The zebrafish (Danio rerio) has arrived in the pantheon of genetic model organisms. Perched next to the mouse in the vertebrate alcove of the hall of metazoans, the zebrafish looks out fondly at its better-established mentors, Drosophila and Caenorhabditis elegans. Like them, the zebrafish now has proved tractable to large-scale genetic screens. Mutations discovered in these screens already have revealed a logic to components of vertebrate development that could not have been predicted by the study of invertebrates.
And yet, the very nature of the exalted genetic company kept by the zebrafish makes comparisons inevitable and serves to reveal its lack of molecular sophistication. In brief, cloning of the mutations can be hard. The majority of mutations generated to date are single-base-pair changes generated by exposure to the chemical ethylnitrosourea. Mutation cloning requires assignment of chromosomal position to the mutant locus, a process that begins with meiotic mapping, and may then require laborious “walks” along genomic DNA. Dense genetic and physical maps, accompanied by the placement of many known genes along both, are now under construction to facilitate the cloning process. Gene mapping is key to abbreviating mutation cloning time, because such mapping provides candidate genes for the mutations. A combination of genetic and physical maps has been the hallmark of success of the Human Genome Project, facilitating cloning of genes in the other model organisms, including human, and forming the underpinning for the most powerful tool, sequencing of the entire genome. Radiation hybrid (RH) maps, reported by Hukriede et al. (1) in a recent issue of the Proceedings and by Geisler et al. (2), add an important buttress to the cloning infrastructure.
Zebrafish Genetic Screens Establish a Logic for Vertebrate Development.
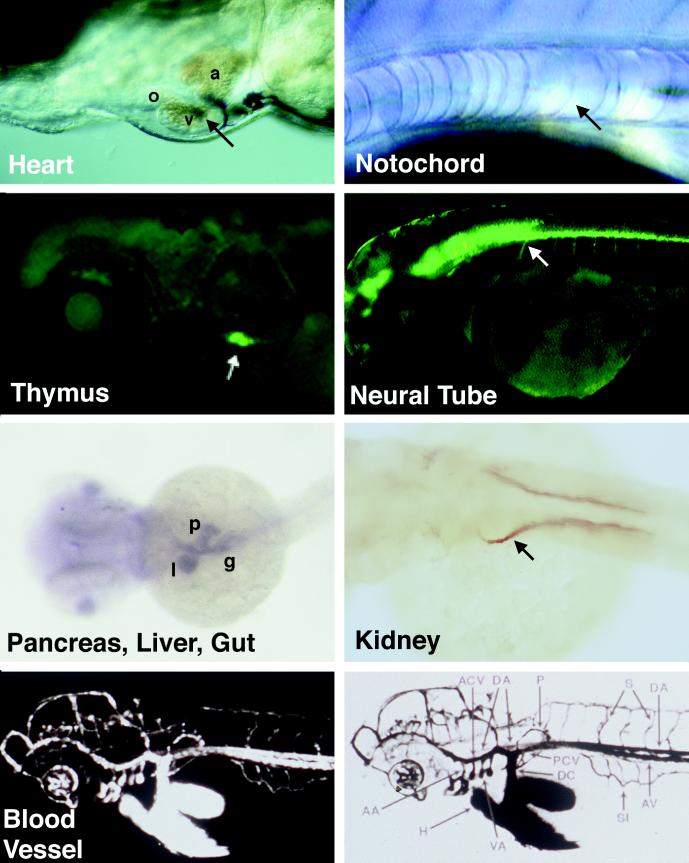
As powerful as the nematode and fruit-fly models are for discovering the logic of cell-fate decisions and pattern formation and for assembling the molecular components of responsible pathways, there are some questions these invertebrates simply cannot answer. The neural crest, multichambered heart, endocrine and exocrine pancreas, notochord, and endothelium are chordate “inventions” (although some have evolved, apparently independently, in invertebrates). By using embryological techniques in many organisms and targeted mutations in mice, much has been learned about elements of these processes, but an overarching set of principles is largely elusive. As exemplified in Fig. 1, by 72 h of development, essentially all of the vertebrate-specific organs have formed in the zebrafish embryo and are directly visible through the relatively transparent skin or may be rendered so by using tissue-specific green fluorescent protein expression, in situ hybridization, immunohistochemistry, or injection of dyes into living embryos in a manner akin to angiography in humans. The first hope for zebrafish screens was that a collection of single gene defects, in its entirety, would begin to provide a logical framework for generation of these vertebrate-specific characteristics (3–5). The second hope was that the cloning of the revealed critical genes would provide a molecular entrance to the pathways. How well has the zebrafish done with regard to the first hope, that we could learn about the unitary steps of vertebrate development from phenotypic analyses even before cloning the genes? Let me mention a few of the many developmental events already established by single gene mutations. Some confirm received wisdom; others are revelations. For example, these mutations have uncovered pathways for cell-fate decisions in the dorsal mesoderm (6, 7), for patterning of the nervous system (8, 9) and of neural-crest-derived pigmentation (10), and for organ morphogenesis (11–13).
