Abstract
During organogenesis, tissues expand in size and eventually acquire consistent ratios of cells with dazzling diversity in morphology and function. During this process progenitor cells exit the cell cycle and execute differentiation programs through extensive genetic reprogramming that involves the silencing of proliferation genes and the activation of differentiation genes in a step-wise temporal manner. Recent years have witnessed expansion in our understanding of the epigenetic mechanisms that contribute to cellular differentiation and maturation during organ development, as this is a crucial step toward advancing regenerative therapy research for many intractable disorders. Among such epigenetic programs, the developmental roles of the polycomb repressive complex 2 (PRC2), a chromatin remodeling complex that mediates silencing of gene expression, have been under intensive examination. This review summarizes recent findings of how PRC2 functions to regulate the transition from proliferation to differentiation during organogenesis and discusses some aspects of the remaining questions associated with its regulation and mechanisms of action.
Keywords: Polycomb, cell fate, histone methyltransferase, differentiation, PRC2, epigenetics
Chromatin remodeling and gene regulation
During embryonic development, the process of organogenesis requires that multipotent progenitor cells respond to developmental cues that drive specific cell fate decisions. These developmental events are orchestrated through significant changes in gene expression that ultimately execute programs of cellular differentiation and maturation. One powerful means of regulating gene expression during development is through control of chromatin structure, which determines accessibility to DNA. Changes in the structure of chromatin are governed in part by post-translational modifications (PTMs) of histones, processes that are mediated by complexes that bind and covalently modify the amino acid side chains of histone tails that are exposed over the surface of the nucleosome. Histone modifications are diverse in nature and include acetylation of lysines, methylation of arginines and lysines, and phosphorylation of serines and threonines, among others (Berger, 2007). Mechanistically, a histone tail may simultaneously harbor several modifications that collectively form a unique docking site that promotes the recruitment of distinct protein complexes that subsequently affect chromatin structure and gene expression (Berger, 2007; Turner, 2007).
The correlation between histone modifications and transcription states has been the subject of focused investigation. The current view is that under certain signaling conditions, positive- or negative-acting histone PTMs are established on gene promoters which in turn can facilitate recruitment of activators or repressors of gene expression, respectively (Berger, 2007). For instance, trimethylation of lysine 4 in histone 3 (H3K4me3) is enriched on the 5′ end of open reading frames and correlates well with transcription activation, and is thus considered an activating mark (Berger, 2007; Chi et al., 2010; Turner, 2007). On the other hand, enrichment in modifications such as H3K9me3 and H3K27me3 is associated with silenced genes (Boyer et al., 2006; Lee et al., 2006; Snowden et al., 2002). However, how well histone PTMs can be predictive of the state of transcription remains unclear. Nevertheless, it is increasingly clear that modulating chromatin structure through histone modification is an important means of regulating the expression of large groups of developmental and signaling genes, and that this is a central mechanism for coordinating developmental transitions during organogenesis.
One important class of chromatin modifiers are the Polycomb group (PcG) genes, which encode highly conserved factors that mediate gene silencing. They were initially identified in Drosophila as repressors of Hox genes during developmental patterning (Alexander et al., 2009; Sparmann and van Lohuizen, 2006). Mutations of PcG members in Drosophila embryos disrupt the correct spatial and temporal expression pattern of Hox genes in body segmentation, leading to embryonic posteriorization (Ringrose and Paro, 2004). This function is also conserved in vertebrates where mutations in several polycomb factors lead to skeletal malformations as a result of disruption of Hox gene expression (Akasaka et al., 1996; del Mar Lorente et al., 2000).
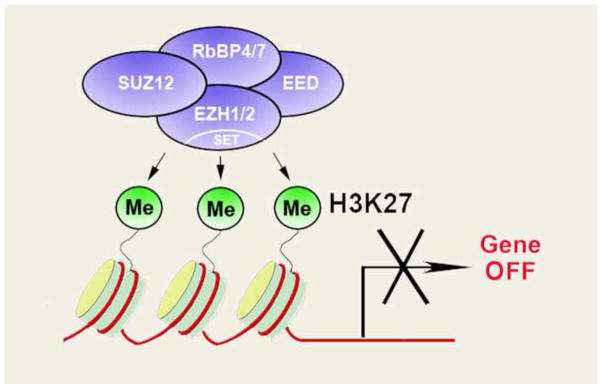
The mechanisms underlying Polycomb-mediated repression are still under intensive study. Several biochemically and functionally distinct complexes, termed Polycomb Repressive Complexes (PRCs), have been purified including, PRC1 and PRC2 (Akizu et al., 2010; Martinez and Cavalli, 2006). PRC1 catalyzes the monoubiquitylation of lysine 119 of histone H2A (H2A119ub) while PRC2 has methyltransferase activities and is primarily responsible for histone3 lysine 27 di-/tri-methylation (H3K27me2/3) (Fig. 1) (Kuzmichev et al., 2002; Sawarkar and Paro, 2010). Interestingly, PRC1 binds the PRC2-mediated mark H3K27me3, and shares occupancy with many of its target genes, providing a functional link between both complexes (Fischle et al., 2003). The addition of H3K27me3 by PRC2 has been proposed to facilitate gene repression by recruiting PRC1 to the methylated region (Cao et al., 2005; Spivakov and Fisher, 2007). However this particular recruitment order (PRC2 then PRC1) has not been firmly established (Margueron and Reinberg, 2011), and there is also evidence that PRC1 and PRC2 do not always occupy the same genomic loci (Ku et al., 2008). Notably, in embryonic stem (ES) cells, PRC1 and PRC2 act redundantly to regulate the ability of these cells to differentiate, since they both repress common developmental regulators, and both PRC1 and PRC2 must be eliminated to prevent ES cell differentiation (Leeb et al., 2010). Thus it is likely that PRC1 and PRC2 have overlapping as well as distinct roles (Richly et al., 2011; Simon and Kingston, 2009).
