Abstract
Mucosal epithelium M cells are dispersed across Peyer’s patch follicle associated epithelium (PPFAE) with minimal clustering. Since Notch signaling can influence patterning in epithelia, we examined its influence on PPFAE M cell distribution. Conditional deletion of Notch1 in intestinal epithelium increased PPFAE M cells and also increased M cell clustering, implying a role for Notch in both M cell numbers and lateral inhibition. By contrast, conditional deletion of the ligand Jagged1 also increased M cell clustering, but with a paradoxical decrease in M cell density. In vitro, inhibition of Notch signaling reduced expression of an M cell associated gene CD137, consistent with cis-promoting effects on M cell development. Thus, Jagged1 may have a cis-promoting role in committed M cells, but a trans- inhibitory effect on neighboring cells. In sum, Jagged1-Notch signaling may edit the pattern of M cells across the PPFAE, which may help optimize mucosal immune surveillance.
Keywords: mucosal immunity, cytokines, lymphoid organs
1. Introduction
Notch receptors are transmembrane receptors that, when activated by one of several known ligands (Delta-like/Serrate or Jagged), undergo proteolytic processing and nuclear localization to directly activate expression of gene targets (1–3). In the immune system, Notch signaling regulates the development and effector cell induction of several cell types including T and B lymphocytes and dendritic cells; the expression of Notch receptors and ligands is distributed among many cell types with many of the direct interactions still only incompletely understood (4–7). However, known functions of Notch also extend to more basic developmental processes, where cell fates and tissue patterning are regulated. These decisions are made as a consequence of direct cell-cell signaling, where a cell expressing a Notch ligand influences the fate of an adjacent cell expressing a Notch receptor. Thus, tissue patterns can be established or reinforced by the directional interactions between cells with regulated expression of Notch ligands and receptors. These interactions can result in lateral inhibition or lateral activation, with Notch activation inhibiting or inducing the development of a particular phenotype. These effects can be used to limit the production of specialized cells along a default pathway, or to help establish tissue boundaries.
Notch and its ligands can be used to develop rather complex patterns of specialized cell types, and this is especially notable in the development of sensory organs, such as the Drosophila eye disc, where Notch signaling insures regular spacing of photoreceptor cells (8). In the mammalian inner ear, Notch signaling appears to insure the orderly arrangement of hair cells (9–12). This appears to be in part by lateral inhibition mediated by Delta-like 1, but in these studies it has also been suggested that there is also an (as yet unproven) inductive signal provided by Jagged1. Thus, the possibility has been raised that a single Notch ligand may simultaneously provide both trans-inhibitory and cis-inductive signals, depending on the cellular context.
In the intestine, Notch has an important role in regulating intestinal epithelium lineage specification; Notch signaling suppresses the development of secretory cell types such as goblet cells (13–17). The production of secretory cells is not associated with sensory function, but the production of another specialized intestinal epithelial cell, the M cell, does fit the sensory organ pattern. M cells are mainly found in Peyer’s patch follicle associated epithelium (PPFAE), and are responsible for the capture of lumenal particles such as bacteria and viruses, and transcytosis across the epithelial barrier to underlying dendritic cells, triggering mucosal immune responses (18–20). The distribution of M cells in the PPFAE appears to be highly regulated, with a distributed checkerboard pattern (21–24). In addition, goblet cells are less frequent in PPFAE than in neighboring villi, with correspondingly less mucus over the PPFAE epithelium. Since the localized assembly of M cells in PPFAE comprises a mucosal immune surveillance unit, their organized pattern may be beneficial to their function.
We recently reported that in a cell culture model of M cell function, the expression of the Notch ligand Jagged1 was increased in M cell-like cells (25), raising the possibility that its expression and interaction with Notch receptors may influence development of M cells in the PPFAE. However, in a survey of Notch and Notch ligand expression in the gut (26), Jagged1 expression was primarily detected in the intestinal crypt, suggesting that if Jagged1 is indeed influencing M cell development, it may be primarily at the earliest stages in lineage decisions (14–17). Here we report the results of studies on the requirements for Notch and Jagged1 in M cell development and distribution in PPFAE. Our results are consistent with the notion that M cell expression of Jagged1 and Notch may have an editing effect on the production and distribution of M cells across the PPFAE, while also having a slight inductive influence on committed M cells.
2. Material and Methods
2.1. Animals
VilCre mice (Jax #4586, expressing Cre recombinase under the Villin promoter), FloxNotch1 mice (Jax #6951), and FloxJag1 mice (Jax #10618), all on the C57BL/6 background, were purchased from Jackson Labs (Bar Harbor, ME, USA) and bred in the UC Riverside vivarium under SPF conditions. All mice were genotyped according to Jackson Lab website protocols. Conditional Notch1 KO mice were generated by crossing VilCre with FloxNotch1; conditional knockouts were homozygous for FloxNotch1, while controls were heterozygous. The same strategy was used to generate conditional knockout Jagged1 KO mice. All mice were used around 8 weeks of age. Mice were handled according to institutional IACUC and NIH guidelines.
2.2. Cell line and tissue culture
Caco-2BBe cells were obtained from ATCC and cultured with ADMEM with 10%FBS, 1.5% penicillin/streptomycin, and 10mM HEPES. For qPCR analysis, 500,000 cells were plated in 12 well plates for 24 hours. Cytokines were added at the time of plating at the concentration of 100ng/ml for TNFα (Peprotech, Rocky Hill, NJ, USA) and 5ug/ml of LTβR agonist (R&D Systems, Minneapolis, MN, USA). Conditions for cytokine induction were developed and reported by Wang et al. (27). Jagged1 peptide (Anaspec, Fremont, CA, USA) was used at 4 or 40uM in culture, added at the time of plating and continued culture for 24 hours. For DAPT (Tocris Bioscience, Minneapolis, MN, USA) treatment, DAPT was added to the culture at the time of plating at the concentration of 10uM and 100uM and followed by combined cytokine treatment 4 hours after plating. DAPT treated samples were compared with control samples treated with DMSO at the same concentration. The data for cytokine induction of CD137 and Jagged1, and CD137 inhibition by DAPT shown in the figures is the mean “fold-increase” compared to control non-cytokine treated cultures, determined from three independent biological replicate experiments (shown as the mean and SEM of the three experiments together), with each individual experiment showing the same trends.
