fig. 1 C). In outgrowth cultures of E3.5 embryos, we found that Ns−/− mutants also gradually degenerated and produced nothing resembling the ICM or TE (Fig. 1 D). In addition, BrdU incorporation and TUNEL staining demonstrated a decrease in proliferation and a concomitant increase in apoptosis in E3.5 stage Ns−/− embryos (Fig. 1, E and F, respectively), correlating with the failure of Ns−/− embryos to develop into blastocysts and the subsequent death of the embryos. Collectively, these data indicate that the early death of Ns−/− embryos can be attributed to failure of the morula to survive beyond compaction. Ns heterozygous embryos were indistinguishable from wild-type controls (Fig. S1, F–I) and heterozygous mice were normal in gross appearance and were fertile (unpublished data). Our observations are consistent with a previous report that homozygous Ns-null mice abort embryogenesis before blastulation (Zhu et al., 2006).
Ns is necessary for the maintenance of ESC self-renewal
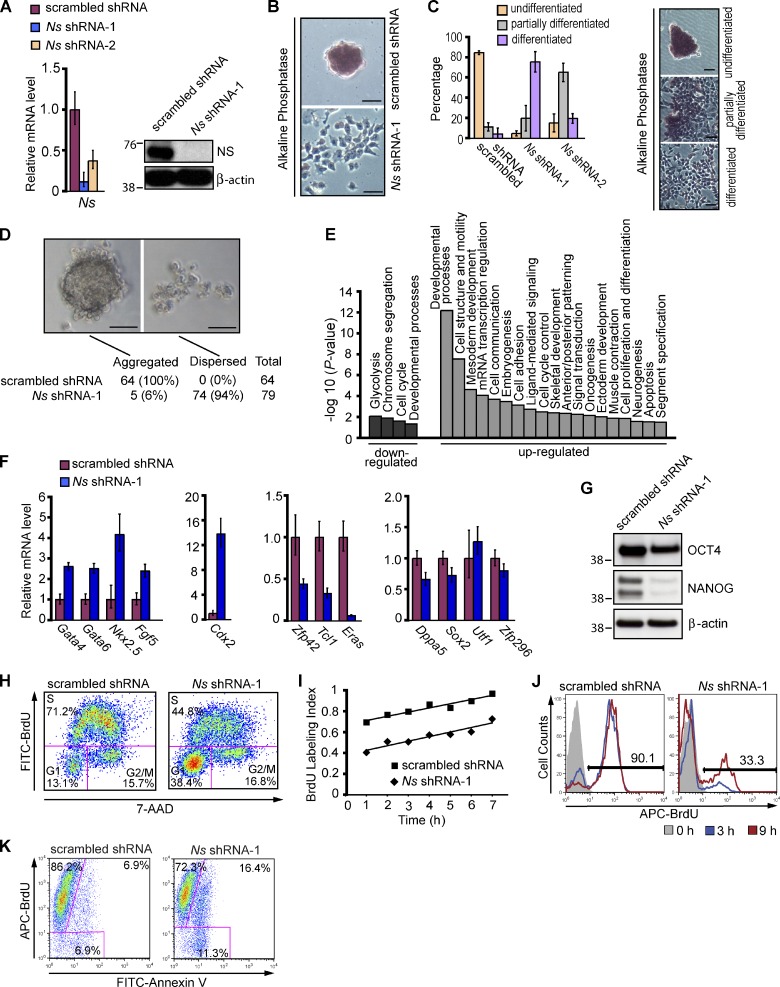
We next turned our attention to the role of Ns in ESCs. Because no cells of the ICM or later stage embryos could be obtained with Ns−/− mice, we studied mouse ESCs in which we depleted NS by the use of RNA interference. We tested 10 different short hairpin RNA (shRNA) expression constructs or small interfering RNA oligonucleotides that specifically target different regions of the Ns transcript. One shRNA (shRNA-1) reduced Ns expression to nearly undetectable levels in Western blot analysis and to <10% of controls with scrambled shRNA by QPCR analysis of Ns RNA (Fig. 2 A). By 4 d after transfection, NS knockdown (KD) cells had become flat, and had lost the colony morphology and strong alkaline phosphatase (AP) staining that are typical of ESCs (Fig. 2, B and C), suggesting that the cells had initiated differentiation in response to the depletion of NS. Consistent with the changes in morphology, NS KD cells had a defect in the formation of embryoid bodies (EBs), a distinct characteristic of ESCs (Fig. 2 D). When placed in hanging drop cultures, control ESCs formed EBs in almost every drop within 24 h, whereas NS KD cells generally remained dispersed. The few EBs that formed from ESCs subjected to NS KD were much smaller than controls after 3 d of culture (unpublished data). The response of ESCs to KD of NS could not be attributed to off-target effects of the shRNA (Fig. S2).
Figure 2.
Nucleostemin is essential for maintaining the self-renewal of ESCs. (A) Depletion of NS in ESCs. For A–K, the E14 line of mouse ESCs was transfected with vectors expressing the puromycin resistance gene and either an shRNA targeting Ns or scrambled shRNA. Puromycin selection was initiated 24 h after transfection and continued for 3 d before cells were harvested for subsequent analysis. Untransfected cells were eliminated by 2 d of drug selection. Left, QPCR analysis of Ns mRNA level. Right, Western analysis of NS protein with β-actin as a loading control. The RNA data in A and F were normalized to the expression of Gapdh and are represented as mean ± SEM with n = 3. The data are displayed relative to results with scrambled shRNA transfected controls. The positions of molecular mass standards (in kilodaltons) are shown in A and G. (B) Representative images showing differentiation of ESCs upon NS KD. 4 d after the indicated transfections, NS KD cells became flattened and lost the stereotypical colony morphology and AP staining. Under the same culture conditions, a normal undifferentiated phenotype with distinct colonies strongly expressing AP was maintained in controls with scrambled shRNA. (C) Quantification of undifferentiated, partially differentiated, and fully differentiated ESC colonies 5 d after the indicated transfections. The data in the left panel represent the results of two independent experiments with error bars indicating standard deviations. Approximately 100 colonies were counted for each transfection. The right panel illustrates representative morphology of the three types of colonies used to score the extent of differentiation, as described in Materials and methods. The KD of NS by shRNA-2 was less efficient than that by shRNA-1 (see Fig. S2, C and D for more information). (D) Failure of NS KD cells to form embryoid bodies. 3 d after transfection with shRNAs, ESCs were placed in hanging drops and examined 24–72 h later. The percentage values represent the fraction of hanging drops that formed EBs or remained dispersed after 24 h of culture. (E) Categories of genes whose expression changed in response to KD of NS by Ns shRNA-1 in ESCs. Data were obtained from whole-genome expression analysis of four biological replicates of NS KD cells and scrambled shRNA transfected controls. See Materials and methods and Table 1 for details. (F) QPCR analysis of RNAs specific for endoderm (Gata4 and Gata6), mesoderm (Nkx2.5), ectoderm (Fgf5), TE (Cdx2), and ESCs (Zfp42, Tcl1, Eras, Dppa5, Sox2, Utf1, and Zfp296) in NS KD cells and controls. Analysis by QPCR was performed 4 d after the indicated transfections. (G) Western analysis of ESC markers OCT4 and NANOG 4 d after NS KD in ESCs, with β-actin as a loading control. (H) Cell cycle profiles of asynchronously growing ESCs 4 d after transfection with shRNAs. Cells were pulsed with BrdU for 20 min, then fixed and stained with anti-BrdU antibodies and 7-AAD followed by FACS analysis. The percentages of cells in the various phases of the cell cycle are shown. (I) Cumulative BrdU labeling curves. 4 d after the indicated transfections, cells were fed with fresh media containing BrdU every 2 h, and the BrdU labeling index was determined by FACS at the indicated time points. The time necessary to reach the maximum labeling index corresponds to the total cell cycle length minus the length of the S phase (TG2+M+G1; Nowakowski et al., 1989) and was extrapolated by linear regression analysis. One of three independent experiments with comparable results is shown. (J) Cell cycle reentry analysis. Cells with the indicated transfections were synchronized at the G2/M transition at 4 d after transfection with shRNAs, then released to enter S phase, as described in Materials and methods. At subsequent time points, cells were pulsed with BrdU for 20 min and subjected to FACS analysis to measure entry into S phase. Analyses were performed at the time of release from the cell cycle block and 3 h and 9 h afterward. Frequencies indicate percentages of BrdU-positive cells at 9 h after release. (K) DNA replication and apoptosis. 4 d after transfection of ESCs with shRNAs, cells were incubated with BrdU for 22 h, then fixed and stained with anti-BrdU antibodies and Annexin V for subsequent FACS analysis. In total, 93.1% (86.2% + 6.9%) of control transfectants and 88.7% (72.3% + 16.4%) of NS KD cells were positive for BrdU incorporation. Cells that have cycled through the S phase and have become apoptotic during the BrdU labeling period are positive for both BrdU incorporation and Annexin V (6.9% of controls vs. 16.4% of NS KD cells). Bars, 50 µm.