Figure 1.
The polycomb complex PRC2 functions as a histone methyltransferase. PRC2 contains four core components: EZH1/2, SUZ12, EED and RbBP4/7. PRC2 recruitment to gene promoters leads to deposition of H3K27me3, which is associated with gene repression.
PcGs can mediate silencing of a broad range of genes, and are associated with important biological contexts such as maintenance and differentiation of ES cells, as well as cancer progression (Boyer et al., 2006; Lee et al., 2006; Schwartz et al., 2006). While much has been learned about the biochemical roles of PcGs (Margueron and Reinberg, 2011; Simon and Kingston, 2009), only recently are we gaining an appreciation for their fundamental roles as developmental regulators. While both PRC1 and PRC2 likely function to regulate key aspects of development, recently there has been a particular focus on PRC2, with increasing evidence that this complex plays a critical role in regulating differentiation decisions during vertebrate embryogenesis. Thus, in this review we specifically highlight what is known about the developmental roles of PRC2 function during tissue development.
The Polycomb Repressive Complex PRC2
PRC2 consists of four core subunits: SUZ12 (the mammalian orthologue of Suppressor of Zeste Su(z) 12), EZH2 (the mammalian orthologue of Enhancer of Zeste (E(z)), EED (the mammalian orthologue of Extra Sex Combs ESC) and Retinoblastoma Associated Protein RbAP46/48 (also known as RbBP4/7; the mammalian orthologue of P55) (Fig. 1) (Kuzmichev et al., 2002; Margueron and Reinberg, 2011). These components encompass a diverse cohort of functional activities. SUZ12, for instance, contains a zinc finger motif and is required for EZH2 catalytic activities (Pasini et al., 2004). EZH2 bears histone methyltransferase activity through its highly conserved SET domain: mutations in the SET domain cause loss of H3K27me3 in Drosophila as well as in vertebrates (Kuzmichev et al., 2002; Muller et al., 2002; Su et al., 2003). Interestingly, recent studies have identified a version of PRC2 that contains the EZH2 homolog, EZH1, which can also mediate trimethylation of H3K27 (Margueron et al., 2008; Shen et al., 2008). The third component, EED, is a WD-40 repeat protein that interacts with EZH2 and is required for the EZH2 methyltransferase activity (Ketel et al., 2005; Kuzmichev et al., 2005). EED also plays an important role in the maintenance and propagation of H3K27me3 during cell division since it binds H3K27me3 through its C-terminal domain (Margueron et al., 2009). Together, EZH2, EED and SUZ12 constitute the minimal PRC2 subunits required for catalytic activity and subsequent initiation of gene repression (Ketel et al., 2005; Sparmann and van Lohuizen, 2006). The fourth core component, RbBP4/7, is required for association of PRC2 with the histone tail (Kuzmichev et al., 2002).
Beside the core subunits, PRC2 contains other factors such as JARID2, AEBP2, and PCL that have been shown to occupy most PRC2 target genes (Margueron and Reinberg, 2011; Nekrasov et al., 2007; Peng et al., 2009; Shen et al., 2009). The exact function of these components is not well understood, but evidence suggests that while they are not essential for PRC2-mediated catalytic function per se, their presence modulates PRC2 enzymatic activities and promotes DNA binding (Margueron and Reinberg, 2011).
Roles of PRC2 in differentiation and cell fate commitment
Invertebrate studies support the concept that PcGs play important roles as regulators of developmental gene expression, but only recently is a detailed picture emerging for vertebrate models (Fig. 2). In mouse, mutants of Suz12, Ezh2 and Eeddisplay developmental and proliferative abnormalities and are lethal at early postimplantation stages (Faust et al., 1998; O’Carroll et al., 2001; Pasini et al., 2004). Although these mutations demonstrate how essential PRC2 is for vertebrate development, they shed no light on the tissue-specific roles that PRC2 might play in differentiation and cell fate acquisition.
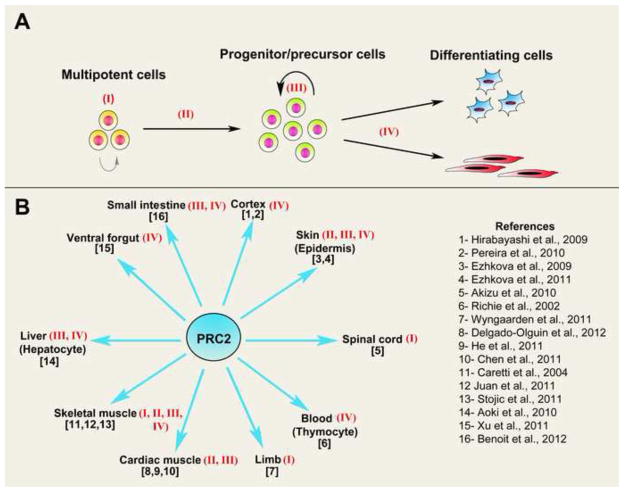
Figure 2. Roles of PRC2 during tissue differentiation.
(A) Schematic figure showing major developmental transitions at which PRC2 functions, including (I) multipotent cell identity, (II) lineage commitment, (III) progenitor expansion, (IV) differentiation/cell fate choice.
(B) Reported tissues that are under regulation by PRC2 during development. Roman numbers represent steps from panel A that have been shown to be regulated by PRC2 while numbers refer to related citations on the reference list to the right. See text for details.
