Abstract
RECQL5 is one of the five human RecQ helicases, involved in the maintenance of genomic integrity. While much insight has been gained into the function of the Werner (WRN) and Bloom syndrome proteins (BLM), little is known about RECQL5. We have analyzed the recruitment and retention dynamics of RECQL5 at laser-induced DNA double strand breaks (DSBs) relative to other human RecQ helicases. RECQL5-depleted cells accumulate persistent 53BP1 foci followed by γ-irradiation, indicating a potential role of RECQL5 in the processing of DSBs. Real time imaging of live cells using confocal laser microscopy shows that RECQL5 is recruited early to laser-induced DSBs and remains for a shorter duration than BLM and WRN, but persist longer than RECQL4. These studies illustrate the differential involvement of RecQ helicases in the DSB repair process. Mapping of domains within RECQL5 that are necessary for recruitment to DSBs revealed that both the helicase and KIX domains are required for DNA damage recognition and stable association of RECQL5 to the DSB sites. Previous studies have shown that MRE11 is essential for the recruitment of RECQL5 to the DSB sites. Here we show that the recruitment of RECQL5 does not depend on the exonuclease activity of MRE11 or on active transcription by RNA polymerase II, one of the prominent interacting partners of RECQL5. Also, the recruitment of RECQL5 to laser-induced damage sites is independent of the presence of other DNA damage signaling and repair proteins BLM, WRN and ATM.
Keywords: RECQL5, micro-irradiation, confocal laser microscopy
1. INTRODUCTION
RecQ helicases are a conserved group of proteins that play significant roles in the maintenance of genomic stability. There are five RecQ helicases in humans: RECQL1, BLM, WRN, RECQL4 and RECQL5; in contrast, single cell eukaryotes and prokaryotes express only one RecQ helicase. RecQ helicases are proposed to play important roles in various DNA metabolic processes including DNA replication, recombination, base excision repair and telomere maintenance [1–3]. Mutations in BLM, WRN, and RECQL4 are associated with Bloom syndrome (BS), Werner syndrome (WS), and Rothmund-Thomson syndrome (RTS), respectively. These syndromes are characterized by carcinogenesis and/or premature aging [4–6]. Although no clinical syndrome has been associated with defects in RECQL5, recent studies demonstrate that RECQL5-deficient mice are highly cancer prone and undergo premature aging [7], suggesting that RECQL5 may also function as a tumor suppressor.
RECQL5 exists in three isoforms generated by alternative splicing [8, 9]. RECQL5α and RECQL5γ isoforms contain 410 and 435 amino acids, respectively, and are predominantly cytoplasmic [9]. The larger RECQL5β isoform, which contains 991 amino acids, is nuclear and has an extended C-terminal region [9]. Although all three isoforms contain the core helicase domain, only RECQL5β has demonstrable helicase activity in vitro [9]. Biochemical studies indicate that RECQL5β unwinds a variety of DNA structures, albeit with low processivity [10, 11]. The conserved Zn2+ binding motif modulates RECQL5 helicase activity and is required for DNA binding [12]. The C-terminal half of RECQL5β has intrinsic DNA strand annealing activity, which is inhibited by ATP [10]. RECQL5β has been studied more than any other human RECQL5 isoform, and is the focus of the present study.
RECQL5 is implicated in DNA replication, transcription and repair processes. It promotes strand exchange at stalled replication forks, associates with DNA replication factories in S phase nuclei [11] and also plays an important role in cell survival after camptothecin treatment [13]. RECQL5 interacts with, and inhibits RNA polymerase II during transcription initiation and elongation, indicating a potential role in transcription [14, 15]. Recent studies reveal that both the KIX and SRI domains in the C-terminus of RECQL5 are required for binding to RNA Pol II [16, 17] and that the SRI domain plays a role in transcription-associated DNA double strand break (DSB) repair [18]. RECQL5 interacts with several proteins involved in DNA replication and recombination including Rad51 [19], PCNA [11], FEN1 [20], MRN [21], TopoIIIα and TopoIIIβ [9]. Our recent studies indicate potential co-operation of RECQL5 with Topoisomerase IIα in DNA decatenation and cell cycle progression [22].
In mice, RECQL5 deficiency induces genomic instability [7, 13]. Embryonic stem cells and primary embryonic fibroblasts derived from RECQL5−/− mice show a high level of sister chromatid exchanges (SCEs), and accumulate spontaneous Rad51 and γ-H2AX foci, indicating accumulation of DSBs [7]. Consistent with this observation, spontaneous DNA breaks also accumulate in Drosophila melanogaster RECQL5 mutant larvae [23]. RECQL5 plays an important role in homologous recombination (HR) by displacing Rad51 from nucleoprotein filaments [7]. Collectively, these observations implicate a potential role for RECQL5 in DSB repair.
In this study, we investigated the cellular dynamics of RECQL5 in living cells and monitored its recruitment to laser-induced DSB sites using confocal laser microscopy. The recruitment kinetics of RECQL5 to DSB sites is similar with that of other human RecQ helicases like BLM, WRN and RECQL4. Our retention studies indicate that RECQL5 associates longer than RECQL4 but shorter than BLM and WRN at the laser-induced DSB sites. We demonstrate that both the N-terminal helicase domain and the C-terminal KIX domain are essential for the recruitment and retention of RECQL5 to the DSB sites. Further, we show that the recruitment of RECQL5 to laser-induced DSB sites is independent of functional activities of its interacting partners, MRE11 and RNA polymerase II. To further investigate any potential involvement of other DNA damage signaling and repair proteins, the recruitment of RECQL5 was examined in BLM, WRN and ATM null fibroblasts.
2. MATERIALS AND METHODS
2.1 Cell lines and Transfection
HeLa cells, SV40-transformed normal (GM637), WRN (AG11395), and BLM (GM08505) fibroblasts cell lines were obtained from Coriell cell repositories. HeLa cells were cultured in DMEM media supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Pen-Strep) at 37°C in a humidified atmosphere with 5% CO2. SV40-transformed normal GM637, WRN and BLM fibroblasts were cultured in MEM media supplemented with 10% FBS, 1% Pen–Strep along with 1X non-essential and essential amino acids, vitamins and 2 mM glutamine. For live-cell experiments, cells were plated 24 hrs before transfection in 2 cm glass bottom plates (MatTek Corporation, Ashland, MA, USA) at 60–70% confluency. Lipofectamine LTX was used for transfection, following the manufacturer’s instructions (Invitrogen, Life Technologies, Grand Island, NY).
