Abstract
Mutations in the TMPRSS3 gene are known to cause autosomal recessive nonsyndromic hearing impairment (ARNSHI). After undergoing a genome scan, ten consanguineous Pakistani families with ARNSHI were found to have significant or suggestive evidence of linkage to the TMPRSS3 region. In order to elucidate if the TMPRSS3 gene is responsible for ARNSHI in these families, the gene was sequenced using DNA samples from these families. Six TMPRSS3 variants were found to co-segregate in ten families. None of these variants were detected in 500 control chromosomes. Four novel variants, three of which are missense [c.310G>A (p.Glu104Lys), c.767C>T (p.Ala256Val) and c.1273T>C (p.Cys425Arg)] and one nonsense [c.310G>T (p.Glu104Stop)], were identified. The pathogenicity of novel missense variants was investigated through bioinformatics analyses. Additionally, the previously reported deletion c.208delC (p.His70ThrfsX19) was identified in one family and the known mutation c.1219T>C (p.Cys407Arg) was found in five families, which makes c.1219T>C (p.Cys407Arg) the most common TMPRSS3 mutation within the Pakistani population. Identification of these novel variants lends support to the importance of elements within the low-density lipoprotein receptor A (LDLRA) and serine protease domains in structural stability, ligand binding and proteolytic activity for proper TMPRSS3 function within the inner ear.
Keywords: autosomal recessive nonsyndromic hearing impairment, LDLRA domain, Pakistan, serine protease domain, TMPRSS3
INTRODUCTION
Hearing impairment (HI) is the most common sensory defect in humans, affecting at least 278 million people worldwide (1). Eighty percent of the burden of HI is borne by low- and middle-income countries (1) such as Pakistan, where the prevalence of HI among children has been estimated to be as high as 7.9% (2). Of 1-2 per 1000 neonates with pre-lingual HI, about 50% have a genetic etiology for HI (3). Of the genetic HI cases, 70% are non-syndromic; for nonsyndromic (NS) HI, 77% are of autosomal recessive (AR) inheritance (4). For ARNSHI alone, >90 loci have been mapped and about 40 genes have been identified (Hereditary Hearing Loss Homepage).
The TMPRSS3 gene (MIM 605511) on 21q22.3 encodes a transmembrane serine protease that is hypothesized to target the epithelial amiloride-sensitive sodium channel (ENaC), which mediates sodium reabsorption in the endolymph (5). In individuals of European descent, TMPRSS3 variants cause <0.5% of ARNSHI (6). TMPRSS3 variants have also been observed in a small number of Tunisian and Turkish families with ARNSHI (7,8). On the other hand, 3% of Pakistani families with ARNSHI segregate TMPRSS3 variants (6,9), and this prevalence rate is lower compared to the prevalence rates within the Pakistani population of five other ARNSHI genes, namely SLC26A4, GJB2, HGF, TMC1 and MYO15A (10-13). Sixteen TMPRSS3 variants, which are found in three of the four protein domains, have been described (Table 1). Most TMPRSS3 variants have been shown to affect proteolytic cleavage and ENaC activation in functional studies (5,8,14), although the presence of an unknown TMPRSS3 substrate aside from ENaC has been suggested (15). The knowledge of how HI mutations affect function is important to the development of therapeutic strategies, while the knowledge of population-specific prevalence of HI genes and variants could improve HI genetic screening and counseling services.
Table 1.
Previously published TMPRSS3 a mutations
| Variant b | Exon/ Intron |
Protein Domain c | Population | Reference |
|---|---|---|---|---|
| c.208delC (p.His70ThrfsX19) d | Exon 4 | LDLRA (predicted to delete SRCR, serine protease) |
Spanish, Greek, Pakistani, Newfoundlander |
6,15 |
| c.268G>A (p.Ala90Thr) e | Exon 4 | LDLRA | UK Caucasian | 16 |
| c.308A>G (p.Asp103Gly) | Exon 4 | LDLRA | Greek | 6 |
| c.323 −6G>A f | Intron 4 | LDLRA (predicted to delete part of SRCR) |
Pakistani | 17 |
| c.325C>T (p.Arg109Trp) | Exon 5 | LDLRA | Pakistani | 9 |
| c.413C>A (p.Ala138Glu) | Exon 5 | SRCR | UK Caucasian | 16 |
| c.581G>T (p.Cys194Phe) | Exon 7 | SRCR | Pakistani | 9,15 |
| c.646C>T (p.Arg216Cys) | Exon 8 | Serine protease | German | 18 |
| c.647G>T (p.Arg216Leu) | Exon 8 | Serine protease | Turkish | 8 |
| c.753G>C (p.Trp251Cys) | Exon 8 | Serine protease | Tunisian | 7 |
| c.782 +8insT g | Intron 8 | Serine protease | Newfoundlander | 15 |
| c.916G>A (p.Ala306Thr) | Exon 9 | Serine protease | German | 18 |
| c.1180_1187del8ins68 h | Exon 11 | Serine protease | Palestinian | 17 |
| c.1192C>T (p.Gln398Stop) | Exon 11 | Serine protease | Turkish | 8 |
| c.1211C>T (p. Pro404Leu) i | Exon 12 | Serine protease | Turkish, Tunisian | 7,8 |
| c.1219T>C (p.Cys407Arg) | Exon 12 | Serine protease | Pakistani | 9,15 |
NCBI Reference Sequence: NM_024022.2 (isoform a). Isoform a is the most abundantly and widely expressed isoform, including in fetal cochlea (17)
Variant names were modified to follow updated nomenclature rules. None of these variants are in 1000 Genomes.