Important insights into the roles of PRC2 in development came from studies on ES cells. Recent excellent reviews have covered this topic (Surface et al., 2010) and we shall only discuss it briefly. Genome wide analysis of PRC2 targets in ES cells revealed that PRC2 and its mark H3K27me3 occupy inactive promoters of key developmental regulators, suggesting a role in the maintenance of ES cell pluripotency (Boyer et al., 2006; Lee et al., 2006). However, this role has been questioned in more recent studies (Chamberlain et al., 2008; Shen et al., 2008; Surface et al., 2010). For example, ES cells can be established from PRC2 core subunit mutants, and, in the case of the Eed mutant, they contribute robustly to multiple lineages in vivo, suggesting that PRC2 is not strictly required for pluripotency (Chamberlain et al., 2008; Pasini et al., 2007; Shen et al., 2008). Rather, PRC2 has a prominent role in the proper differentiation of ES cells since ES cells lacking PRC2 components fail to differentiate in culture conditions (Pasini et al., 2007; Pietersen and van Lohuizen, 2008; Shen et al., 2008). These findings have led to consideration of PRC2 as a regulator of cellular transitions and differentiation decisions.
Consistent with a role for PRC2 in regulating differentiation, in ES cells many of the genes involved in differentiation are co-occupied by the repressive mark H3K27me3 and the activating mark H3K4me3, forming a unique status of “bivalent domain” (Bernstein et al., 2006). Upon differentiation, PRC2 occupancy is lost and H3K27me3 is removed while H3K4me3 is maintained, permitting expression of differentiation genes (Boyer et al., 2006; Lee et al., 2006). Thus the PRC2-mediated repression of the developmental gene promoters occupied with the bivalent domain is transient and seems to prime ES cells for subsequent lineage commitment and cell fate decisions (Landeira et al., 2010; Pietersen and van Lohuizen, 2008; Surface et al., 2010). Recently, it has been shown that the presence of H3K4me3 allosterically inhibits PRC2 catalytic function, raising interesting questions about how bivalent domains are established in ES cells (Schmitges et al., 2011).
In principle, if PRC2 regulates aspects of embryonic stem cell proliferation and differentiation, it may also do so during organ development. The discovery that H3K27me3 can be actively removed by specific demethylases has further potentiated interest in the involvement of PRC2 during tissue development because such function implies that this mark can be transiently utilized to control gene expression, and thus has the potential to play important roles in organogenesis (Lan et al., 2007). If tissue differentiation is executed according to an ES cell culture differentiation paradigm, this predicts that tissue-specific inactivation of PRC2 core components during organogenesis should lead to suppression of differentiation and cell fate acquisition (Margueron and Reinberg, 2011). However, observed outcomes from studying the effect of PRC2 mutations on tissue development suggest that PRC2 function is context-specific and depends on selective targeting of gene expression. What is clear is that PRC2 functions to regulate cellular transitions during development, acting to either promote or block differentiation, to fine-tune cell fate acquisition and/or to preserve proper cell identity during progression from proliferation to differentiation (Fig. 2).
Consistent with what has been observed in ES cells, in some contexts PRC2 is required for tissue differentiation. For example, a mouse mutation in Eed causes a partial block in thymocyte differentiation, and inactivation of Ezh2 in adipose tissue impairs adipocyte differentiation due to an abnormal activation of canonical Wnt signaling, a major inhibitor of adipogenesis (Richie et al., 2002; Wang et al., 2010). In contrast, in some contexts PRC2 is required to constrain differentiation. For instance, inhibition of Ezh2 function in mouse epidermal progenitors results in accelerated skin development likely as the result of precocious recruitment of the transcription factor AP-l, which directs a late epidermal differentiation program, to terminal differentiation gene promoters (Ezhkova et al., 2009; Pirrotta, 2009).. Similarly, loss of Ezh2 enhances hepatogenesis and accelerates hepatic maturation in cultured uncommitted hepatic cells although the mechanism is poorly understood (Aoki et al., 2010). Moreover, knockdown of Suz12 in intestinal epithelial cells results in a precocious differentiation due to selective upregulation of terminal differentiation genes (Benoit et al., 2012). Together these findings reveal more complexity than initially apparent from ES cell studies, which do not recapitulate the spatial and temporal aspects of in vivo tissue development, nor the influence of environmental factors or tissue interactions.
Analysis of PRC2 function at multiple stages of development within a given tissue demonstrates additional complexity to PRC2 function. For example, during limb development, Ezh2 is required at early stages for establishing the limb anterior-posterior axis, while at later stages it is required for cell survival and digit elongation, in part through changes in the regulation of Hox gene expression (Wyngaarden et al., 2011). Importantly, the authors found that during late limb bud stages Ezh2 is required for cells to switch from plasticity to a determined state, since Ezh2 mutant but not wild type cells could be respecified in the presence of new positional cues (Wyngaarden et al., 2011). This parallels work in C. elegans embryos showing that the Polycomb complex protein MES-2/E(Z) is required for transition from a developmentally plastic state to the onset of differentiation (Yuzyuk et al., 2009).
There is also evidence that in some contexts PRC2 prevents inappropriate expression of genes from alternate lineages. For example, Ezh2 knockouts cause heart defects as a result of a disruption of normal gene expression profile in cardiomyoctes, including an upregulation of noncardiomyocyte genes, such as Six1, which promotes activation of skeletal muscle genes in differentiating cardiac muscle (Chen et al., 2012; Delgado-Olguin et al., 2012; He et al., 2011). Ezh2 also regulates terminal cell fate choices during the differentiation of the ventral foregut endoderm by suppressing the pancreatic cell fate gene Pdx1 to allow cells to adopt a liver cell fate (Xu et al, 2011). Together these data underscore the importance of in vivo analysis in diverse lineages for defining the role of PRC2 in differentiation decisions.