Figure 1.
The transparency of the zebrafish embryo permits visualization of embryonic organs in the living embryo (Heart and Notochord), when enhanced by fluorescence after green fluorescent protein transgenesis (Thymus and Neural Tube), or after injection of fluorescent dextran to fill the vascular tree (Blood Vessel and Bottom Right). Screens may also be done by using in situ hybridization (Pancreas, Liver, Gut) or immunohistochemistry (Kidney). Heart and Notochord images were modified from refs. 35 and 36, respectively; green fluorescent protein images were provided by S. Lin (Medical College of Georgia, Augusta, GA); the Pancreas, Liver, Gut image was provided by J.-N. Chen (Massachusetts General Hospital); the Kidney image was provided by F. Serluca (Massachusetts General Hospital); and angiograms were modified from ref. 14.
As determined by phenotypic analysis, there are mutations that resemble complex human disorders, ranging from vascular malformations (14) to common adult diseases, including heart failure and arrhythmias (11, 15). In two cases, phenotypic resemblance has been confirmed by molecular definition: mutations in δ-aminolevulinate synthase cause congenital sideroblastic anemia in humans and the sauternes mutation in fish (16). Uroporphyrinogen decarboxylase mutations in humans cause human hepatoerythropoietic porphyria and the yquem mutation in fish (17).
Cloning Is Expedited by Candidate Genes.
How about the second hope, cloning the mutations? If cloning is the primary goal and the particular biological target is less of an issue, nothing today beats insertional mutagenesis, chiefly because of the efforts of Hopkins and her colleagues (18). There have been 12 mutations cloned as a result of retrovirus insertional screens (ref. 19 and N. Hopkins, personal communication). The lesser efficiency of mutagenesis by insertion, however, makes it unlikely that insertional methods will replace chemical methods for many purposes. Ethylnitrosourea screens provide the flexibility to select, in any lab, new morphological or functional targets and, in a matter of months, to do a screen and examine a thousand or more genomes. Thus, the insertional and chemical mutagenesis strategies are complementary. About two dozen ethylnitrosourea-induced mutations have been cloned. The canonical approach begins with meiotic mapping. Then, if there is a candidate gene, map positions are compared between candidate and mutation loci. How genes become candidates is not standard. Some examples show the investigators’ ingenuity: gastrulation defects suggested consideration of genes in Bone Morphogenetic Protein signaling pathways (20), the roles of which were known through extensive embryological work, especially in Xenopus. The spatiotemporal pattern of dharma expression in the yolk syncytial layer plus its induction by LiCl made it a good candidate for the bozozok mutation (21). The combination of reduced pectoral fins and somite patterning defects in sonic-you (syu) made sonic hedgehog a good candidate (22). The knowledge that tail formation depends on Brachyury led to the search for mutation in another fibroblast growth factor-dependent T box gene in the trunk-defective spadetail mutant (23). The yoo-too mutation, phenotypically similar to syu, mapped to a zebrafish linkage group with enough synteny conservation to human chromosome 2 to suggest that a hedgehog pathway component, Gli2, should be considered as a candidate (24).
one-eyed pinhead, by contrast, was identified through positional cloning, starting with meiotic markers and closing the interval by use of bacterial artificial chromosomes and P1-derived artificial chromosomes and then fine mapping the interval, taking advantage of the ease of generating thousands of mutant embryos in the fish. The gene hunt was done by using bacterial artificial chromosomes to select cDNAs, and causality was proven by the pattern of expression and rescue (25). A similarly arduous approach was needed to clone the sauternes gene (16). Although positional cloning can demand a substantial effort, this strategy makes no a priori assumptions about the identity of the gene of interest and thus is applicable to any mapped mutation.
RH Maps Facilitate Cloning.