2.3. Real Time-PCR
For quantitative PCR analysis of gene expression in Caco-2BBe cells, RNA was harvested after 24 hours of culture with TRIZOL (Invitrogen, Grand Island, NY, USA); next, 2μg of total RNA was made into cDNA using Superscript III first-strand synthesis system (Invitrogen). Quantitative PCR was performed using a CFX96 Real-Time PCR detection system (BioRad, Hercules, CA, USA) using SYBR Green for quantification of PCR product. All samples were calibrated for relative expression using GAPDH in parallel reactions as the reference housekeeping gene. All PCR assays were done in triplicate in 96 well plates with at least 3 replicate experiments with similar results; error bars shown reflect the variation in three independent biological replicate experiments. Relative mRNA expression was calculated using the ΔΔCT method. Primers used for Real-time PCR (all sequences are 5’ to 3’) were: GAPDH, For- CATGAGAAGTATGACAACAGCCT, Rev- AGTCCTTCCACGATACCAAAGT; CD137, For- AGGTGTTTTCAGGACCAGGAAGGA, Rev- GTCGACAGATGCCACGTTTCTGAT; Jagged1, For- TACACTGCCTGCCTTAAGTGAGGA, Rev- CACGGTCTCAATGGTGAACCAACA.
2.4. Immunohistochemistry and confocal microscopy
For whole mount Peyer’s patch microscopy, freshly dissected Peyer’s Patches from the small intestine (typically 6 to 8 Peyer’s patches recovered from stomach to cecum) were washed briefly in PBS then kept in 4% paraformaldehyde in PBS/ 30% sucrose for 30 minutes. Samples were then washed with 0.1% Tween in PBS twice and blocked with Casein 0.1% Tween for another 30 minutes. For primary antibody staining, Rhodamine conjugated UEA-1 (Vector Laboratories, Burlingame, CA, USA) was used. Whole mount Peyer’s patches were then cleaned and mounted after 10 minutes of 4% PFA post-fix. Samples were washed with 3 times PBS 0.1% Tween and followed by secondary staining (Streptavidin Alexa 647 (Invitrogen)). For goblet cell staining, intestines (also from the small intestine between stomach and cecum) were kept on ice in 4% paraformaldehyde/PBS/30% sucrose for 3 hours before freezing. Cryostat sections were stained with Alcian blue (Sigma-Aldrich, St. Louis, MO, USA) for 1 minute and cleaned using tap water until washes were clean. Images were taken using bright field microscopy. Staining of Caco-2BBe cells for CD137 and Jagged1 was performed as follows: 50,000 Caco-2BBe cells were plated in chamber slides (BD Biosciences, San Jose, CA) with the same cytokine concentrations as for qPCR culture for 48 hours before staining. Staining was done using Jagged1 rabbit antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and CD137 goat antibody (Santa Cruz), using donkey anti-goat Alexa 488 and donkey anti-rabbit Alexa 647 (Invitrogen) as secondary reagents.
2.5. Goblet cell count and M cell density analysis
Goblet cell counts was assessed by counting the number of goblet cells over the distance on the basement membrane obtained from stained intestinal cryostat sections. Each data point was the analysis from a single confocal z-stack image. For M cell quantification, mice were used at 8 weeks of age. Images were taken from whole mount Peyer’s patches through confocal microscopy and analyzed using Volocity 5 software (PerkinElmer, San Jose, CA, USA). M cell counts were counted based on UEA-1 staining, which distinguishes goblet cells and M cells by differences of brightness and shapes. Adjacent cell percentages were calculated by the number of M cells with contiguous edges in direct contact over a length of >3 μm, divided by the total M cells counted from the same follicle. Each data point was the analysis from a single image, and data was accumulated from multiple Peyer’s patches from at least three different mice for each genotype. Statistical tests were performed using Prism software (GraphPad, La Jolla, CA, USA). We used a two-tailed t-test for M cell and goblet cell density counts, and Mann-Whitney for percentage clustering analysis, though similar results were obtained using either method. For quantitative PCR analysis, three independent biological replicate cultures and each associated PCR assay result (in fold induction) was combined for statistical analysis.
3. Results
3.1. Removal of epithelial Notch increases both goblet cell and M cell lineages in the intestine
Notch signaling has a critical role in intestinal epithelium lineage fate decisions; blocking Notch signaling resulted in the nearly exclusive production of goblet cells at the expense of other cell types (16; 17). However, in the epithelium overlying intestinal Peyer’s patches, the influence of cytokines from lymphoid cells including lymphoid tissue inducer cells (LTi) changes the local context, dramatically altering patterns of gene regulation (28). For example, in contrast to neighboring villous epithelium, PPFAE show expression of genes such as CCL20 (29; 30). The development of M cells is even more complex, since local conditions only induce a subset of epithelial cells to the M cell lineage; the regulation of this selective induction remains to be explained. If Notch influences M cell lineage decisions in the same way it affects goblet cells, then an increase in M cell numbers in mice lacking epithelial Notch expression might be evidence for Notch regulation of M cell production.