In many tissues a marked reduction in cell proliferation is also observed upon loss of Ezh2, in part due to an abnormal upregulation of the tumor suppressor gene p16(Ink4A), a major target for PRC2 repression (Aoki et al., 2010; Chen et al., 2009; Ezhkova et al., 2009; He et al., 2011; Juan et al., 2011). This suggests an additional role of PRC2 in controlling the balance between proliferation and differentiation and is consistent with data from cancer studies where upregulation of Ezh2 has been linked to different types of malignancies, and thus used as a marker for several types of aggressive tumors (Fullgrabe et al., 2011; Kleer et al., 2003; Varambally et al., 2002). For instance, Ezh2 is highly expressed in certain types of gliomas and in glioma stem-like cells, and is required for glioma cell proliferation (Orzan et al., 2011). Interestingly, recent studies reported recurrent somatic mutations in lysine27 of histone variants H3.3 and H3.1 in pediatric brain gliomas, further underscoring the importance of epigenetic mechanisms in the regulation of cancer malignancies and providing potential avenues for diagnosis and treatment of cancer (Schwartzentruber et al., 2012; Wu et al., 2012).
The roles of PRC2 and H3K27me3 during myogenesis
The magnitude of the complexity of PRC2 function during organogenesis can be demonstrated by briefly considering its multiple stage-specific roles during skeletal muscle differentiation. Ezh2 was initially found to be expressed in dividing myoblasts of the mouse embryo. In fibroblast reporter assays, Ezh2 could inhibit the activation of transcription mediated by the myogenic factor MYOD, and in undifferentiated cultured myoblasts it was required to restrict the expression of muscle differentiation genes, suggesting that PRC2 is necessary to prevent premature differentiation (Caretti et al., 2004; Prezioso and Orlando, 2011). However, myoblast differentiation proceeded normally upon Ezh2 knockdown in cell cultures, and Ezh2 conditional knockout in mice produced no obvious muscle defects during embryonic development (Juan et al., 2011; Stojic et al., 2011). Rather, Ezh2 seems to be required for postnatal muscle growth and regeneration, acting to maintain identity of postnatal muscle stem cells by constraining the expression of genes irrelevant to muscle development rather than suppressing muscle-specific transcription (Juan et al., 2011). It seems that Ezh1, which is expressed in differentiating myoblasts, plays a more prominent role during embryonic muscle development in the progression from proliferation to differentiation. Upon knockdown of Ezh1, but not Ezh2, cultured myoblasts failed to differentiate properly and exhibited a delay in expression of the muscle-specific bHLH gene myogenin due to reduced recruitment of MYOD to the myogenin promoter (Stojic et al., 2011). This example highlights how the composition of PRC2 subunits can be an important factor in the regulation of multiple steps in the differentiation process.
The roles of PRC2 and H3K27me3 during neurogenesis
The first glimpse of possible functions of PRC2 in neural differentiation came from ES cell studies where it was shown that many of the genes involved in neurogenesis are targets for PRC2-mediated deposition of H3K27me3 (Boyer et al., 2006; Lee et al., 2006). However, ES cells lacking Suz12 do not overproduce neurons but rather suffer failure in executing a proper neural differentiation program under differentiation conditions, presumably due to loss of H3K27me3 (Pasini et al., 2007). Interestingly, sustained maintenance of H3K27me3 by knocking down the H3K27me3-specific demethylase Jmjd3 is also detrimental to ES cell neural differentiation, further suggesting that transient H3K27me3 deposition is essential for the proper execution of the neural differentiation program (Burgold et al., 2008; Sen et al., 2008).
In agreement with data from non-neural tissue development, the function of PRC2 during neural development is context-dependent and does not necessarily follow the ES cell differentiation model. For instance, in mammalian neocortex, either shRNA-mediated knockdown of Eed, or tamoxifen-induced Ezh2 conditional inactivation in neural precursor cells in culture caused a delay in the switch in neural precursor cells from generating neurons to astrocytes, resulting in increased production of neurons (Hirabayashi et al., 2009). Tamoxifen-induced disruption of Ezh2 in vivo during the neurogenic period under the control of ERT2-Cre had a similar effect by extending the neurogenic phase in the developing cortex. In this tissue PRC2 cooperates with PRC1 to restrict the ability of neural progenitors to generate neurons by repressing expression of the proneural bHLH factor neurogenin (Ngn1) during the late phase of neocortical development when the time is proper for astrocyte production (Hirabayashi and Gotoh, 2010; Hirabayashi et al., 2009). In a separate study, conditional inactivation of Ezh2 using Emx1-Cre, which inactivates Ezh2 before the onset of neurogenesis, results in a shift from self renewal towards differentiation, and accelerates the developmental timing for both cortical neurogenesis and gliogenesis (Pereira et al., 2010). This is in contrast to extension of the neurogenic period and delay of gliogenesis reported by Hirabayashi and colleagues (Hirabayashi et al., 2009). The differences in these findings could be due to differences in Cre drivers and timing of inactivation, which warrants further investigation (Testa, 2011). Nevertheless, these studies reinforce the general concept that in many contexts PRC2 functions to regulate the timing of developmental transitions.
Interestingly, PRC1 and PRC2 appear to play similar roles in regulating neocortical development, since tamoxifen-induced inactivation of the PRC1 component Ring1b in neural progenitors in vivo using ERT2-Cre phenocopies the Ezh2 mutant (Hirabayashi and Gotoh, 2010; Hirabayashi et al., 2009). However, there are likely additional functions for PRC1 components since the PRC1 component Bmi-1 is required for neural stem cell self-renewal, in part through repression of the cell cycle inhibitors p16, p19, and p21 (Fasano et al., 2007; Molofsky et al., 2005; Molofsky et al., 2003). Analysis of PRC1/PRC2 double mutants will be important for assessing the degree of functional overlap for these complexes during neural development.
