Abstract
The cell extract of a cultured terrestrial Nostoc sp. (UIC 10062), obtained from a sample collected at Grand Mere State Park in Michigan, displayed antiproliferative activity against the HT-29 human colon cancer cell line. Bioactivity-guided fractionation of the cell extract, combined with LC-MS analysis, led to the isolation of two cyclophanes, named merocyclophanes A and B (1 and 2). Their structures were determined by various spectroscopic techniques including HRESIMS, and 1D and 2D NMR analyses. The stereoconfiguration was assigned on the basis of X-ray crystallographic and CD analyses. The structures of merocyclophanes A and B (1 and 2) established a hitherto unknown [7.7]paracyclophane skeleton in nature, as characterized by α-branched methyls at C-1/14. Merocyclophanes A and B (1 and 2) displayed antiproliferative activity against the HT-29 human colon cancer cell line with IC50 values of 3.3 and 1.7 µM, respectively.
Keywords: Nostoc sp., Cyanobacterium, Antiproliferative activity, HT-29 human colon cancer cells, [7.7]paracyclophanes
1. Introduction
Cyanobacteria (blue-green algae) have been shown to be prolific producers of bioactive secondary metabolites (Tan, 2007; Wagoner et al., 2004; Harada, 2004). Several terrestrial cyanobacterial species belonging to the order Nostocales have been reported to produce naturally occurring paracyclophanes. Of these, nostocyclyne A, an acetylenic [14]paracyclophane, was isolated from the natural bloom of Nostoc sp. and displayed antimicrobial activity against Gram-positive bacteria S. aureus and B. subtilis (Ploutno et al., 2000). Additionally, cylindrocyclophanes and nostocyclophanes, the two classes of cyanobacterial [7.7]paracyclophanes isolated from the cultured Nostoc sp. and Cylindrospermum sp., have exhibited a broad spectrum of biological activities, including antibacterial, antifungal and cytotoxic activities (Moore et al., 1990; Chen et al., 1991; Moore et al., 1992; Bui et al., 2007; Chlipala et al., 2010). As regards their formation, the polyketide biosynthetic pathway to these natural cyclophanes was determined by an isotope precursor administration experiment, and involves dimerization of two acetate-derived nonaketides and subsequent modification by chlorination, oxidation and/or methylation, resulting in diverse chemical structures (Bobzin et al., 1993).
Recently, several cyclindrocyclophanes were reported from a terrestrial Nostoc sp. (UIC 10022A) obtained from the material collected in the city of Chicago. These compounds displayed inhibitory activity against the 20S proteasome and were found to be cytotoxic against cancer cell lines, including HT-29, NCI-H460, SF268 and MCF7 cells (Chlipala et al., 2010). Herein, structure elucidation and biological activity of two additional cyclophanes, named merocyclophanes A and B (1 and 2), that were isolated from a second Nostoc sp. (UIC 10062) are reported. This strain was obtained from a sample collected at Grand Mere State Park in Michigan, and the merocyclophanes were named in recognition of the collection site. Their structures were determined using various spectroscopic methods including HRESIMS, and 1D and 2D NMR analyses. The stereoconfiguration was assigned by a combination of X-ray crystallographic and CD analyses, and the structures of merocyclophanes A and B (1 and 2) established a new [7.7]paracyclophane carbon skeleton, as characterized by the presence of α-branched methyls at C-1/14.
2. Results and Discussion
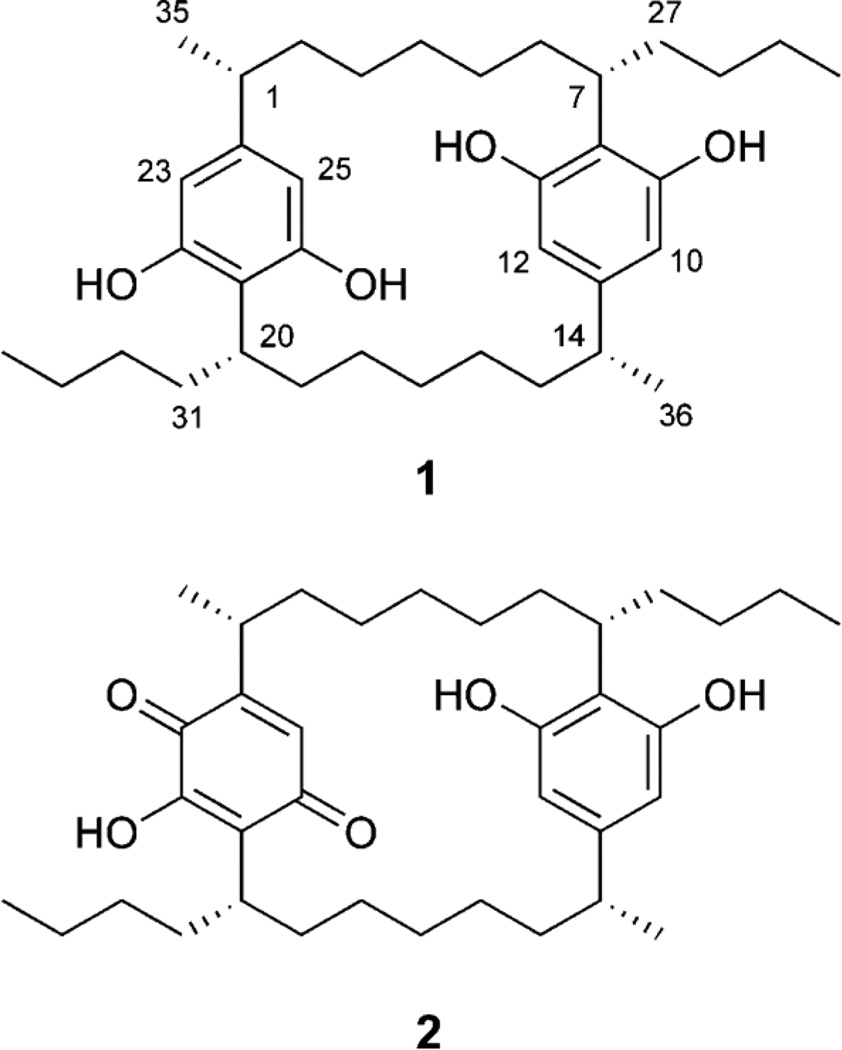
Nostoc sp. (UIC 10062) was isolated from a sample collected at Grand Mere State Park in Michigan, and cultured in Z media (Falch et al., 1995). The freeze-dried cells were extracted with a mixture of CH2Cl2 and MeOH (1:1, v/v) and dried in vacuo, with the resulting extract (156.6 mg) subjected to bioassay-guided fractionation. This cell extract showed antiproliferative activity against the HT-29 human colon cancer cell line (IC50 13.1 µg/mL), and was fractionated using Diaion HP-20 resin and an increasing amount of iPrOH in H2O. Fractions eluting at 70 (5.0 mg) and 80 (6.5 mg) % iPrOH were found to be active, with IC50 values of 0.9 and 1.2 µg/ml, respectively. LC-MS and UV analyses of the active fractions also indicated the presence of two cyclophanes with molecular weights of 552 and 566. Reversed-phase HPLC of these fractions yielded merocyclophanes A (1, 2.4 mg, 0.11%) and B (2, 0.9 mg, 0.04%) as minor compounds (Fig. 1).