LDLRA: low-density lipoprotein receptor A; SRCR: scavenger-receptor cysteine rich. No variant has been found in the first transmembrane domain
This variant was originally reported as 207delC (6)
This variant was found in heterozygosity in two probands but a second mutation was not found. Although the c.268G>A variant was predicted to be pathogenic and was not found in 165 controls, it is catalogued in dbSNP as rs45598239 and the variant is present in 5% of the CEU panel.
Originally reported as IVS4 −6G>A
Originally reported as IVS8 +8insT
This is the same as the Ins(βsat)+del mutation
This variant is catalogued in dbSNP as rs28939084 but no population frequency data is available
Within a collection of Pakistani families with ARNSHI, six different TMPRSS3 variants were found to co-segregate with HI and are not found in 250 control individuals from Pakistan. Of these variants, one nonsense and three missense substitutions are novel. The novel variants were found within the low-density lipoprotein receptor A (LDLRA) and serine protease domains, which further strengthens the importance of these domains to TMPRSS3 function within the inner ear.
MATERIALS AND METHODS
Ascertainment and phenotyping
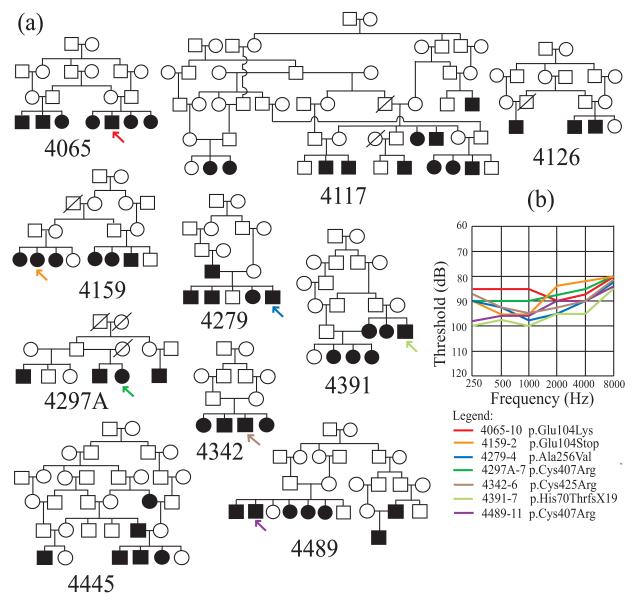
The study was approved by the Institutional Review Boards of the Quaid-I-Azam University and the Baylor College of Medicine and Affiliated Hospitals. Informed consent was obtained from all family members who participated in the study. Eight unrelated families segregating ARNSHI for which TMPRSS3 gene mutations were identified were ascertained from Punjab province, Pakistan, with each family speaking either Punjabi or Sairiki, while two families 4159 and 4489 hail from Sindh province and speak Sindhi (Fig.1a). Family members were interviewed for comprehensive clinical history in order to rule out perinatal, ototoxic, infectious and traumatic factors that could lead to HI. Physical examination included careful assessment for syndromic and vestibular features. Air-conduction thresholds were tested at frequencies 250-8,000 Hz using a portable audiometer. The audiograms demonstrate HI that is bilateral and severe-to-profound at all tested frequencies (Fig.1b). In addition, 250 unrelated hearing individuals from Pakistan without a family history of HI were recruited as control individuals.
Fig. 1.
(a) Pedigree drawings of ten families with ARNSHI segregating TMPRSS3 mutations.