There are additional potential roles for Ezh2 in the developing nervous system. In neurosphere culture of cells isolated from the mouse telencephalon at E14, Ezh2 controls the cell fate choice between oligodendrocytes and astrocytes, with downregulation of Ezh2 expression being required to promote the production of astrocytes (Fasano et al., 2007; Sher et al., 2008). Whether Ezh2 plays a role in the development of oligodendrocytes in vivo remains to be determined. In the chick spinal cord, EZH2 activity is not required for neuroblast proliferation or for neural differentiation, but is required for dorsoventral patterning through regulation of Noggin expression and dorsal BMP signaling (Akizu et al., 2010). This is consistent with previously described roles for PRC2 in consolidating positional identity of progenitors in development (Sparmann and van Lohuizen, 2006). Additionally, there is preliminary evidence that PRC2 may be involved in the regulation of neural crest cells. PRC2 components are expressed in neural crest derivatives, and the PRC2 binding partner Aebp2 is required for mouse neural crest derivatives (Aldiri and Vetter, 2009; Kim et al., 2011).
In general, the role of PRC2 in the development of various tissues is complex, and likely to be stage-specific. Nevertheless, a clear picture emerges of PRC2 as a regulator of developmental transitions as cells progress from being multipotent, developmentally plastic progenitors to lineage committed precursors and ultimately terminally differentiated cells. During this process PRC2 can act to either promote or block these transitions, and can also consolidate or preserve proper cell identity (Fig. 2).
Complementary roles of Ezh1 and Ezh2 during development
Ezh1 and Ezh2 are partially redundant in establishing H3K27me3 and can occupy similar target genes, and in some cases have been proposed to play redundant roles. In ES cells, Ezh1 is required for differentiation and for repression of developmental genes, similar to Ezh2 (Margueron et al., 2008; Shen et al., 2008). In skin, both Ezh1 and Ezh2 are expressed, and target similar epidermal differentiation genes (Ezhkova et al., 2009). Furthermore, disruption of Ezh2 results in an incomplete loss of H2K27me3 in skin, suggesting compensation by Ezh1. Ezh1 is dispensable for epidermal differentiation during development, however double inactivation of Ezh1 and Ezh2 leads to arrest in hair follicle morphogenesis and impairs skin regeneration in postnatal mice demonstrating functional redundancy in this tissue (Ezhkova et al., 2011).
Nevertheless, it is also clear that the developmental roles of Ezh1 and Ezh2 can also be distinct and context dependent. During organogenesis in many tissues Ezh2 expression is mainly confined to embryonic tissues while Ezh1 persists postnatally (Ezhkova et al., 2009; Margueron et al., 2008). Meanwhile, Ezh1, but not Ezh2, is required for myoblast differentiation in mouse, and Ezh1 regulates left-right asymmetry in medaka through silencing of Nodal (Arai et al., 2010; Stojic et al., 2011). These data highlight the importance of studying the consequences of inactivation Ezh1 alone or in combination with Ezh2 to dissect the contribution of these enzymes to organ formation. Additionally, if Ezh1 can truly compensate for the absence of Ezh2, as many studies have proposed, then knocking Ezh1 into the Ezh2 locus in mice should restore organ defects observed upon loss of Ezh2. Such experiments might be necessary to reveal the extent to which Ezh1 and Ezh2 can act redundantly during organogenesis.
H3K27me3 deposition during development
Understanding the mechanism by which PRC2 regulates progression from proliferation to differentiation has relied heavily on identifying target genes occupied by H3K27me3 and characterizing the pattern of this mark during development using chromatin immunoprecipitation coupled with microarray analysis (ChIP–chip) and ChIP-sequencing analyses. It has been found that H3K27me3 deposition is dynamic and particularly selective in a tissue-specific manner during organogenesis. For example H3K27me3 mark is enriched on the promoter of the bHLH factor Ngn1 to suppress neurogenesis as progenitors transition to the generation of astrocytes in the developing neocortex (Hirabayashi et al., 2009). Conversely, H3K27me3 deposition contributes to postnatal olfactory bulb neurogenesis by repressing the expression of the neurogenic gene Dlx2 in neural stem cells residing in the subventricular zone (SVZ), thus preserving their potential to produce astrocytes and oligodendrocytes (Lim et al., 2009). In addition, during skin development H3K27me3 occupancy is maintained on terminal differentiation genes in basal epidermal cells and is progressively lost as development proceeds toward terminal differentiation (Ezhkova et al., 2009). These examples underscore how a single molecular mechanism can be utilized in a tissue- and stage-specific manner to achieve differential roles during the progression from proliferation to differentiation and final fate acquisition in organ development.
Notably, H3K27me3 enrichment is not limited only to tissue-specific genes during organogenesis, suggesting that PRC2 loss of function may cause a global de-repression of genes associated with multiple lineages, as was observed in ES cells (Boyer et al., 2006; Lee et al., 2006). However, inactivation of PRC2 in vivo leads to upregulation of only a minority of those genes, and the overall effect of PRC2 conditional mutants on organ development is relatively mild (Ezhkova et al., 2009; He et al., 2011; Hirabayashi et al., 2009; Wyngaarden et al., 2011). Hence, it is unlikely that the H3K27me3-mediated repression is the sole mechanism that acts to constrain gene expression during organ formation.