Fig. 1.
Structures of merocyclophanes A (1) and B (2).
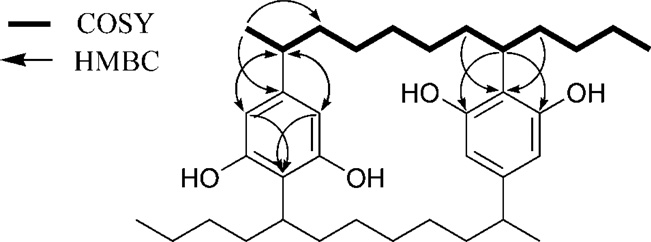
Merocyclophane A (1) was obtained as white amorphous powder, and the molecular formula was determined as C36H56O4 by HRESIMS analysis (m/z 551.4170 [M-H]•). The total numbers of proton and carbon signals, determined by analysis of 1H and 2D NMR spectra, were only half of those required by the molecular formula, thus indicating the presence of C2 axis of symmetry in 1. The structure of 1 was elucidated by analysis of 2D NMR spectra including COSY, HSQC and HMBC (Fig. 2). The appearance of only two aromatic singlets (H-10/23, δH 6.04 and H-12/25, δH 6.00) indicated the presence of two tetrasubstituted aromatic rings. The carbon chemical shifts of C-9/22 (δC 158.5) and C-13/26 (δC 156.9), together with HMBC correlations from H-7/20 (δH 3.10) to C-9/22 (δC 158.5) and C-13/26 (δC 156.9), identified these partial structures as two 2,5-dialkylresorcinol moieties. Sequential COSY correlations from H1/14 to H7/20 combined with HMBC correlations from H-10/23 (δH 6.04) and H-12/25 (δH 6.00) to C-1/14 (δC 41.8) and from H-7/20 (δH 3.10) to C-9/22 (δC 158.5) and C-13/26 (δC 156.9) completed the [7.7]paracyclophane core. Additional sequential COSY correlations from the H-7/20 to the triplet methyls H3-30/34 via three methylenes (H2-27/31, H2-28/32 and H2-29/33) further expanded the carbon chains. Structure determination was completed by an HMBC correlation from the doublet methyl (H3-35/36, δH 1.15) to the aromatic carbon (C-11/24, δC 146.6) combined with a COSY correlation between H-1/14 (δH 2.30) and H-35/36 (δH 1.15), placing methyl groups at C-1/14.
Fig. 2.
Key 2D correlations used for structure determination of 1
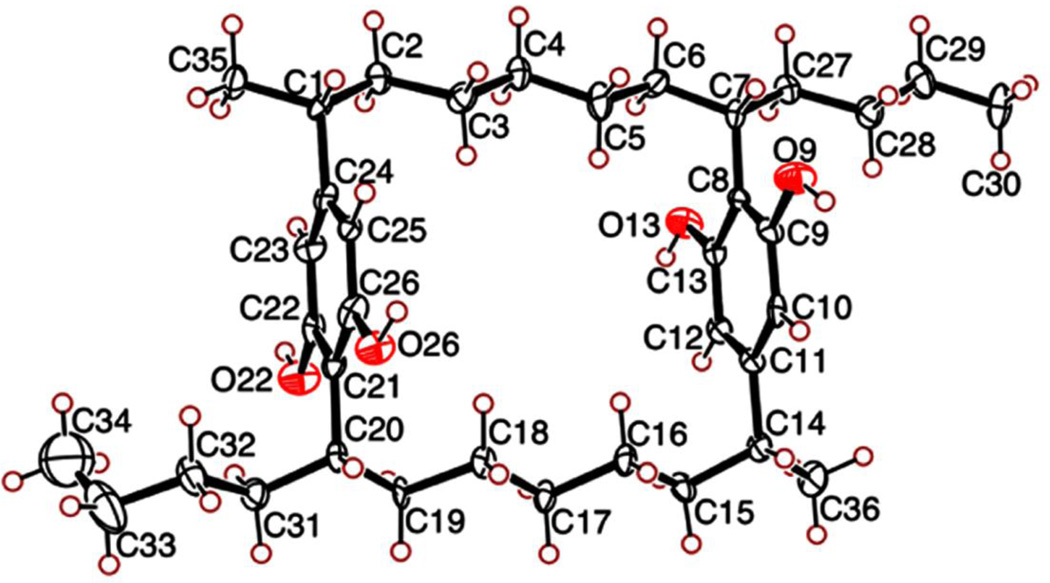
Structure 1 established the presence of four stereogenic centers, whose stereoconfiguration was determined by a combination of X-ray crystallographic and CD spectral analyses. Merocyclophane A (1) was crystallized from CH3CN. Single-crystal X-ray analysis established the relative configuration of 1 as shown in Fig. 3. The absolute configurations at C-1/14 and C-7/20 were established by comparison of the CD spectrum of 1 with those reported for the nostocyclophanes (Chen et al., 1991). Cotton effects observed between 220 ~ 230 nm and between 270 ~ 280 nm arise from π - π * transitions of a benzene chromophore. According to the benzene sector rule, the chlorine-bearing stereogenic carbons in the nostocyclophanes, three carbons away from the benzene chromophore, should not affect Cotton effects in these regions (Smith, 1998). The CD spectrum of 1 exhibited negative Cotton effects at 227 (Δε, −4.35) and 277 (Δε, −2.73), similar to those observed for the nostocyclophanes, suggesting the same absolute configuration. Therefore, the absolute configurations of the four stereogenic carbons C-1/14 and C-7/20 in 1 were assigned as “R” and “S”, respectively.