Filled symbols are hearing-impaired individuals, while clear symbols denote unaffected family members. The position within the pedigree of each individual with an audiogram is marked by an arrow. Families 4117, 4126 and 4445 (without arrows) segregate the known variant c.1219T>C (p.Cys407Arg) (b) Air-conduction thresholds averaged for both ears for each of seven hearing-impaired individuals with TMPRSS3 mutation. Each line color denotes an individual per family: red, individual 4065-10 with c.310G>A (p.Glu104Lys) mutation; orange, 4159-2 with c.310G>T (p.Glu104Stop); blue, 4279-4 with c.767C>T (p.Ala256Val); dark green, 4297A-7 with c.1219T>C (p.Cys407Arg); brown, 4342-6 with c.1273T>C (p.Cys425Arg); light green, 4391-7 with c.208delC(p.His70ThrfsX19); violet, 4489-11 with c.1219T>C (p.Cys407Arg). All seven individuals have severe-to-profound hearing impairment at tested frequencies.
Genome scan and linkage analysis
Ten families provided venous blood for DNA isolation. Samples from these families were screened for GJB2 (MIM 121011) mutations using sequencing and were found to be negative. DNA samples from three families (4065, 4117 and 4126) underwent a genome scan using ~395 microsatellite markers at the Center for Inherited Disease Research (CIDR). Additionally seven families (4159, 4279, 4297A, 4342, 4391, 4445 and 4489) were genotyped at CIDR using the HumanLinkage-12 panel which contains 6,090 SNP markers.
Multipoint linkage analysis was performed using Allegro1.2c (19). An AR mode of inheritance with complete penetrance and a disease allele frequency of 0.001 were used. Marker allele frequencies were estimated from observed and reconstructed genotypes of the founders from families in the same genome scan. Genetic map distances were derived from the Rutgers combined linkage-physical map of the human genome (20).
TMPRSS3 sequencing and bioinformatics analyses
Primers were designed for all exons of the TMPRSS3 gene (NM_024022.2). From each of ten families, DNA samples from one hearing and two HI individuals were initially selected for sequencing. After standard PCR and purification of PCR products, TMPRSS3 sequencing was performed with the BigDye Terminator v3.1 Cycle Sequencing Kit and Applied Biosystems 3730 DNA Analyzer (Applied Biosystems Inc, Foster City, CA USA). DNA sequences were assembled and analyzed through Sequencher software V4.9 (Gene Codes Corp., Ann Arbor, MI USA). If a variant is identified and is not found in dbSNP or the 1000 Genomes database, DNA samples from remaining family members were sequenced in order to verify that the variant co-segregates with HI status. The identified mutations were named using Mutalyzer (21). TMPRSS3 exons 4, 8 and 12 where variants were identified were sequenced in 250 control individuals from Pakistan.
The possible deleterious effects of the identified missense variants were predicted using SIFT (22) and PolyPhen-2 (23). For assessment of evolutionary conservation of specific amino acid residues and nucleotides, ConSeq (24) and PhyloP (25) scores were calculated. The PROSITE database was scanned for protein motifs (26). For novel variants, protein modeling was performed using the SWISS-MODEL Workspace (27) and model figures were created using Swiss-Pdb Viewer Deep View v4.0 (28).
RESULTS
Of ten families, five families had significant multipoint LOD scores of >3.3 in the TMPRSS3 region, while the remaining families had suggestive LOD scores (1.8<LOD<3.3) (Table 2). Six causal variants [c.208delC(p.His70ThrfsX19), c.310G>A (p.Glu104Lys), c.310G>T (p.Glu104Stop), c.767C>T (p.Ala256Val), c.1219T>C (p.Cys407Arg), c.1273T>C (p.Cys425Arg)] were identified in these families. For each family, only one TMPRSS3 variant was found to co-segregate with HI. None of the six causal variants were found in 250 control individuals from Pakistan.
Table 2.