In principle, the expression of PRC2 subunits and its mark H3K27me3 should extensively overlap during tissue development. Paradoxically, in several tissues while Ezh2 is mainly enriched in dividing cells, the global level of the H3K27me3 mark is maintained or increased concomitant with differentiation, including in mouse retina, heart, limb, skin and chick spinal cord (Akizu et al., 2010; Ezhkova et al., 2009; He et al., 2011; Rao et al., 2010; Wyngaarden et al., 2011). The apparent inverse correlation between the expression of Ezh2 and H3K27me3 deposition is counterintuitive, however, studies have revealed that the EZH2 homologue, EZH1, can be responsible for the addition of the H3K27me3 in differentiating cells (Akizu et al., 2010; Ezhkova et al., 2009; Margueron et al., 2008; Stojic et al., 2011). Since the catalytic function of EZH1 also requires the presence of the core subunits SUZ12 and EED, it will be important to characterize the expression of these components after the initiation of differentiation in detail. Indeed, a recent study has shown that while the protein levels of EZH2 diminish with differentiation, EED and SUZ12 are maintained, albeit at low levels, and in association with EZH1 are required for myoblast differentiation (Stojic et al., 2011). The biological significance of the presence of PRC2 complexes that contain EZH1 instead of the canonical EZH2 is poorly understood, but may reflect a potential role in target selectivity (Ho and Crabtree, 2008; Margueron et al., 2008; Stojic et al., 2011). Similarly, why H3K27me3 is enriched in fully differentiated cells remains unclear, but it is possible that it is used to stabilize terminal cell fate decisions by permanently suppressing the expression of all genes that are not related to the maintenance of the fully differentiated cells.
Given that H3K27me3 occupancy can be transient during differentiation, it is unclear how H3K27me3 is removed during this process. Histone demethylases are important class of chromatin remodeling factors and have increasingly been found to have essential functions during development and diseases (reviewed in (Pedersen and Helin, 2010). H3K27me3 specific demethylases, UTX and JMJD3, have been identified and implicated in neural commitment and the differentiation of muscle and skin in culture (Burgold et al., 2008; Lan et al., 2007; Seenundun et al., 2010; Sen et al., 2008). Recently, in vivo analysis has demonstrated that UTX is essential for heart development, and acts as a developmental switch in the cardiac lineage to induce expression of cardiac genes in association with core cardiac transcription factors (Lee et al., 2012). Since UTX is broadly expressed there is much to be learned about the roles of H3K27me3 demethylases, and how their functions are coordinated with PRC2 activities during embryonic development.
Regulation of PRC2 function during development
1- Regulation of PRC2 subunit expression
The enrichment of PRC2 core subunit expression in proliferating cells suggests the presence of a regulatory mechanism that tightly controls the induction/maintenance of PRC2 transcription in progenitors while shutting it off upon initiation of differentiation (Akizu et al., 2010; Ezhkova et al., 2009; He et al., 2011; Stojic et al., 2011). Given that the function of PRC2 is context-dependent, this mechanism is likely to be tissue-specific as well. Additionally, since EZH1 and EZH2 show differential expression patterns, it is likely that these two subunits are regulated by distinct mechanisms. We propose that early transcription factors or signaling pathways that drive tissue specification and differentiation may control PRC2 expression: factors that control cell proliferation and self renewal could be involved in maintaining high expression of PRC2 components, while factors that promote cellular differentiation could function to constrain PRC2 transcription as part of their differentiation program. While there is no direct in vivo evidence to support this model, information from tissue culture, ES cells, and cancer studies may provide insight into possible mechanisms. For example, the microRNA miR-214, which drives muscle specification, is involved in a negative feedback loop to inhibit the translation of Ezh2 in skeletal muscle cells and ES cells (Juan et al., 2009). Further, it has been shown that the transcription factor E2F induces the expression of the PRC2 core subunits Eed and Ezh2 in tumor cells and in fibroblasts (Bracken et al., 2003; Muller et al., 2001). More recently, Myc family members were found to be necessary and sufficient to promote the expression of PRC2 components in ES cells (Neri et al., 2011). Whether any of these factors is part of the regulatory mechanism governing PRC2 expression during organogenesis remains to be tested.
2- Posttranslational modifications
There is mounting evidence that PRC2 proteins are targeted for sumoylation and phosphorylation (Margueron and Reinberg, 2011; Riising et al., 2008). While the functional significance of sumoylation remains unclear, EZH2 phosphorylation has been particularly studied, and been shown to modulate PRC2 binding and catalytic activities in a site-dependent manner. While phosphorylation of particular sites inhibits catalytic activities and interfere with EZH2 binding, other sites seem to promote EZH2 function (reviewed in(Caretti et al., 2011; Chou et al., 2011). The responsible kinases have been identified and shown to be the cell cycle regulators CDK1 and CDK2 (Chen et al., 2010; Kaneko et al., 2010; Zeng et al., 2011). Hence, regulation of EZH2 phosphorylation can provide an additional mechanism to modulate PRC2 activities in a spatial and temporal manner during the transition from proliferation to differentiation. For example, the CDK1-mediated phosphorylation of human EZH2 at Thr 487 inhibits its catalytic function, promoting osteogenesis in mesenchymal stem cells (Wei et al., 2011). EZH2 can also be phosphorylated by AKT signaling, which opens the door for investigating the link between environmental cues and regulation of PRC2 function during organ formation (Cha et al., 2005). Further, whether EZH1 activity is subject to regulation by posttranslational modification in a similar manner to EZH2 remains to be fully explored.