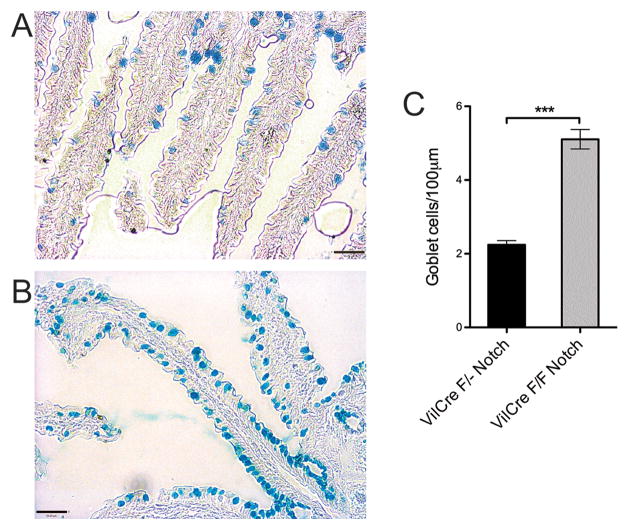
Intestinal epithelium may use Notch1 or Notch2 to mediate signaling; here, we used a conditional deletion of Notch1 in intestinal epithelium. A floxed Notch1 allele was crossed to a transgene expressing Cre recombinase driven by the Villin promoter. This transgene is expressed early in the intestinal epithelium during development (31; 32), and appears to be specific to intestinal epithelium. As confirmation of this strategy, mice homozygous for the floxed Notch1 allele and carrying the Vil-Cre transgene showed approximately two-fold increased numbers of goblet cells throughout the intestinal villi as compared to mice heterozygous for the floxed allele (Figure 1A-C). Consistent with previous effects of a complete block in Notch signaling, these results confirm the major influence of Notch1 signaling on intestinal epithelium lineage fate.
Figure 1. Goblet cells on small intestine villi are greatly increased in the absence of Notch1.
A. Control villi (single floxed Notch1) stained for goblet cells using Alcian Blue, showing normal dispersed distribution and density of goblet cells on intestinal villi. (Scale bar: 50 um)
B. FloxNotch/VilCre villi stained with Alcian Blue. Here, there is greatly increased goblet cell density, with many goblet cells directly adjacent to each other. (Scale bar: 50 um)
C. Quantitation of goblet cell density in wild type and Notch1 conditional knockout villi. Goblet cell density was measured by goblet cell counts per villous basement membrane length. The Notch1 conditional knockout villi showed approximately double the density of goblet cells versus control villi. (single flox Notch N=22; FloxNotch N=15; ***, P<0.0005)
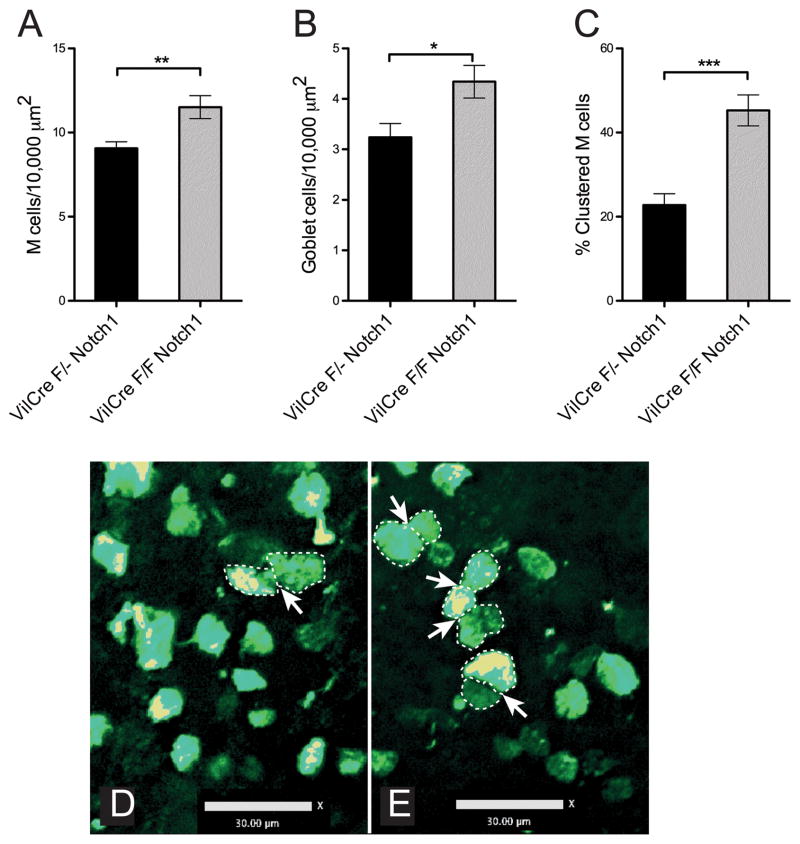
In the PPFAE, we assayed the production of M cells, using staining with the fucose-binding lectin Ulex Europeus Agglutinin-1 (UEA-1) (33). Goblet cells can also bind to UEA-1; though their numbers are much lower in PPFAE compared to villi, we used analysis of z-stack images to rule out goblet cells in our counts by using the distinctive 3-dimensional morphology of the different cells. We found that the density of M cells was significantly increased by about 25% (Figure 2A). Moreover, we also observed a significant increase in PPFAE goblet cell density (Figure 2B), leaving the M cell/goblet cell ratio unchanged around a value of 3. It is conceivable that changes in Notch signaling might affect M cell morphology relative to goblet cells; however, the coordinated changes in the numbers of both M cells and goblet cells in PPFAE argue against such an effect. Notch1 may influence both lineage fate decisions as well as M cell patterning through lateral inhibition. In support of this mechanism, we also found that the percentage of M cells showing clustering (defined by adjacent M cells with more than 3 microns in direct contiguous contact) was doubled (Figure 2C-E). Thus, our data supports the hypothesis that the both the numbers and distribution of M cells across the PPFAE are influenced by Notch.