2.2 Laser micro-irradiation and confocal microscopy
Confocal laser scanning microscopy coupled with micro-irradiation has previously been used to study kinetics of protein-protein and protein-DNA interactions in living cells [24–26]. We and others have applied this technique to study recruitment of RecQ helicases to laser-induced DSBs [27–29]. A Nikon Eclipse 2000E spinning disk confocal microscope was employed with five laser imaging modules and a CCD camera (Hamamatsu, Tokyo, Japan). The set-up integrated a Stanford Research Systems (SRS) NL100 nitrogen laser by Micro point ablation system (Photonics Instruments, St. Charles, IL, USA). Site specific DNA damage was induced using the SRS NL100 nitrogen laser that was adjusted to emit at 435 nm. The power of the laser was attenuated using Improvision’s Volocity software version 5.0 (Improvision/PerkinElmer, Waltham, MA) in terms of percent intensity to induce single strand or DSBs. Positions internal to the nuclei of either live HeLa cells or HeLa cells transfected with GFP-tagged plasmids were targeted using a 40X oil objective lens. Laser intensity at or above 7% was optimized to induce DSBs, while laser intensities lower than 3–4% induce single strand breaks. The cells were transfected with GFP-tagged proteins, targeted at 7% laser intensity and images were captured at various time points and analyzed using Volocity version 5.0 build 6. The recruitment kinetics was calculated by plotting an increase in fluorescence intensity with time. Retention experiments were performed using a special environmental chamber attached to the microscope stage to maintain the physiological conditions (i.e. 37°C, 5% CO2).
2.3 Immunofluorescence and antibodies
Polyclonal rabbit anti-RECQL5 antibody was generated against the C-terminal portion of RECQL5 (amino acids 813–963) and affinity purified as previously described [22]. For endogenous recruitment experiments, HeLa cells were grown in 2 cm gridded glass plates (MatTek Corporation, Ashland, MA) and subjected to micro-irradiation. Following micro-irradiation, cells were fixed with 3.7% formaldehyde in PBS for 10 min at room temperature. The cells were stained with mouse monoclonal γ-H2AX antibody (1:200, Upstate biotechnology, NY) and affinity purified rabbit polyclonal RECQL5 antibody, (1:200) overnight at 4°C. γ-H2AX was used as a marker for DSB sites. The cells were then visualized by incubating with secondary antibodies, Alexa FITC (488)–conjugated donkey anti-mouse, Alexa 647–conjugated donkey anti-rabbit (Invitrogen, Life Technologies, Grand Island, NY). Images were captured using the confocal scanning microscope. For recruitment experiments with Flag-tagged or Omni-tagged RECQL5 plasmids, the cells were fixed 5 min after micro-irradiation and then stained with rabbit polyclonal antibody against Flag (Sigma, St Louis, MO) and mouse monoclonal antibody against Omni (Santa Cruz Biotechnology Inc, Santa Cruz, CA) respectively. Expression vectors for both wild type and ATPase dead Omni-tagged RECQL5 plasmids (pJP 136 RECQL5) were a kind gift from Dr. Pavel Janscak (Zurich, Switzerland).
2.4 Generation of lentivirus and stable RECQL5 knockdown cells
Stable RECQL5-depleted cells were generated previously by using lentiviral shRNA [22]. The pLKO.1 vector harboring a short hairpin, shRNA construct targeting human RECQL5 was obtained from Sigma Aldrich. shRNA targeting the coding region of RECQL5 gene; 5′-CCGGCCCTAAAGGTACGAGTAAGTTCTCGAGAACTTACTCGTACCTTTAGGGT TTTTG- 3′ was used. shRNA construct expressing scrambled sequence was purchased from Addgene, (Plasmid # 1864, CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG, deposited by Sabatini lab). Second-generation VSV-G pseudotyped lenti viruses were generated by transient co-transfection of HEK293T cells with a three-plasmid combination as described previously [22]. For lentiviral transduction, 2 × 105 cells were seeded in 10 cm culture plates and transfected the following day with appropriate lentivirus. Cells were split 48 hours after transfection and selected in the presence of 2 μg/ml puromycin.
2.5 Analysis of 53BP1 foci after γ-irradiation
Both scrambled and RECQL5-depleted cells (25,000 cells each) were seeded on 4 chambered slides (MatTek corporation), grown overnight and then treated with 2 Gy γ-irradiation. Cells were then fixed 0, 1, 8 or 16 hrs, followed by γ-irradiation, and immuno-stained with rabbit polyclonal 53BP1 antibody (1:200, Abcam, Cambridge, MA, USA), visualized with Alexa donkey anti-rabbit 647 (Invitrogen, Carlsbad, CA, USA). An average of ten images representing about 15–20 cells per experiment were taken and the data was analyzed using Volocity software version 5.0 build 6. Average number of 53 BP1 foci was counted per each cell and a graph was plotted against time (in hrs).
2.6 Cloning of GFP tagged RECQL5 fragments
GFP-RECQL5 plasmid was constructed as follows. RECQL5 gene was PCR-amplified from pPG10 plasmid [10], using primers: RQ5-Met-KpnI-F, 5′-GAGATTGGTACCTATGAGCAGCCACCATACAACCT-3′ and RQ5-nostop-XbaI-R, 5′-GAGATTTCTAGATCTCTGGGGGCCACACAG-3′. PCR products were digested and ligated between KpnI and XbaI sites of the pcDNA3.1-CT-GFP plasmid (Invitrogen) generating RQ5/pcDNA3.1-CT-GFP construct. The RECQL5 fragments 1-240, 1-500, 501-650 and 853-991 were amplified by PCR and cloned by using the Topo pcDNA3.1 C-terminal GFP vector (Cat # K4820-01, Invitrogen, Life technologies). Further expression vectors of Flag-tagged full-length RECQL5 and deletion mutants were previously cloned in the pCMV24-3xFLAG (N-terminal flag tag) and C-terminal MYC-tagged vector [30]. SV40 nuclear localization signal (SV40 NLS) was cloned into these truncated flag and GFP-RECQL5 fragments to facilitate nuclear localization.
3. RESULTS
3.1 RECQL5-depleted cells accumulate persistent 53BP1 foci followed by γ-irradiation
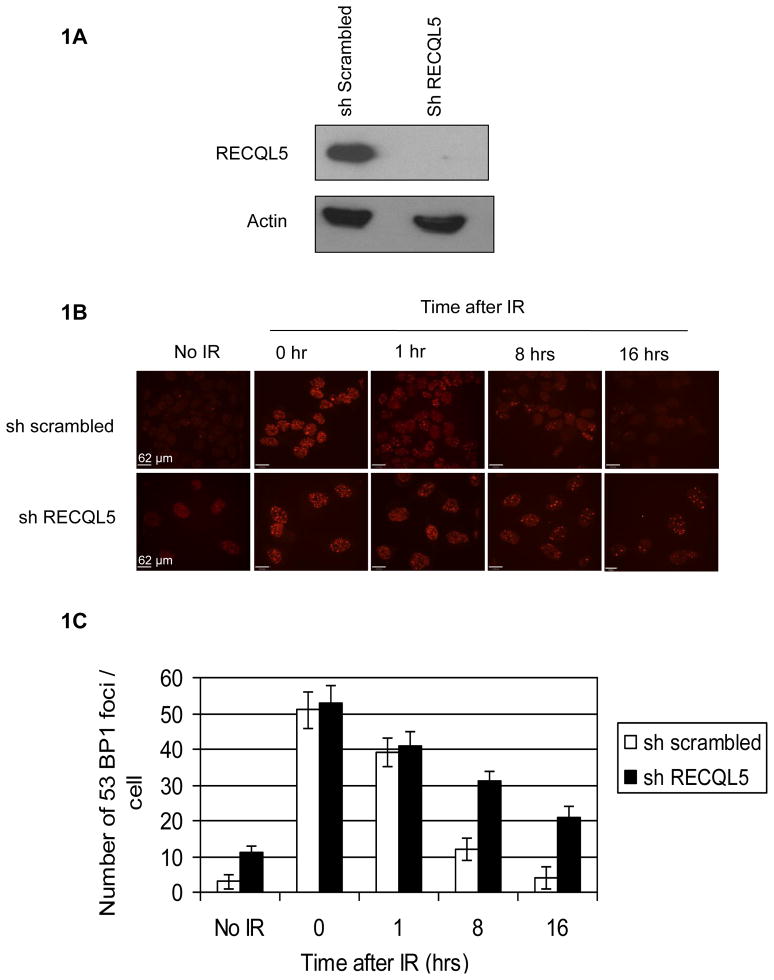
53BP1 foci are widely used as a marker for the accumulation of DSBs [31]. RECQL5 was stably knocked-down in HeLa cells using lenti-viral shRNA. We first verified the RECQL5 depletion using a western blot analysis (Fig. 1A). The efficiency of RecQL5 knockdown (KD) is ~ 90–95% on an average. Both scrambled and RECQL5-depleted cells were irradiated with 2 Gy of γ-irradiation and analyzed for DSBs by quantifying 53BP1 foci (Fig. 1B). RECQL5-depleted cells accumulate relatively more 53BP1 foci than scrambled cells, indicating spontaneous accumulation of DSBs in the absence of irradiation. However, after γ-irradiation, RECQL5-depleted cells accumulate about ~3-fold and 5-fold more 53BP1 foci after 8 hrs and 16 hrs, respectively, compared to scrambled cells (Fig. 1C). The plot shows the average number of 53BP1 foci per cell plotted against time after γ-irradiation and the foci disappeared more quickly in scrambled cells than in RECQL5-depleted cells (Fig 1C).