Fig. 3.
ORTEP drawing of merocyclophane A (1)
Merocyclophane B (2) was obtained as purple amorphous powder, and the HRESIMS data (m/z 565.3954 [M-H]•) established the molecular formula as C36H54O5. The 1H NMR spectrum of 2 closely resembled 1, except for one of the two aromatic rings. This resulted in an unsymmetric structure. The presence of a hydroxyquinone in 2 was suggested by analysis of the UV spectrum. A chromophore corresponding to the quinone absorption was observed at 521 nm, which shifted to 413 nm in the presence of acid (0.04 v/v % TFA in MeOH). This was consistent with the observed color change from deep purple to yellow upon addition of TFA. The appearance of the down-fielded carbon chemical shift of C-23 (δC 132.6 compared to δC 104.5 in 1) further supported the presence of the hydroxyquinone moiety in 2. However, significant line broadening of NMR signals was observed in the hydroxyquinone moiety, causing a number of expected correlations to be absent in the HMBC spectrum, even at the elevated temperature (343 K). Thus, the carbon chemical shifts of C-21, C-22, C-25 and C-26 could not be detected. The presence of the unsymmetrical hydroxyquinone moiety in 2 raised the possibility of two conformations arising from hindered rotation around the C1–C24 and C20–C21 bonds, resulting in the formation of two diastereotopic atropisomers. A torsion energy scan carried out using MM3 force-field calculation showed high rotational energy barriers between the two possible atropisomers (see Supplementary data). However, a slow conversion of 1 into 2 was observed during prolonged storage, indicating 2 to be an oxidation artifact of 1. This non-enzymatic oxidation suggests that both atropisomers are present in a nearly equal amount, which is supported by the identical CD spectrum observed between 1 and 2. The stereoconfiguration of 2 (C-1/14, C-7/20) was determined by comparison of the CD spectrum with that of 1. Negative Cotton effects observed at 227 (Δε, −2.22) and 274 (Δε, −2.03) nm indicated the same absolute configuration of 2 as determined for 1.
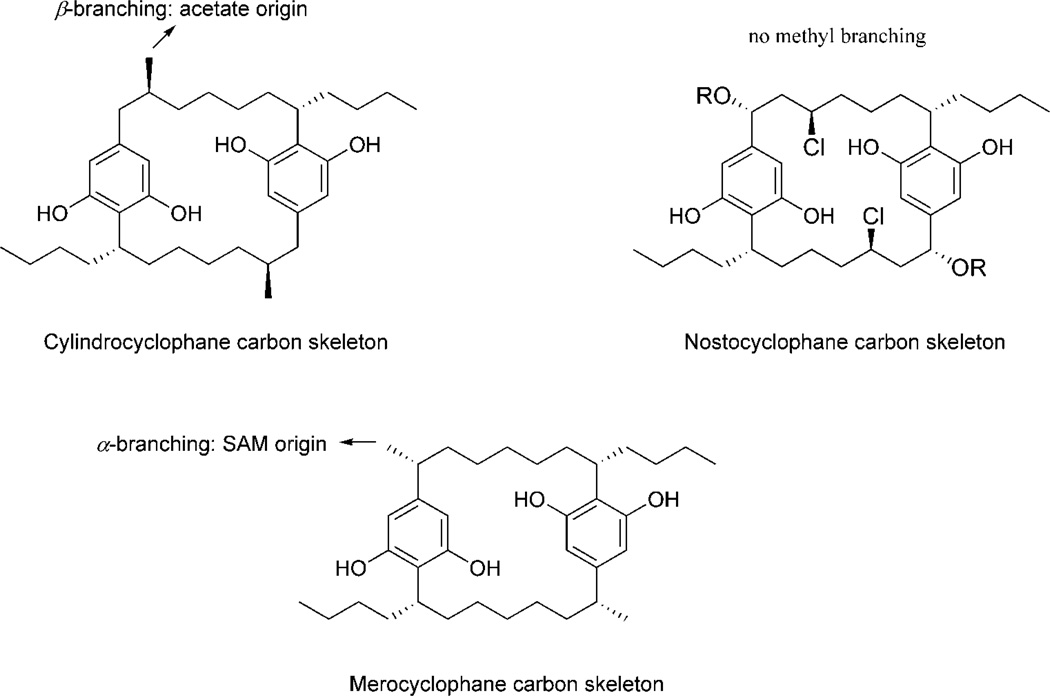
The structures of merocyclophanes A and B (1 and 2) differ from the previously described cylindrocyclophanes and nostocyclophanes by the presence of α-branched methyls at C-1/14 as shown in Fig. 4, whereas cylindrocyclophanes have branched methyls at C-2/15 (β-branching). Isotope precursor administration experiments performed by Bobzin et al. indicated the acetate origin of these β-branched methyl groups (Bobzin et al., 1993). However, the C-methylation in the merocyclophanes occurred at α-position, suggesting that these methyl groups are likely derived from SAM (S-adenosyl methionine) (Jones et al, 2010).