Figure 2. Peyer’s patch M cells show increased numbers and clustering in floxNotch mice.
A. Quantitation of M cell density across Peyer’s patch epithelium, measured by direct counting of M cells per Peyer’s patch follicle epithelium surface area. Notch1 conditional knockout Peyer’s patches contained a higher density of M cells. (single flox Notch N=11; FloxNotch N=10; **, P<0.005)
B. Quantitation of goblet cell density across Peyer’s patch epithelium. In Notch1 conditional knockout mice, goblet cell density was increased in Peyer’s patch in similar fashion to that seen in intestinal villi, and in parallel to the increase seen for M cells. (single flox Notch N=11; FloxNotch N=10; *, P<0.05)
C. Calculation of M cell clustering across Peyer’s patch epithelium. Adjacent cells with more than 3 microns in direct contiguous contact are considered clustered, and counts show the proportion of M cells in clustered arrangements. (single flox Notch N=11; FloxNotch N=10; ***, P<0.0005)
D. Confocal image of Peyer’s patch whole mount showing dispersal of M cells in control Peyer’s patch and occasional M cell cluster (arrow). (Scale bar: 30 um)
E. Image showing increased clustering in FloxNotch Peyer’s patch. Clustered M cells with increased contacts along sides (outlined by dotted lines) are identified with arrows. (Scale bar: 30 um)
3.2. Deletion of epithelial Jagged1 reduces PPFAE M cell numbers while increasing M cell clustering
Goblet cell lineage commitment is determined in the intestinal crypt, regulated in part by expression of Delta-like 1 (Dll1) expression (13; 15; 26). Interestingly, Dll1 may have both a lateral inhibition effect on Notch-expressing cells, and a positive induction effect that may be Notch-independent; unfortunately, details on this mechanism are limited, since Dll1 expression is only transiently evident in the crypt cells (13; 15). In the case of PPFAE M cells, a similar challenge is present for deciphering any potential role of Jagged1, which we identified in a cell culture model as a candidate gene in M cell development (25). As noted earlier, Jagged1 expression is mainly limited to the lower crypt, so any influence of Jagged1 expression may be limited to the early stages in the crypt followed by reduced Jagged1 expression thereafter. In addition, we previously reported evidence that early lineage decisions toward M cell commitment occur prior to expression of other M cell associated genes such as CD137, gp2, and PGRP-S (24; 34), so for Jagged1 to influence M cell development, it should also be at an early stage in lineage commitment.
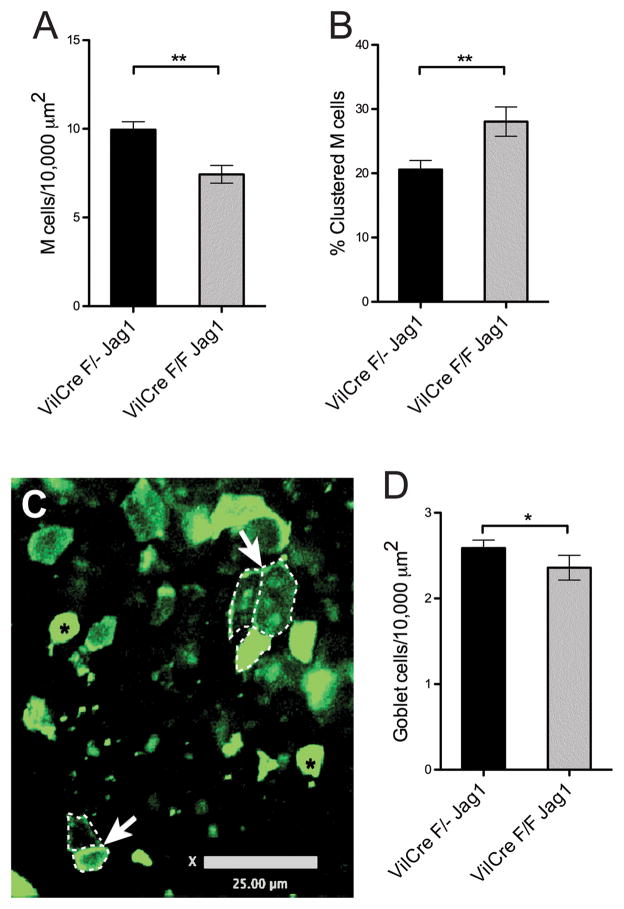
We examined the development of M cells in mice homozygous for a floxed Jagged1 gene plus the villin-Cre transgene, so that Jagged1 was specifically eliminated only in the intestinal epithelium. As with the floxed Notch mice, we assayed for M cell numbers and distribution. In contrast to the floxed Notch mice, M cell numbers were reduced by about 25% (Figure 3A). However, despite this reduction the proportion of clustered M cells was actually increased (Figure 3B,C), consistent with loss of lateral inhibition. Interestingly, PPFAE goblet cell numbers were also decreased (Figure 3D). Here too, because of parallel decreases in both M cells and goblet cells, it seems unlikely that changes in M cell numbers due to loss of Jagged1 signaling can be explained by alterations in M cell morphology. Thus, the expression of Jagged1 in PPFAE appears to be involved in the control of M cell numbers with additional effects on goblet cells, and may also mediate lateral inhibition effects to limit M cell clustering.
Figure 3. FloxJagged1 Peyer’s patches have fewer M cells but increased clustering.