Fig 1. RECQL5-depleted cells accumulate persistent 53BP1 foci.
(A) The efficiency of RECQL5 knockdown using lentiviral shRNA. (B) Accumulation of 53BP1 foci in RECQL5-depleted cells followed by γ-irradiation. Both scrambled and RECQL5-depleted cells were irradiated with 2 Gy of γ-irradiation and fixed 0, 1, 8 and 16 hrs after IR treatment and stained for 53BP1. (C) The plot showing the average number of 53BP1 foci/cell plotted against time (in hrs) after γ-irradiation. The data points represent the mean of three independent experiments.
3.2 Laser micro-irradiation and recruitment of RECQL5 to laser-induced DSBs
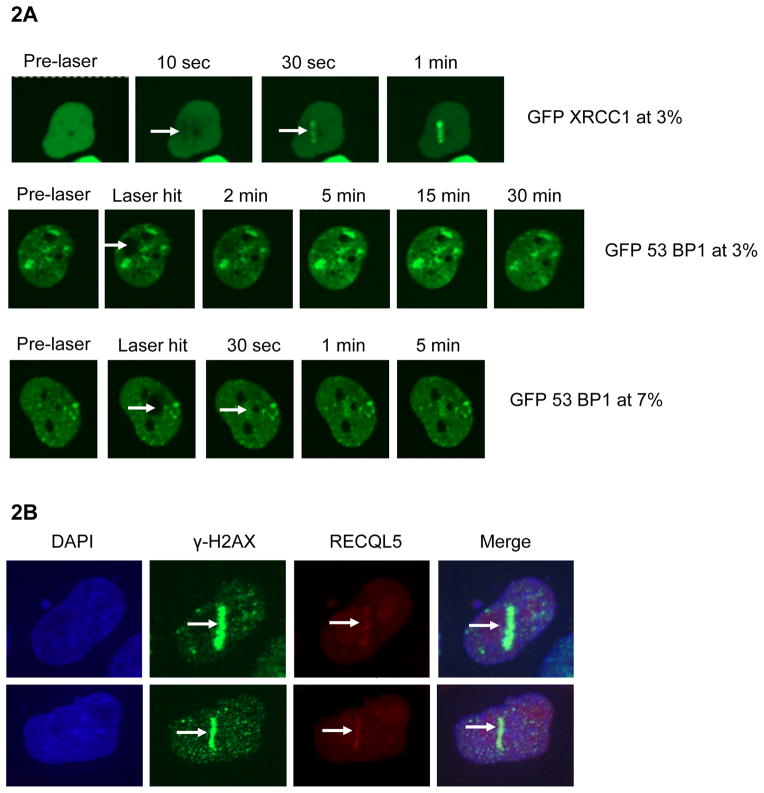
Laser micro-irradiation produces a variety of DNA lesions (such as single strand breaks (SSBs), DSBs and oxidative base modifications) and their frequency depends upon factors like laser intensity and wavelength [32]. DSBs were induced using a 435 nm laser [27]. We established our experimental conditions for selectively generating SSBs (≤ 3% laser intensity, marked by recruitment of XRCC1, Fig. 2A) and DSBs (≥ 7% laser intensity, marked by recruitment of 53BP1 and γ-H2AX, Fig. 2A, B). At lower laser intensities (~ 3%), GFP-XRCC1 was recruited spontaneously within 10 seconds of micro-irradiation (Fig. 2A). But the recruitment of GFP-53BP1 was not observed even 30 min after micro-irradiation at 3% laser intensity, indicating no accumulation of DSBs. At higher laser intensity (~ 7%), we observed efficient recruitment of GFP-53BP1 within 30 seconds of micro-irradiation (Fig. 2A). We then assayed the recruitment of endogenous RECQL5 at the ~7% laser intensity. HeLa cells were fixed 5 min after micro-irradiation and then immuno-stained with RECQL5 and γ-H2AX antibodies. Our results indicate that endogenous RECQL5 co-localizes with γ-H2AX at the laser sites within 5 min of micro-irradiation (Fig. 2B). γ-H2AX was used as a marker for DSBs.
Fig 2. Optimization of laser micro-irradiation and recruitment of endogenous RECQL5 to laser-induced DSB sites.
Confirmation of laser intensity necessary for DSB induction: (A) GFP-XRCC1 is recruited to single strand breaks using 3% laser intensity, while GFP-53BP1 is not. GFP-53BP1 is recruited to DSBs at or above 7 % laser intensity. The white arrow indicates the site of the laser hit. (B) Recruitment of endogenous RECQL5 to laser-induced DSBs at 7% laser intensity. HeLa cells were plated in small 2 cm gridded glass plates and the cells were then fixed and immuno-stained 5 min after micro-irradiation. The arrows in white indicates the site of laser hit and recruitment of γ-H2AX (in green), and RECQL5 (in red) were observed.