To characterize further the extent to which ESCs differentiated in response to depletion of NS, we compared the gene expression profile of NS KD cells that had been subjected to Ns shRNA-1 with that of ESCs expressing scrambled shRNA 4 d after transfection of the shRNAs. Out of the 41,174 probe sets examined in genome-wide expression profiling, 1,935 genes were differentially expressed, including 1,122 up-regulated genes and 813 down-regulated genes in NS KD cells (adjusted P < 0.05; Tables S1 and S2). Among 386 genes with at least fourfold change, down-regulated genes were enriched for cell cycle regulators and metabolic genes, whereas up-regulated genes were enriched for several categories related to embryogenesis and lineage commitment (Fig. 2 E and Table 1), consistent with the differentiated phenotype of NS KD cells. The activation of representative markers for multiple somatic lineages and down-regulation of ESC markers was also confirmed by QPCR analysis (Fig. 2 F and Table S2). Up-regulated genes included Gata4, Gata6, and Sox17 (endoderm); Nkx2.5, T, and Msx1 (mesoderm); Fgf5, Ednra, and Gap43 (ectoderm); and Cdx2, Krt8, and Krt18 (TE). Down-regulated genes included archetypal molecular markers for ESCs, such as Zfp42, Tcl1, and Eras measured as mRNA (Fig. 2 F and Table S2), and Nanog measured as protein (Fig. 2 G). The reduction in Oct4 expression after NS KD was less than that of Nanog (Fig. 2 G). The residual expression of Oct4 after KD of NS in ESCs is consistent with recent reports that Oct4 expression is not restricted to undifferentiated ESCs (Fazzio et al., 2008; Thomson et al., 2011; Yang et al., 2011). Similarly, the levels of mRNAs for the ESC markers Dppa5, Sox2, Utf1, and Zfp296 changed by less than twofold (Fig. 2 F, right-hand panel). Thus, KD of NS shifted the transcriptional program of ESCs in the direction of differentiation, but the shift was not complete.
Table 1.
Summary of categories of biological processes that are significantly overrepresented by genes misregulated in NS knockdown cells (with at least fourfold change and adjusted P-value < 0.05) when compared with controls with scrambled shRNA (Ctl)
| NS KD vs. Ctl ESCs | Biological Process | Count | Expected Count | P-value |
| Down-regulated | Glycolysis | 4 | 0.2 | 8.07E–03 |
| Chromosome segregation | 5 | 0.41 | 1.21E–02 | |
| Cell cycle | 11 | 3.49 | 2.40E–02 | |
| Developmental processes | 18 | 8.29 | 4.42E–02 | |
| Up-regulated | Developmental processes | 60 | 20.25 | 6.30E–13 |
| Cell structure and motility | 33 | 9.58 | 2.67E–08 | |
| Cell structure | 25 | 5.76 | 1.83E–07 | |
| Mesoderm development | 20 | 4.89 | 2.28E–05 | |
| mRNA transcription regulation | 36 | 14.32 | 7.97E–05 | |
| Cell communication | 29 | 10.67 | 1.94E–04 | |
| Embryogenesis | 10 | 1.42 | 3.19E–04 | |
| mRNA transcription | 41 | 19.32 | 6.14E–04 | |
| Cell adhesion | 18 | 5.71 | 6.99E–04 | |
| Ligand-mediated signaling | 14 | 3.35 | 1.82E–03 | |
| Cell cycle control | 14 | 3.61 | 3.05E–03 | |
| Skeletal development | 8 | 1.11 | 3.97E–03 | |
| Anterior/posterior patterning | 6 | 0.58 | 4.34E–03 | |
| Signal transduction | 60 | 37.94 | 5.43E–03 | |
| Oncogenesis | 13 | 3.97 | 6.84E–03 | |
| Ectoderm development | 18 | 6.19 | 9.23E–03 | |
| Muscle contraction | 8 | 1.73 | 1.27E–02 | |
| Cell proliferation and differentiation | 20 | 8.52 | 1.32E–02 | |
| Cell cycle | 20 | 8.53 | 1.34E–02 | |
| Neurogenesis | 16 | 5.4 | 2.61E–02 | |
| Apoptosis | 13 | 4.62 | 2.76E–02 | |
| Segment specification | 6 | 0.82 | 2.98E–02 |
Analysis was performed with the Panther classification system (http://www.pantherdb.org/tools/genexAnalysis.jsp). The biological process terms of the Panther classification system are similar to those of the Gene Ontology (GO), but abbreviated and simplified to facilitate high-throughput analyses. The set of all genes in the mouse genome was used as a reference list. Included in the table are the number of misregulated genes found in each category (count), the expected number of genes for each category among misregulated genes based on the reference list (expected count), and the P-value of enrichment as calculated by the binomial statistic, which is the probability that the number of genes observed in each category occurred by chance as determined by the reference list. Ns shRNA-1 was used for depletion of NS.
We conclude that Ns is essential for the maintenance of ESC self-renewal. The mixed nature of the transcriptional program elicited by NS KD suggests that the cells progressed to an intermediate state of differentiation, similar to that reported previously in response to KD of other ESC maintenance genes (Fazzio et al., 2008). A recent report implicates another nucleolar protein, nucleolin, in the maintenance of ESC self-renewal (Yang et al., 2011).
Ns contributes to the maintenance of ESC self-renewal by sustaining rapid transit through the G1 phase of the cell cycle
Cells generally make the decision to self-renew, differentiate, or die during the G1 phase of the cell division cycle (Massagué, 2004). The cell cycle of mouse ESCs differs from that of somatic cells by having an abbreviated G1 phase, constitutively active cyclin E–Cdk2, and virtually undetectable cyclin D–Cdk4 activity (Savatier et al., 1996; Stead et al., 2002). It has been proposed that brevity of the G1 phase is essential to the maintenance of ESC self-renewal (Orford and Scadden, 2008; Singh and Dalton, 2009). We explored this possibility by analyzing changes in the cell cycle elicited by Ns shRNA-1 in ESCs. Notably, KD of NS elicited an increase in the G1 population and a concomitant decrease in the percentage of S-phase cells (Fig. 2 H). This might indicate either prolongation of the G1 phase or cell cycle arrest in the G1 phase. We explored these possibilities in three ways. First, we used cumulative BrdU labeling to demonstrate that KD of NS in ESCs increased the combined time spent in the G2, M, and G1 phases by twofold compared with controls with scrambled shRNA (∼8 h for control transfectants vs. ∼16.5 h for NS KD cells; Fig. 2 I). Second, after being synchronized at the G2/M transition and then released from the cycle arrest, NS KD cells progressed into S phase in a delayed manner in comparison with control transfectants (Fig. 2 J). Third, 4 d after depletion of NS we labeled cells with BrdU for 22 h, which is roughly the time required for two cell divisions by normal mouse ESCs. We found that ∼90% of NS KD cells were able to progress into S phase during the BrdU labeling period (Fig. 2 K). We conclude that the increased population of cells in the G1 phase resulted from elongation of G1 rather than arrest of the cycle, although it is conceivable that the cells might also pause in a G0-like state during their transit of G1. Ns has been implicated previously in the proliferation of ESCs (Nomura et al., 2009), but the restructuring of the cell cycle effected by NS KD described here has not been reported previously. Ns is required for the survival of both somatic and pluripotent cells (Tsai and McKay, 2002; Nomura et al., 2009). Accordingly, we found that KD of NS in ESCs increased apoptosis (Fig. 2 K; unpublished data).
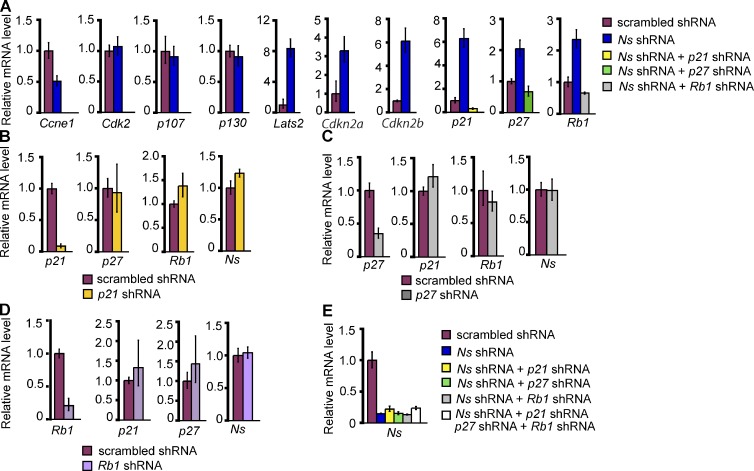
In an effort to explain the reconfiguration of the cell cycle elicited by NS KD, we used QPCR to analyze the expression of cell cycle regulators in NS KD cells. Expression of p107 (Rbl1) and p130 (Rbl2) was not affected by the depletion of NS (Fig. 3 A). In contrast, there was decreased expression of Ccne1 (cyclin-E) and up-regulation of inhibitors of the cyclin E–Cdk2 complex, including p21 (Cdkn1a), p27 (Cdkn1b), Rb1, and Lats2 (Fig. 3 A); these changes suggest a decrease in cyclin E–Cdk2 activity. Expression of p21 was also elevated in Ns-null embryos (Fig. S1 D). Consistent with a fundamental change in the cell cycle control of NS KD cells, we also observed up-regulation of Cdkn2a and Cdkn2b (Fig. 3 A), both members of the INK/ARF family that repress the activity of cyclin D–Cdk4/6 complexes and are thought to be associated with lineage specification during development (Zindy et al., 1997; Faast et al., 2004). In summary, the findings with cell cycle regulators in NS KD cells are in accord with the loss of the characteristic cell cycle structure of ESCs and the differentiated phenotype of the cells.
Figure 3.