The continuing development of maps, libraries, and other genomic resources facilitates molecular analysis by the candidate-gene approach and positional cloning. The zebrafish genome is 1.7 × 109 bp (about half the size of the mouse genome), corresponding to about 2,400 centimorgans. There is some evidence that at least part of the genome in zebrafish underwent an additional round of duplication. In theory, this redundancy could render many mutations phenotypically silent, were the duplicates functionally identical. However, the large number of phenotypes discovered through mutagenesis suggests that one copy of many of the duplicated genes became dysfunctional or that the two genes divided the jobs of one ancestral gene (26). Classical genetic maps have been built for zebrafish based on meiotic recombination frequency by using markers polymorphic between two strains. Several types of markers are used. The microsatellite map (27) uses anonymous sequences surrounding CA repeats. It currently has about one marker per centimorgan, on average, and is a useful tool to compare mutation and candidate-gene positions and to serve as the anchor to initiate positional cloning by walks with large-insert genomic libraries. Inclusion of genes on meiotic maps (28, 29) permits comparison between mutation and candidate position. However, sequence conservation is high within genes; thus, polymorphism detection can be arduous.
RH mapping of genes, on the other hand, does not require polymorphism. The substrate for RH mapping is a panel of clonal somatic cell hybrid cell lines (30). The hybrids are generated between cell lines of two different species. The donor cells are lethally irradiated, causing double-stranded breaks in the chromosomes. Cell fusion contributes different collections of pieces of the donor genome to each recipient cell. As shown first by Goss and Harris (31), loci in proximity on a donor chromosome tend to be retained together. Map distances are inferred from the pattern of retention across many clonal lines.
The resolution of the RH map is determined by the degree of fragment retention and the size of fragments retained in the hybrid cells (32). Higher resolution, needed for positional cloning, is achieved with higher radiation doses but is accompanied by reduction in long-range continuity of the map. Not all chromosomal fragments are retained equally. It seems, for example, that centromeric regions are retained in hybrids to a greater degree (30). Practically speaking, with a marker-retention frequency of about 20%, about 100 lines are needed to generate a map that is useful for cloning without introducing too many gaps. Such a map has been generated for several species.
There are two RH panels for zebrafish, both made by using donor irradiated AB9 cells (grown from fin), fused with either mouse (1) or hamster (33) recipients. Both panels are being widely used in the zebrafish research community. The former was used for the map reported by Hukriede et al. It is composed predominantly of microsatellite markers, along with some genes and expressed sequence tags. There is overall agreement between the positions of markers on the meiotic and RH maps, with some gaps and inversions, as is expected until the RH map becomes denser. Intermarker distances are expressed in centiRays, where 1 centiRay corresponds to a 1% frequency of breakage between two markers. For this map, 1 centiRay = 148 kilobases. The potential resolution of about 500 kilobases is comparable to that of RH maps generated for other species. The panel used for the map reported by Geisler et al. (2) provides better resolution, with some loss of continuity (R. Geisler and C. Nusslein-Volhard, personal communication). The two maps are complementary.
The key contribution of these RH maps is that any DNA sequence may be mapped, without the need to determine polymorphism. Thus, RH panels are proving important for mutation cloning, because they permit comparison of map positions between mutations and candidate genes or expressed sequence tags. Even if walking remains necessary, yeast artificial chromosome ends may be directly mapped on the RH map, establishing their order. RH mapping of sequenc tagged sites from yeast artificial chromosomes or bacterial artificial chromosomes also helps to begin the integration of meiotic and physical maps. As markers are added, the refinement of the maps will continue, a process facilitated by the universal use of the same reagents, DNA generated in bulk from the RH panel clonal cell lines.
There are some technical considerations to keep in mind when using RH maps. For example, PCR will not amplify all markers to an equal degree. Fragments are retained at different concentrations among cell lines; as such, scoring for marker retention requires care. Distances between markers are arrived at by statistical means. They are not directly proportional to either physical or genetic distance. However, given these caveats, the zebrafish RH maps are proving quite valuable for mutation cloning.
Future Arrivals.
Dense maps of the zebrafish genome therefore will facilitate gene discovery from the screens. The next generation of screens will be refined to focus on additional vertebrate-specific questions, such as those concerned with integrative organ system physiology; mechanisms of homeostasis; and the molecular bases of behavior, learning, and memory. In the genetic discovery gyre, from gene to function and back, high-throughput sequencing by the Human Genome Project has proved breathtakingly rapid in gene discovery but slow at putting function to gene. Phenotype-first genetic screens, as in zebrafish, provide elements of function at a rapid pace. With the building of the necessary genomic infrastructure, gene cloning will become facile, and the gyre will spin more quickly from function to gene.
