Abstract
Background
The DARC (Duffy blood group, chemokine receptor) gene encodes for a transmembrane glycoprotein that functions as a chemokine transporter, is a receptor for Plasmodium vivax and knowlesi, and expresses the Duffy blood group antigens (Fy). The Fy(a−b−) phenotype found in people of African descent is typically associated with a −67t>c mutation in the 5′ untranslated region (UTR), which prevents red blood cells being invaded by Plasmodium vivax and knowlesi. The aim of this study was to establish DARC allele frequencies in an African American blood donor cohort, determine a phylogenetic tree for DARC, and compare human and Neandertal DARC genes.
Methods
The DARC nucleotide sequence of 54 African American blood donors was determined from genomic DNA. Heterozygous substitutions were resolved by sequencing of haplotype specific amplifications. A phylogenetic tree for DARC was established using the neighbor-joining method with Pan troglodytes as root.
Results
108 haplotypes of the DARC gene could be unambiguously determined from nucleotide position −300 in the 5′ UTR to +300 in the 3′ UTR. 11 different alleles were found, including the clinically relevant FY*A, FY*B, FY*B-67C, FY*B298A, and FY*X alleles. All phenotype predictions based on genotypes matched exactly the serologically determined phenotypes: 52% Fy(a−b−), 28% Fy(a−b+), and 20% Fy(a+b−).
Conclusions
The nucleotide sequencing approach using one amplicon is a practical genotyping method for DARC and allows the determination of haplotypes even in heterozygous constellations. We developed a phylogenetic tree for DARC alleles and postulated a distinct FY*B allele as ancestral for the extant DARC alleles in humans.
Introduction
The DARC (Duffy blood group, chemokine receptor) gene encodes for a trans-membranous glycoprotein expressing the Duffy blood group antigens (Fy) that functions as a chemokine transporter and as a receptor for the malaria parasites Plasmodium vivax and knowlesi.1-4 DARC is located on chromosome 1 (1q21-q22) and transcribed in two mRNA variants leading to two different protein isoforms. The initially described mRNA variant has one exon and encodes for a protein with 338 amino acids.5 The later described mRNA variant has two exons, the intron encompassing the initial part of the first mRNA variant, and encodes for a protein with 336 amino acids.6,7 Despite encoding for a shorter protein, the second mRNA variant is longer than the first because of a longer 5′ untranslated region (UTR). DARC proteins are expressed on red blood cells (RBC) and in various tissues such as endothelium, brain, heart, kidney and pancreas.7 While some DARC mRNA is expressed by mesenchymal stem cells, Fy antigens cannot be detected.8
Antithetical antigens, Fya and Fyb, are encoded by co-dominant allele groups FY*A and FY*B, which differ by a single nucleotide polymorphism (SNP) 125G>A with an amino acid substitution Gly42Asp.5,6 A homozygous single nucleotide substitution in the 5′ UTR, −67t>c, also called GATA box mutation, leads to a lack of DARC protein in red blood cells (RBC), serologically detected as Fy(a−b−), which prevents invasion by P. vivax and P. knowlesi.2,3,9,10 These FY*B-67C alleles have become the most prevalent DARC alleles in populations living in regions with endemic malaria.11,12 In contrast to null alleles, which are rare causes of Fy(a−b−) phenotypes,13 the FY*B-67C alleles lead to an expression of the Duffy glycoprotein in non-erythroid tissues.5,9,14 These FY*B-67C alleles remained an effective escape mechanism from infections by certain malaria parasites for a long time. They also represent a striking example for evolution and expansion of advantageous alleles under selective pressure.11 However, infections of Fy(a−b−) individuals by certain P. vivax strains have recently been reported.15
While genotype and allele frequencies for DARC are known for various populations, a nucleotide sequencing approach had not been applied to the African American population. Typically, specific known polymorphisms are used for genotype screening. Alternatively, serologic findings may trigger a search for novel mutations. We chose a random approach and nucleotide sequencing of the DARC gene for the current study. The aim of this study was to establish frequencies of DARC alleles at high resolution. We screened for novel mutations and gained more detailed haplotype information with the intention of determining the phylogenetic tree for the DARC alleles.