3- Recruitment of PRC2
One of the least understood aspects of PRC2 function is how it achieves target specificity during the transition state from proliferation to differentiation. The core PRC2 binds DNA with low affinity, indicating the presence of a recruiting mechanism that directs PRC2 to its intended targets. Additional PRC2 cofactors such as JARID2 promote binding of PRC2 to the DNA (Landeira et al., 2010; Li et al., 2010; Peng et al., 2009). However, since JARID2 is a bona fide partner of PRC2, it is still unclear why recruitment is particularly selective (Landeira and Fisher, 2010; Margueron and Reinberg, 2011). In principle, PRC2 recruitment can be facilitated by the presence of unique DNA elements in the targeted promoters. Indeed, such unique sequences, termed Polycomb Response Elements (PREs), have been previously identified in Drosophila and, to a certain extent, mouse and shown to bind PRC2 via association with PRC1 (Bracken and Helin, 2009; Sing et al., 2009). More importantly, a model that suggests the involvement of transcription factors in the regulation of PcG recruitment was proposed (Bracken and Helin, 2009). According to this model, factors that drive cell fate specification can promote recruitment or dissociation of PRC2 during differentiation in a tissue-specific manner (Bracken and Helin, 2009). In support to this model, a recent study elegantly demonstrates that the homeoprotein MSX1, which regulates myoblast differentiation and limb formation, physically interacts with EZH2 and forces it to relocalize to the nuclear periphery (Wang et al., 2011). This relocalization of EZH2 leads to the redistribution of H3K27me3 to the nuclear lamina and subsequent repression of MSX1 target genes in myoblasts. Hence, we expect that performing tissue-specific pull-down experiments may identify additional tissue-specific PRC2 binding partners. However, it should be taken into consideration that the association between PRC2 and these factors could be transient and depend upon posttranslational modifications of PRC2 components (Palacios et al., 2010; Singh and Dilworth, 2011).
Additionally, long non-coding RNAs (ncRNAs) have been implicated in the recruitment of PRC2 (Bracken and Helin, 2009; Margueron and Reinberg, 2011; Ng et al., 2012). For instance, the ncRNA HOTAIR associates with PRC2 and promotes its recruitment to HOXD locus for subsequent repression in trans (Rinn et al., 2007). Several long ncRNAs have been identified and shown to have tissue-specific expression, suggesting possible PRC2-dependent roles in organogenesis (Pauli et al., 2011). For example, the lncRNA Six3OS is specifically expressed in the developing retina and hypothalamus, is involved in retinal cell fate decisions and interacts with Ezh2 as well as Eya family members (Rapicavoli et al., 2011). Recently, Margueron and Reinberg proposed a model stipulating that the collective step-wise weak interactions of PRC2 core and auxiliary components with histone, DNA, and H3K27me3, and its association with long ncRNA, provides a sufficient platform for PRC2 recruitment to its targets (Margueron and Reinberg, 2011). Validating this model during organogenesis awaits further experimentation.
Perspectives and Future Directions
Recent years have witnessed tremendous progress in our understanding of the contribution of PRC2 to differentiation and cell fate specification, yet much remains to be explored. We expect that additional tissues will be added to the list of organs regulated by PRC2, and more details about PRC2 mechanism of action will be revealed. Most studies have focused on the roles of nuclear PRC2 in catalyzing the addition of H3K27me3 and repressing gene expression during organogenesis. However, the full spectrum of PRC2 alternative roles has not been explored. For instance, PRC2 can localize to the cytoplasm where it promotes actin polymerization through its methyltransferase activities (Bryant et al., 2008; Su et al., 2005). Regulation of actin polymerization is essential for proper cell morphogenesis during organogenesis, suggesting that PRC2 might be involved in this process. Strikingly, in breast cancer cells EZH2 binds the Wnt effector β-catenin and promotes transcriptional activation of genes under estrogen control, independent of its methyltransferase activities, and in the absence of other PRC2 core subunits (Li et al., 2009; Shi et al., 2007). This indicates that EZH2 function (and perhaps other PRC2 components) can be uncoupled from PRC2 enzymatic activities, and can act as a transcriptional switch under certain conditions. These studies highlight the need for a reexamination of the subcellular localization of PRC2 components, and for a proper dissection of its functional activities during the progression from proliferation to differentiation.
Highlights.
Polycomb factors function in chromatin-remodeling complexes to silence genes.
PRC2 regulates developmental transitions during organogenesis.
PRC2 functions are context-dependent and tissue-specific.
H3K27me3 is dynamic and selective during tissue development.
PRC2 subunit expression and function is likely regulated by tissue-specific factors.
Acknowledgments
We are grateful to Drs. Alejandro Sánchez Alvarado, Richard Dorsky and Kathryn Moore for their helpful comments on this manuscript. This work was supported by NIH grant EY012274 to MLV.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akasaka T, Kanno M, Balling R, Mieza MA, Taniguchi M, Koseki H. A role for mel-18, a Polycomb group-related vertebrate gene, during theanteroposterior specification of the axial skeleton. Development. 1996;122:1513–1522. doi: 10.1242/dev.122.5.1513. [DOI] [PubMed] [Google Scholar]
- Akizu N, Estaras C, Guerrero L, Marti E, Martinez-Balbas MA. H3K27me3 regulates BMP activity in developing spinal cord. Development. 2010;137:2915–2925. doi: 10.1242/dev.049395. [DOI] [PubMed] [Google Scholar]
- Aldiri I, Vetter ML. Characterization of the expression pattern of the PRC2 core subunit Suz12 during embryonic development of Xenopus laevis. Dev Dyn. 2009;238:3185–3192. doi: 10.1002/dvdy.22120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander T, Nolte C, Krumlauf R. Hox genes and segmentation of the hindbrain and axial skeleton. Annu Rev Cell Dev Biol. 2009;25:431–456. doi: 10.1146/annurev.cellbio.042308.113423. [DOI] [PubMed] [Google Scholar]