Analysis of cell cycle regulators in NS knockdown cells. (A) QPCR analysis of expression of Ccne1, Cdk2, and inhibitors of the cyclin E–Cdk2 and cyclin D–Cdk4/6 complexes after NS KD in ESCs. For the three right panels in A and the panel in E, samples for co-depletion of NS and a cell cycle inhibitor were transfected with two plasmids, one encoding both the puromycin resistance gene and an shRNA for Ns, the other encoding both the blasticidin resistance gene and an shRNA for a cell cycle inhibitor. For KD of NS alone, cells were transfected with two plasmids, one containing Ns shRNA and the puromycin resistance gene, the other expressing a scrambled shRNA and the blasticidin resistance gene. The controls were transfected with two plasmids, one encoding the puromycin resistance gene and scrambled shRNA, the other encoding the blasticidin resistance gene and the same scrambled shRNA. For the seven left panels in A and all panels in B–D, single-depletion samples were transfected with plasmids encoding the puromycin resistance gene and either a scrambled shRNA, an shRNA for Ns, or an shRNA for a cell cycle inhibitor. Selection with puromycin and blasticidin was initiated 24 h after transfection and continued for 3 d before cells were analyzed. Ns shRNA-1 was used in all experiments for KD of NS. For A–E, the data were normalized to the expression of Gapdh and are represented as mean ± SEM with n = 3. The data are displayed relative to results with scrambled shRNA-transfected controls. B, C, and D show QPCR analysis of the effect of shRNA constructs targeting p21, p27, and Rb1, respectively, in ESCs. (E) QPCR analysis of Ns mRNA in ESCs that were co-depleted of NS and the indicated cell cycle inhibitors.
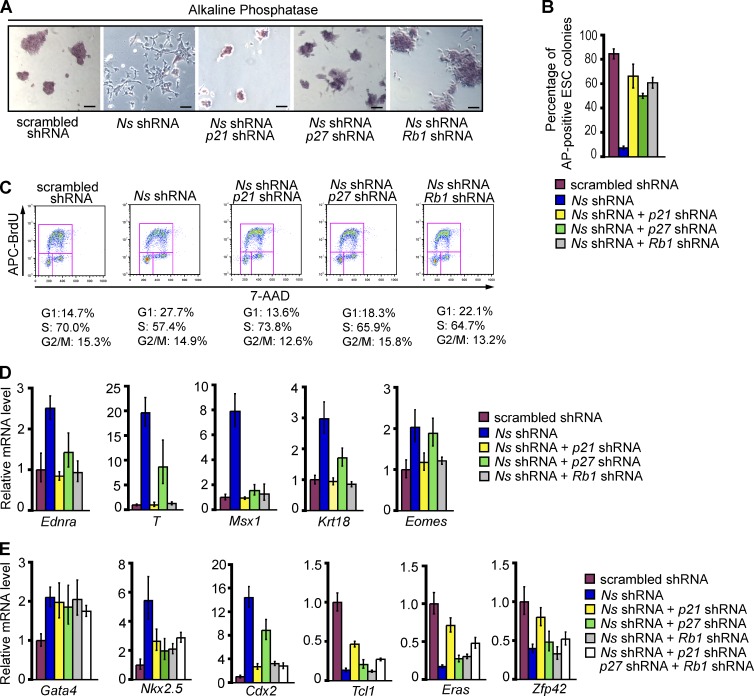
We next examined whether impedance to the G1/S transition of ESCs played any causal role in the loss of self-renewal elicited by KD of NS. We used shRNAs to co-deplete cell cycle inhibitors with NS (Fig. 3, A and E), then examined the effect of the co-depletions on the response of ESCs to the KD of NS (Fig. 4). In each instance, the co-depletions returned the respective inhibitors to or below the constitutive level observed in the absence of NS KD (defined by treatment with scrambled shRNA alone), without affecting the KD of NS (Fig. 3, A and E). KDs of either p21, p27, or RB1 did not affect the levels of the other two inhibitors (Fig. 3, B–D). Remarkably, co-depletion of either p21, p27, or RB1 with NS substantially impeded the differentiation that would normally result from depletion of NS. 4 d after any of the co-depletions, ESC self-renewal appeared to have been at least partially rescued (Fig. 4, A–E). The rescue was apparent in colony morphology and AP staining (Fig. 4, A and B). The extent of rescue for the cell cycle varied among the several KDs of cycle inhibitors, but was virtually complete with the KD of p21 (Fig. 4 C).
Figure 4.
Depleting inhibitors of the cyclin E–Cdk2 complex restores ESC properties in NS knockdown cells. ESCs were transfected with the indicated constructs as described for co-depletion samples in Fig. 3 A, using Ns shRNA-1 for KD of NS. (A) Representative morphology and AP staining of ESCs after the indicated transfections. Bars, 100 µm. (B) Quantification of undifferentiated, AP-positive ESC colonies (as defined in Fig. 2 C) after the indicated transfections. Shown are the results in two independent experiments with error bars indicating standard deviations. (C) Cell cycle profiles of the cells represented in A. (D) QPCR analysis of differentiation markers Ednra (ectoderm), T and Msx1 (mesoderm), and Krt18 and Eomes (TE) in ESCs after the indicated transfections. For D and E, the RNA data were normalized to the expression of Gapdh and are represented as mean ± SEM with n = 3. The data are displayed relative to results with scrambled shRNA-transfected controls. (E) QPCR analysis of markers specific for endoderm (Gata4), mesoderm (Nkx2.5), TE (Cdx2), and ESCs (Tcl1, Eras, and Zfp42) in ESCs after the indicated transfections.
The transcriptional response to co-depletions was mixed. Expression of several markers for differentiation, including Ednra (ectoderm), T and Msx1 (mesoderm), and Krt18 and Eomes (TE), was restored to their normal low levels in ESCs by the KD of p21 or Rb1 (Fig. 4 D). On the other hand, there was persistent up-regulation of the differentiation marker Gata4 (endoderm), reduced yet still elevated expression of the differentiation markers Nkx2.5 (mesoderm) and Cdx2 (TE), and increased but still relatively low expression of the ESC markers Tcl1, Eras, and Zfp42 (Fig. 4 E), leading us to conclude that the effect of NS KD had not been completely reversed. Even simultaneous depletion of p21, p27, and RB1 failed to fully reverse either the induction of Gata4, Nkx2.5, and Cdx2 or the repression of Tcl1, Eras, and Zfp42 elicited by NS KD (Fig. 4 E). These results raise the possibility that restraining the expression of cell cycle inhibitors is not the sole manner in which Ns influences ESC identity. Nevertheless, it appears that mitigation of the S-phase entry block imposed by NS KD can substantially prevent the loss of ESC self-renewal. We conclude that Ns contributes to the undifferentiated state of ESCs, at least in part by sustaining rapid transit through G1, and that elongation of G1 contributes to the differentiation of ESCs elicited by NS KD.
It has been suggested that prolongation of G1 creates a “window of opportunity” that allows ESCs to respond to signals responsible for differentiation (Orford and Scadden, 2008; Singh and Dalton, 2009). A major example of these signals is provided by a pathway that includes the protein kinases MEK and ERK (Burdon et al., 1999; Kunath et al., 2007; Stavridis et al., 2007). Pharmacological inhibition of MEK can sustain the self-renewal of ESCs in the absence of otherwise essential external factors if glycogen synthase kinase-3 (GSK3) is also inhibited (Ying et al., 2008). We hypothesized that inhibition of these two kinases might counter the effect of NS KD on ESCs. We tested this hypothesis by using inhibitors of MEK (PD0325901, or PD) and GSK3 (CHIR99021, or CH), applying them either separately or together to ESCs that had been transfected with Ns shRNA-1 (Fig. 5). The inhibitor treatments were performed with the cells in conventional ESC media supplemented with serum and leukemia inhibitory factor (LIF). The KD of NS was roughly 90% in all three instances (unpublished data).
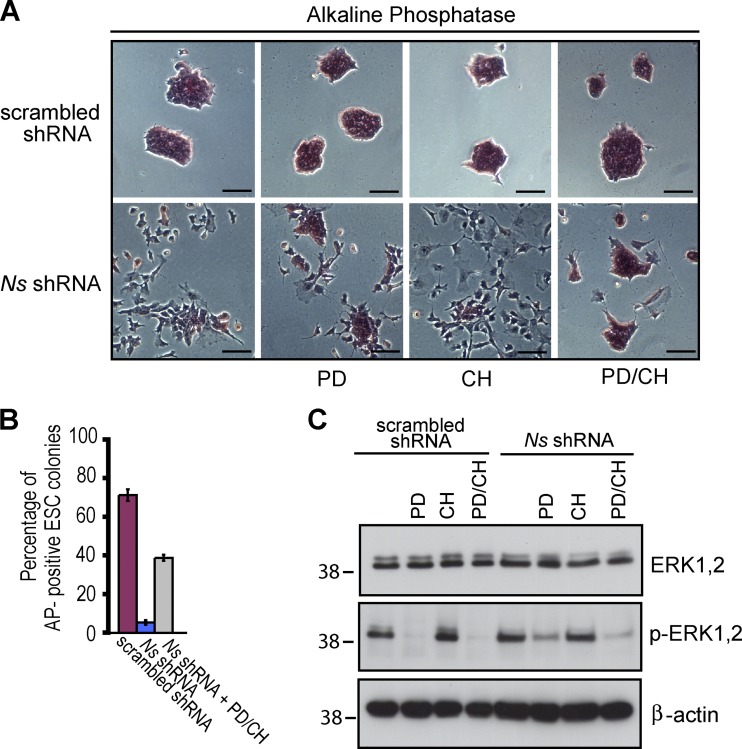
Figure 5.