Materials and Methods
Blood samples and Fy phenotype
EDTA blood samples were drawn from 54 African American blood donors at the NIH Blood Bank from October 2009 to March 2010 after obtaining written informed consent. They were random with respect to the Duffy (Fy) phenotype determined by indirect antiglobulin tube method with FDA licensed polyclonal reagents (derived from pools of human sera; Ortho, Raritan, NJ). We used fresh RBC or thawed RBC that were cryopreserved with a sucrose/dextrose freezing solution and maintained in liquid nitrogen for less than 3 months.16
DARC nucleotide sequencing
DNA was extracted (Qiagen EZ1 DNA blood kit on the BioRobot EZ1; Qiagen, Valencia, CA) and sequenced by a method similar to one previously published for RHD.17 One amplification reaction covering the complete DARC gene was run for each sample (total volume 50 μl): 41.75 dH2O, 5 μl 10x buffer, 1 μl dNTP (10 mM), 0.5 μl each of FyAF and FyAR primers (10 μM; Table 1; Eurofins MWG Operon, Huntsville, AL), 0.25 μl Taq enzyme (FastStart High Fidelity PCR System, dNTPack; Roche Applied Science, Indianapolis, IN), and 1 μl DNA (~100 ng/μl). Thermocycler (Bio-Rad C1000; Bio-Rad, Hercules, CA) conditions were 94°C for 3 min; 10 cycles with 94°C for 30 sec, 60°C for 30 sec, 68°C for 5 min; 35 cycles with 94°C for 30 sec, 60°C for 30 sec, 68°C for 5:20 min and extended by 20 sec/cycle. PCR cleanup of the 2493 bp long amplicon was accomplished by QIAquick PCR purification kit (Qiagen) with an elution volume of 40 μl.
Table 1.
Primer sequences
| Name | Use | Location | Nucleotide sequence (5′ to 3′) |
|---|---|---|---|
| FyAF | Amplification | 5′ UTR | cttttgaactgcctttccttgg |
| FyAR | Amplification | 3′ UTR | cttccccaccactgtcctaatc |
| Fy-67Tf | Amplification | 5′ UTR GATA box specific T | cctcattagtccttggctcttat |
| Fy-67Cf | Amplification | 5′ UTR GATA box specific C | cctcattagtccttggctcttac |
| FySF1 | Sequencing | 5′ UTR | gatggaggagcagtgagagtc |
| FySF2 | Sequencing | Intron 1 | atccctatgcccctcatttc |
| FySF3 | Sequencing | Intron 1 | cctgctttgtccttttccac |
| FySF4 | Sequencing | Exon 2 | AGTGCCCTCTTCAGCATTGT |
| FySF5 | Sequencing | Exon 2 | TGGGCCTGGTTTATTTTCTG |
| FySR1 | Sequencing | Intron 1 | ccaagagaccaggatggaac |
| FySR2 | Sequencing | Exon 2 | CGTGCTGTATATCAGGGTGC |
| FySR3 | Sequencing | 3′ UTR | atgactcccctcatgctctg |
Eight sequencing reactions were run for each amplicon using 1 μl amplicon, 1.25 μl primer (Table 1), 1.8 μl BigDye v3.1 Terminatory Mix (Applied Biosystems, Carlsbad, CA), and 16 μl dH2O. Thermocycler conditions were 25 cycles of 96°C for 15 sec, 58°C for 10 sec, 60° for 4 min. Unincorporated dye was removed using DyeEx 96 well plates (Qiagen), the sequences dehydrated (Savant SPD 2010 SpeedVac Concentrator; ThermoScientific, Wilmington, DE) and resuspended in 10 μl formamide (Hi-Di; Applied Biosystems) before chromatography analysis on an AB 3730 DNA analyzer (Applied Biosystems). Nucleotide sequences were aligned (CodonCode Aligner; CodonCode, Dedham, MA) to NCBI RefSeq NG_011626.1 and nucleotide positions defined using the first nucleotide of the coding sequence (CDS) of the NM_002036.2 isoform as nucleotide position 1.
Haplotype specific sequence analysis
Heterozygous mutations were resolved by haplotype specific amplifications (Fy-67T or Fy-67C with FyAR primers; modified PCR program applying an annealing temperature of 63°C and extension of 72°C for 4 min in all cycles) followed by sequencing reactions of the haplotype amplicons as described above.
Terminology
We propose and apply in this study a terminology that is based on prior nomenclatures,13,18 but additionally allows the specification of alleles at high resolution as obtained by haplotype specific sequence analysis. Numbers after a colon, assigned according to the allele frequency, are used to distinguish between alleles with identical exon and GATA box sequences.