3.3 The recruitment kinetics of RECQL5 to the laser-induced DSB sites is similar to that of other human RecQ helicases
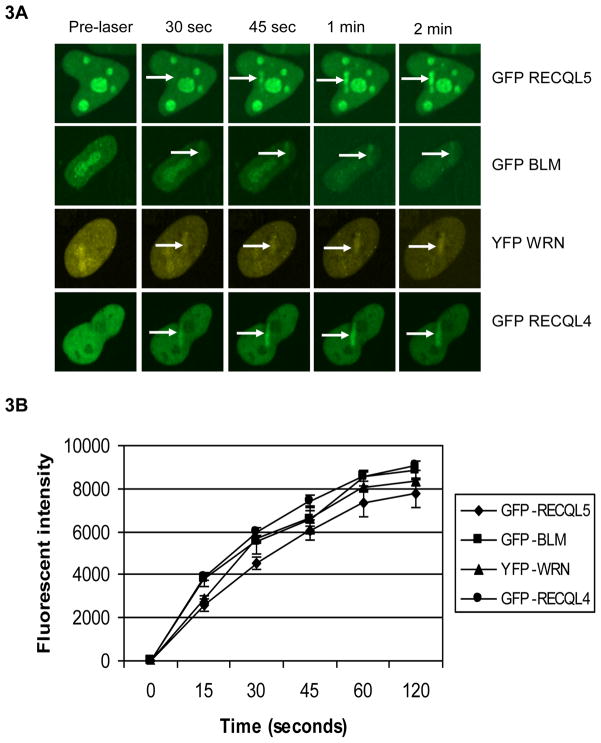
We next used GFP-tagged RECQL5 to investigate its recruitment kinetics to the laser-induced DSB sites and compare with that of other human RecQ helicases. Kinetic studies were performed in transiently-transfected HeLa cells expressing GFP-RECQL4 [27], GFP-BLM [28] YFP-WRN [33] or GFP-RECQL5. Our results indicate that all the proteins were recruited with similar kinetics within 30 seconds of micro-irradiation (Fig. 3A). The plot (Fig. 3B) compares the recruitment kinetics and is similar for all the RecQ helicases over a period of 2 min. It is interesting to note that BLM, WRN and RECQL5 are preferentially localized to the nucleoli while RECQL4 is nuclear. A similar localization of BLM to the nucleolus, mediated by its C-terminal domain, was previously reported and shown to be more important than its localization to PML bodies in maintaining genomic stability [34]. Both BLM and WRN are mobilized from the nucleolus to the nucleus following micro-irradiation, while a major portion of RECQL5 is still retained in the nucleolus after micro-irradiation (Fig. 3A).
Fig 3. Recruitment kinetics of GFP-RECQL5 is comparable with that of other RecQ helicases.
(A) HeLa cells were transfected with GFP-BLM, YFP-WRN, GFP-RECQL4 and GFP-RECQL5, targeted with 7% laser and imaged at the indicated time points. The arrows in white indicate the site of the laser irradiation and recruitment of these RecQ helicases. (B) The plot comparing the recruitment kinetics of different RecQ helicases. The data points represent the mean of three independent experiments (about 6–8 cells per each experiment).
3.4 Retention of GFP-RECQL5 at the laser-induced damage sites is longer than RECQL4, but shorter than BLM and WRN
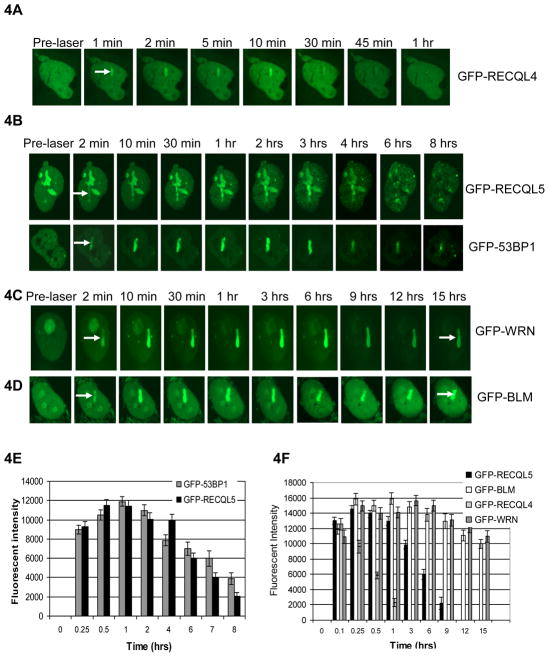
DNA repair efficiency can depend on the dynamics of the DNA repair proteins at the sites of DNA damage. The dynamics include the recruitment and retention of repair enzymes at the DSB sites. While there have been many studies on recruitment there is much less information about the retention of DNA repair proteins at the DSB sites. Our studies show that GFP-RECQL4 associates at the laser-induced DSB sites for a maximum of 45 min-1 hr in human GM637 fibroblasts (Fig. 4A, [27]). Here, we compare the retention dynamics of RECQL5 with that of other human RecQ helicases such as BLM and WRN. GM637 fibroblasts were transiently transfected with GFP-RECQL5, micro-irradiated at 7% laser intensity and live cell imaging was performed over several hours in a specially designed environmental chamber. Our retention studies indicate that GFP-RECQL5 associates at the laser-induced damage sites for a maximum of about 7–8 hrs in GM637 fibroblasts (Fig. 4B, upper panel). The DSB repair efficiency was estimated based on the retention of GFP-53BP1 (Fig. 4B, lower panel). The plot (Fig. 4E) compares the retention kinetics of GFP-53BP1, and shows that it is similar to that of GFP-RECQL5 at the laser-induced DSB sites. The fluorescent intensity of GFP-53BP1 (although not completely lost) decreased about 3–4 fold in 8 hrs after micro-irradiation (Fig. 4E). Our results are consistent with the previous observations by Jensen et al indicating that GFP-53BP1 was lost from the DSB tracks between 7 and 12 hrs followed by micro-irradiation [35]. Similar analysis of endogenous 53BP1 was also performed followed by γ-irradiation. GM637 fibroblasts were irradiated with 2 Gy of γ-irradiation and then analyzed for DSBs by quantifying 53BP1 foci. After γ-irradiation, cells were fixed either immediately (0 hr) or after 1, 8 or 16 hrs and then immuno-stained with 53BP1 antibody and analyzed by confocal microscopy. Our results indicate a direct correlation between the number of DSBs (53BP1 foci) and the kinetics of RECQL5 association at the damage sites and show that the number of 53BP1 foci were reduced by 4-fold within 8 hrs (Supplementary Fig. 1), indicating that more than ~ 75% of the DSBs were repaired during the presence of RECQL5. The foci had almost completely disappeared by 16 hrs. To compare the retention dynamics of RECQL5 relative to that of other human RecQ helicases, retention experiments were performed with GFP-BLM (Fig. 4C) and GFP-WRN (Fig. 4D) under similar conditions. Our retention studies indicate that both GFP-BLM and GFP-WRN can associate longer than 15 hrs. The plot (Fig. 4F) compares the retention kinetics of RECQL5 with that of RECQL4, BLM and WRN. The florescent intensity of GFP-RECQL5 was almost completely gone after 8 hrs, which is longer than that of GFP-RECQL4 (~ 1 hr), but shorter than those of GFP-BLM and GFP-WRN (> 15 hrs).
Fig 4. Retention of GFP-RECQL5, compared with that of other human RecQ helicases in GM637 fibroblasts.
Comparison of retention of GFP-RECQL4 (A) GFP-WRN (C) GFP-BLM (D) in GM637 fibroblasts. (B) Retention of GFP-RECQL5 in GM637 fibroblasts, compared with the DSB repair marker GFP-53BP1. (E) Plot comparing retention of GFP-RECQL5 with GFP-53BP1 in GM637 fibroblasts. (F) Plot comparing the retention of RecQ helicases at the laser-induced DSB sites in GM637 fibroblasts. The data points represent the mean of three independent experiments (about 6–8 cells per each experiment).