TMPRSS3 variants identified in Pakistani families with significant or suggestive LOD scores
| Family |
N Affected with DNA |
N Unaffected with DNA |
Max. Multipoint LOD Score a |
Exon | Variant |
|---|---|---|---|---|---|
| 4065 | 7 | 3 | 5.71 | 4 | c.310G>A (p.Glu104Lys)b |
| 4117 | 10 | 13 | 9.16 | 12 | c.1219T>C (p.Cys407Arg) c |
| 4126 | 3 | 3 | 3.44 | 12 | c.1219T>C (p.Cys407Arg) c |
| 4159 | 6 | 5 | 2.53 | 4 | c.310G>T (p.Glu104Stop)b |
| 4279 | 5 | 2 | 2.41 | 8 | c.767C>T (p.Ala256Val)b |
| 4297A | 4 | 3 | 1.76 | 12 | c.1219T>C (p.Cys407Arg) c |
| 4342 | 4 | 2 | 3.01 | 12 | c.1273T>C (p.Cys425Arg)b |
| 4391 | 6 | 3 | 2.99 | 4 | c.208delC (p.His70ThrfsX19)d |
| 4445 | 5 | 7 | 4.77 | 12 | c.1219T>C (p.Cys407Arg) c |
| 4489 | 7 | 7 | 5.79 | 12 | c.1219T>C (p.Cys407Arg) c |
Three novel missense variants [c.310G>A (p.Glu104Lys), c.767C>T (p.Ala256Val), c.1273T>C (p.Cys425Arg)] and one novel nonsense variant [c.310G>T (p.Glu104Stop)] were identified. Two variants were identified within the LDLRA domain at position 104 which has a glutamic acid residue. The p.Glu104Lys variant was labeled as probably damaging or functional by SIFT and PolyPhen-2 (Table 3). The nonsense mutation p.Glu104Stop is predicted to result in truncated protein with removal of two domains, the scavenger-receptor cysteine rich (SRCR) and serine protease domains. The novel missense variants p.Ala256Val and p.Cys425Arg and the known variant p.Cys407Arg, which all occur within the serine protease domain, were also deemed to affect protein function or to be probably damaging by SIFT and PolyPhen-2 (Table 3). PROSITE predicts that the three novel missense variants plus the known variant p.Cys407Arg occur within active sites of TMPRSS3 (Table 3).
Table 3.
Functional prediction for TMPRSS3 missense variants found in Pakistani families in this study
| Variant | SIFT | PolyPhen-2 | PhyloP a | ConSeq b | PROSITE |
|---|---|---|---|---|---|
| c.310G>A (p.Glu104Lys) |
Affect protein function |
Probably damaging |
4.41 | 9 (f) | LDLRA domain signature, tyrosine kinase phosphorylation site |
| c.767C>T (p.Ala256Val) |
Affect protein function |
Probably damaging |
6.26 | 9 (s) | Histidine active site, adjacent to charge relay system |
| c.1219T>C (p.Cys407Arg) c |
Affect protein function |
Probably damaging |
4.92 | 7 (b) | Disulfide bridge, adjacent to serine active site |
| c.1273T>C (p.Cys425Arg) |
Affect protein function |
Probably damaging |
4.92 | 9 (s) | Disulfide bridge, N-myristoylation site |
Vertebrate Basewise Conservation for 44 species by PhyloP (phyloP44wayAll) from the UCSC Genome Browser, Build 36. Positive scores are assigned for nucleotides that are predicted to be conserved and represent −log p-values under a null hypothesis of neutral evolution.
ConSeq conservation scale ranges from 9 for conserved to 1 for variable. e = exposed; b = buried; f = functional (highly conserved and exposed); s = structural (highly conserved and buried)
The p.Cys407Arg variant was first reported by Ben-Yosef et al. (9)
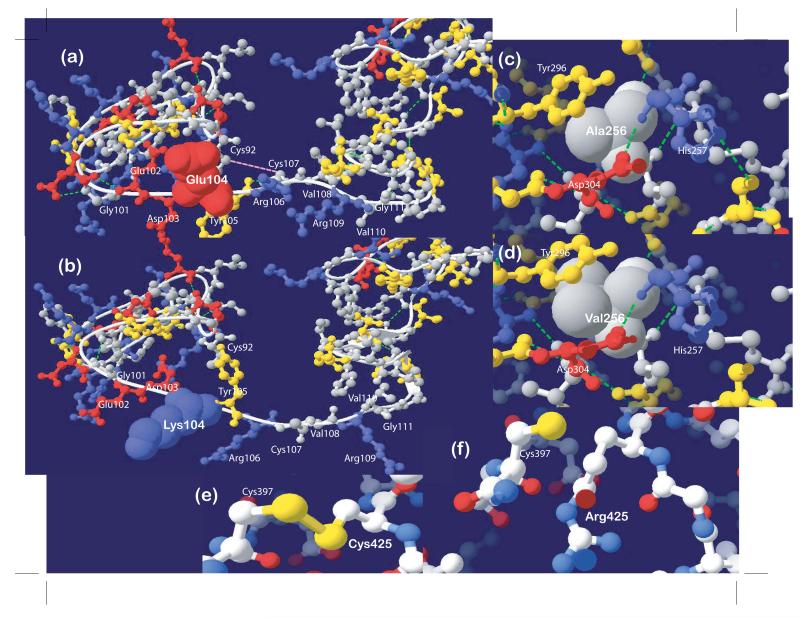