A. Direct quantitation of M cell density across Peyer’s patch epithelium showed that, in contrast to Notch1 conditional knockout mice, there were reduced numbers in Jagged1 conditional knockout mice compared to wild type. (single flox Jag1 N=32; FloxJag1 N=32;**, P<0.005)
B. Quantitation of M cell clustering (measured as described for Notch1 conditional knockout). Despite the decrease in M cell density, Jagged1 conditional knockout Peyer’s patch epithelium showed increased M cell clustering. (single flox Jag1 N=32; FloxJag1 N=32; **, P<0.005)
C. Confocal image of FloxJagged1 Peyer’s patch whole mount showing M cell clustering (outlined by dotted lines, arrows). Two goblet cells are identified with black asterisks. (Scale bar: 25 μm)
D. Goblet cell density counts showed a slight reduction in Jagged1 conditional knockout Peyer’s patch compare to wild type, though not as strong as the reduction in M cell density. (single flox Jag1 N=32; FloxJag1 N=32; *, P<0.05)
3.3. Jagged1 and CD137 are coordinately regulated in a cell culture model of M cell gene expression
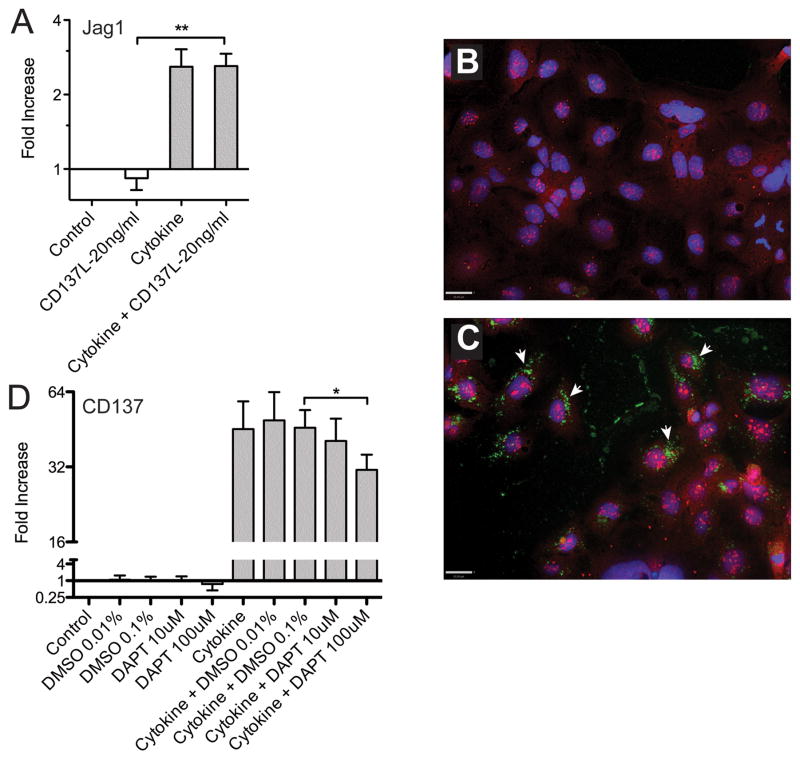
Our studies in vivo suggested that while Notch signaling has an inhibitory effect on M cell numbers and clustering, Jagged1 has paradoxical inhibitory effects on clustering but positive effects on M cell numbers. These results raised the possibility that Jagged1 has both cis and trans activity, so we examined possible gene interactions in a cell culture model of M cell associated gene regulation. In earlier studies on a Caco-2 co-culture model of M cell-like induction, we found that Jagged1 transcripts were induced (25), so we also studied Jagged1 expression in a more recent study on the induction of M cell associated genes. We recently reported that a combination of agonists for the TNFα receptors and the LTβR induced upregulation of PPFAE and M cell associated genes in the intestinal epithelium cell line Caco-2BBe (27). Among the induced genes was CD137, a member of the TNFR superfamily gene CD137 (27; 34), which proved to be required for M cell functional development but not lineage commitment in vivo. In this context, we also found a consistent 2–3-fold increase in Jagged1 expression similar to the level of induction in the Caco-2 coculture studies (Figure 4A). Under similar conditions, strong induction of CD137 was also evident (Figure 4B-D). Jagged2 induction was less than 1.5-fold (not shown). In immunohistochemical analysis of the Caco-2BBe cells (Figure 4B,C), Jagged1 protein was already evident in untreated cells, so upregulation was subtle. It should be noted that expression of Jagged1 in Caco-BBe cells is consistent with studies suggesting that freshly passaged Caco-2 cells resemble crypt cells both in terms of their initial lack of brush border microvilli and patterns of gene expression (35–37). The staining for Jagged1 was distributed in the nucleus, cytoplasm and in part also on the cell membrane, while CD137 was found in cytoplasmic vesicles as previously reported (27). Both Jagged1 and CD137 were detected in the same cells, consistent with cis interactions; however, CD137 was found in cytoplasmic vesicles that did not co-localize with Jagged1.
Figure 4. Studies on Caco-2BBe cell gene expression show coordinate regulation of M cell genes Jagged1 and CD137.
A. TNFα plus LTβR agonist (“Cytokine”: 100ng/ml TNFα and 5μg/ml of LTβR agonist antibody) induced upregulation of Jagged1 with no additional influence of soluble CD137L agonist. (t-test analysis; **, P<0.005)
B. Confocal images of untreated Caco-2BBe cells stained for CD137 (green) and Jagged1 (red). Nuclei are blue. (Scale bar: 30 μm)
C. Confocal images of cytokine treated Caco-2BBe cells shows upregulation of CD137 (arrows), but there is no co-localization of CD137 (green) with Jagged1 (red). (Scale bar: 30 μm)
D. Cytokine treatment of Caco-2BBe cells induces expression of CD137, but expression shows a dose-dependent inhibition by the Notch signaling inhibitor DAPT. One tailed paired t- test comparing DMSO control to 100μm DAPT was significant (P<0.05).