Neandertal genome
The published data of the Neandertal genome19 was analyzed using the SAMtools.20 Alignments to the human genome (hg18)21 in the region chr1:157,441,074-157,443,164 were used for analysis of the summary 1x Neandertal genome sequence and the 3 fossils with highest genome coverage: Vi33.16 (54.1%), Vi33.25 (46.6%), and Vi33.26 (45.2%). The other 3 fossils reported to date were analyzed with ≤ 2% coverage, lacked nucleotide sequences for DARC in the UCSC Genome Browser, and were excluded from analysis.19,21
Phylogenetic tree
The topologic associations between the various alleles were analyzed using a neighbor joining clustering method (CodonCode Aligner).22 Each single nucleotide substitution was counted as one event. The DARC sequence from chimpanzee (Pan troglodytes, NC_006468.2, nucleotides 138460591 to 138471719) was used for external rooting, as previously described for RHD.23
Statistics
95% confidence intervals (CI) for allele frequencies using the Poisson distribution and χ2 test were calculated with MedCalc (MedCalc Software, Mariakerke, Belgium).
Results
Genotype
We developed a nucleotide sequencing method for DARC based on one amplicon (Table 1). Nucleotide sequence analysis of DARC in 54 African American blood donors revealed 10 single nucleotide polymorphisms in the range of −300 nucleotides in the 5′ UTR to +300 nucleotides in the 3′ UTR (see genotype details in the supplementary Table S1). One polymorphism occurred in the GATA box in the 5′ UTR, 4 in the intron, 3 in exon 2, and 2 in the 3′ UTR. An intron nucleotide substitution (IVS1+54t>c) was found in all samples and also in the chimpanzee and Neandertal reference sequences indicating a likely error in the currently used reference sequence NG_011626.1.
All haplotypes could be unambiguously determined revealing 11 different alleles in the studied population (Table 2). Based on the number of analyzed samples, alleles not found in this study may have a population frequency of < 3.4% (upper limit of 95% CI, Poisson distribution). For comparison to previous studies, we clustered the alleles into 5 clinically relevant, i.e. phenotypically differing or potentially differing, allele groups based on exon and GATA box mutations (Table 3).
Table 2.
DARC allele distribution in African American blood donors
| Nucleotide substitution (position) * |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele † |
5′ UTR |
Intron (IVS1) |
Exon 2 |
3′ UTR |
Population frequency |
||||||||||
| Designation | Trivial name | −67 | +54 | +115 | +150 | −243 | −58 | 125 | 265 | 298 | +250 | +268 | Observed (n) | Mean | 95% CI ‡ |
| FY*01:1 | FY*A:1 | t | c | t | c | t | a | G | C | G | c | a | 9 | 8% | 4% – 16% |
| FY*01:2 | FY*A:2 | t | c | t | c | – | a | G | C | G | c | a | 1 | 1% | 0.02% – 5% |
| FY*01:3 | FY*A:3 | t | c | t | c | – | a | G | C | G | t | a | 1 | 1% | 0.02% – 5% |
| FY*02:1 | FY*B:1 | t | c | c | c | – | g | A | C | G | c | a | 4 | 4% | 1% – 9% |
| FY*02:2 | FY*B:2 | t | c | t | t | – | a | A | C | G | c | a | 3 | 3% | 0.5% – 8% |
| FY*02:3 | FY*B:3 | t | c | c | c | – | a | A | C | G | c | a | 2 | 2% | 0.2% – 7% |
| FY*02:4 | FY*B:4 | t | c | t | t | – | a | A | C | G | c | g | 1 | 1% | 0.02% – 5% |
| FY*03:1 | FY*B-67C:1 | c | c | t | c | – | a | A | C | G | c | a | 72 | 67% | 52% – 84% |
| FY*03:2 | FY*B-67C:2 | c | c | c | c | – | a | A | C | G | c | a | 9 | 8% | 4% – 16% |
| FY*04:1 | FY*B298A:1 | t | c | t | t | – | a | A | C | A | c | g | 5 | 5% | 2% – 11% |
| FY*05:1 | FY*X:1 | t | c | t | t | – | a | A | T | A | c | g | 1 | 1% | 0.02% – 5% |
| Total | 108 | ||||||||||||||
Nucleotide substitutions are shown relative to the reference sequence (NG_011626.1) used for analysis. Nucleotide positions are defined using the first nucleotide of the coding sequence (CDS) of the NM_002036.2 isoform as nucleotide position 1.