The Protein Data Bank was searched for experimentally derived three-dimensional structures that are similar to TMPRSS3. The LDLRA domain is modeled after the LDLR protein 1N7D (29) which is comprised of cysteine-rich repeats that contain two loops connected by three disulfide bonds. The missense substitution p.Glu104Lys changes the acidic residue to a basic residue, which is expected to affect not only the calcium and ligand-binding affinity of the domain but also the conformation of residues at positions 101 to 111 (Fig.2a-b). Predicted conformational changes include the following: (1) Gly101 and Glu102 side chains are moved from non-exposed to exposed relative to solvent; (2) the side chain of Lys104 is moved away from the calcium-binding site relative to native Glu104; (3) the oxygen-bearing side chains of adjacent residues Asp103 and Tyr105 are moved central to calcium-binding site, which does not conform with the octahedral conformation of calcium-stabilizing side chains (30); (4) Cys107 is prevented from forming a disulfide bridge with Cys92; (5) multiple hydrogen bonds are lost; and (6) the conformation of linker residues Val108-Arg109-Val110-Gly111 are changed. Although the linker region is expected to be flexible (31), changes in position of the cysteine-rich repeat regions relative to each other should affect ligand specificity. Overall the conformational changes that occur with p.Glu104Lys are predicted to result in decreased calcium-binding affinity, ligand specificity and structural stability of the LDLRA domain.
Fig. 2.
(a-b) The p.Glu104Lys mutation is predicted to result in conformational change of the linker region from positions 101 to 111. Multiple hydrogen bonds (dotted green lines) and the disulfide bond between cysteine residues at positions 92 and 107 (pink dotted line) would have been lost. (c-d) The extra side chains of valine compared to alanine at position 256 are expected to interfere with proteolysis at the catalytic center which is comprised of three residues, His257, Tyr296 and Asp304. (e-f) The p.Cys425Arg mutation is predicted to remove the disulfide bond (yellow stick connecting residues) between cysteine residues at positions 397 and 425.
The serine protease domain was most similar to those of other members of the serine protease family, bovine enteropeptidase 1EKB (32) and human hepsin 1Z8G or TMPRSS1 (33). The alanine at position 256 is adjacent to His257, which is one of three signature structural elements that comprise the catalytic center of the serine protease domain or the catalytic triad (32). Valine has two additional non-polar side chains compared to alanine, which may result in direct overlap of the electron density map of residue 256 with the electron density maps of the polar side chain of Tyr296 and of catalytic residues His257 and Asp304 (Fig.2c-d), thus it is expected that Val256 fits poorly within the catalytic center. The novel variant p.Cys425Arg within the serine protease domain occurs at the loop between β sheets 11-12. The Cys425 residue forms a disulfide bond with Cys397 (Fig.2e), which is required to stabilize the serine protease domain. The substitution at position 425 (Fig.2f) is therefore predicted to cause destabilization of the catalytic domain.
Additionally, five of the ten families have the known variant p.Cys407Arg, while one family segregates the previously reported p.H70TfsX19 (c.208delC) (Table 2). Bioinformatic predictions of functional effects of previously reported missense substitutions that were not identified in this study are also reported in Table 4.
Table 4.