To determine whether CD137 and Jagged1-Notch signaling are connected, we tested the importance of Notch signaling in cytokine treated Caco-2BBe cells (Figure 4D). Inhibition of Notch signaling by the use of the gamma-secretase inhibitor DAPT resulted in a slight dose-dependent decrease in CD137 induction by cytokines. Thus, it appears that at least in the context of cytokine-dependent induction of M cell associated genes, Notch signaling promotes rather than inhibits the M cell phenotype. It is possible that constitutive Jagged1 expression by these cells drives persistent cis-activation of Notch and boosts the cytokine-induced CD137 expression; this contribution was only revealed by the DAPT inhibition of Notch. Indeed, treatment with soluble Jagged1 did not induce additional CD137 expression (not shown). By contrast, treatment of cytokine-treated cells with CD137L showed no consistent effect on Jagged1 expression (not shown). Thus, Notch signaling appears to have an influence on M cell-associated gene expression in these homogeneous cultures.
4. Discussion
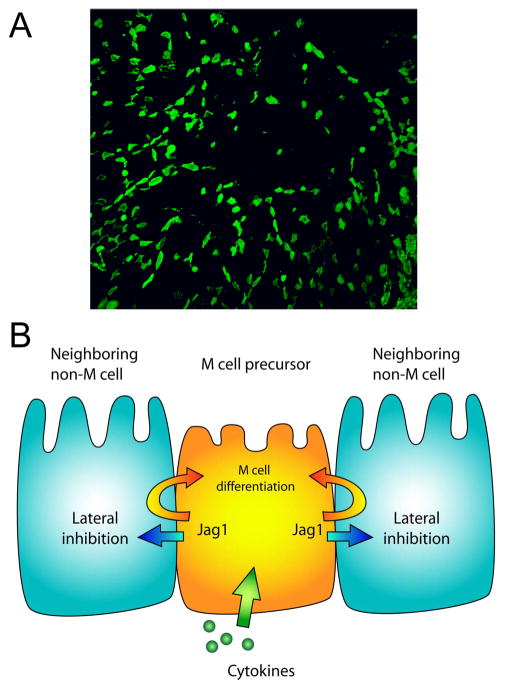
Our studies provide evidence that Jagged1 and Notch influence PPFAE M cell numbers and distribution by regulating M cell development at an early stage within the crypts adjacent to the Peyer’s patch follicle. While it is unclear what factors cause the initial commitment of crypt stem cells to M cell versus enterocyte phenotypes, the present data suggest that the eventual output of M cells from the crypt is subject to editing through signals such as Jagged1-Notch interactions. The effect of Notch signaling appears to be complex and context-dependent, as the loss of Jagged1 suggests the possibility of both trans-inhibitory and cis-inducing effects on M cells. Consistent with this dual role, preliminary analysis of mice with intestinal epithelium expression of a constitutively active human Notch cytoplasmic domain showed no significant effect on PPFAE M cell numbers (not shown); here it is likely that the Notch signaling was both inhibitory on some cells yet reinforcing in others, resulting in a balanced effect on total M cell numbers. The possibility of simultaneous trans-inhibitory and cis-inducing functions of Jagged1 in the editing of PPFAE M cells is consistent with studies on other Notch ligands; for example, cell-autonomous Delta-Notch signaling has been implicated in Drosophila hair bristle formation (38). Considered in aggregate, the effects of Notch signaling appear to insure the scattered distribution of M cells across the PPFAE (Figure 5), a necessarily dynamic function in the face of continuous regeneration of the short-lived Peyer’s patch epithelial cells.
Figure 5. Model of Jagged-Notch interactions and M cell patterns in Peyer’s patch epithelium.
A. A low magnification view of the overall distribution of M cells (green) across Peyer’s patch follicle epithelium. Note that at this low magnification, dispersion of M cells derived from crypt stem cell progenitors is evident, though the rare M cell clusters are not as visible.
B. Simplified cartoon model of M cell editing by Jagged1 and Notch interactions, indicating a possible role for Jagged1 in both lateral inhibition of M cell development and cis-reinforcement of M cell lineage decisions.
If we view the distributed array of M cells across the PPFAE as a type of sensory organ with a defined tissue pattern (Figure 5A), then Jagged1 and Notch are appropriate candidates for regulating intestinal crypt production of M cells. A regulated M cell distribution could have several benefits. First, the full surface area of the follicle epithelium would be used to optimum efficiency, with optimum distribution of M cell-specific capture receptors such as gp2 (39). In addition, the dendritic cells underlying the follicle epithelium would all have similar opportunity to take up antigens transcytosed by the M cells and present them to nearby interfollicular zone T lymphocytes. Second, because M cells have a basolateral pocket containing B lymphocytes, the dispersal of M cells may minimize the disadvantages of epithelial cells with reduced basement membrane contacts and potential for loss of epithelial integrity and barrier function.
A third potential benefit of dispersed M cells was raised in our recent studies on particle uptake by Nasal Associated Lymphoid Tissue M cells (40). We found that the ionic strength of the dispersion buffer affected M cell-dependent uptake, suggesting a role for electrostatic forces in M cell function. Since cell membranes and biological particles (e.g., bacteria and viruses) are nearly always negatively charged, electrostatic repulsion between the membranes and particles would minimize direct interactions. However, the smooth (“microfold”) apical membranes of M cells may have lower surface charge relative to adjacent enterocytes with extensive microvilli, so electrostatic forces might drive particles toward the M cell membranes. Thus, dispersed M cells surrounded by microvilli-covered enterocytes may be most effective in taking advantage of both long range electrostatic forces and short range interactions between capture receptors and target ligands.
The contrast between intestinal villus and Peyer’s patch epithelium organization of specialized cell types is striking in view of the common contribution of crypt stem cells to both. We found that while Notch signaling clearly regulates the production of both goblet cells and M cells, it is the local environment (villus vs PPFAE) that determines whether the main non-enterocyte produced is a goblet cell or M cell. That is, the proximity to the Peyer’s patch provides the context that promotes the generation of M cells rather than goblet cells. In addition, cis-signaling may provide yet additional specificity in a binary choice between goblet versus M cell phenotype; a speculative hypothesis is that Jagged1 helps support the M cell lineage while Delta-like 1 provides cis-signaling for nascent goblet cells. In pathological settings such as inflammatory bowel disease, these context-dependent contrasts may be important determinants of whether the local crypts are induced to provide additional goblet cells or M cells.