Retention of GFP-RECQL5 was also investigated in HeLa cells. Dissociation of GFP-RECQL5 from DSBs occurred over approximately 3 hrs in HeLa cells (Fig. 5A). Our observations indicate that retention of RECQL5 at the damage sites may be cell type specific. Our previous studies showed that BLM and WRN are rapidly recruited to DSBs and remained for longer times (~ 6 and 16 hrs) in HeLa cells, while GFP-RECQL4 dissociated more rapidly within 1 hr after micro-irradiation [27]. Thus the association of RECQL5 at the damage sites is consistently longer than RECQL4 but shorter than BLM and WRN.
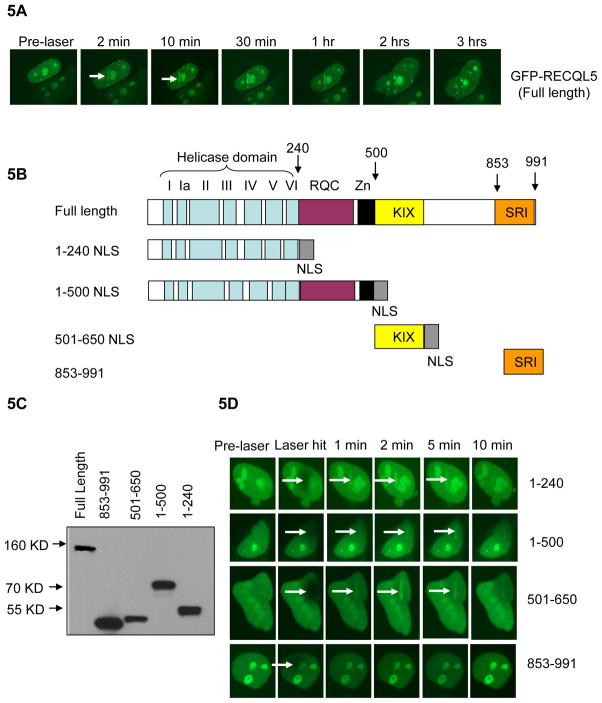
Fig 5. Domain mapping of RECQL5, responsible for its recruitment to DSB sites.
(A) Retention of full length RECQL5 at the laser-induced DSBs in HeLa cells. (B) Construction of GFP-RECQL5 fragments: 1-240, 1-500, 501-650, and 853-991 in pcDNA3.1 C-terminal GFP vector. The fragments 1-240, 1-500 and 501-650 include an artificial NLS at the C-terminus. (C) Expression of GFP-RECQL5 fragments. (D) GFP-RECQL5 fragments were transiently transfected into HeLa cells and micro-irradiated at 9% laser intensity. The arrows in white indicate the site of the laser hit and recruitment of these GFP-RECQL5 fragments.
3.5 RECQL5 helicase activity is not necessary for its recruitment to DSBs
To test whether a functional helicase activity is essential for recruitment of RECQL5 to laser-induced DSBs, Omni-tagged wild type and K58R RECQL5 (ATPase/helicase-dead RECQL5) plasmids were transiently-transfected into HeLa cells and recruitment to laser-induced DSBs was analyzed. The results show similar recruitment of both wild type and RECQL5 K58R to DSBs indicating that the helicase activity is not required (Supplementary Fig. 2A). Although, the helicase dead K58R RECQL5 was recruited within 5 min after micro-irradiation (similar to the wild type RECQL5), it dissociates quickly, and was not observed at the laser stripes 30 min after micro-irradiation (Supplementary Fig. 2B), whereas the wildtype RECQL5 was observed after 2 hrs at the laser-induced DNA damage sites (Supplementary Fig. 2C).
3.6 The helicase and KIX domains are required for recruitment of RECQL5 to DSB sites
To determine which domains of RECQL5 are required for its recruitment to DSBs, constructs were designed to express four GFP-tagged fragments of RECQL5: 1-240 (the helicase domain alone), 1-500 (helicase and RQC domains), 501-650 (the KIX domain), and 853-991 (the SRI domain) (Fig. 5B). The expression of these fragments was confirmed by a Western blot (Fig. 5C). HeLa cells were transiently-transfected with GFP-RECQL5 fragments and live cell imaging was performed followed by micro-irradiation. The results show that the fragments 1-240, 1-500 and 501-650 were recruited to DSBs, but the C-terminal fragment 853-991 (SRI domain) was not, indicating that the helicase and KIX domains can independently participate in DNA damage recognition and aid in the recruitment of RECQL5 to the DSB sites (Fig. 5D). The recruitment efficiency of these fragments appeared slightly lower than that of full length RECQL5, as higher laser intensity was used to detect them (~ 9%, compared with 7% for the full length protein). These GFP-RECQL5 fragments were also more labile and dissociated from DSBs within 10–15 min (Fig. 5D), while the full length protein was retained at DSBs for approximately 3 hrs (Fig. 5A). These results suggest that the presence of both helicase and KIX domains could be essential for efficient retention of RECQL5 at DSB sites.
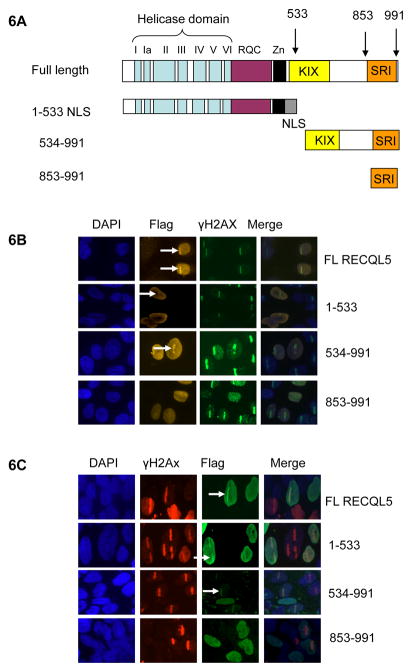
Similar mapping results were also obtained using flag-tagged RECQL5 fragments 1-534, 534-991 and 853-991 (Fig. 6A). Both HeLa cells (Fig. 6B) and U2OS cells (Fig. 6C) were transfected with these flag-tagged RECQL5 fragments and analyzed as described. The short C-terminal SRI fragment, 853-991, was not recruited to DSBs in either cell line, while the remaining fragments 1-534 and 534-991 were recruited efficiently. The results further confirm that both the helicase and KIX domains can independently be responsible for the recruitment of RECQL5 to the DSB sites.
Fig 6. Recruitment of flag tagged RECQL5 fragments to the laser-induced DSB sites.
(A) Graphic image of Flag tagged RECQL5 fragments. The fragments 1-533 and 534-991 include an artificial NLS at the C-terminus. The full length and flag-tagged RECQL5 fragments; 1-533, 534-991 and 853-991 were transfected in HeLa cells (B) and U2OS cells (C) and fixed 5 min after micro-irradiation at 9% laser intensity. The arrows in white indicate the site of the laser hit and recruitment of these flag-tagged RECQL5 fragments. The fragment 853-991 is excluded from the recruitment. The fragments containing the helicase and KIX domains are recruited.