This allele terminology is proposed as an extension of previous nomenclatures13,18 to reflect the high resolution of the study results (see text for details). The ISBT Working Party on Red Cell Immunogenetics and Blood Group Terminology30,31 recommended using the ISBT symbol FY when referring to serologically defined alleles or molecularly defined alleles that represent a serologically defined antigen and using the Human Genome Organization (HUGO) Gene Nomenclature Committee (HGNC) symbol DARC in all other circumstances. The allele sequences have been deposited as GenBank accession numbers JN251907 to JN251917.
95% confidence interval (CI) based on Poisson distribution
Table 3.
Frequencies of clinically relevant DARC allele groups in African Americans
| Proposed allele group name * |
Population frequency |
|||||||
|---|---|---|---|---|---|---|---|---|
| Designation | Trivial | Alleles † | ISBT working draft ‡ |
Traditional terminology § |
Associated Phenotype |
Observed (n) | Mean | 95% CI ∥ |
| FY*01 | FY*A | FY*01:1 to FY*01:3 | FY*01 or FY*A | FY*A | Fy(a+b−) | 11 | 10% | 5% – 18% |
| FY*02 | FY*B | FY*02:1 to FY*02:4 | FY*02 or FY*B | FY*B | Fy(a−b+) | 10 | 9% | 4% – 17% |
| FY*03 | FY*B-67C | FY*03:1 to FY*03:2 | FY*02N.01 | FY*B-33¶ | Fy(a−b−) ** | 81 | 75% | 60% – 93% |
| FY*04 | FY*B298A | FY*04:1 | n/a | FY*B-298A | Fy(a−b+) | 5 | 5% | 2% – 11% |
| FY*05 | FY*X | FY*05:1 | FY*02M.01 | FY*X | Fy(a−b+w) | 1 | 1% | 0.02% – 5% |
| Total | 108 | |||||||
The alleles were grouped based on the nucleotide sequence in the exons and GATA box as detailed in the text. Allele terminology using FY instead of DARC according to the ISBT Working Party on Red Cell Immunogenetics and Blood Group Terminology (see footnote to Table 2).30,31
Allele designation as defined in Table 2
Current working draft of the new proposed ISBT terminology for blood group alleles18
Commonly used terminology as summarized by Castilho13
95% confidence interval (CI) based on Poisson distribution
This allele, which is not a DARC null allele, has also been designated Fy,2 FY,9 Fy−,10 FY*Fy,32 FY*0,33 FY*silent,34 FY*−33C,35 FY*−67C,36 FY*BES,37 FY*Bnull,38 DARC-null39 using various relative nucleotide positions to indicate the t>c polymorphism (−46,9 −365,10 −33,13 −6718) while the position −67 used in the new terminology is based on the fixed start of the CDS of NM_002036.2.
In contrast to rare DARC null alleles,13 the Fyb antigen is expressed in non-erythroid tissues.
Phenotype
All phenotype predictions based on the genotype matched the serologically determined phenotype. Twenty-eight donors were Fy(a−b−), 15 Fy(a−b+), and 11 Fy(a+b−), in accordance with reference Duffy phenotype occurrences in African Americans.24 The observed phenotype distribution did not differ significantly from the expected distribution predicted by the observed alleles (p = 0.49, χ2 test; supplementary Table S2). We found no evidence that any of the detected intron polymorphisms affected the phenotypes that were predicted by the exon and GATA box polymorphisms.
DARC alleles in Neandertals
We analyzed the currently available Neandertal nucleotide sequence19 (Table 4), which covers 80% of DARC. Among the 11 SNP described in the current study (Table 2), no difference was found between the predicted ancestral human and the Neandertal alleles (6 equal to and 5 without sequence data in the Neandertal genome). Among the 13 SNP differing between P. troglodytes and H. sapiens, 10 nucleotides in the Neandertal genome were identical to the human genome, 1 was heterozygous for the human and chimpanzee nucleotide, and 2 lacked data.
Table 4.