Fig. 4.
Comparison of carbon skeletons between the cylindrocyclophanes, the nostocyclophanes and merocyclophane A
Merocyclophanes A and B (1 and 2) were tested for their antiproliferative activity against the HT-29 human colon cancer cell line. Both 1 and 2 displayed antiproliferative activity with IC50 values of 3.3 µM and 1.7 µM, respectively. Merocyclophanes A and B (1 and 2) possessed similar levels of antiproliferative activity as those reported for cylindrocyclophanes A–F against various cell lines (0.5 – 5 µg/mL). A SAR study on cyclindrocyclophane A suggested that the 2,5-dialkylresorcinol moiety was the pharmacophore needed for the antiproliferative activity of cylindrocyclophanes, and that the activity was significantly enhanced by the presence of [7.7]paracyclophane core structure (Yamakoshi et al, 2009). The structure of merocyclophane A (1) differs from that of cyclindrocyclophane A by the position of methyl groups and the lack of alkyl hydroxy groups; however, both compounds showed a similar level of activity. This indicates that replacement of the hydroxyl groups at C-1/14 or C-2/15 by methyl groups does not significantly change the antiproliferative activity of natural [7.7]paracyclophanes.
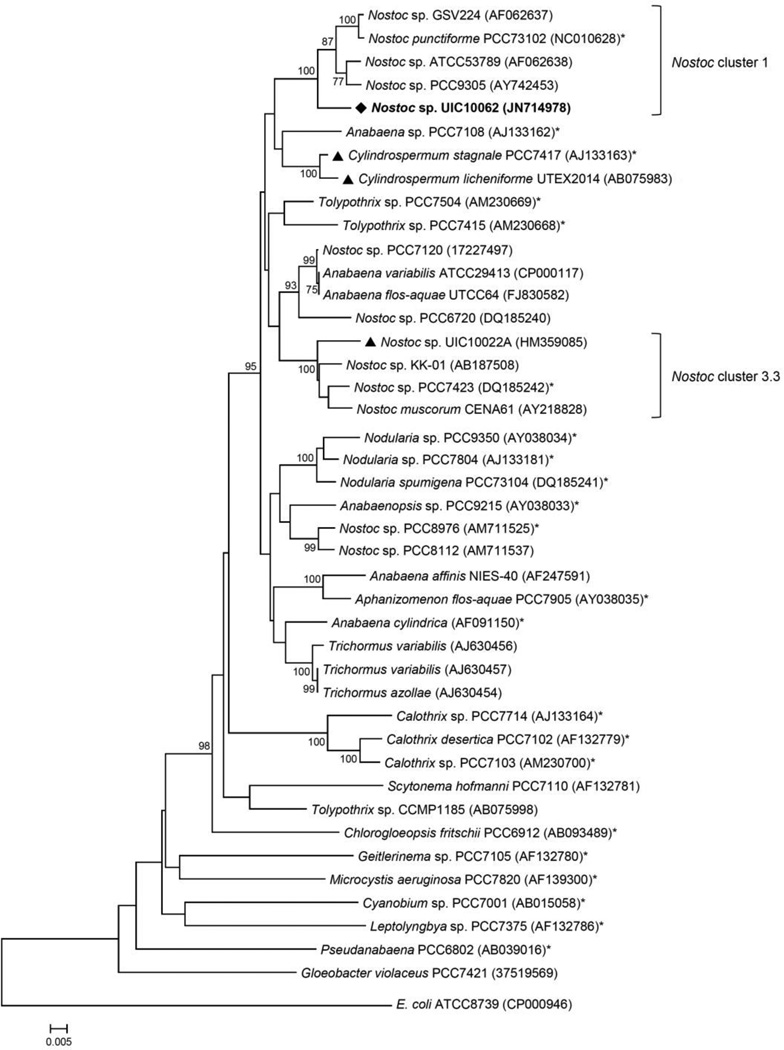
Taxonomic identification of the strain UIC 10062 was conducted on the basis of microscopic examination and phylogenetic analysis using a 16S rRNA gene sequence. Microscopic observation of the cultured UIC 10062 suggested this strain to be a Nostoc sp. (see Supplementary data) (Wehr et al., 2003; Castenholz et al., 2001). For phylogenetic analysis, a nearly 1.3 kb sequence of the partial PCR-amplified 16S rRNA gene of UIC 10062 was aligned with the 16S rRNA gene sequences of Bergey’s reference strains and other related species retrieved from GenBank. Phylogenetic trees were constructed using three different methods (neighbor-joining, minimum evolution and maximum parsimony) and showed nearly identical topology, particularly in the clade that includes UIC 10062, with very similar bootstrap values (the neighbor-joining tree shown in Fig. 5). The resulting phylogenetic tree indicated that UIC 10062 formed a monophyletic clade with Nostoc sp. GSV224, Nostoc sp. ATCC53789, Nostoc sp. PCC9305 and Nostoc punctiforme PCC73102 (the Bergey’s reference strain for Nostoc cluster 1), and thus UIC 10062 was designated within this cluster. The comparison of 16S rRNA gene sequences among three [7.7]paracyclophane-producing cyanobacteria (Nostoc sp. UIC 10062, Nostoc sp. UIC 10022A and Cylindrospermum licheniforme UTEX 2014) showed that variation in 16S rRNA gene sequences did not correlate with chemical variability. The two genetically quite distant strains of Nostoc sp. UIC 10022A and C. licheniforme UTEX 2014 (5% sequence divergence) have been identified to produce cylindrocyclophanes. However, the 16S rRNA gene sequence of C. licheniforme UTEX 2014 showed a higher sequence homology to Nostoc sp. UIC 10062 (96%) than to UIC 10022A (95%). Merocyclophanes produced by Nostoc sp. UIC 10062 have α-branched methyls at C-1/14, which are biosynthetically distinct from the β-branched methyls found in the cyclindrocyclophanes. Also, among these three strains, only Nostoc sp. UIC 10022A was known to produce chlorinated cyclophanes. Considering the quite distant phylogenetic relationships of 16S rRNA genes among cyclophane-producing species, it can be hypothesized that there has been a potential horizontal gene transfer between species, and these genes have evolved to have different modification patterns due to different environmental conditions.