3.7 Recruitment of RECQL5 to the laser-induced damage sites does not depend upon the presence of BLM, WRN and is independent of the functional activities of its interacting partners, MRE11 and RNA polymerase II
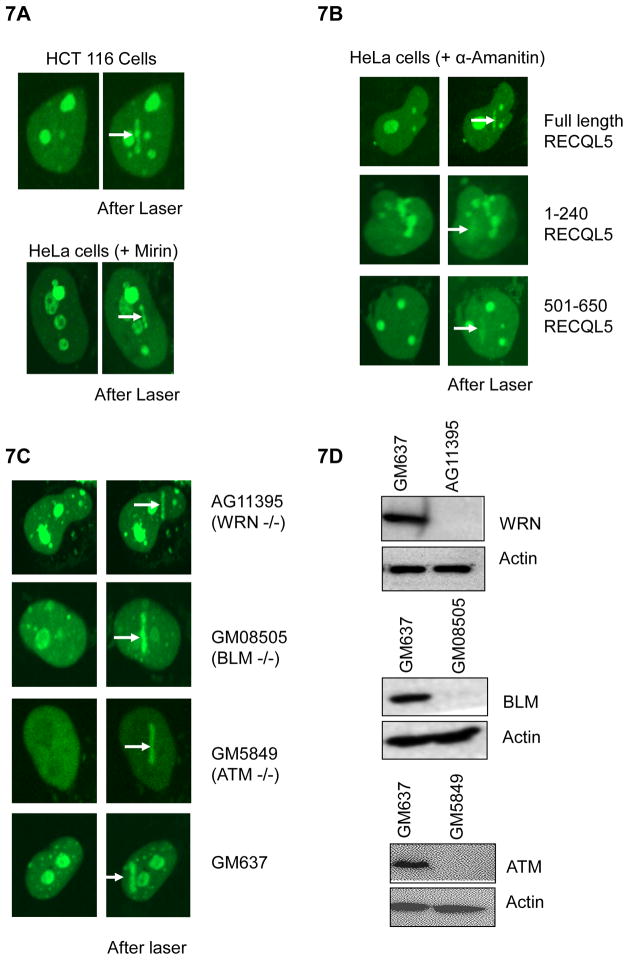
Previous studies indicate that RECQL5 interacts with the MRN complex and that MRE11 is essential for recruitment of RECQL5 to DSBs [21]. We examined the recruitment of GFP-RECQL5 in HCT116 cells which express an exonuclease-defective mutant allele of MRE11 [36]. MRE11 exonuclease activity was not required for recruitment of RECQL5 to DSBs (Fig. 7A, upper panel). Similar results were obtained in HeLa cells treated with Mirin (100 μg/ml) (Fig. 7A, lower panel), which inhibits MRE11 exonuclease [37]. These results indicate that the recruitment of RECQL5 does not depend on the strand resection activity of MRE11.
Fig 7. Recruitment of RECQL5 is independent of the presence of other human RecQ helicases or functional activities of its interacting partners, MRE11 and RNA polymerase II.
(A) Recruitment of GFP-RECQL5 in HCT116 cells (upper panel). Recruitment of GFP-RECQL5 in HeLa cells treated with Mirin for 1 hr prior to laser micro-irradiation (Tocris Bioscience, 100 μM) (lower panel). (B) Recruitment of GFP RECQL5 in HeLa cells, treated with α-Amanitin (Sigma, 50 μg/ml) for 2 hrs prior to laser micro-irradiation. (C) Recruitment of RECQL5 in WRN- (AG11395), BLM- (GM8505), ATM-deficient (GM5849) fibroblasts, compared with the normal GM637 fibroblasts. (D) Western analyses of these fibroblasts confirming their deficiency in WRN, BLM and ATM respectively.
RNA polymerase II was previously shown to be a prominent and specific interacting partner of RECQL5 (relative to that of other human RecQ helicases) [14]. The KIX domain, one of the domains responsible for recruitment of RECQL5, was proposed to bind both initiation (Pol IIa) and elongation (Pol IIo) forms of the RNA polymerase II [16]. To test if a transcribing RNA polymerase is essential for the recruitment of GFP-RECQL5, HeLa cells were transfected with GFP-RECQL5, and then treated with α-Amanitin (50 μg/ml), which specifically blocks RNA Polymerase II [38]. The recruitment experiments were performed as described above, and the results indicate that active transcription by RNA polymerase II is not required for recruitment of RECQL5 to DSBs (Fig. 7B). We also observed that the recruitment of the GFP-RECQL5 helicase and KIX domains was not affected by treatment with α-Amanitin, indicating that active transcription is not required for the recruitment of either domains to the DSB sites (Fig. 7B).
To identify additional proteins that might be required for the recruitment of RECQL5, similar recruitment studies were performed in WRN (AG11395), BLM (GM08505) and ATM-deficient (GM5849) patient fibroblasts (Fig. 7C). SV40-transformed GM637 were used as control fibroblasts. The fibroblasts were assayed by western blot and they were deficient of the respective proteins (Fig. 7D). Recruitment of GFP-RECQL5 to the sites of laser micro-irradiation was proficient in these cell lines and thus independent of the presence of BLM, WRN and other DNA damage signaling proteins such as ATM (Fig. 7C).
4. DISCUSSION
Although previous studies have implicated RECQL5 in DSB repair [7, 11, 23], there has been no report on the association dynamics of RECQL5 at DSB sites and how it differs from that of other RecQ helicases in both recognition and association at the damage sites. RECQL5-depleted cells accumulate persistent 53BP1 foci after γ-irradiation, indicating accumulation of DSBs and direct involvement of RECQL5 in DSB repair. We employed laser micro-irradiation to image the kinetic behavior of RecQ helicases in response to laser-induced DNA damage in living cells. As previously shown for BLM, WRN and RECQL4 [27–29], GFP-RECQL5 is recruited rapidly to the site of laser-induced DNA damage. The endogenous RECQL5 is also recruited to DSBs, where it co-localizes with γ-H2AX at the micro-irradiated site.
Our studies provide evidence that although these human RecQ helicases are recruited to DSBs with similar kinetics, there is a difference in their retention times. The retention pattern of GFP-RECQL5 was similar to that of GFP-53BP1; more than about 75% of DSBs were repaired during the presence of RECQL5, indicating its potential role in DSB repair. We show that RECQL5 associates longer than RECQL4 at the DSB sites, but disassociates faster than BLM and WRN. Thus, these human RecQ helicases appear to have different intrinsic capacities for retention at the DSB sites. This is a novel observation, and could be attributed to diverse and differential roles of these human RecQ helicases, such as their upstream and downstream roles both in DNA damage signaling and repair processes. Consistent with our observations, RECQL5 was shown to have a prominent role in early homologous recombination (HR), like disruption of Rad51 nucleo-protein filaments [7], but to our knowledge no downstream roles of RECQL5 in the DNA repair processes have been described. On the other hand, consistent with the longer retention, both BLM and WRN were proposed to play significant upstream and downstream roles in both DNA damage signaling and repair processes such as HR and NHEJ [1, 39–42]. In addition, DNA repair proteins like MRN, Rad51 and γH2AX have been reported to persist at the damage sites for more than 24 hrs, and this was attributed to their potential role in DNA damage surveillance [43]. BLM and WRN might have a similar potential role in the surveillance of genomic integrity.