Comparison of DARC in human, Neandertal and chimpanzee genomes
| Nucleotides position * |
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5′ UTR |
IVS 1 |
Exon 2 |
3′ UTR |
|||||||||||||||||||||
| Species † | −214 | −135 | −67 | −29 | +54 | +115 | +123 | +150 | −243 | −58 | −19 | 24 | 123 | 125 | 265 | 298 | 343 | 573 | 807 | +88 | +219 | +250 | +268 | +283 |
| H. sapiens | c | g | t | c | c | c | t | c | - | a | c | G | T | A | C | G | G | T | G | g | c | c | g | a |
| Neandertal | c | g | ? | c | ? | c | t | ? | - | ? | c | AG | ? | ? | C | G | G | T | G | ? | c | c | g | a |
| P. troglodytes | t | t | t | g | c | c | - | c | - | a | t | A | C | A | C | G | A | A | C | t | t | c | g | c |
Nucleotide positions are shown according to the human reference sequence (NG_011626.1) and defined using the first nucleotide of the coding sequence (CDS) of the NM_002036.2 isoform as nucleotide position 1. The following single nucleotide polymorphisms (SNP) have been used for this analysis: all 11 SNP found in sequence analysis of 54 African Americans (Table S1) and all 13 SNP in which the predicted ancestral human DARC alleles differed from P. troglodytes in the range of −300 nucleotides in the 5′ UTR to +300 nucleotides in the 3′ UTR. Additional 65 nucleotide substitutions were found in the Neandertal genome sequence, which differ from both the human and the chimpanzee nucleotides and were not considered for this analysis.
Used DARC nucleotide sequences for analysis: The postulated ancestral human DARC allele (Fig. 1), NC_006468.2 for P. troglodytes, and the sequence of the Neandertal Genome project.19
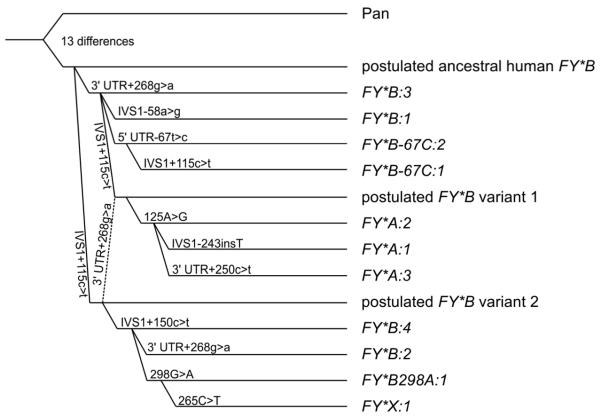
Phylogenetic tree of DARC alleles
Using the DARC sequence of the chimpanzee for external rooting, a distinct FY*B allele could be determined to be ancestral to all known DARC alleles (Fig. 1). Other alleles are younger, in particular all alleles with the GATA box mutation −67t>c and also all alleles that lead to Fya expression. Characteristic nucleotides of this predicted ancestral human FY*B allele (IVS1+115c and 3′ UTR+268g) were also found in the Neandertal genome (Table 4).
Fig. 1. Phylogeny of DARC alleles in humans.
A phylogenetic tree of DARC is shown for the 11 alleles found in this study using the DARC sequence from Pan troglodytes (NC_006468.2) for external rooting. Clustering of the described DARC alleles is based on neighbor-joining method.22 For each evolutionary step, the event is indicated; the depicted distances of the alleles are arbitrary, as previously described for RHD.23
Discussion
The aim of this study was to determine DARC alleles at high resolution and their frequencies in an African American population. For the present study, all haplotypes could be determined unambiguously in the range of −300 nucleotides at 5′ UTR to +300 nucleotides at 3′ UTR. This DNA stretch covered all nucleotides of the two published mRNA variants.5-7
The frequencies of the DARC allele groups (Table 3) were comparable to the results of earlier studies.13,14 This detailed study of complete haplotypes allowed us to determine the phylogenetic tree of DARC alleles. Using the reference sequence for the chimpanzee DARC gene as root, a distinct ancestral human DARC allele can be predicted, which represents a FY*B allele (Fig. 1). Our results confirm previous studies25,26 that a FY*B allele is the ancestral allele. A recent claim that FY*A is ancestral11 was explained to be a typographical error and should have stated that FY*B is the ancestral allele (written communication, Dr Philip W Hedrick, July 27, 2011). Our conclusion is also supported by comparison of this ancestral human FY*B allele with the Neandertal DARC gene (Table 4).
Future studies on DARC alleles at such a high resolution will reveal additional alleles and allow even more differentiation of the phylogenetic tree. In the current study we focused on alleles occurring in individuals of African descent, because knowledge of human evolution from the African continent27 may give us a better chance of revealing ancestral alleles.
Despite the limitation of an 80% coverage of the DARC gene locus in the Neandertal genome, the comparison of 24 characteristic nucleotides revealed a close similarity with the predicted human DARC allele: 16 nucleotides were identical to the human genome, 1 was heterozygous for the human and chimpanzee genomes, while information for 7 nucleotides was lacking. This reflects the similarity of human and Neandertal genomes with common ancestors within the last 500,000 years and potential interbreeding,19 whereas common ancestors of humans and chimpanzees lived around 6.1 million years ago.28 While the current Neandertal genome data lacks the nucleotides indicative for the Fya/Fyb and GATA box polymorphisms, two nucleotides (IVS1+115c and 3′ UTR+268g) were identical to polymorphisms found in human FY*B but not in FY*A alleles, which indicates that the Neandertal individuals studied19 may have had a Fy(a−b+) phenotype.