Dual inhibition of the MEK and GSK3 kinases partially rescues ESCs from the differentiation imposed by depletion of NS. ESCs were transfected with the indicated constructs as described in Fig. 2, using Ns shRNA-1 for KD of NS. Treatment with PD0325901 (PD) and CHIR99021 (CH) was initiated 24 h later. After an additional 3 d, the cells were harvested for analysis. Throughout the experiments, cells were maintained in conventional ESC media (supplemented with serum and LIF). (A) Representative morphology and AP staining of controls and NS KD cells treated with the indicated chemical inhibitors. Bars, 100 µm. (B) Quantification of undifferentiated, AP-positive ESC colonies (as defined in Fig. 2 C) with the indicated treatment. Shown are the results in two independent experiments with error bars indicating standard deviations. (C) Western analysis of cells with the indicated treatment with antibodies to ERK, phosphorylated-ERK, and β-actin. The positions of molecular mass standards (in kilodaltons) are shown.
When applied separately, PD partially blocked differentiation, whereas the effect of CH was marginal (Fig. 5, A and B). In contrast, a combination of the two inhibitors (PD/CH) greatly impeded the differentiation of ESCs normally elicited by KD of NS (Fig. 5, A and B). By 4 d after NS KD, cells treated with PD/CH largely maintained ESC-like colony morphology and strong AP staining, although many of the colonies appeared relatively flat, had rough edges indicative of differentiation, and lacked the dome shape with well-defined borders that is characteristic of undifferentiated ESC colonies (Fig. 5, A and B). The activity of the MEK/ERK pathway in these experiments was evaluated by assessing the phosphorylated (activated) forms of ERK (Fig. 5 C). In the absence of inhibitors, roughly equivalent basal levels of activated ERK were readily detected in ESCs transfected with either scrambled shRNA or Ns shRNA-1. Treatment with either PD/CH or PD alone greatly reduced the phospho-ERK levels, whereas application of CH alone had no effect. Thus, the activity of ERK was essential for differentiation, but was effective only after depletion of NS and the ensuing prolongation of G1. These findings are consistent with the possibility that the prolongation of G1 in response to KD of NS creates a window of opportunity that allows ESCs to respond to the signaling.
Intrinsic Ns is required for the reprogramming of somatic cells to pluripotency by ectopic factors
Having found that Ns is essential for the maintenance of ESC self-renewal, we asked whether Ns might play a role in the reprogramming of somatic cells to iPS cells. We chose female adult mammary epithelial cells (MECs) for study, with the hope that successful reprogramming of these cells would prove useful in future studies of both the normal and diseased breast. It has been reported recently that MECs can be reprogrammed to iPS cells by retroviral transduction of OCT4, SOX2, MYC, and KLF4 (OSMK; Li et al., 2010), the canonical factors that were first used to reprogram a variety of mouse and human somatic cells (Takahashi and Yamanaka, 2006; Takahashi et al., 2007). We have confirmed that finding, using human cDNAs as reprogramming factors in order to permit discrimination between ectopic and intrinsic versions of the corresponding genes (Fig. S3, A–C).
The expression of endogenous NS protein in OSMK iPS cells was markedly up-regulated, reaching levels similar to those in normal ESCs (see Fig. 6 C), as previously reported for iPS cells derived from porcine fetal fibroblasts (Ezashi et al., 2009). These data prompted us to examine whether Ns is required during the reprogramming process elicited by OSMK. We transduced MECs with OSMK in combination with either Ns shRNA-1 or scrambled shRNA. KD of NS in cells transduced with OSMK led to decreased expansion in cell number (Fig. S4 A), increased apoptosis (Fig. S4 B), and failure to form AP-positive, ESC-like colonies, when compared with cells transduced with OSMK and scrambled shRNA (Fig. S4 C). These results are in accord with previous reports that NS is required for both cellular proliferation and survival (Tsai and McKay, 2002; Nomura et al., 2009), which could at least partially explain the requirement for NS during reprogramming. It is notable that the four reprogramming factors cannot overcome the requirement for NS.
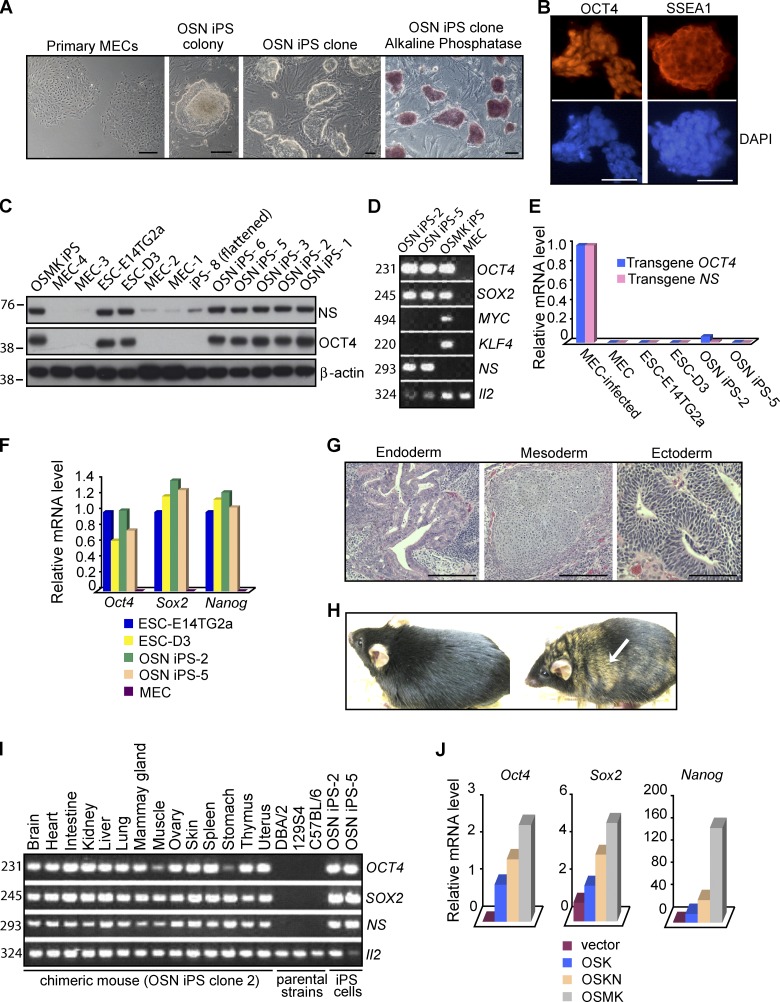
Figure 6.
Induction of pluripotent stem cells from mouse mammary epithelial cells by a combination of OCT4, SOX2, and NS. (A) Morphology of primary MECs, primary OSN iPS colony 4 wk after transduction, and established OSN iPS clones, the last displaying both characteristic ESC colony morphology and strong staining for AP activity. (B) Expression of pluripotency markers (OCT4 and SSEA1) in OSN iPS cells, as assessed by immunofluorescence microscopy. Nuclei were counterstained with DAPI. The antibodies used in B and C could recognize both human and mouse homologues, but the data likely represent only endogenous mouse proteins due to silencing of the retroviral vectors. (C) Western analysis of NS and OCT4 proteins in OSN iPS cells, OSMK iPS cells, normal ESCs (E14TG2a and D3), and the MECs used to generate the iPS cells. β-Actin was used as a loading control. The positions of molecular mass standards (in kilodaltons) are shown. (D) Detection of transgenes in iPS cells. Genomic DNA extracted from OSN iPS cells, OSMK iPS cells, and MECs used to generate the iPS cells was analyzed by PCR with a forward primer specific for viral vector sequences and a reverse primer for cDNA sequences of human transgenes. Il-2 was used as an internal control. The sizes of the PCR products in D and I are indicated in base pairs. (E) Silencing of transduced genes in iPS cell lines. Gene expression was assessed by QPCR, using primers specific for the transduced human genes. Analyses were performed on uninfected MECs, MECs 48 h after infection with the retroviral vectors, two lines of ESCs (E14TG2a and D3), and two OSN iPS cell lines. The data were normalized to the expression of Gapdh and represent the average of triplicate QPCR analyses. The data are displayed relative to results with newly infected MECs. For E, F, and J, one of three independent experiments with comparable results is shown. (F) Reactivation of endogenous mouse Oct4, Sox2, and Nanog in OSN iPS cells. Shown is QPCR analysis with primers detecting transcripts from the respective endogenous mouse loci as opposed to the transduced human cDNAs in OSN iPS cells, two lines of ESCs (E14Tg2a and D3), and the MECs used to generate the iPS cells. Results were normalized to the expression of Gapdh and were the average of triplicate QPCR analyses. The data are shown relative to results with E14Tg2a. (G) Hematoxylin and eosin staining of teratomas generated from OSN iPS cells. Shown is a teratoma containing endoderm (gut-like epithelium), mesoderm (cartilage), and ectoderm (neural tissue). (H) Adult chimeric mouse derived from OSN iPS cells by injection into B6XB6D2 F1 (black) 8-cell-stage embryos and transplantation into pseudopregnant mice. The agouti coat color (arrow) originated from OSN iPS cells derived from129S4 mice. A normal C57BL/6 mouse is shown on the left. (I) Tissue distribution of OSN iPS cells in chimeras. Genomic DNA isolated from indicated organs from an adult chimera derived from OSN iPS cells was analyzed by PCR for the presence of transduced genes as in D. (J) QPCR analysis of endogenous pluripotency markers in early reprogramming cells 12 d after the indicated transductions. The data were normalized to the expression of Gapdh and represent the average of triplicate QPCR analyses. For Sox2 and Nanog, the data are displayed relative to results with MECs transduced with empty vector. For Oct4, the data are displayed relative to results with MECs transduced with OSK. Bars, 100 µm.