RECQL5 is structurally unique and distinct from other RecQ helicases, in that, it does not possess the C-terminal HRDC domain which is present in both BLM and WRN, nor does it have any homology with the longer N-terminal domain of RECQL4. Previous studies indicate that both BLM and WRN are rapidly recruited to laser-induced DSBs via the HRDC domain [28, 29]. Further, the domain of RECQL4 responsible for its DNA damage localization has been mapped to its unique N-terminus domain (363–492), which shares no homology to the recruitment domains of BLM and WRN [27]. Mapping of domains within RECQL5 reveal that both the N-terminal helicase domain and the C-terminal KIX domain are essential for its recruitment. Our results are consistent using two independent flag-tagged and GFP-tagged RECQL5 fragments. All of the above observations indicate that RecQ helicases might use different domains in order to localize to the damage sites. Previous studies by Islam et al., indicates that RECQL5 requires both the helicase and KIX domains for correction of sister chromatid exchanges and resistance to camptothecin [16]. We propose that both the helicase and KIX domains are the primary DNA damage recognition sites and are also essential for stable association of RECQL5 to the DSB sites. Although both domains get recruited independently to the DSB sites, they are labile and disassociate within 10 min compared with full length RECQL5, which associates for about 3 hrs in HeLa cells. This suggests that RECQL5 requires both domains in order to be retained efficiently at the DSB sites and facilitate effective DNA repair. Interestingly, the helicase dead K58R RECQL5 also dissociates quickly compared to the wildtype RECQL5, indicating the helicase dead RECQL5 might not have an effective role in the DNA repair process, compared with that of the wildtype protein, and hence disperse quickly from the site of the DNA damage.
Interestingly, the region of RECQL5 spanning the amino acids 561–651, within the KIX domain, was shown to be responsible for unwinding the lagging-strand arm and to promote strand exchange at a stalled replication fork [11]. The KIX domain is also a part of the RNA polymerase II (RNA pol II) interacting domain and a mutation in KIX domain completely disrupts the interaction. Primary sequence alignment revealed a high degree of similarity between RecQL5-KIX and the KIX domains from several Pol II transcriptional regulators, CBP, p300, and Med15 [16]. RNA pol II has recently been shown to be recruited to the sites of laser micro-irradiation [44]. Therefore, we tested whether RECQL5 recruitment required active RNA polymerase II transcription. Our results showed proficient recruitment of full length GFP-RECQL5 and both the helicase and KIX domains after treatment with α-Amanitin, suggesting that active transcription by RNA pol II is not essential for the recruitment or loading of RECQL5 to DSB sites. We speculate that the role of RECQL5 in DSB repair could be independent of its role in transcription.
Previous studies by Zheng et al indicate that MRE11 is essential for recruitment of RECQL5 to DSB sites. Here we show that the recruitment of RECQL5 is independent of the exonuclease activity of MRE11 and that RECQL5 does not require strand resection by MRE11. However, the interaction between the two proteins indicates that MRE11 may function in physically loading RECQL5 on to the DSB sites. MRE11 is a 3′-5′ exonuclease and produces 5′-overhangs. RECQL5 might not favor loading on to the 5′-overhangs, as it belongs to the 3′-5′ translocating family of RecQ helicases.
Our study thus provides an initial overview of the association of RECQL5 with the damage sites in comparison with that of other human RecQ helicases. Although the recruitment of RECQL5 is independent of BLM and WRN, it would be interesting to observe if the retention of RECQL5 is affected in their absence. Such studies would help us understand the overlapping roles or a possible functional interplay among these RecQ helicases in the maintenance of genomic stability. Previous studies indicate that RECQL5 has overlapping functions with that of BLM in homologous recombination [7, 45]. It would be of further interest to determine functional co-operations among these RecQ helicases. Future studies are being undertaken to explore this interesting avenue.
Supplementary Material
Highlights.
DNA double strand breaks (DSBs) were specifically generated using micro-irradiation.
RECQL5 depleted cells accumulate persistent DSBs following γ-irradiation.
We provide an insight into the recruitment and retention dynamics of RECQL5 at DSBs.
Both helicase and KIX domains of RECQL5 confer its recruitment and retention at DSBs.
Human RecQ helicases have different inherent capacities to be retained at DSB sites.
Acknowledgments
We would like to thank Dr. Leslie Ferrari and Dr. Chandrika Canugovi for critical reading of the manuscript. This work was supported by funds from the Intramural Research Program of the National Institute on Aging, NIH, AG000726-20.
ABBREVIATIONS
- DSBs
Double strand breaks
- SSBs
Single strand breaks
- BLM
Bloom syndrome protein
- WRN
Werner syndrome protein
- HR
Homologous recombination
- BS
Bloom syndrome
- WS
Werner syndrome
- RTS
Rothmund-Thomson syndrome
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh DK, Ahn B, Bohr VA. Roles of RECQ helicases in recombination based DNA repair, genomic stability and aging. Biogerontology. 2009;10:235–252. doi: 10.1007/s10522-008-9205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh DK, Ghosh AK, Croteau DL, Bohr VA. RecQ helicases in DNA double strand break repair and telomere maintenance. Mutat Res. 2011 doi: 10.1016/j.mrfmmm.2011.06.002. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hickson ID. RecQ helicases: caretakers of the genome. Nat Rev Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 5.Ichikawa K, Noda T, Furuichi Y. Preparation of the gene targeted knockout mice for human premature aging diseases, Werner syndrome, and Rothmund-Thomson syndrome caused by the mutation of DNA helicases. Nihon Yakurigaku Zasshi. 2002;119:219–226. doi: 10.1254/fpj.119.219. [DOI] [PubMed] [Google Scholar]
- 6.Lindor NM, Furuichi Y, Kitao S, Shimamoto A, Arndt C, Jalal S. Rothmund-Thomson syndrome due to RECQ4 helicase mutations: report and clinical and molecular comparisons with Bloom syndrome and Werner syndrome. Am J Med Genet. 2000;90:223–228. doi: 10.1002/(sici)1096-8628(20000131)90:3<223::aid-ajmg7>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 7.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, Jasin M, Vogel H, Sung P, Luo G. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sekelsky JJ, Brodsky MH, Rubin GM, Hawley RS. Drosophila and human RecQ5 exist in different isoforms generated by alternative splicing. Nucleic Acids Res. 1999;27:3762–3769. doi: 10.1093/nar/27.18.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimamoto A, Nishikawa K, Kitao S, Furuichi Y. Human RecQ5beta, a large isomer of RecQ5 DNA helicase, localizes in the nucleoplasm and interacts with topoisomerases 3alpha and 3beta. Nucleic Acids Res. 2000;28:1647–1655. doi: 10.1093/nar/28.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren H, Dou SX, Zhang XD, Wang PY, Kanagaraj R, Liu JL, Janscak P, Hu JS, Xi XG. The zinc-binding motif of human RECQ5beta suppresses the intrinsic strand-annealing activity of its DExH helicase domain and is essential for the helicase activity of the enzyme. Biochem J. 2008;412:425–433. doi: 10.1042/BJ20071150. [DOI] [PubMed] [Google Scholar]