Acknowledgments
I thank R. Geisler, C. Nusslein-Volhard, and N. Hopkins for sharing data before publication; S. Lin, J.-N. Chen, and F. Serluca for unpublished images; and W. Talbot, J.-N. Chen, and D. Jackson for comments on the manuscript. Both title and theme, to some degree, borrow from V. S. Naipul’s book, The Enigma of Arrival (34).
Footnotes
The companion to this commentary begins on page 9745 in issue 17 of volume 96.
References
- 1.Hukriede N A, Joly L, Tsang M, Miles J, Tellis P, Epstein J A, Barbazuk W B, Li F N, Paw B, Postlethwait J H, et al. Proc Natl Acad Sci USA. 1999;96:9745–9750. doi: 10.1073/pnas.96.17.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geisler R, Rauch G-J, Baier H, van Bebber F, Bross L, Dekens M P S, Finger K, Fricke C, Gates M A, Geiger G, et al. Nat Genet. 1999;23:86–89. doi: 10.1038/12692. [DOI] [PubMed] [Google Scholar]
- 3.Streisinger G, Walker C, Dower N, Knauber D, Singer F. Nature (London) 1981;291:293–296. doi: 10.1038/291293a0. [DOI] [PubMed] [Google Scholar]
- 4.Nüssslein-Volhard C. Science. 1994;266:572–574. doi: 10.1126/science.7939708. [DOI] [PubMed] [Google Scholar]
- 5.Driever W, Fishman M C. J Clin Invest. 1996;97:1788–1794. doi: 10.1172/JCI118608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halpern M E, Thisse C, Ho R K, Thisse B, Riggleman B, Trevarrow B, Weinberg E S, Postlewait J H, Kimmel C B. Development (Cambridge, UK) 1995;121:4257–4264. doi: 10.1242/dev.121.12.4257. [DOI] [PubMed] [Google Scholar]
- 7.Melby A E, Warga R M, Kimmel C B. Development (Cambridge, UK) 1996;122:2225–2537. doi: 10.1242/dev.122.7.2225. [DOI] [PubMed] [Google Scholar]
- 8.Moens C B, Yan Y-L, Apel B, Force A G, Kimmel C B. Development (Cambridge, UK) 1996;122:3981–3990. doi: 10.1242/dev.122.12.3981. [DOI] [PubMed] [Google Scholar]
- 9.Trowe T, Klostermann S, Baier H, Granato M, Crawford A D, Grunewald B, Hoffmann H, Karlstrom R O, Meyer S, Muller B, et al. Development (Cambridge, UK) 1996;123:439–450. doi: 10.1242/dev.123.1.439. [DOI] [PubMed] [Google Scholar]
- 10.Kelsh R N, Brand M, Jiang Y-J, Heisenberg C-P, Lin S, Haffter P, Odenthal J, Mullins M C, van-Eeden F J M, Furutani-Seiki M, et al. Development (Cambridge, UK) 1996;123:369–389. doi: 10.1242/dev.123.1.369. [DOI] [PubMed] [Google Scholar]
- 11.Stainier D Y R, Fouquet B, Chen J, Warren K S, Weinstein B M, Meiler S, Mohideen M P K, Neuhauss S C F, Solnica-Krezel L, Schier A F, et al. Development (Cambridge, UK) 1996;123:285–292. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- 12.Pack M, Solnica-Krezel L, Malicki J, Neuhauss S C F, Schier A F, Stemple D L, Driever W, Fishman M. Development (Cambridge, UK) 1996;123:321–328. doi: 10.1242/dev.123.1.321. [DOI] [PubMed] [Google Scholar]
- 13.Schilling T F, Walker C, Kimmel C B. Development (Cambridge, UK) 1996;122:1417–1426. doi: 10.1242/dev.122.5.1417. [DOI] [PubMed] [Google Scholar]
- 14.Weinstein B M, Stemple D L, Driever W, Fishman M C. Nat Med. 1995;1:1143–1147. doi: 10.1038/nm1195-1143. [DOI] [PubMed] [Google Scholar]
- 15.Baker K, Warren K S, Yellen G, Fishman M C. Proc Natl Acad Sci USA. 1997;94:4554–4559. doi: 10.1073/pnas.94.9.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brownlie A, Donovan A, Pratt S J, Paw B H, Oates A C, Brugnara C, Witkowska H E, Sassa S, Zon L I. Nat Genet. 1998;20:244–250. doi: 10.1038/3049. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Long Q, Marty S D, Sassa S, Lin S. Nat Genet. 1998;20:239–243. doi: 10.1038/3041. [DOI] [PubMed] [Google Scholar]