NS can cooperate with OCT4 and SOX2 in the reprogramming of somatic cells to pluripotency
Having demonstrated that intrinsic Ns was required for the reprogramming of MECs, we examined whether ectopic human NS could contribute to reprogramming by substituting it for one or more of the four canonical factors (OSMK) in the induction of pluripotency in MECs. We first asked whether NS could replace either OCT4, SOX2, KLF4, or MYC in this set of four factors. We found that combinations containing either human OCT4, SOX2, KLF4, and NS (OSKN, without MYC) or OCT4, SOX2, MYC, and NS (OSMN, without KLF4) were capable of generating iPS cell lines (Table S3). Using the yield of established iPS cell lines as a measure, we found that the reprogramming efficiencies with OSKN, OSMN, AND OSMK were all ∼0.05%. This conforms to previous reports for OSMK (Takahashi and Yamanaka, 2006; Takahashi et al., 2007; Jaenisch and Young, 2008; Hochedlinger and Plath, 2009), although ESC-like colonies induced by the combinations including NS were slower to emerge than those elicited by OSMK (Table S3).
We then performed a more decisive test by asking whether NS alone could cooperate with OS in reprogramming, by comparing the activities of O, OS, and OSN. Only OSN induced iPS cells, although it did so at a lower frequency than any of the four factor combinations (Table S3). Typically, 1–5 independent OSN iPS cell lines could be established from roughly 3 × 104 triply-transduced MECs, although in approximately half of the experiments no iPS cell lines were obtained. No evidence of reprogramming was obtained with any factor combination that did not include OCT4 and SOX2, or from any two-factor combination within the set of OSMKN (Table S3). The failure of O and OS alone to produce iPS cells affirmed the view that NS made an essential contribution to reprogramming by OSN.
The presence of transduced transgenes for all four factors could be detected in OSMK iPS cells, whereas OSN iPS cells contained only the OCT4, SOX2, and NS transgenes (Fig. 6 D). Thus, reprogramming by OSN could not be attributed to accidental contamination by either KLF4 or MYC. Expression of transduced genes was readily detected within 48 h after infection of MECs, but diminished over the course of iPS cell selection and was nearly undetectable in established iPS cell lines (Fig. 6 E), indicating silencing of the retroviral vectors.
The OSN iPS cells were phenotypically and functionally similar to regular ESCs. They could be maintained in standard ESC media and grew as compact, three-dimensional colonies with well-defined borders (Fig. 6 A), a clonal morphology that is characteristic of iPS cells and ESCs. The OSN iPS cells uniformly expressed the pluripotency markers AP (Fig. 6 A), OCT4, and SSEA1 (Fig. 6 B). QPCR analysis with primers specific for transcripts of the endogenous mouse genes demonstrated up-regulation of intrinsic Oct4, Sox2, and Nanog in OSN iPS cells to levels found in ESCs (Fig. 6 F), indicating reactivation of the stem cell transcriptional program. It is notable that as expression of ectopic NS was silenced, there was a compensatory rise in expression of intrinsic Ns (Fig. 6 C), presumably reflecting a continuing requirement for NS in the reprogramming process and the resulting iPS cells. This observation is consistent with our previous conclusion that Ns is required for the maintenance of ESC self-renewal.
The OSN iPS cells demonstrated pluripotency by forming teratomas containing various cell types representing all three germ layers (Fig. 6 G). We also obtained adult chimeric mice (Fig. 6 H) from two OSN iPS clones by injection of cells into 8-cell-stage embryos and transplantation into pseudopregnant mice. The OSN chimeric mice appeared grossly normal over 4 mo of observation. In contrast, progeny derived from OSMK iPS cells often succumbed to lethal tumors as early as 100 d after birth, presumably due to reactivation of the transduced MYC (Okita et al., 2007; Nakagawa et al., 2008). PCR analysis of tissues from chimeras revealed widespread contributions from the OSN iPS cells (Fig. 6 I). We conclude that the OSN iPS cells have a developmental potential similar to that of regular ESCs.
Previous studies of reprogramming mouse embryonic fibroblasts (MEFs) with OSMK indicated that expression of the ectopic genes is required for ∼10 d, after which cells enter a self-sustaining pluripotent state (Stadtfeld et al., 2008). To better understand the contribution of ectopic NS during the reprogramming process, we examined MECs relatively early in reprogramming by either OSK, OSKN, or OSMK (Fig. 6 J). Expression of endogenous Oct4, Sox2, and Nanog was low or undetectable 8 d after transduction of OSMK (unpublished data), but became vigorous by 12 d (Fig. 6 J). Expression of these endogenous genes at 12 d was substantially lower in OSK cells, but appeared to be augmented by the presence of NS in OSKN cells (Fig. 6 J). One possible explanation for the augmentation would be the large increase in cell number that follows transduction of MECs with OSKN. Two points argue against this explanation. First, the analyses of expression were normalized to an internal standard (Gapdh), so cell number should play no role in the results. Second, the accumulation of cells was not affected by the addition of NS to either OS or OSK (Fig. S4 D), yet expression of Oct4, Sox2, and Nanog was augmented by the presence of NS in OSKN. These data also support the view that the contribution of NS to reprogramming is not restricted to effects on cellular survival and proliferation: although addition of NS to OS had no effect on cell number, it enabled reprogramming. In summary, we conclude that NS contributes to reprogramming not only by sustaining cellular proliferation and survival, but also by enhancing the expression of crucial endogenous ESC regulators during the early stages of reprogramming. The mechanism of this enhancement is not known.
Much of the prior work on reprogramming has been conducted with MEFs, rather than the MECs used here. We found that OSN could reprogram MEFs to iPS cells (Fig. S3, D–F), albeit at a 10-fold lower efficiency than what we observed with MECs (unpublished data), which is consistent with a previous report that epithelial cells such as MECs permit more efficient reprogramming than mesenchymal cells such as MEFs (Li et al., 2010).
Discussion
Ns is required for the maintenance of self-renewal by ESCs
The central finding of this work is that by virtue of its effect on the cell cycle, Ns is essential to the maintenance of ESC self-renewal. We have concluded that the requirement for Ns in self-renewal is due largely to its role in sustaining the brevity of G1 in ESCs, which in turn shields the cells from signals for differentiation. The evidence for our conclusion is of three sorts. First, depletion of NS lengthened G1 and led to partial differentiation of ESCs. Second, depletion of any or all of the cell cycle inhibitors p21, p27, and RB1 in ESCs mitigated the effect of NS depletion on the cell cycle and rescued various other features of self-renewal. It appears that reconfiguration of the cell cycle may be the prime mover in the loss of self-renewal upon depletion of NS, and the principal change in that reconfiguration is a prolongation of G1. Third, it has been suggested that prolongation of G1 creates a window of opportunity during which ESCs can respond to signals responsible for differentiation (Orford and Scadden, 2008; Singh and Dalton, 2009). We have obtained evidence that is consistent with this view by examining the MEK/ERK signaling pathway, an established trigger for differentiation (Burdon et al., 1999; Kunath et al., 2007; Stavridis et al., 2007; Ying et al., 2008). The pathway was constitutively active in control ESCs without effect on self-renewal. The pathway remained equally active after depletion of NS, but now the cells differentiated. The differentiation could be blocked by inhibiting MEK. Thus, the signaling from MEK/ERK is required for differentiation of ESCs, but it is effective only when presented with a window of opportunity provided by a protracted G1.
These findings demonstrate that Ns is required for the maintenance of ESC self-renewal, provide an explanation for how Ns might contribute to self-renewal, and dramatize the importance of rapid transit through G1 to stemness. It seems unlikely, however, that the change in the cell cycle elicited by NS KD was the sole trigger for differentiation because a number of differentiation genes remained active and a few ESC markers remained low even when depletion of p21 completely rescued the ESC form of the cell cycle from NS KD.
Ns as a reprogramming factor
We have found that ectopic NS can cooperate with OCT4 and SOX2 to produce lines of iPS cells, albeit less efficiently than the canonical OSMK. OS alone produced no iPS cells, affirming that NS made an essential contribution to reprogramming by OSN. The ability of NS to serve as surrogate for either MYC or KLF4 in the cooperation with OS is notable because MYC and KLF4 are thought to play at least partially distinctive roles in reprogramming (Sridharan et al., 2009; Plath and Lowry, 2011). In this view, MYC mainly regulates genes involved in cellular proliferation and metabolism, whereas KLF4 joins OCT4, SOX2, and NANOG in controlling genes involved in pluripotency and differentiation (Chen et al., 2008; Kim et al., 2008, 2010; Sridharan et al., 2009). Ns is required to maintain the expression of both classes of genes in the stem-like state (see Table 1), which might account for its ability to substitute for either MYC or KLF4 in the cooperation with OS to reprogram somatic cells.