Fig. 5.
Phylogenetic relationships of 16S rRNA genes from cyanobacteria. Evolutionary distances were determined using the neighbor-joining method with 1000 replicate bootstrap resampling to construct the phylogenetic tree. Strains were obtained from NCBI with the accession number given in parentheses. Strains marked with a (*) were obtained as Bergey’s reference strains. Cyanobacterial strains previously reported to produce cylindrocyclophanes are denoted by a triangle (▴). Only bootstrap values greater than or equal to 75% are displayed.
3. Conclusion
Chemical investigation of Nostoc sp. (UIC 10062), guided by antiproliferative assay against the HT-29 human colon cancer cell line, led to isolation of two new [7.7]cyclophanes, named merocyclophanes A (1) and B (2). Their structure determinations established a new carbon skeleton, as characterized by the presence of α-branched methyls at C1/14. Both merocyclophanes A and B displayed antiproliferative activity against the HT-29 human colon cancer cell line. The strain UIC 10062 was taxonomically classified to a Nostoc sp. based on analysis of the microscopic morphology and 16S rRNA gene sequence.
4. Experimental
4.1. General Experimental Procedures
Optical rotations were measured using a Perkin-Elmer 241 polarimeter. UV spectra were measured on a Shimadzu UV spectrometer UV2401 and scanned from 190 to 360 nm. CD spectra were recorded on a JASCO J-710 CD spectrometer. IR spectra were acquired using a Jasco FTIR-410 Fourier transform infrared spectrometer. 1D and 2D NMR spectra were obtained on a Bruker Avance DRX 600 MHz NMR spectrometer with a 5 mm CPTXI Z-gradient. 1H and 13C NMR chemical shifts were referenced to the solvent signals (MeOH-d4 and DMSO-d6). The HMBC spectrum was recorded with the average 3JCH of 8 Hz and the HSQC spectrum was measured with the average 1JCH of 140 Hz. Low- and high-resolution ESI mass spectra were obtained using a Shimadzu IT-TOF LC mass spectrometer.
4.2. Biological Material
Nostoc sp. (UIC 10062) was isolated from a sample collected at the north pond of Grand Mere State Park in Michigan in 2007 (N 42°00.679´, W 86°32.417´). The unialgal strain (UIC 10062) was produced through micropipette isolation techniques (Chlipala et al., 2009). The strain was cultured in a 22 L glass flask containing Z media (18 L) with sterile aeration (Falch et al., 1995). Cultures were illuminated with fluorescent lamps at 1.03 klx with an 18/6 h light/dark cycle. The temperature of the culture room was maintained at 22 °C. After 6 weeks, the biomass of cyanobacteria was harvested by centrifugation and then freeze-dried.
4.3. Strain Identification
Morphological studies were performed using a cultivated cyanobacterium UIC 10062 (See biological material) with microscopic observation for morphological characterization conducted using a Zeiss Axiostar Plus light microscope equipped with a Canon PowerShot A620 camera. The following parameters were selected to characterize its morphology: thallus morphology, shape of trichome, morphology of terminal cells, shape and arrangement of vegetative cells, presence and arrangement of heterocystes and akinetes (see Supplementary data). Taxonomic identification of the cyanobacterial specimen was made in accordance with the modern taxonomic system (Komárek et al., 2003).
4.4. DNA Extraction, PCR Amplification and Sequencing
Cell mass (258 mg), obtained from a static culture of Nostoc sp. (UIC 10062), was centrifuged at 14,000 × g for 5 min, and pretreated with lysozyme and proteinase K as follows prior to using the Wizard Genomic DNA purification kit. The cell pellet was re-suspended in lysis buffer (2.5 mL, 10 mM Tris, 0.1M EDTA, 0.5% w/v SDS, 20 µg/mL pancreatic RNase, pH 8.0) containing lysozyme (1 mg/mL) and incubated at 37°C for 1 hr. To this mixture, proteinase K was added to a final concentration of 100 µg/mL, and incubated at 50°C for 1 h. After incubation, the cell material was recovered by centrifugation at 14,000 × g for 3 min. DNA was extracted from this pretreated cell material using the Wizard Genomic DNA purification kit (Promega), where the protocol was slightly modified to include mechanical disruption after the addition of the nuclei lysis solution (step 6). A portion of the 16S rRNA gene was PCR-amplified using the cyanobacteria-specific primers 106F and 1509R (Martínez-Murcia et al., 1995). For a total volume of 50 µL, the reaction mixture contained DNA (5 µL, approximately 90 ng), Phusion HF Buffer (10 µL, 5x), dNTP mix (1 µL, 10 mM), each primer (1 µL, 10 µM), Phusion high-fidelity DNA polymerase (0.5 µL), and H2O (31.5 µL). The reaction was performed in a Bio-Rad C1000 thermal cycler as following reaction program: initial denaturation for 30 s at 98°C, 35 amplification cycles of 10 s at 98°C, 30 s at 53°C and 30s at 72°C, and a final extension for 10 min at 72°C. PCR products were purified using a MinElute PCR purification kit (Qiagen) and sequenced using the cyanobacteria-specific primers 106F and 1509R as well as the internal primers, 359F and 781R (Nübel et al., 1997). The resulting 16S rRNA gene sequence was deposited in the NCBI GenBank under the accession number JN714978.