- 18.Gaiano N, Allende M, Amsterdam A, Kawakami K, Hopkins N. Proc Natl Acad Sci USA. 1996;93:7777–7782. doi: 10.1073/pnas.93.15.7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allende M L, Amsterdam A, Becker T, Kawakami K, Gaiano N, Hopkins N. Genes Dev. 1996;10:3141–3155. doi: 10.1101/gad.10.24.3141. [DOI] [PubMed] [Google Scholar]
- 20.Schulte-Merker S, Lee K J, McMahon A P, Hammerschmidt M. Nature (London) 1997;387:862–863. doi: 10.1038/43092. [DOI] [PubMed] [Google Scholar]
- 21.Fekany K, Yamanaka Y, Leung T C, Sirotkin H I, Topczewski J, Gates M A, Hibi M, Renucci A, Stemple D, Radbill A, et al. Development (Cambridge, UK) 1999;126:1427–1438. doi: 10.1242/dev.126.7.1427. [DOI] [PubMed] [Google Scholar]
- 22.Schauerte H E, van Eeden F J M, Fricke C, Odenthal J, Strähle U, Haffter P. Development (Cambridge, UK) 1998;125:2983–2993. doi: 10.1242/dev.125.15.2983. [DOI] [PubMed] [Google Scholar]
- 23.Griffin K J P, Amacher S L, Kimmel C B, Kimelman D. Development (Cambridge, UK) 1998;125:3379–3388. doi: 10.1242/dev.125.17.3379. [DOI] [PubMed] [Google Scholar]
- 24.Karlstrom R O, Talbot W S, Schier A F. Genes Dev. 1999;13:388–393. doi: 10.1101/gad.13.4.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Talbot W S, Schier A F. Cell. 1998;92:241–251. doi: 10.1016/s0092-8674(00)80918-6. [DOI] [PubMed] [Google Scholar]
- 26.Force A, Lynch M, Pickett F B, Amores A, Yan Y L, Postlethwait J. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimoda N, Knapik E W, Ziniti J, Sim C, Yamada E, Kaplan S, Jackson D, deSauvage F, Jacob H, Fishman M C. Genomics. 1999;58:219–282. doi: 10.1006/geno.1999.5824. [DOI] [PubMed] [Google Scholar]
- 28.Postlethwait J H, Yan Y L, Gates M A, Horne S, Amores A, Brownlie A, Donovan A, Egan E S, Force A, Gong Z, et al. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- 29.Gates M A, Kim L, Egan E S, Cardozo T, Sirotkin H I, Dougan S T, Lashkari D, Abagyan R, Schier A F, Talbot W S. Genome Res. 1999;9:334–347. [PubMed] [Google Scholar]
- 30.Walter, M. A. & Goodfellow, P. N. (1993) 9, 352–356. [DOI] [PubMed]
- 31.Goss S J, Harris H. Nature (London) 1975;255:680–684. doi: 10.1038/255680a0. [DOI] [PubMed] [Google Scholar]
- 32.Walter, M. A., Spillett, D. J., Thomas, P., Weissenbach, J. & Goodfellow, P. N. (1994) 7, 22–28. [DOI] [PubMed]
- 33.Kwok C, Korn R M, David M E, Burt D W, Critcher R, McCarthy L, Paw B H, Zong L I, Goodfellow P N, Schmitt K. Nucleic Acids Res. 1998;26:3562–3566. doi: 10.1093/nar/26.15.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naipul V S. The Enigma of Arrival. New York: Random House; 1988. [Google Scholar]
- 35.Chen J-N, Haffter P, Odenthal J, Vogelsang E, Brand M, van-Eeden F J M, Furutani-Seiki M, Granato M, Hammerschmidt M, Heisenberg C-P, et al. Development (Cambridge, UK) 1996;123:293–302. doi: 10.1242/dev.123.1.293. [DOI] [PubMed] [Google Scholar]
- 36.Odenthal J, Haffter P, Vogelsang E, Brand M, van-Eeden F J M, Furutani-Seiki M, Granato M, Hammerschmidt M, Heisenberg C-P, Jiang Y-J, et al. Development (Cambridge, UK) 1996;123:103–115. doi: 10.1242/dev.123.1.103. [DOI] [PubMed] [Google Scholar]