Ns is a transcriptional target of the MYC protein in both somatic cells and ESCs (http://www.myccancergene.org; Chen et al., 2008; Kim et al., 2008; Sridharan et al., 2009), and its expression can be induced by ectopic expression of MYC in the MECs that we have used for reprogramming (unpublished data). This relationship could account for the ability of NS to substitute for MYC in reprogramming, although it seems likely that the highly pleiotropic MYC also governs other genes that contribute to stem cell identity (Chen et al., 2008; Kim et al., 2008, 2010; Sridharan et al., 2009). In contrast, Ns functionally resembles Klf4 in the maintenance of normal levels of Nanog, as well as in the repression of differentiation markers and the cell cycle inhibitor p21 in ESCs (Chen et al., 2008; Ema et al., 2008; Kim et al., 2008; Sridharan et al., 2009). This resemblance could in turn account for the ability of NS to substitute for KLF4 in reprogramming. The mechanism by which a nucleolar protein might substitute for functions of both MYC and KLF4 is not readily apparent.
Materials and methods
Generation of Ns mutant mice
The S4-4B1 clone of ESCs (derived from the129S4 strain of mice) containing a gene trap mutation in the Ns locus was previously generated by using the ROSAFARY vector (Chen et al., 2004) and provided by P. Soriano (Mount Sinai School of Medicine, New York, NY). This gene trap vector contains a promoter trap module (SAβgeo*pA), a poly-A trap module (PGKhygroSD) that is flanked by FRT sites and can be removed by Flp recombinase, and the long terminal repeats (LTR) of the retroviral vector (Fig. S1 A; Chen et al., 2004). We further mapped the genomic structure of the disrupted Ns locus in this ESC clone by amplifying and sequencing a PCR fragment using a primer in intron 1 of Ns and a primer in the 5′ end of the vector. The gene trap insertion was detected at 860 bp downstream of exon 1 in the intron 1 region of the Ns locus. Chimeras were generated at the UCSF Transgenic Core according to standard protocols. In brief, S4-4B1 ESCs were injected into C57BL/6 blastocysts and transplanted into pseudopregnant mice. Chimeric male mice displaying >50% coat color chimerism were bred to C57BL/6 females to obtain germline transmission. Several backcross generations were performed on the C57BL/6 genetic background before setting up heterozygous intercrosses. Animal experiments were conducted in accordance with all local and federal guidelines and were approved by the UCSF Institutional Animal Care and Use Committee (IACUC).
Embryo collection, culture, and manipulation
Mouse embryos at different stages of pre- and post-implantation development were isolated from uteri dissected out from pregnant female mice that were prepared by timed natural matings following standard protocols (Nagy et al., 2003; Qu et al., 2003). Preimplantation embryos were isolated by flushing oviducts and uteri with M2 medium (Sigma-Aldrich). Postimplantation embryos were dissected from deciduas. To observe developmental potential of embryos in vitro, embryos were incubated in single drops of M2 medium overlaid with mineral oil in a humidified CO2 chamber at 37°C for 72–96 h, and photographed every 12 h. Embryos were transferred to M16 medium (Sigma-Aldrich) when reaching morula stage. For blastocyst outgrowth analyses, E3.5 embryos were placed in single drops of ESC media containing DME (Invitrogen) supplemented with 15% heat-inactivated fetal bovine serum (FBS), 0.055 mM β-mercaptoethanol, 2 mM l-glutamine, 0.1 mM MEM nonessential amino acids, 100 U/ml penicillin/streptomycin, and 1,000 U/ml LIF (Millipore). Embryos or explants were harvested for PCR genotyping at the end of observation or when signs of degeneration were apparent.
Genotyping of mice and embryos
Ns genotyping was performed by PCR analysis of genomic DNA isolated from mouse tail biopsies or fetal tissues of whole embryos as described previously (Qu et al., 2003). We used a competitive PCR method with three primer sets that included a common forward primer (5′-ATCTCTTGAGGGCCAGGAATATGG-3′) and two reverse primers specific for the wild-type (5′-ATGGCAGGTCATACGTTTACTCGC-3′) and mutant (5′-CATCAAGGAAACCCTGGACTACTG-3′) alleles. To genotype preimplantation embryos and blastocyst outgrowths, genomic DNA was extracted with the Pico Pure DNA Extraction kit (Thermo Fisher Scientific), and analyzed by two rounds of PCR amplification using nested PCR primers. The primers for the first round of PCR reaction were identical to those described above. The second round of PCR used 1 µl of the first round of PCR products and nested primers (5′-GATAGGTAGAACTGACTCCTGGAA-3′; 5′-GAGAAAGACTGCACACATGAGCAG-3′; and 5′-TCGAGGTCGACGGTATCGATTAGT-3′).
Microscopy
All samples were photographed at room temperature and all images compared were taken under identical conditions. Fixed embryos were photographed using a stereomicroscope (MZ FLIII; Leica) equipped with standard filter sets suitable for FITC and TMR red. Bright-field images of live embryos, outgrowth cultures, or cells were photographed with an inverted microscope (Axiovert 25; Carl Zeiss). Immunostaining images of cells were captured using a fluorescent microscope (Axiophot; Carl Zeiss) with 20x/0.50 and 40x/0.75 Plan Neofluar objectives. All images were acquired with a camera (DFC 420 C; Leica) and FireCam software (Leica). Photoshop (Adobe) was used for linear adjustments and pseudocoloring of images.
Detection of LacZ gene expression in embryos
Embryos isolated from intercrosses of Ns+/− with Ns+/+ mice were fixed in phosphate buffer solution (PBS) containing 2% paraformaldehyde (PFA) and 0.2% glutaraldehyde on ice for 5 min. After three washes with PBS, embryos were stained overnight at 37°C in 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) solution, containing 1 mg/ml X-Gal, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 2 mM MgCl2, 0.01% sodium deoxycholate, and 0.02% nonidet P-40 (NP-40) in PBS. After staining, embryos were washed in PBS containing 2 mM MgCl2, 0.01% sodium deoxycholate, and 0.02% NP-40, and examined under a stereomicroscope. The littermate wild-type embryos demonstrated negative staining and were used as a control.
BrdU incorporation assays of embryos
Preimplantation embryos derived from Ns+/− intercrosses were cultured for 16 h in M16 medium containing 50 µM BrdU (BrdU labeling and detection kit I; Roche) before fixation in 4% PFA. The BrdU-positive cells were identified by indirect immunofluorescence according to the manufacturer’s directions. After photography, embryos were genotyped.
TUNEL staining
Embryos derived from Ns+/− intercrosses were fixed in 4% PFA in PBS for 5 min at room temperature. After three washes with PBS for 5 min, embryos were processed for TUNEL staining according to the manufacturer’s instructions (In situ Cell Death Detection kit, TMR red; Roche). Embryos were analyzed by fluorescence microscopy and genotyped.
Plasmids
The pLKO.1-shRNA plasmids targeting Ns and containing the puromycin resistance gene were obtained from the Broad-TRC collection (Thermo Fisher Scientific). TRC number for the hairpin that most efficiently depletes NS is TRCN0000096885. The shRNA expression plasmids used for targeting p21, p27, and Rb1 were derived from TRCN0000042583, TRCN0000071063, and TRCN0000042543, respectively, with the puromycin resistance gene replaced with the blasticidin resistance gene. The shRNA expression vector targeting Trp53 (pSicoR-p53) was from Addgene, with the EGFP gene replaced with the blasticidin resistance gene. The human cDNAs for OCT4, SOX2, and KLF4 in the pMig vectors were obtained from Addgene. The human cDNAs for MYC and NS were cloned into the pMig and pMSCV-puro vectors (Takara Bio Inc.), respectively.
ESC culture and analysis
E14 feeder-free mouse ESCs were cultured at 37°C with 5% CO2 and were maintained on gelatin-coated dishes in ESC media as described above. Transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. ESCs were selected with puromycin (2 µg/ml) or puromycin plus blasticidin S (20 µg/ml) for cotransfection 1 d after transfection. The transfected cells were fed with fresh media every day. Stable ESC lines expressing human NS were generated by electroporation of plasmids and subsequent selection of ESCs with puromycin (1 µg/ml) for 2 wk. Individual colonies were selected and analyzed for NS expression. Detection of AP as a marker for the undifferentiated state of ESCs was performed using the ESC characterization kit according to the manufacturer’s protocol (Millipore). Drawing upon previous reports (Ivanova et al., 2006; Ema et al., 2008), we used both ESC colony morphology and AP staining to assess the differentiation state of ESCs. We divided ESCs into three categories: undifferentiated (ESC colony morphology with defined borders and strong AP staining), partially differentiated (poor ESC colony morphology with differentiated cells at periphery and weak AP staining, and flattened cells with weak AP staining), and differentiated (flattened cells with nondetectable AP staining). These are illustrated in Fig. 2 C.
The inhibitor treatments (1 µM PD0325901; 3 µM CHIR99021; Stemgent) were performed with the cells in conventional ESC media as described above. Differentiation of ESCs as EBs was performed using the hanging drop culture method. ESCs were seeded onto the lids of bacterial culture dishes at 400 cells/droplet in ESC media without LIF, which were then placed onto the bacterial dishes half-filled with water to assist in the maintenance of humidity. For synchronization experiments, cells were blocked at G2/M transition with nocodazole (Sigma-Aldrich) treatment at 50 ng/ml for 12 h. Mitotic cells were collected and released to normal ESC media to enter S phase. For cell cycle profile analysis, cells were pulse-labeled with 10 µM BrdU for 20 min, then fixed with PFA and stained with anti-BrdU antibodies and 7-AAD according to the manufacturer’s protocol (BD). For cumulative BrdU labeling, cells were fed with fresh media containing BrdU every 2 h during the course of analyses. BrdU-positive cells were quantified by FACS. The extrapolation of the time necessary to reach the maximum labeling index corresponds to the time in G2, M, and G1 (TG2+M+G1; Nowakowski et al., 1989). For detection of apoptosis, cells were stained with Annexin V following the manufacturer’s recommendation (BD). FACS analysis was performed using a FACSCalibur (BD) and Flowjo software (Tree Star).