Functional prediction for TMPRSS3 missense variants from previous publications that were not identified in this study
| Variant | SIFT | PolyPhen-2 | PhyloP a | ConSeq b | PROSITE |
|---|---|---|---|---|---|
| c.268G>A (p.Ala90Thr) c |
Tolerated | Probably damaging |
2.85 | 5 (e) | Within LDLRA domain |
| c.308A>G (p.Asp103Gly) |
Affect protein function |
Probably damaging |
3.89 | 9 (f) | Tyrosine kinase phosphorylation site |
| c.325C>T (p.Arg109Trp) |
Affect protein function |
Probably damaging |
2.15 | 8 (f) | SRCR domain signature |
| c.413C>A (p.Ala138Glu) |
Affect protein function |
Possibly damaging |
3.22 | 7 (b) | Within SRCR domain |
| c.581G>T (p.Cys194Phe) |
Affect protein function |
Probably damaging |
4.72 | 8 (b) | Disulfide bridge, N-myristoylation site |
| c.646C>T (p.Arg216Cys) |
Affect protein function |
Probably damaging |
6.15 | 9 (f) | Protein kinase C phosphorylation site |
| c.647G>T (p.Arg216Leu) |
Affect protein function |
Probably damaging |
5.13 | 9 (f) | Protein kinase C phosphorylation site |
| c.753G>C (p.Trp251Cys) |
Affect protein function |
Probably damaging |
3.60 | 8 (b) | Close to histidine active site |
| c.916G>A (p.Ala306Thr) |
Tolerated | Probably damaging |
4.85 | 9 (s) | N-myristoylation site |
| c.1211C>T (p. Pro404Leu) d |
Affect protein function |
Probably damaging |
6.19 | 9 (f) | Serine active site |
Vertebrate Basewise Conservation for 44 species by PhyloP (phyloP44wayAll) from the UCSC Genome Browser, Build 36. Positive scores are assigned for nucleotides that are predicted to be conserved and represent −log p-values under a null hypothesis of neutral evolution.
ConSeq conservation scale ranges from 9 for conserved to 1 for variable. e = exposed; b = buried; f = functional (highly conserved and exposed); s = structural (highly conserved and buried)
This variant is catalogued in dbSNP as rs45598239 and the A allele is present in 5% of the CEU panel
This variant is catalogued in dbSNP as rs28939084 but no population frequency data is available
DISCUSSION
We observed 10 families with TMPRSS3 variants in our collection of 353 ARNSHI Pakistani families, which is consistent with the previously reported prevalence rate for the Pakistani population (6, 9). The known variant p.Cys407Arg has been identified in three out of eight Pakistani families with TMPRSS3-related HI (9, 15 and 17). In this study, five of ten families carry the p.Cys407Arg, which makes this variant the most common TMPRSS3 mutation among Pakistanis. The deletion c.208delC (previously called c.207delC) was reported in two out of four Pakistani families with TMPRSS3 mutations (15). However the c.208delC mutation was found in only one family from this study.
Two novel missense variants were identified within the serine protease domain of TMPRSS3. Database searches, evolutionary conservation and protein modeling point towards the p.Ala256Val and p.Cys425Arg variants as pathologic. Failure of proteolysis in the presence of variants at adjacent residues (e.g. Trp251Cys, Ala426Thr) also lends support to the pathogenicity of these novel variants at the serine protease domain (14).
The glutamic acid residue at position 104 lies within the LDLRA domain, which is recognized by means of its cysteine-rich repeats that are important for proper protein folding (29). The c.310G>T variant results in a stop codon at position 104, and is expected to truncate the protein at the LDLRA domain, while deleting the entire SRCR and serine protease domains. The SRCR domain is hypothesized to have binding properties (17), while the serine protease domain serves the catalytic function of TMPRSS3, thus elimination of both domains is predicted to render the protein non-functional.
For the missense variant p.Glu104Lys, bioinformatic prediction through evolutionary conservation and its position within the cluster of highly conserved acidic residues lead to the hypothesis of its pathogenic nature, either by structure destabilization and/or non-binding of calcium and ligand. Interestingly position 104 of TMPRSS3 aligns with the LDLR gene (MIM 606945) at which the familial hypercholesterolemia variant p.Glu207Lys occurs (34). Specifically the LDLRA domain of TMPRSS3 aligns with ligand-binding repeats (LR) 5 and 6 of the LDLR protein, and LR5, which aligns with the consensus sequence that includes Glu104, is considered a basic element that is required to bind ligand (35). Based on known LDLR protein structure, Glu104, together with three aspartic acid residues at positions 93, 100 and 103, participates in calcium binding (30). It has been shown that calcium binding confers stability to disulfide bonds (i.e. Cys79-Cys98 and Cys92-Cys107) and subsequently to protein folding and ligand binding within the LDLRA domain (35,36). In functional experiments, LDLRA domain mutations (i.e. p.Asp103Gly and p.Arg109Trp) resulted in reduced proteolytic activity (5,14).
Identification of novel TMPRSS3 variants lends support to the importance of elements within the low-density lipoprotein receptor A (LDLRA) and serine protease domains in structural stability, ligand binding and proteolytic activity for proper TMPRSS3 function within the inner ear.
ACKNOWLEDGEMENTS
We thank the families for their participation in the study. This work was made possible through grants from the Higher Education Commission, Pakistan (to W.A.) and the National Institutes of Health (NIH) - National Institute of Deafness and Other Communication Disorders Grant DC03594 (to S.M.L.). Genotyping services were provided by the Center for Inherited Disease Research through a fully funded federal contract from the NIH to The Johns Hopkins University, Contract Number N01-HG-65403.