4.5. Phylogenetic Analysis
Phylogenetic and molecular evolutionary analyses were conducted using MEGA 5.0 (Tamura et al., 2011). The resulting sequence chromatograms were visually inspected, and the total sequence of 1,290 nucleotides was aligned with 40 cyanobacterial species retrieved from GenBank as well as Gloeobacter violaceus PCC7421 as an outgroup. Cyanobacterial reference strains were firstly selected from Bergey’s Manual (Castenholz, 2001), and only sequences of at least 1 kb were retrieved from GenBank. Multiple sequence alignment was performed with ClustalW in MEGA 5.0 with standard gap opening and extension penalties. The evolutionary history was inferred using the neighbor-joining (NJ), minimum evolution (ME) and maximum parsimony (MP) methods. One thousand replicates were used to evaluate the robustness of branches in the inferred trees generated. Subgroups with greater than 75% consistency in the tree are labeled at the respective nodes.
4.6. Extraction and Isolation
Freeze-dried biomass (2.2 g) from 18 L of culture was harvested and extracted with CH2Cl2-MeOH (250 mL, 1:1) three times and concentrated in vacuo to yield an extract (0.2 g), which was fractionated using Diaion HP-20 resin with iPrOH-H2O (50 mL) to generate 8 sub-fractions (0:10, 2:8, 4:6, 6:4, 7:3, 8:2, 9:1 and 10:0 v/v). Fractions eluting with iPrOH-H2O 7:3 v/v, (5.0 mg) and 8:2 v/v, (6.5 mg) were found to be active in the antiproliferative assay against HT-29 cells with IC50 values of 0.9 and 1.2 µg/ml, respectively. LC-MS analysis of these fractions indicated the presence of new compounds, and were subjected to reversed-phase HPLC (Varian C8 semi-preparative column, 10 mm × 250 mm, 3 mL/min) eluting with gradient of MeOH-H2O (7:3 to 9:1 v/v for 45 min). Merocyclophanes A and B (1 and 2) were eluted at 37.9 min (1, 2.4 mg, 0.11%) and 31.6 min (2, 0.9 mg, 0.04%), respectively.
4.6.1. Merocyclophane A (1)
White amorphous powder; [α]25D – 29 (c 0.024, MeOH); UV (MeOH) λmax (log ε) 207 (4.55), 221 (4.02), 272 (3.24) nm; CD (MeOH) λmax (Δε) 213 (−6.40), 227 (−4.35), 277 (−2.73); IR (neat) νmax 2954, 2920, 2851, 1650, 1541, 1507 cm−1; for 1H and 13C NMR spectroscopic data, see Table 1; HRESIMS m/z 551.4170 [M-H]• (cacld for C36H55O4, 551.4100)
Table 1.
NMR spectroscopic data of merocyclophane A (1) in MeOH-d4 (600 MHz)
| NO. | Merocyclophane A (1) | ||||
|---|---|---|---|---|---|
| δCa | δH | mult. (J in Hz) |
COSY | HMBC | |
| 1 / 14 | 41.8 | 2.30 | dqd (10.8, 6.9, 3.8) | 2/15, 35/36 | 35/36, 10/23, 12/25, 11/24 |
| 2 / 15 | 40.5 | 1.32 | m | 1/14, 3/16 | 1/14, 3/16, 11/24, 35/36 |
| 1.45 | m | ||||
| 3 / 16 | 30.7 | 0.65 | m | 2/15, 4/17 | 1/14, 2/15, 5/18 |
| 0.93 | m | ||||
| 4 / 17 | 32.4 | 0.88 | m | 3/16, 5/18 | |
| 1.27 | m | ||||
| 5 / 18 | 30.6 | 0.67 | m | 4/17, 6/19 | |
| 0.98 | m | ||||
| 6 / 19 | 35.1 | 1.31 | m | 5/18, 7/20 | 7/20 |
| 1.97 | qd (12.4, 4.0) | ||||
| 7 / 20 | 36.7 | 3.10 | m | 6/19, 27/31 | 5/18, 6/19, 8/21, 9/22, 13/26, 27/31, 28/32 |
| 8 / 21 | 116.1 | ||||
| 9 / 22 | 158.5 | ||||
| 10 / 23 | 104.5 | 6.04 | s | 14/1, 8/21, 12/25, 9/22 | |
| 11 / 24 | 146.6 | ||||
| 12 / 25 | 109.1 | 6.00 | s | 14/1, 8/21, 10/23, 13/26 | |
| 13 / 26 | 156.9 | ||||
| 27 / 31 | 34.7 | 1.49 | m | 7/20, 28/32 | 28/32, 29/33, 7/20 |
| 1.92 | m | ||||
| 28 / 32 | 31.6 | 1.11 | m | 27/31, 29/33 | |
| 1.19 | m | ||||
| 29 / 33 | 23.8 | 1.25 | m | 28/32, 30/34 | |
| 1.29 | m | ||||
| 30 / 34 | 14.4 | 0.83 | t (7.1) | 29 / 33 | 28/32, 29/33 |
| 35 / 36 | 23.5 | 1.15 | d (6.9) | 1 / 7 | 1/14, 2/15, 11/24 |
assigned from the HSQC and HMBC spectra
4.6.2. Merocyclophane B (2)
Purple amorphous powder; [α]25D – 19 (c 0.070, MeOH); UV (MeOH) λmax (log ε) 207 (4.25), 221 (3.98), 273 (3.57), 324 (2.76), 521 (2.59) nm; CD (MeOH) λmax (Δε) 211 (−3.66), 227 (−2.22), 274 (−2.03) nm; IR (neat) νmax 2955, 2924, 2855, 1664, 1635, 1595, 1527 cm−1; for 1H and 13C NMR spectroscopic data, see Table 2; HRESIMS m/z 565.3954 [M-H]• (cacld for C36H54O5, 565.3893)
Table 2.