Quantitative real-time PCR (QPCR)
Total RNA was extracted from cells with the RNeasy kit (QIAGEN). RNA was treated with DNase I (Invitrogen) for 15 min before reverse transcription with the SuperScript III First-Strand Synthesis kit (Invitrogen). cDNA synthesis was performed with 1 µg of total RNA and subsequently diluted 10 times. Total RNA from individual preimplantation embryos was obtained and analyzed using the Taqman PreAmp Cells-to-CT kit (Applied Biosystems). QPCR was performed on an ABI Prism 7300 machine (Applied Biosystems) with inventoried Taqman probes for each gene of interest (Applied Biosystems) and analyzed using the SDS software. Full information regarding individual probes is available upon request. Each QPCR was performed on at least three different experimental samples and each reaction was performed in triplicate.
Whole-genome expression analysis
Total RNA from ESCs was isolated using Trizol reagent combined with PureLink Micro-to-Midi columns (Invitrogen). Subsequent sample preparation, labeling, and array hybridizations were performed according to standard protocols from the UCSF Shared Microarray Core Facilities (http://www.arrays.ucsf.edu) and Agilent Technologies (http://www.agilent.com). In brief, total RNA quality was assessed using a Pico Chip on an Agilent 2100 Bioanalyzer (Agilent Technologies). RNA was amplified and labeled with Cy3-CTP using the Agilent low RNA input fluorescent linear amplification kit following the manufacturer’s protocol. Labeled cRNA was assessed using a spectrophotometer (ND-100; Nanodrop Technologies), and equal amounts of Cy3 labeled target were hybridized to Agilent whole-mouse genome 4 × 44K ink-jet arrays. Hybridizations were performed for 14 h, according to the manufacturer’s protocol. Arrays were scanned using the Agilent microarray scanner and raw signal intensities were extracted with Feature Extraction v9.1 software. Two group t-tests were formulated to examine differences between NS KD and scrambled shRNA transfected control groups in four biological replicates. Moderated t-statistic, B statistic, false discovery rate, P-value, and adjusted P-value for each probe were obtained. All procedures were performed using functions in the R package limma in Bioconductor. We considered genes to be differentially expressed if at least one probe for the gene had an adjusted P-value < 0.05.
Western blot analysis
Cell lysates were subjected to Western blot analysis with standard protocols. Primary antibodies were incubated overnight at 4°C and included rabbit anti-NS, which was generated against a peptide corresponding to the mouse Ns codons 32–49 (Tsai and McKay, 2002) in conjunction with Open Biosystems, goat anti-OCT4 (N-19; Santa Cruz Biotechnology, Inc.), rabbit anti-p53 (CM5; Vector Laboratories), rabbit anti-NANOG (ab21603; Abcam), rabbit anti-ERK1/2 (9102; Cell Signaling Technology), rabbit anti–phospho-ERK1/2 (9101; Cell Signaling Technology), rabbit anti-HA (Y-11; Santa Cruz Biotechnology, Inc.), and mouse anti–β-actin (A5441; Sigma-Aldrich). After incubation with the appropriate secondary antibodies, detection was performed by ECL (GE Healthcare).
Immunofluorescence
Cells were fixed in 4% PFA in PBS for 10 min at room temperature and subjected to immunofluorescence using standard protocols. The fixed cells were incubated with primary antibodies overnight at 4°C, which included rabbit anti-OCT4 (ab19857; Abcam), rabbit anti-NANOG (ab21603; Abcam), and mouse-anti-SSEA1 (SCR001; Millipore). This was followed by staining with the appropriate secondary antibodies conjugated to Alexa Fluor 594 or 488 (Invitrogen). Nuclei were counterstained with DAPI.
Generation of iPS cells
MEFs were derived from wild-type E13.5 embryos as described previously (Qu et al., 2003). In brief, embryos with heads, livers, and gonads removed were minced, trypsinized, and cultured in DME supplemented with 10% FBS, 2 mM glutamine, and 100 U/ml penicillin/streptomycin. Low passage MEFs (up to passage 4) were used for reprogramming studies. Adult MECs were derived according to standard protocols (Edwards et al., 1996). In brief, mammary glands free of adjacent tissues were dissected from 3–4-mo-old female wild-type mice, minced into small pieces (<1 mm), and digested for 1 h at 37°C in DME/F12 medium supplemented with 2.5% FBS and 2 mg/ml collagenase. The resultant organoids were recovered by centrifugation at 1,500 rpm for 10 min, resuspended in DME/F12, and separated from red blood cells and fibroblasts by 8–10 times of pulse-centrifugation. MECs were subsequently cultured in growth media (DME/F-12 supplemented with 10% FBS, 5 µg/ml insulin, 0.5 µg/ml hydrocortisone, 10 ng/ml EGF, 1 mM glutamine, 50 µg/ml gentamicin, and 100 U/ml penicillin/streptomycin) for 2–3 d, and replated as single cells the day before transduction.
Retroviral vectors were separately cotransfected with helper plasmids into BOSC 23 cells using the Effectene transfection reagent (QIAGEN). Viral supernatants were collected 48 h after transfection and filtered through 0.45-µm filters. MECs or MEFs were spin infected (1,800 rpm, room temperature, 1 h) with combinations of viral supernatants (equal amount of each desired factor), supplemented with 5 µg/ml polybrene. The transduced cells were maintained in standard ESC media with LIF for 1 d, replated on mitomycin C–inactivated MEF feeders, and 3 d later switched to Knockout-DME (Invitrogen) supplemented with 15% knockout serum replacement (Invitrogen), β-mercaptoethanol, l-glutamine, MEM nonessential amino acids, penicillin/streptomycin, and LIF without any further selection. iPS colonies were mechanically isolated. Individual cells were dissociated, replated onto feeders, and maintained in standard ESC media with LIF. All lentiviruses were generated using a third generation approach, with lentiviral plasmids cotransfected with packaging constructs MDL, REV, and VSV-G into 293T cells using the Effectene transfection reagent. Viral supernatants were harvested 48 h after transfection and filtered through 0.45-µm filters. MECs were infected with viral supernatants overnight and fed with fresh media for subsequent experiments.
Calculation of reprogramming efficiency
We used the expression of GFP in the pMig control virus to estimate the transduction efficiencies of MECs as ∼50% for individual retroviruses, and therefore ∼25% (0.52) of cells were infected with two factors, ∼12.5% (0.53) of cells were infected with three factors, and ∼6.25% (0.54) of cells were infected with four factors. The reprogramming efficiency was then calculated as the number of established iPS cell lines divided by the number of cells infected with desired combinations of factors (Takahashi and Yamanaka, 2006; Takahashi et al., 2007).
Genotyping of iPS cells
Genotyping was performed on genomic DNA isolated from iPS cells using the QIAamp DNA mini kit (QIAGEN), and analyzed by PCR with a forward primer specific for viral vector sequences and a reverse primer for cDNA sequences of human NS, OCT4, SOX2, MYC, or KLF4. Il-2 was used as an internal control as described by The Jackson Laboratory. The primer sequences were: Vector-F: 5′-CCCTTGAACCTCCTCGTTCGACC-3′; NS-R: 5′-CCTTAGCTCAGCTTCCCTAAGA-3′; OCT4-R: 5′-GCTTAGCCAGGTCCGAGGATC-3′; SOX2-R: 5′-TTGACGCGGTCCGGGCTGTTT-3′; MYC-R: 5′-TTTGATGAAGGTCTCGTCGTCC-3′; KLF4-R: 5′-GGTTATTCGGGGCACCTGCTT-3′.
Teratoma formation
One to two million cells for each iPS cell line were injected subcutaneously into the dorsal flank of Nude mice. Teratomas were recovered 4 wk after injection, fixed overnight in 10% formalin, paraffin embedded, and processed for hematoxylin and eosin staining according to standard protocols.
Chimera production from iPS cells
Chimeras were generated at the UCSF Transgenic Core according to standard protocols. In brief, iPS cells generated from MECs derived from 129S4 mice were injected into B6XB6D2 F1 8-cell-stage mouse embryos and transplanted into pseudopregnant mice. Adult female chimeric mice with agouti coat color were obtained and analyzed.
Online supplemental material
Fig. S1 shows disruption of the Ns gene by means of gene trap mutation. Fig. S2 shows evaluation of the specificity of Ns shRNA. Fig. S3 shows reprogramming of mouse MECs and MEFs. Fig. S4 shows that intrinsic Ns is required for the reprogramming of somatic cells to pluripotency by ectopic factors. Table S1 is an Excel file containing the complete expression array dataset for the NS KD cells vs. controls with scrambled shRNA (Ctl) comparison. Table S2 is an Excel file including all genes found to be significantly up-regulated or down-regulated by expression array analysis in NS KD cells (adjusted P-value < 0.05). Table S3 is a summary of data on reprogramming of MECs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201103071/DC1.