We describe a practical nucleotide sequencing system that allows haplotype specific determination of DARC alleles from genomic DNA with one amplicon. The described approach encompassed the nucleotide sequence that is known to be relevant for blood group genotyping (from −67 in the 5′ UTR to the end of the CDS) and the complete nucleotide sequences of the two published mRNA variants5-7 known at the time of study design (NM_001122951.2 and NM_002036.2). However, an update of the mRNA reference sequence NM_002036.3 after 11 November 2010 presents a 5′ UTR that is not completely covered by our approach. If this longer mRNA, which is based on a cDNA nucleotide sequence (AK291593.1) found in placenta, is representative for prevalent DARC transcripts, future studies should encompass the nucleotide sequence of this longer mRNA as well.
Terminology of DARC alleles, genotypes, and Fy phenotypes have evolved over time reflecting the increasing depth of resolution and detail of information. Currently used terminologies have the limitation that they cannot discriminate the detail of our genotyping approach covering the range of −300 nucleotides in the 5′ UTR to +300 nucleotides in the 3′ UTR. Hence, we propose an extension of the current terminologies (Table 2). The alleles can be clustered in biologically and clinically relevant allele groups (Table 3), and the group names may be used when genotyping methods with lower resolution than nucleotide sequencing are applied.
Genotyping reports should not only describe the genotype and predicted phenotype for RBC and non-erythroid tissues (if applicable), but also document the applied genotyping approach, including method/system and version, its quality/resolution, and particularly the range of the effectively analyzed nucleotide sequence. This range can be a few nucleotides in single nucleotide polymorphism (SNP) typing or up to several hundred or thousand base pairs in sequence based typing (SBT). Current information technology software is capable of handling this kind of data in nested clusters. This suggested approach for DARC would also be useful for other blood group systems.
Supplementary Material
Acknowledgments
We are very grateful to Karen M. Byrne, Lorraine G. Caruccio, Elizabeth J. Furlong, Supatta M. Lucas, Traci D. Paige, and A. Hallie Lee-Stroka for expert technical support. We thank the staff of the Blood Donor Section for sample collection and of the HLA Laboratory for DNA extraction. Part of this study was completed by Kanaeko R. Ravenell as her project work in the SBB program at the NIH (year of graduation, class of 2010).29 This research was supported by the Intramural Research Program of the NIH Clinical Center.
Footnotes
Conflict of interest disclosure: None.
Statement of Disclaimer: The views expressed do not necessarily represent the view of the National Institutes of Health, the Department of Health and Human Services, or the U.S. Federal Government.
Authorship contribution: Study design: PS and WAF. Sequencing methods: PS. Sample collection and preparation: KRR, SLS. Experiment execution: PS, KRR. Data analysis: PS, KRR, WAF. Phylogenetic tree and terminology: PS, WAF. Manuscript writing: PS, WAF. All authors read and approved the final manuscript.