- 13.Hu Y, Lu X, Zhou G, Barnes EL, Luo G. Recql5 plays an important role in DNA replication and cell survival after camptothecin treatment. Mol Biol Cell. 2009;20:114–123. doi: 10.1091/mbc.E08-06-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aygun O, Svejstrup J, Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc Natl Acad Sci U S A. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aygun O, Xu X, Liu Y, Takahashi H, Kong SE, Conaway RC, Conaway JW, Svejstrup JQ. Direct inhibition of RNA polymerase II transcription by RECQL5. J Biol Chem. 2009;284:23197–23203. doi: 10.1074/jbc.M109.015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Islam MN, Fox D, III, Guo R, Enomoto T, Wang W. RecQL5 promotes genome stabilization through two parallel mechanisms--interacting with RNA polymerase II and acting as a helicase. Mol Cell Biol. 2010;30:2460–2472. doi: 10.1128/MCB.01583-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanagaraj R, Huehn D, MacKellar A, Menigatti M, Zheng L, Urban V, Shevelev I, Greenleaf AL, Janscak P. RECQ5 helicase associates with the C-terminal repeat domain of RNA polymerase II during productive elongation phase of transcription. Nucleic Acids Res. 2010;38:8131–8140. doi: 10.1093/nar/gkq697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Xu X, Liu Y. The SET2-RPB1 interaction domain of human RECQ5 is important for transcription-associated genome stability. Mol Cell Biol. 2011;31:2090–2099. doi: 10.1128/MCB.01137-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwendener S, Raynard S, Paliwal S, Cheng A, Kanagaraj R, Shevelev I, Stark JM, Sung P, Janscak P. Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J Biol Chem. 2010;285:15739–15745. doi: 10.1074/jbc.M110.110478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Speina E, Dawut L, Hedayati M, Wang Z, May A, Schwendener S, Janscak P, Croteau DL, Bohr VA. Human RECQL5beta stimulates flap endonuclease 1. Nucleic Acids Res. 2010;38:2904–2916. doi: 10.1093/nar/gkp1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng L, Kanagaraj R, Mihaljevic B, Schwendener S, Sartori AA, Gerrits B, Shevelev I, Janscak P. MRE11 complex links RECQ5 helicase to sites of DNA damage. Nucleic Acids Res. 2009;37:2645–2657. doi: 10.1093/nar/gkp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramamoorthy M, Tadokoro T, Rybanska I, Ghosh AK, Wersto R, May A, Kulikowicz T, Sykora P, Croteau DL, Bohr VA. RECQL5 cooperates with Topoisomerase II alpha in DNA decatenation and cell cycle progression. Nucleic Acids Res (2011) 2011;40:1621–1635. doi: 10.1093/nar/gkr844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama M, Yamaguchi S, Sagisu Y, Sakurai H, Ito F, Kawasaki K. Loss of RecQ5 leads to spontaneous mitotic defects and chromosomal aberrations in Drosophila melanogaster. DNA Repair (Amst) 2009;8:232–241. doi: 10.1016/j.dnarep.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 24.Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 25.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 26.Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008;9:91–96. doi: 10.1038/sj.embor.7401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh DK, Karmakar P, Aamann M, Schurman SH, May A, Croteau DL, Burks L, Plon SE, Bohr VA. The involvement of human RECQL4 in DNA double-strand break repair. Aging Cell. 2010;9:358–371. doi: 10.1111/j.1474-9726.2010.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karmakar P, Seki M, Kanamori M, Hashiguchi K, Ohtsuki M, Murata E, Inoue E, Tada S, Lan L, Yasui A, Enomoto T. BLM is an early responder to DNA double-strand breaks. Biochem Biophys Res Commun. 2006;348:62–69. doi: 10.1016/j.bbrc.2006.07.037. [DOI] [PubMed] [Google Scholar]
- 29.Lan L, Nakajima S, Komatsu K, Nussenzweig A, Shimamoto A, Oshima J, Yasui A. Accumulation of Werner protein at DNA double-strand breaks in human cells. J Cell Sci. 2005;118:4153–4162. doi: 10.1242/jcs.02544. [DOI] [PubMed] [Google Scholar]
- 30.Popuri V, Croteau DL, Bohr VA. Substrate specific stimulation of NEIL1 by WRN but not the other human RecQ helicases. DNA Repair (Amst) 2010;9:636–642. doi: 10.1016/j.dnarep.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dinant C, de JM, Essers J, van Cappellen WA, Kanaar R, Houtsmuller AB, Vermeulen W. Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. J Cell Sci. 2007;120:2731–2740. doi: 10.1242/jcs.004523. [DOI] [PubMed] [Google Scholar]
- 33.Liu FJ, Barchowsky A, Opresko PL. The Werner syndrome protein functions in repair of Cr(VI)-induced replication-associated DNA damage. Toxicol Sci. 2009;110:307–318. doi: 10.1093/toxsci/kfp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yankiwski V, Noonan JP, Neff NF. The C-terminal domain of the Bloom syndrome DNA helicase is essential for genomic stability. BMC Cell Biol. 2001;2:11. doi: 10.1186/1471-2121-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J Cell Biol. 2005;170:201–211. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wen Q, Scorah J, Phear G, Rodgers G, Rodgers S, Meuth M. A mutant allele of MRE11 found in mismatch repair-deficient tumor cells suppresses the cellular response to DNA replication fork stress in a dominant negative manner. Mol Biol Cell. 2008;19:1693–1705. doi: 10.1091/mbc.E07-09-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML, Kopelovich L, Jasin M, Baer R, Paull TT, Gautier J. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol. 2008;4:119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gong XQ, Nedialkov YA, Burton ZF. Alpha-amanitin blocks translocation by human RNA polymerase II. J Biol Chem. 2004;279:27422–27427. doi: 10.1074/jbc.M402163200. [DOI] [PubMed] [Google Scholar]
- 39.Karmakar P, Piotrowski J, Brosh RM, Jr, Sommers JA, Miller SP, Cheng WH, Snowden CM, Ramsden DA, Bohr VA. Werner protein is a target of DNA-dependent protein kinase in vivo and in vitro, and its catalytic activities are regulated by phosphorylation. J Biol Chem. 2002;277:18291–18302. doi: 10.1074/jbc.M111523200. [DOI] [PubMed] [Google Scholar]
- 40.Patro BS, Frohlich R, Bohr VA, Stevnsner T. WRN helicase regulates the ATR-CHK1-induced S-phase checkpoint pathway in response to topoisomerase-I-DNA covalent complexes. J Cell Sci. 2011;124:3967–3979. doi: 10.1242/jcs.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr Homologous recombination resolution defect in werner syndrome. Mol Cell Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu L, Chan KL, Ralf C, Bernstein DA, Garcia PL, Bohr VA, Vindigni A, Janscak P, Keck JL, Hickson ID. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 2005;24:2679–2687. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim JS, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J Cell Biol. 2005;170:341–347. doi: 10.1083/jcb.200411083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci USA. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu Y, Lu X, Barnes E, Yan M, Lou H, Luo G. Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol Cell Biol. 2005;25:3431–3442. doi: 10.1128/MCB.25.9.3431-3442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.