Supplementary Material
Acknowledgments
We thank J. Meneses for advice and assistance with the isolation and culture of preimplantation embryos; L. Urisman for assistance with animal husbandry; members of the Bishop laboratory for helpful discussions; and R. Blelloch, T. Fazzio, G. Martin, and B. Panning for critical comments on the manuscript. Microarray data collection and preliminary analyses were performed by R. Barbeau, Y. Xiao, A. Barczak, and D. Erle at the UCSF Sandler Asthma Basic Research (SABRE) Center Functional Genomics Core Facility. The gene trap S4-4B1 clone of ESCs was kindly provided by P. Soriano. We thank N. Killeen and members of the UCSF Transgenic Core for the production of chimeric mice; G. Daley for providing the human cDNAs for OCT4, SOX2, and KLF4 in the pMig vectors; and T. Jacks for the pSicoR-p53 plasmid through Addgene.
This work was supported by funds from the George W. Hooper Research Foundation, UCSF (J.M. Bishop), and the National Institutes of Health grants R35 CA044338 and T32 CA09043 (J.M. Bishop). J. Qu was a Special Fellow of the Leukemia and Lymphoma Society.
Author contributions: J.Qu and J.M. Bishop conceived and designed the experiments. J. Qu performed the experiments. J. Qu and J.M. Bishop analyzed the data and wrote the paper.
Footnotes
Abbreviations used in this paper:
- EB
- embryoid body
- ESC
- embryonic stem cell
- gal
- galactosidase
- ICM
- inner cell mass
- iPS
- induced pluripotent stem
- KD
- knockdown
- LIF
- leukemia inhibitory factor
- MEC
- mammary epithelial cell
- MEF
- mouse embryonic fibroblast
- PD/CH
- PD0325901 plus CHIR99021
- PI
- propidium iodide
- QPCR
- quantitative real time PCR
- shRNA
- short hairpin RNA
- TE
- trophectoderm
References
- Beekman C., Nichane M., De Clercq S., Maetens M., Floss T., Wurst W., Bellefroid E., Marine J.C. 2006. Evolutionarily conserved role of nucleostemin: controlling proliferation of stem/progenitor cells during early vertebrate development. Mol. Cell. Biol. 26:9291–9301 10.1128/MCB.01183-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon T., Stracey C., Chambers I., Nichols J., Smith A. 1999. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev. Biol. 210:30–43 10.1006/dbio.1999.9265 [DOI] [PubMed] [Google Scholar]
- Chen W.V., Delrow J., Corrin P.D., Frazier J.P., Soriano P. 2004. Identification and validation of PDGF transcriptional targets by microarray-coupled gene-trap mutagenesis. Nat. Genet. 36:304–312 10.1038/ng1306 [DOI] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J., et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 133:1106–1117 10.1016/j.cell.2008.04.043 [DOI] [PubMed] [Google Scholar]
- Edwards P.A., Abram C.L., Bradbury J.M. 1996. Genetic manipulation of mammary epithelium by transplantation. J. Mammary Gland Biol. Neoplasia. 1:75–89 10.1007/BF02096304 [DOI] [PubMed] [Google Scholar]
- Ema M., Mori D., Niwa H., Hasegawa Y., Yamanaka Y., Hitoshi S., Mimura J., Kawabe Y., Hosoya T., Morita M., et al. 2008. Krüppel-like factor 5 is essential for blastocyst development and the normal self-renewal of mouse ESCs. Cell Stem Cell. 3:555–567 10.1016/j.stem.2008.09.003 [DOI] [PubMed] [Google Scholar]
- Ezashi T., Telugu B.P., Alexenko A.P., Sachdev S., Sinha S., Roberts R.M. 2009. Derivation of induced pluripotent stem cells from pig somatic cells. Proc. Natl. Acad. Sci. USA. 106:10993–10998 10.1073/pnas.0905284106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faast R., White J., Cartwright P., Crocker L., Sarcevic B., Dalton S. 2004. Cdk6-cyclin D3 activity in murine ES cells is resistant to inhibition by p16(INK4a). Oncogene. 23:491–502 10.1038/sj.onc.1207133 [DOI] [PubMed] [Google Scholar]
- Fazzio T.G., Huff J.T., Panning B. 2008. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 134:162–174 10.1016/j.cell.2008.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K., Plath K. 2009. Epigenetic reprogramming and induced pluripotency. Development. 136:509–523 10.1242/dev.020867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I.R. 2006. Dissecting self-renewal in stem cells with RNA interference. Nature. 442:533–538 10.1038/nature04915 [DOI] [PubMed] [Google Scholar]
- Jaenisch R., Young R. 2008. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 132:567–582 10.1016/j.cell.2008.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Chu J., Shen X., Wang J., Orkin S.H. 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 132:1049–1061 10.1016/j.cell.2008.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Woo A.J., Chu J., Snow J.W., Fujiwara Y., Kim C.G., Cantor A.B., Orkin S.H. 2010. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 143:313–324 10.1016/j.cell.2010.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T., Saba-El-Leil M.K., Almousailleakh M., Wray J., Meloche S., Smith A. 2007. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 134:2895–2902 10.1242/dev.02880 [DOI] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q., et al. 2010. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 7:51–63 10.1016/j.stem.2010.04.014 [DOI] [PubMed] [Google Scholar]
- Massagué J. 2004. G1 cell-cycle control and cancer. Nature. 432:298–306 10.1038/nature03094 [DOI] [PubMed] [Google Scholar]
- Meng L., Lin T., Tsai R.Y. 2008. Nucleoplasmic mobilization of nucleostemin stabilizes MDM2 and promotes G2-M progression and cell survival. J. Cell Sci. 121:4037–4046 10.1242/jcs.037952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Gertsenstein M., Vintersten K., Behringer R. 2003. Manipulating the mouse embryo: a laboratory manual. Third Edition Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 764 pp [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. 2008. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 26:101–106 10.1038/nbt1374 [DOI] [PubMed] [Google Scholar]
- Nomura J., Maruyama M., Katano M., Kato H., Zhang J., Masui S., Mizuno Y., Okazaki Y., Nishimoto M., Okuda A. 2009. Differential requirement for nucleostemin in embryonic stem cell and neural stem cell viability. Stem Cells. 27:1066–1076 10.1002/stem.44 [DOI] [PubMed] [Google Scholar]
- Nowakowski R.S., Lewin S.B., Miller M.W. 1989. Bromodeoxyuridine immunohistochemical determination of the lengths of the cell cycle and the DNA-synthetic phase for an anatomically defined population. J. Neurocytol. 18:311–318 10.1007/BF01190834 [DOI] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S. 2007. Generation of germline-competent induced pluripotent stem cells. Nature. 448:313–317 10.1038/nature05934 [DOI] [PubMed] [Google Scholar]
- Orford K.W., Scadden D.T. 2008. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet. 9:115–128 10.1038/nrg2269 [DOI] [PubMed] [Google Scholar]
- Plath K., Lowry W.E. 2011. Progress in understanding reprogramming to the induced pluripotent state. Nat. Rev. Genet. 12:253–265 10.1038/nrg2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J., Li X., Novitch B.G., Zheng Y., Kohn M., Xie J.M., Kozinn S., Bronson R., Beg A.A., Minden A. 2003. PAK4 kinase is essential for embryonic viability and for proper neuronal development. Mol. Cell. Biol. 23:7122–7133 10.1128/MCB.23.20.7122-7133.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savatier P., Lapillonne H., van Grunsven L.A., Rudkin B.B., Samarut J. 1996. Withdrawal of differentiation inhibitory activity/leukemia inhibitory factor up-regulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene. 12:309–322 [PubMed] [Google Scholar]
- Singh A.M., Dalton S. 2009. The cell cycle and Myc intersect with mechanisms that regulate pluripotency and reprogramming. Cell Stem Cell. 5:141–149 10.1016/j.stem.2009.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R., Tchieu J., Mason M.J., Yachechko R., Kuoy E., Horvath S., Zhou Q., Plath K. 2009. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 136:364–377 10.1016/j.cell.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Maherali N., Breault D.T., Hochedlinger K. 2008. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2:230–240 10.1016/j.stem.2008.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavridis M.P., Lunn J.S., Collins B.J., Storey K.G. 2007. A discrete period of FGF-induced Erk1/2 signalling is required for vertebrate neural specification. Development. 134:2889–2894 10.1242/dev.02858 [DOI] [PubMed] [Google Scholar]
- Stead E., White J., Faast R., Conn S., Goldstone S., Rathjen J., Dhingra U., Rathjen P., Walker D., Dalton S. 2002. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene. 21:8320–8333 10.1038/sj.onc.1206015 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 126:663–676 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 131:861–872 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Thomson M., Liu S.J., Zou L.N., Smith Z., Meissner A., Ramanathan S. 2011. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 145:875–889 10.1016/j.cell.2011.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R.Y., McKay R.D. 2002. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 16:2991–3003 10.1101/gad.55671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A., Shi G., Zhou C., Lu R., Li H., Sun L., Jin Y. 2011. Nucleolin maintains embryonic stem cell self-renewal by suppression of p53 protein-dependent pathway. J. Biol. Chem. 286:43370–43382 10.1074/jbc.M111.225185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. 2008. The ground state of embryonic stem cell self-renewal. Nature. 453:519–523 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Yasumoto H., Tsai R.Y. 2006. Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol. Cell. Biol. 26:9279–9290 10.1128/MCB.00724-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F., Quelle D.E., Roussel M.F., Sherr C.J. 1997. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 15:203–211 10.1038/sj.onc.1201178 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.