References
- 1.Dvorak JA, Miller LH, Whitehouse WC, Shiroishi T. Invasion of erythrocytes by malaria merozoites. Science. 1975;187:748–50. doi: 10.1126/science.803712. [DOI] [PubMed] [Google Scholar]
- 2.Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science. 1975;189:561–3. doi: 10.1126/science.1145213. [DOI] [PubMed] [Google Scholar]
- 3.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–4. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 4.Hadley TJ, Peiper SC. From malaria to chemokine receptor: the emerging physiologic role of the Duffy blood group antigen. Blood. 1997;89:3077–91. [PubMed] [Google Scholar]
- 5.Chaudhuri A, Polyakova J, Zbrzezna V, Williams K, Gulati S, Pogo AO. Cloning of the glycoprotein D cDNA, which encodes the major subunit of the Duffy bllod group system and the receptor for the Plasmodium vivax malaria parasite. Proc Natl Acad Sci U S A. 1993;90:10793–7. doi: 10.1073/pnas.90.22.10793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwamoto S, Li J, Omi T, Ikemoto S, Kajii E. Identification of a novel exon and spliced form of Duffy mRNA that is the predominant transcript in both erythroid and postcapillary venule endothelium. Blood. 1996;87:378–85. [PubMed] [Google Scholar]
- 7.Le Van Kim C, Tournamille C, Kroviarski Y, Cartron JP, Colin Y. The 1.35-kb Duffy mRNA isoforms are differently regulated in various regions of brain, differ by the length of their 5′ untraslated sequence, but encode the same polypeptide. Blood. 1997;90:2851–3. [PubMed] [Google Scholar]
- 8.Schäfer R, Schnaidt M, Klaffschenkel RA, Siegel G, Schüle M, Rädlein MA, Hermanutz-Klein U, Ayturan M, Buadze M, Gassner C, Danielyan L, Kluba T, Northoff H, Flegel WA. Expression of blood group genes by mesenchymal stem cells. Br J Haematol. 2011;153:520–8. doi: 10.1111/j.1365-2141.2011.08652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tournamille C, Colin Y, Cartron JP, Le Van KC. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10:224–8. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- 10.Iwamoto S, Li J, Sugimoto N, Okuda H, Kajii E. Characterization of the Duffy gene promoter: evidence for tissue-specific abolishment of expression in Fy(a−b−) of black individuals. Biochem Biophys Res Commun. 1996;222:852–9. doi: 10.1006/bbrc.1996.0833. [DOI] [PubMed] [Google Scholar]
- 11.Hedrick PW. Population genetics of malaria resistance in humans. Heredity. 2011 doi: 10.1038/hdy.2011.16. doi:10.1038/hdy.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howes RE, Patil AP, Piel FB, Nyangiri OA, Kabaria CW, Gething PW, Zimmerman PA, Barnadas C, Beall CM, Gebremedhin A, Menard D, Williams TN, Weatherall DJ, Hay SI. The global distribution of the Duffy blood group. Nat Commun. 2011;2:266. doi: 10.1038/ncomms1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castilho L. The value of DNA analysis for antigens in the Duffy blood group system. Transfusion. 2007;47(1 Suppl):28S–31S. doi: 10.1111/j.1537-2995.2007.01307.x. [DOI] [PubMed] [Google Scholar]
- 14.Moulds JM, Hayes S, Wells TD. DNA analysis of Duffy genes in American blacks. Vox Sang. 1998;74:248–52. [PubMed] [Google Scholar]
- 15.Mendes C, Dias F, Figueiredo J, Mora VG, Cano J, de SB, do Rosario VE, Benito A, Berzosa P, Arez AP. Duffy Negative Antigen Is No Longer a Barrier to Plasmodium vivax - Molecular Evidences from the African West Coast (Angola and Equatorial Guinea) PLoS Negl Trop Dis. 2011;5:e1192. doi: 10.1371/journal.pntd.0001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmid P, Huvard MJ, Lee-Stroka AH, Lee JY, Byrne KM, Flegel WA. Red blood cell preservation by droplet freezing with polyvinyl pyrrolidone or sucrose/dextrose and by bulk freezing with glycerol. Transfusion. 2011 doi: 10.1111/j.1537-2995.2011.03258.x. doi:10.1111/j.1537-2995.2011.03258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner FF, Gassner C, Müller TH, Schönitzer D, Schunter F, Flegel WA. Molecular basis of weak D phenotypes. Blood. 1999;93:385–93. [PubMed] [Google Scholar]
- 18.Terminology for blood group alleles (working draft) ISBT Red Cell Immunogenetics and Terminology Working Party ; 2010. [accessed June 20, 2011]. [Internet] http://ibgrl.blood.co.uk/ISBTPages/AlleleTerminology/allele-terminology.htm . [Google Scholar]
- 19.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, Hansen NF, Durand EY, Malaspinas AS, Jensen JD, Marques-Bonet T, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–22. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nei M, Kumar S. Molecular Evolution and Phylogenetics. 1st ed Oxford University Press; USA: 2000. [Google Scholar]