NMR spectroscopic data of merocyclophane B (2) in DMSO-d6 (600 MHz)
| No. | Merocyclophane B (2) | ||
|---|---|---|---|
| δCa | δH | mult. (J in Hz) | |
| 1 | 28.7 | 2.86 | m |
| 2 | 37.6 | 1.22 | m |
| 1.44 | m | ||
| 3 | 29.0 | 0.62 | m |
| 0.94 | m | ||
| 4 | 30.3 | 0.84 | m |
| 1.24 | m | ||
| 5 | 28.9 | 0.66 | m |
| 0.94 | m | ||
| 6 | 33.0 | 1.27 | m |
| 1.89 | m | ||
| 7 | 34.3 | 3.06 | m |
| 8 | 113.8 | ||
| 9 | 155.4 | ||
| 10 | 102.5 | 5.97 | s |
| 11 | 144.3 | ||
| 12 | 107.3 | 5.95 | s |
| 13 | 156.7 | ||
| 14 | 39.6 | 2.26 | |
| 15 | 38.6 | 1.21 | m |
| 1.40 | m | ||
| 16 | 29.0 | 0.62 | m |
| 0.94 | m | ||
| 17 | 30.3 | 0.84 | m |
| 1.24 | m | ||
| 18 | 28.9 | 0.69 | m |
| 0.95 | m | ||
| 19 | 33.1 | 1.32 | m |
| 1.63 | m | ||
| 20 | 35.5 | 2.80 | br m |
| 21 | ndb | ||
| 22 | ndb | ||
| 23 | 132.6 | 6.30 | br s |
| 24 | 149.1 | ||
| 25 | ndb | ||
| 26 | ndb | ||
| 27 | 32.8 | 1.45 | m |
| 1.80 | m | ||
| 28 | 29.7 | 1.05 | m |
| 1.12 | m | ||
| 29 | 21.9 | 1.19 | m |
| 30 | 13.8 | 0.79 | t (7.3) |
| 31 | 32.9 | 1.40 | m |
| 1.70 | m | ||
| 32 | 29.7 | 1.05 | m |
| 1.12 | m | ||
| 33 | 21.9 | 1.19 | m |
| 34 | 13.8 | 0.79 | t (7.3) |
| 35 | 20.0 | 1.01 | br d |
| 36 | 22.4 | 1.08 | d (6.9) |
| 9-OH | 8.53 | br s | |
| 13-OH | 8.54 | br s | |
| 26-OH | 10.24 | br s | |
assigned from the HSQC and HMBC spectra,
ndb no signals observed at 25°C presumably due to signal broadening
4.7. X-ray Crystallographic Analysis of Merocyclophane A (1)
Crystals for X-ray analysis were grown from CH3CN. A small single crystal, roughly 30 × 30 × 30 µm, was selected for data collection at sector-22-BM, and encased in Paratone-N oil and cooled to 100K to minimize crystal degradation and X-ray radiation damage. The MAR 225 CCD detector was set at a distance of 85 mm from the crystal using a wavelength of 0.80Å to collect data near a resolution of 0.86 Å. 36 images were collected, each with a rotation sweep of 10°. The images were indexed, and the intensities were integrated and scaled with XDS (Kabsch, 1993). The WinGX package was used for structure solution and refinement. The space group was identified as C2 with one molecule in the asymmetric unit (Farrugia, 1999). SHELX was used for structure solution and refinement on F2. Six CH3CN molecules were found in the asymmetric unit, with two of them sitting on 2-fold axes. Four CH3CN molecules are each involved in hydrogen bonding with the four aromatic hydroxyl groups; the remaining two CH3CN molecules reside in channels formed at the interface of the larger macrocycles as they pack together in the unit cell. Crystal data: C36H56O4·5CH3CN, MW = 758.1, monoclinic, space group C2 (5); a = 14.735 (2) Å, b = 10.840 (5) Å, c = 29.842 (3) Å, β = 95.226 (4)°, V = 4747 (2) Å3; Z = 4, Dc = 0.057 mg/m3; µ = 0.057 mm−1; F000 = 1656. Reflections collected / unique = 13719 / 5724 (Rint = 0.0815); final R1 = 0.0882, wR2 = 0.2285 for reflections with I > 2σ (I); R1 = 0.1311, wR2 = 0.2572 for all unique data. Crystallographic data (ID No. 818958) have been deposited with the Cambridge crystallographic Data Centre. Copies of the data can be obtained, free of charge, on application to the Director, CCDC, 12 Union Road, Cambridge CB2 1EZ, UK (deposit@ccdc.cam.ac.uk).
4.8. HT-29 Antiproliferative Assay
The antiproliferative activity of 1 and 2 against the HT-29 cancer cell line was evaluated according to a previously established protocol (Seo et al., 2001).
Supplementary Material
Highlights.
-
-
Two [7.7] paracyclophanes named merocyclophanes A and B were obtained.
-
-
Both displayed antiproliferative activity against the HT-29 cancer cell line.