Highlights.
Notch and Jagged1 genes were floxed in intestinal epithelium
Floxed Notch1 caused a doubling of villous goblet cell numbers
Floxed Notch1 increased both Peyer’s patch M cell numbers and clustering
Floxed Jagged1 decreased M cell numbers but increased clustering
In vitro, M cell genes Jagged1 and CD137 were coordinately induced
Acknowledgments
The authors thank Andrea Saraswati for assistance with histology. This work was supported by the National Institutes of Health (R01 grant AI063426 and R21 grant AI073689 to DDL)
ABBREVIATIONS
- PPFAE
Peyer’s patch follicle associated epithelium
- Dll1
Delta-like 1
- UEA-1
Ulex Europeus Agglutinin-1
- PGRP-S
Peptidoglycan Recognition Protein-S
Footnotes
Conflicts: The authors have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 3.Gazave E, Lapebie P, Richards G, Brunet F, Ereskovsky A, Degnan B, Borchiellini C, Vervoort M, Renard E. Origin and evolution of the Notch signalling pathway: an overview from eukaryotic genomes. BMC Evolutionary Biology. 2009;9:249. doi: 10.1186/1471-2148-9-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng P, Zhou J, Gabrilovich D. Regulation of dendritic cell differentiation and function by Notch and Wnt pathways. Immunol Rev. 2010;234:105–119. doi: 10.1111/j.0105-2896.2009.00871.x. [DOI] [PubMed] [Google Scholar]
- 5.Wong GW, Zúñiga-Pflücker JC. gammadelta and alphabeta T cell lineage choice: resolution by a stronger sense of being. Semin Immunol. 2010;22:228–236. doi: 10.1016/j.smim.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi E, Chiba S, Kumano K, Kunisato A, Takahashi T, Takahashi T, Hirai H. Expression of Notch ligands, Jagged1, 2 and Delta1 in antigen presenting cells in mice. Immunol Lett. 2002;81:59–64. doi: 10.1016/s0165-2478(01)00326-1. [DOI] [PubMed] [Google Scholar]
- 7.Yuan JS, Kousis PC, Suliman S, Visan I, Guidos CJ. Functions of notch signaling in the immune system: consensus and controversies. Annu Rev Immunol. 2010;28:343–365. doi: 10.1146/annurev.immunol.021908.132719. [DOI] [PubMed] [Google Scholar]
- 8.Roignant J-Y, Treisman JE. Pattern formation in the Drosophila eye disc. Int J Dev Biol. 2009;53:795–804. doi: 10.1387/ijdb.072483jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133:1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- 10.Eddison M, Le Roux I, Lewis J. Notch signaling in the development of the inner ear: lessons from Drosophila. Proc Natl Acad Sci USA. 2000;97:11692–11699. doi: 10.1073/pnas.97.22.11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiernan AE, Ahituv N, Fuchs H, Balling R, Avraham KB, Steel KP, Hrabé de Angelis M. The Notch ligand Jagged1 is required for inner ear sensory development. Proc Natl Acad Sci USA. 2001;98:3873–3878. doi: 10.1073/pnas.071496998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiernan AE, Xu J, Gridley T. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet. 2006;2(1):e4. doi: 10.1371/journal.pgen.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akiyama J, Okamoto R, Iwasaki M, Zheng X, Yui S, Tsuchiya K, Nakamura T, Watanabe M. Delta-like 1 expression promotes goblet cell differentiation in Notch-inactivated human colonic epithelial cells. Biochem Biophys Res Commun. 2010;393:662–667. doi: 10.1016/j.bbrc.2010.02.048. [DOI] [PubMed] [Google Scholar]
- 14.Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 15.Stamataki D, Holder M, Hodgetts C, Jeffery R, Nye E, Spencer-Dene B, Winton DJ, Lewis J. Delta1 Expression, Cell Cycle Exit, and Commitment to a Specific Secretory Fate Coincide within a Few Hours in the Mouse Intestinal Stem Cell System. PLoS ONE. 2011;6(9):e24484. doi: 10.1371/journal.pone.0024484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 17.Zecchini V, Domaschenz R, Winton D, Jones P. Notch signaling regulates the differentiation of post-mitotic intestinal epithelial cells. Genes & Development. 2005;19:1686–1691. doi: 10.1101/gad.341705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraehenbuhl JP, Neutra MR. Epithelial M cells: differentiation and function. Annu Rev Cell Dev Biol. 2000;16:301–332. doi: 10.1146/annurev.cellbio.16.1.301. [DOI] [PubMed] [Google Scholar]
- 19.Neutra MR, Frey A, Kraehenbuhl JP. Epithelial M cells: gateways for mucosal infection and immunization. Cell. 1996;86:345–348. doi: 10.1016/s0092-8674(00)80106-3. [DOI] [PubMed] [Google Scholar]
- 20.Neutra MR, Pringault E, Kraehenbuhl JP. Antigen sampling across epithelial barriers and induction of mucosal immune responses. Annu Rev Immunol. 1996;14:275–300. doi: 10.1146/annurev.immunol.14.1.275. [DOI] [PubMed] [Google Scholar]
- 21.Bye WA, Allan CH, Trier JS. Structure, distribution, and origin of M cells in Peyer’s patches of mouse ileum. Gastroenterology. 1984;86:789–801. [PubMed] [Google Scholar]