Footnotes
DECLARATION OF ETHICAL STANDARDS AND CONFLICT OF INTEREST All procedures performed comply with the current laws of Pakistan and the United States. The authors declare that they have no conflict of interest. Granting institutions had no role in study design, data collection, analysis and interpretation, and manuscript preparation for publication.
Electronic Database Information
The following URLs were accessed for data in this article: Hereditary Hearing Loss Homepage (http://hereditaryhearingloss.org)
The Rutgers Combined Linkage-Physical Map of the Human Genome (http://compgen.rutgers.edu/RutgersMap)
UCSC Genome Browser (http://genome.ucsc.edu)
1000 Genomes (http://www.1000genomes.org)
Mutalyzer (http://www.mutalyzer.nl)
SIFT (http://sift.jcvi.org)
PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2)
ConSurf Server (http://consurftest.tau.ac.il)
PROSITE (http://ca.expasy.org/prosite)
SWISS-MODEL Workspace (http://swissmodel.expasy.org/workspace)
Swiss-Pdb Viewer Deep View v 4.0 (http://spdbv.vital-it.ch)
REFERENCES
- 1.WHO/World Health Organization Deafness and hearing impairment. 2006 WHO Fact sheet No300. URL: http://www.who.int/mediacentre/factsheets/fs300.
- 2.Elahi MM, Elahi F, Elahi A, et al. Paediatric hearing loss in rural Pakistan. J Otolaryngol. 1998;27:348–353. [PubMed] [Google Scholar]
- 3.Morton NE. Genetic epidemiology of hearing impairment. Ann N Y Acad Sci. 1991;630:16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- 4.Cohen MM, Gorlin RJ. Chapter 3: Epidemiology, etiology and genetic patterns. In: Gorlin RJ, Toriello HV, Cohen MM, editors. Hereditary hearing loss and its syndromes. Oxford University Press; New York: 1995. pp. 9–12. [Google Scholar]
- 5.Guipponi M, Vuagniaux G, Wattenhofer M, et al. The transmembrane serine protease (TMPRSS3) mutated in deafness DFNB8/10 activates the epithelial sodium channel (ENaC) in vitro. Hum Mol Genet. 2002;11:2829–2836. doi: 10.1093/hmg/11.23.2829. [DOI] [PubMed] [Google Scholar]
- 6.Wattenhofer M, Di Iorio MV, Rabionet R, et al. Mutations in the TMPRSS3 gene are a rare cause of childhood nonsyndromic deafness in Caucasian patients. J Mol Med. 2002;80:124–131. doi: 10.1007/s00109-001-0310-6. [DOI] [PubMed] [Google Scholar]
- 7.Masmoudi S, Antonarakis SE, Schwede T, et al. Novel missense mutations of TMPRSS3 in two consanguineous Tunisian families with non-syndromic autosomal recessive deafness. Hum Mutat. 2001;18:101–108. doi: 10.1002/humu.1159. [DOI] [PubMed] [Google Scholar]
- 8.Wattenhofer M, Sahin-Calapoglu N, Andreasen D, et al. A novel TMPRSS3 missense mutation in a DFNB8/10 family prevents proteolytic activation of the protein. Hum Genet. 2005;117:528–535. doi: 10.1007/s00439-005-1332-x. [DOI] [PubMed] [Google Scholar]
- 9.Ben-Yosef T, Wattenhofer M, Riazuddin S, et al. Novel mutations of TMPRSS3 in four DFNB8/B10 families segregating congenital autosomal recessive deafness. J Med Genet. 2001;38:396–400. doi: 10.1136/jmg.38.6.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anwar S, Riazuddin S, Ahmed ZM, et al. SLC26A4 mutation spectrum associated with DFNB4 deafness and Pendred’s syndrome in Pakistanis. J Hum Genet. 2009;54:266–270. doi: 10.1038/jhg.2009.21. [DOI] [PubMed] [Google Scholar]
- 11.Santos RL, Wajid M, Pham TL, et al. Low prevalence of Connexin 26 (GJB2) variants in Pakistani families with autosomal recessive nonsyndromic hearing impairment. Clin Genet. 2005;67:61–68. doi: 10.1111/j.1399-0004.2005.00379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nal N, Ahmed ZM, Erkal E, et al. Mutational spectrum of MYO15A: the large N-terminal extension of myosin XVA is required for hearing. Hum Mutat. 2007;28:1014–1019. doi: 10.1002/humu.20556. [DOI] [PubMed] [Google Scholar]