- 23.Wagner FF, Ladewig B, Angert KS, Heymann GA, Eicher NI, Flegel WA. The DAU allele cluster of the RHD gene. Blood. 2002;100:306–11. doi: 10.1182/blood-2002-01-0320. [DOI] [PubMed] [Google Scholar]
- 24.Reid M, Lomas-Francis C. The Blood Group Antigen FactsBook. 2nd ed Academic Press; 2004. [Google Scholar]
- 25.Chaudhuri A, Polyakova J, Zbrzezna V, Pogo AO. The coding sequence of Duffy blood group gene in humans and simians: restriction fragment length polymorphism, antibody and malarial parasite specificities, and expression in nonerythroid tissues in Duffy-negative individuals. Blood. 1995;85:615–21. [PubMed] [Google Scholar]
- 26.Li J, Iwamoto S, Sugimoto N, Okuda H, Kajii E. Dinucleotide repeat in the 3′ flanking region provides a clue to the molecular evolution of the Duffy gene. Hum Genet. 1997;99:573–7. doi: 10.1007/s004390050408. [DOI] [PubMed] [Google Scholar]
- 27.Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. abridged pbk. ed Princeton University Press; 1994. [Google Scholar]
- 28.Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 29.Byrne KM, Sheldon SL, Flegel WA. Organization and management of an accredited specialist in blood bank (SBB) technology program. Transfusion. 2010;50(7 Pt 2):1612–7. doi: 10.1111/j.1537-2995.2010.02737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daniels G, Flegel WA, Fletcher A, Garratty G, Levene C, Lomas-Francis C, Moulds JM, Moulds JJ, Olsson ML, Overbeeke MA, Poole J, Reid ME, Rouger P, van der Schoot CE, Scott M, et al. International Society of Blood Transfusion Committee on Terminology for Red Cell Surface Antigens: Cape Town report. Vox Sang. 2007;92:250–3. doi: 10.1111/j.1423-0410.2007.00887.x. [DOI] [PubMed] [Google Scholar]
- 31.Storry JR, Castilho L, Daniels G, Flegel WA, Garratty G, Francis CL, Moulds JM, Moulds JJ, Olsson ML, Poole J, Reid ME, Rouger P, van der Schoot E, Scott M, Smart E, et al. International Society of Blood Transfusion Working Party on red cell immunogenetics and blood group terminology: Berlin report. Vox Sang. 2011;101:77–82. doi: 10.1111/j.1423-0410.2010.01462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tournamille C. Bases moléculaires et relations structure-fonction des antigènes de groupe sanguin Duffy: récepteur de chimiokines et de Plasmodium vivax. Transfus Clin Biol. 2000;7:497–509. doi: 10.1016/s1246-7820(00)80038-5. [DOI] [PubMed] [Google Scholar]
- 33.Hult A, Hellberg A, Wester ES, Olausson P, Storry JR, Olsson ML. Blood group genotype analysis for the quality improvement of reagent test red blood cells. Vox Sang. 2005;88:265–70. doi: 10.1111/j.1423-0410.2005.00623.x. [DOI] [PubMed] [Google Scholar]
- 34.Sellami MH, Kaabi H, Midouni B, Dridi A, Mojaat N, Boukef M, Hmida S. Duffy blood group system genotyping in an urban Tunisian population. Annals of Human Biology. 2008;35:406–15. doi: 10.1080/03014460802082127. [DOI] [PubMed] [Google Scholar]
- 35.Denomme GA, Westhoff CM, Castilho L, Reid ME. Consortium for Blood Group Genes (CBGG): 2008 report. Immunohematology. 2009;25:75–80. [PubMed] [Google Scholar]
- 36.Denomme GA, Westhoff CM, Castilho LM, St-Louis M, Castro V, Reid ME. Consortium for Blood Group Genes (CBGG): 2009 report. Immunohematology. 2010;26:47–50. [PubMed] [Google Scholar]
- 37.Sousa TN, Sanchez BA, Ceravolo IP, Carvalho LH, Brito CF. Real-time multiplex allele-specific polymerase chain reaction for genotyping of the Duffy antigen, the Plasmodium vivax invasion receptor. Vox Sang. 2007;92:373–80. doi: 10.1111/j.1423-0410.2007.00902.x. [DOI] [PubMed] [Google Scholar]
- 38.Maestre A, Muskus C, Duque V, Agudelo O, Liu P, Takagi A, Ntumngia FB, Adams JH, Sim KL, Hoffman SL, Corradin G, Velez ID, Wang R. Acquired antibody responses against Plasmodium vivax infection vary with host genotype for duffy antigen receptor for chemokines (DARC) PLoS One. 2010;5:e11437. doi: 10.1371/journal.pone.0011437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramsuran V, Kulkarni H, He W, Mlisana K, Wright EJ, Werner L, Castiblanco J, Dhanda R, Le T, Dolan MJ, Guan W, Weiss RA, Clark RA, Karim SS, Ahuja SK, et al. Duffy-null-associated low neutrophil counts influence HIV-1 susceptibility in high-risk South African black women. Clin Infect Dis. 2011;52:1248–56. doi: 10.1093/cid/cir119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.