- 22.Gebert A, Posselt W. Glycoconjugate expression defines the origin and differentiation pathway of intestinal M-cells. J Histochem Cytochem. 1997;45:1341–1350. doi: 10.1177/002215549704501003. [DOI] [PubMed] [Google Scholar]
- 23.Lelouard H, Sahuquet A, Reggio H, Montcourrier P. Rabbit M cells and dome enterocytes are distinct cell lineages. J Cell Sci. 2001;114:2077–2083. doi: 10.1242/jcs.114.11.2077. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Gusti V, Saraswati A, Lo DD. Convergent and Divergent Development among M Cell Lineages in Mouse Mucosal Epithelium. J Immunol. 2011;187:5277–5285. doi: 10.4049/jimmunol.1102077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lo D, Tynan W, Dickerson J, Scharf M, Cooper J, Byrne D, Brayden D, Higgins L, Evans C, O’Mahony DJ. Cell culture modeling of specialized tissue: identification of genes expressed specifically by follicle-associated epithelium of Peyer’s patch by expression profiling of Caco-2/Raji co-cultures. Int Immunol. 2004;16:91–99. doi: 10.1093/intimm/dxh011. [DOI] [PubMed] [Google Scholar]
- 26.Sander GR, Powell BC. Expression of notch receptors and ligands in the adult gut. J Histochem Cytochem. 2004;52:509–516. doi: 10.1177/002215540405200409. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Lopez-Fraga M, Rynko A, Lo DD. TNFR and LTbetaR agonists induce follicle-associated epithelium and M cell specific genes in rat and human intestinal epithelial cells. Cytokine. 2009;47:69–76. doi: 10.1016/j.cyto.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roozendaal R, Mebius RE. Stromal cell-immune cell interactions. Annu Rev Immunol. 2011;29:23–43. doi: 10.1146/annurev-immunol-031210-101357. [DOI] [PubMed] [Google Scholar]
- 29.Anderle P, Rumbo M, Sierro F, Mansourian R, Michetti P, Roberts MA, Kraehenbuhl JP. Novel markers of the human follicle-associated epithelium identified by genomic profiling and microdissection. Gastroenterology. 2005;129:321–327. doi: 10.1053/j.gastro.2005.03.044. [DOI] [PubMed] [Google Scholar]
- 30.Rumbo M, Sierro F, Debard N, Kraehenbuhl JP, Finke D. Lymphotoxin beta receptor signaling induces the chemokine CCL20 in intestinal epithelium. Gastroenterology. 2004;127:213–223. doi: 10.1053/j.gastro.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 31.el Marjou F, Janssen KP, Chang BHJ, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. Tissue-specific and inducible Cre- mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 32.Ezzell RM, Chafel MM, Matsudaira PT. Differential localization of villin and fimbrin during development of the mouse visceral endoderm and intestinal epithelium. Development. 1989;106:407–419. doi: 10.1242/dev.106.2.407. [DOI] [PubMed] [Google Scholar]
- 33.Clark MA, Jepson MA, Simmons NL, Booth TA, Hirst BH. Differential expression of lectin-binding sites defines mouse intestinal M-cells. J Histochem Cytochem. 1993;41:1679–1687. doi: 10.1177/41.11.7691933. [DOI] [PubMed] [Google Scholar]
- 34.Hsieh EH, Fernandez X, Wang J, Hamer M, Calvillo S, Croft M, Kwon BS, Lo DD. CD137 Is Required for M Cell Functional Maturation but Not Lineage Commitment. Am J Pathol. 2010;177:666–676. doi: 10.2353/ajpath.2010.090811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mariadason JM, Arango D, Corner GA, Arañes MJ, Hotchkiss KA, Yang W, Augenlicht LH. A gene expression profile that defines colon cell maturation in vitro. Cancer Res. 2002;62:4791–4804. [PubMed] [Google Scholar]
- 36.Tremblay E, Auclair J, Delvin E, Levy E, Ménard D, Pshezhetsky AV, Rivard N, Seidman EG, Sinnett D, Vachon PH, Beaulieu J-F. Gene expression profiles of normal proliferating and differentiating human intestinal epithelial cells: a comparison with the Caco-2 cell model. J Cell Biochem. 2006;99:1175–1186. doi: 10.1002/jcb.21015. [DOI] [PubMed] [Google Scholar]
- 37.Tadjali M, Seidelin JB, Olsen J, Troelsen JT. Transcriptome changes during intestinal cell differentiation. Biochim Biophys Acta. 2002;1589:160–167. doi: 10.1016/s0167-4889(02)00170-2. [DOI] [PubMed] [Google Scholar]
- 38.Jacobsen Tl, Brennan K, Arias AM, Muskavitch MA. Cis-interactions between Delta and Notch modulate neurogenic signalling in Drosophila. Development. 1998;125:4531–4540. doi: 10.1242/dev.125.22.4531. [DOI] [PubMed] [Google Scholar]
- 39.Hase K, Kawano K, Nochi T, Pontes GS, Fukuda S, Ebisawa M, Kadokura K, Tobe T, Fujimura Y, Kawano S, Yabashi A, Waguri S, Nakato G, Kimura S, Murakami T, Iimura M, Hamura K, Fukuoka SI, Lowe AW, Itoh K, Kiyono H, Ohno H. Uptake through glycoprotein 2 of FimH(+) bacteria by M cells initiates mucosal immune response. Nature. 2009;462:226–230. doi: 10.1038/nature08529. [DOI] [PubMed] [Google Scholar]
- 40.Rajapaksa TE, Bennett KM, Hamer M, Lytle C, Rodgers VGJ, Lo DD. Intranasal M cell uptake of nanoparticles is independently influenced by targeting ligands and buffer ionic strength. J Biol Chem. 2010;285:23739–23746. doi: 10.1074/jbc.M110.126359. [DOI] [PMC free article] [PubMed] [Google Scholar]