-
-
Nostoc sp. (UIC 10062) was obtained from Grand Mere State Park in Michigan,
-
-
Nostoc sp. was classified based on microscopic and 16S rRNA gene sequence analysis
Acknowledgments
This research was supported by PO1 CA125066 from NCI/NIH. We acknowledge the use of the Southeast Regional Collaborative Access Team (SER-CAT), beam line 22-BM, for data collection of these small crystals. Use of the Advanced Photon Source at Argonne National Laboratory was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. We also thank Dr. Kirk Hevener and Pin-Chih Su from Center for Pharmaceutical Biotechnology at UIC for performing molecular mechanic study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:
References
- Bobzin SC, Moore RE. Biosynthetic origin of [7.7]paracyclophanes from cyanobacteria. Tetrahedron. 1993;49:7615–7626. [Google Scholar]
- Bui HTN, Jansen R, Pham HTL, Mundt S. Carbamidocyclophanes A-E, chlorinated paracyclophanes with cytotoxic and antibiotic activity from the Vietnamese cyanobacterium Nostoc sp. J. Nat. Prod. 2007;70:499–503. doi: 10.1021/np060324m. [DOI] [PubMed] [Google Scholar]
- Castenholz RW, Phlyum BX. Cyanobacteria. In: Boone DR, Castenholz RW, editors. Bergey’s Manual of Systematic Bacteriology. Vol. 1. New York: Spinger; 2001. pp. 473–599. [Google Scholar]
- Chen JL, Moore RE, Patterson GML. Structures of nostocyclophanes A-D. J. Org. Chem. 1991;56:4360–4364. [Google Scholar]
- Chlipala G, Sturdy M, Krunic A, Lantvit DD, Shen Q, Porter K, Swanson SM, Orjala J. Cyclindrocyclophanes with proteasome inhibitory activity from the cyanobacterium Nostoc sp. J. Nat. Prod. 2010;73:1529–1537. doi: 10.1021/np100352e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falch BS, Konig GM, Wright AD, Sticher O, Angerhofer CK, Pezzuto JM, Bachmann H. Biological-activities of cyanobacteria – Evaluation of extracts and pure compounds. Planta Med. 1995;61:321–328. doi: 10.1055/s-2006-958092. [DOI] [PubMed] [Google Scholar]
- Farrugia LJ. ‘WinGX’ suite for small-molecule single-crystal crystallography. J. Appl. Cryst. 1999;32:837–838. [Google Scholar]
- Harada KI. Production of secondary metabolites by freshwater cyanobacteria. Chem. Pharm. Bull. 2004;52:889–899. doi: 10.1248/cpb.52.889. [DOI] [PubMed] [Google Scholar]
- Jones AC, Monroe EA, Eisman EB, Gerwick L, Sherman DH, Gerwick WH. The unique mechanistic transformations involved in the biosynthesis of modular natural products from marine cyanobacteria. Nat. Prod. Rep. 2010;27:1048–1065. doi: 10.1039/c000535e. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- Komárek J, Komárková J, Kling H. Filamentous Cyanobacteria. In: Wehr JD, Sheath RG, editors. Freshwater Algae of North America. San Diego: Academic Press; 2003. pp. 117–196. [Google Scholar]
- Martínez-Murcia AJ, Acinas SG, Rodriguez-Valera F. Evaluation of prokaryotic diversity by restrictase digestion of 16S rDNA directly amplified from hypersaline environments. FEMS Microbiol. Ecol. 1995;17:247–256. [Google Scholar]
- Moore BS, Chen JL, Patterson GML, Moore RE. [7.7]Paracyclophanes from blue-green algae. J. Am. Chem. Soc. 1990;112:4061–4063. [Google Scholar]
- Moore BS, Chen JL, Patterson GML, Moore RE. Structures of cylindrocyclophanes A-F. Tetrahedron. 1992;48:3001–3006. [Google Scholar]
- Nübel U, GarciaPichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 1997;63:3327–3332. doi: 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploutno A, Carmeli S. Nostocyclyne A, a novel antimicrobial cyclophane from the cyanobacterium Nostoc sp. J. Nat. Prod. 2000;63:1524–1526. doi: 10.1021/np0002334. [DOI] [PubMed] [Google Scholar]
- Rajaniemi P, Hrouzek P, Kastovska K, Willame R, Rantala A, Hoffmann L, Komarek J, Sivonen K. Phylogenetic and morphological evaluation of the genera Anabaena, Aphanizomenon, Trichormus and Nostoc (Nostocales, Cyanobacteria) J. Syst. Evol. Microbiol. 2005;55:11–26. doi: 10.1099/ijs.0.63276-0. [DOI] [PubMed] [Google Scholar]
- Seo E-K, Kim N-C, Mi Q, Chai H, Wall ME, Wani MC, Navarro HA, Burgess JP, Graham JG, Cabieses F, Tan GT, Farnsworth NR, Pezzuto JM, Kinghorn AD. Macharistol, a new cytotoxic cinnamylphenol from the stems of Machaerium aristulatum. J. Nat. Prod. 2001;64:1483–1485. doi: 10.1021/np0103158. [DOI] [PubMed] [Google Scholar]
- Smith HE. Chiroptical properties of the benzene chromophore. A method for the determination of the absolute configurations of benzene compounds by application of the benzene sector and benzene chirality rules. Chem. Rev. 1998;98:1709–1740. doi: 10.1021/cr9410124. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan LT. Bioactive natural products from marine cyanobacteria for drug discovery. Phytochemistry. 2007;68:954–979. doi: 10.1016/j.phytochem.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Wagoner RMV, Drummond AK, Wright JLC. Biogenetic diversity of cyanobacteria metabolites. Adv. Appl. Microbiol. 2007;61:89–217. doi: 10.1016/S0065-2164(06)61004-6. [DOI] [PubMed] [Google Scholar]
- Wehr JD, Sheath RG. Freshwater algae of North America. San Diego: Academic Press; 2003. pp. 117–196. [Google Scholar]
- Yamakoshi H, Ikarashi F, Minami M, Shibuya M, Sugahara T, Kanoh N, Ohori H, Shibata H, Iwabuchi Y. Syntheses of naturally occurring cytotoxic [7.7]paracyclophanes, (−)-cyclindrocyclophane A and its enantiomer, and implications for biological activity. Org. Biomol. Chem. 2009;7:3772–3781. doi: 10.1039/b909646a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.