- 13.Schultz JM, Khan SN, Ahmed ZM, et al. Noncoding mutations of HGF are associated with nonsyndromic hearing loss, DFNB39. Am J Hum Genet. 2009;85:25–39. doi: 10.1016/j.ajhg.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee YJ, Park D, Kim SY, et al. Pathogenic mutations but not polymorphisms in congenital and childhood onset autosomal recessive deafness disrupt the proteolytic activity of TMPRSS3. J Med Genet. 2003;40:629–631. doi: 10.1136/jmg.40.8.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed ZM, Li XC, Powell SD, et al. Characterization of a new full length TMPRSS3 isoform and identification of mutant alleles responsible for nonsyndromic recessive deafness in Newfoundland and Pakistan. BMC Med Genet. 2004;5:24. doi: 10.1186/1471-2350-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchin T, Coy NN, Conlon H, et al. Assessment of the genetic causes of recessive childhood nonsyndromic deafness in the UK – implications for genetic testing. Clin Genet. 2005;68:506–512. doi: 10.1111/j.1399-0004.2005.00539.x. [DOI] [PubMed] [Google Scholar]
- 17.Scott HS, Kudoh J, Wattenhofer M, et al. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat Genet. 2001;27:59–63. doi: 10.1038/83768. [DOI] [PubMed] [Google Scholar]
- 18.Elbracht M, Senderek J, Eggermann T, et al. Autosomal recessive postlingual hearing loss (DFNB8): compound heterozygosity for two novel TMPRSS3 mutations in German siblings. J Med Genet. 2007;44:e81. doi: 10.1136/jmg.2007.049122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gudbjartsson DF, Jonasson K, Frigge ML, et al. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2000;25:12–13. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- 20.Matise TC, Chen F, Chen W, et al. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17:1783–1786. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wildeman M, van Ophuizen E, den Dunnen JT, et al. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum Mutat. 2008;29:6–13. doi: 10.1002/humu.20654. [DOI] [PubMed] [Google Scholar]
- 22.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berezin C, Glaser F, Rosenberg J, et al. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20:1322–1324. doi: 10.1093/bioinformatics/bth070. [DOI] [PubMed] [Google Scholar]
- 25.Pollard KS, Hubisz MJ, Rosenbloom KR, et al. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hulo N, Bairoch A, Bulliard V, et al. The PROSITE database. Nucleic Acids Res. 2006;34:D227–230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold K, Bordoli L, Kopp J, et al. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 28.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 29.Rudenko G, Henry L, Henderson K, et al. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298:2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- 30.Fass D, Blacklow S, Kim PS, et al. Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature. 1997;388:691–693. doi: 10.1038/41798. [DOI] [PubMed] [Google Scholar]
- 31.Beglova N, North CL, Blacklow SC. Backbone dynamics of a module pair from the ligand-binding domain of the LDL receptor. Biochemistry. 2001;40:2808–2815. doi: 10.1021/bi0027276. [DOI] [PubMed] [Google Scholar]
- 32.Lu D, Futterer K, Korolev S, et al. Crystal structure of enteropeptidase light chain complexed with an analog of the trypsinogen activation peptide. J Mol Biol. 1999;292:361–373. doi: 10.1006/jmbi.1999.3089. [DOI] [PubMed] [Google Scholar]
- 33.Herter S, Piper DE, Aaron W, et al. Hepatocyte growth factor is a preferred in vitro substrate for human hepsin, a membrane-anchored serine protease implicated in prostate and ovarian cancers. Biochem J. 2005;390:125–136. doi: 10.1042/BJ20041955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leitersdorf E, Tobin EJ, Davignon J, et al. Common low-density lipoprotein receptor mutations in the French Canadian population. J Clin Invest. 1990;85:1014–1023. doi: 10.1172/JCI114531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fisher C, Abdul-Aziz D, Blacklow SC. A two-module region of the low-density lipoprotein receptor sufficient for formation of complexes with apolipoprotein E ligands. Biochemistry. 2004;43:1037–1044. doi: 10.1021/bi035529y. [DOI] [PubMed] [Google Scholar]
- 36.Blacklow SC, Kim PS. Protein folding and calcium binding defects arising from familial hypercholesterolemia mutations of the LDL receptor. Nat Struct Biol. 1996;3:758–762. doi: 10.1038/nsb0996-758. [DOI] [PubMed] [Google Scholar]