- Aoki R, Chiba T, Miyagi S, Negishi M, Konuma T, Taniguchi H, Ogawa M, Yokosuka O, Iwama A. The polycomb group gene product Ezh2 regulates proliferation and differentiation of murine hepatic stem/progenitor cells. J Hepatol. 2010;52:854–863. doi: 10.1016/j.jhep.2010.01.027. [DOI] [PubMed] [Google Scholar]
- Arai D, Katsura H, Shindo N, Matsumoto M, Higashinakagawa T. Polycomb group protein Ezh1 represses Nodal and maintains the left-right axis. Dev Biol. 2010;341:459–463. doi: 10.1016/j.ydbio.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Benoit YD, Lepage MB, Khalfaoui T, Tremblay E, Basora N, Carrier JC, Gudas LJ, Beaulieu JF. Polycomb repressive complex 2 impedes intestinal cell terminal differentiation. J Cell Sci. 2012 doi: 10.1242/jcs.102061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant RJ, Winder SJ, Cross SS, Hamdy FC, Cunliffe VT. The Polycomb Group protein EZH2 regulates actin polymerization in human prostate cancer cells. Prostate. 2008;68:255–263. doi: 10.1002/pros.20705. [DOI] [PubMed] [Google Scholar]
- Burgold T, Spreafico F, De Santa F, Totaro MG, Prosperini E, Natoli G, Testa G. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PloS One. 2008;3:e3034. doi: 10.1371/journal.pone.0003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti G, Palacios D, Sartorelli V, Puri PL. Phosphoryl-EZH-ion. Cell Stem Cell. 2011;8:262–265. doi: 10.1016/j.stem.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310:306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Gu X, Su IH, Bottino R, Contreras JL, Tarakhovsky A, Kim SK. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 2009;23:975–985. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Ma Y, Kim EY, Yu W, Schwartz RJ, Qian L, Wang J. Conditional ablation of Ezh2 in murine hearts reveals its essential roles in endocardial cushion formation, cardiomyocyte proliferation and survival. PloS One. 2012;7:e31005. doi: 10.1371/journal.pone.0031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Bohrer LR, Rai AN, Pan Y, Gan L, Zhou X, Bagchi A, Simon JA, Huang H. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat Cell Biol. 2010;12:1108–1114. doi: 10.1038/ncb2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou RH, Yu YL, Hung MC. The roles of EZH2 in cell lineage commitment. Am J Transl Res. 2011;3:243–250. [PMC free article] [PubMed] [Google Scholar]
- del Mar Lorente M, Marcos-Gutierrez C, Perez C, Schoorlemmer J, Ramirez A, Magin T, Vidal M. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development. 2000;127:5093–5100. doi: 10.1242/dev.127.23.5093. [DOI] [PubMed] [Google Scholar]
- Delgado-Olguin P, Huang Y, Li X, Christodoulou D, Seidman CE, Seidman JG, Tarakhovsky A, Bruneau BG. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat Genet. 2012;44:343–347. doi: 10.1038/ng.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25:485–498. doi: 10.1101/gad.2019811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Faust C, Lawson KA, Schork NJ, Thiel B, Magnuson T. The Polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development. 1998;125:4495–4506. doi: 10.1242/dev.125.22.4495. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullgrabe J, Kavanagh E, Joseph B. Histone onco-modifications. Oncogene. 2011;30:3391–3403. doi: 10.1038/onc.2011.121. [DOI] [PubMed] [Google Scholar]
- He A, Ma Q, Cao J, von Gise A, Zhou P, Xie H, Zhang B, Hsing M, Christodoulou D, Cahan P, Daley GQ, Kong SW, Orkin SH, Seidman CE, Seidman JG, Pu WT. Polycomb Repressive Complex 2 Regulates Normal Development of the Mouse Heart. Circ Res. 2011;110:406–415. doi: 10.1161/CIRCRESAHA.111.252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y, Gotoh Y. Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci. 2010;11:377–388. doi: 10.1038/nrn2810. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, Koseki H, Vidal M, Gotoh Y. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. An EZ mark to miss. Cell Stem Cell. 2008;3:577–578. doi: 10.1016/j.stem.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan AH, Derfoul A, Feng X, Ryall JG, Dell’Orso S, Pasut A, Zare H, Simone JM, Rudnicki MA, Sartorelli V. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011;25:789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell. 2009;36:61–74. doi: 10.1016/j.molcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketel CS, Andersen EF, Vargas ML, Suh J, Strome S, Simon JA. Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol Cell Biol. 2005;25:6857–6868. doi: 10.1128/MCB.25.16.6857-6868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kang K, Ekram MB, Roh TY, Kim J. Aebp2 as an epigenetic regulator for neural crest cells. PloS One. 2011;6:e25174. doi: 10.1371/journal.pone.0025174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, Adli M, Kasif S, Ptaszek LM, Cowan CA, Lander ES, Koseki H, Bernstein BE. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, Reinberg D. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci U S A. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Landeira D, Fisher AG. Inactive yet indispensable: the tale of Jarid2. Trends Cell Biol. 2010;21:74–80. doi: 10.1016/j.tcb.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, Brookes E, Pombo A, Fisher C, Skarnes WC, Snoek T, Bezstarosti K, Demmers J, Klose RJ, Casanova M, Tavares L, Brockdorff N, Merkenschlager M, Fisher AG. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol. 2010;12:618–624. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee JW, Lee SK. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell. 2012;22:25–37. doi: 10.1016/j.devcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 2010;24:265–276. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gonzalez ME, Toy K, Filzen T, Merajver SD, Kleer CG. Targeted overexpression of EZH2 in the mammary gland disrupts ductal morphogenesis and causes epithelial hyperplasia. Am J Pathol. 2009;175:1246–1254. doi: 10.2353/ajpath.2009.090042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez AM, Cavalli G. The role of polycomb group proteins in cell cycle regulation during development. Cell Cycle. 2006;5:1189–1197. doi: 10.4161/cc.5.11.2781. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Kocher T, Cohen A, Stunnenberg HG, Wilm M, Muller J. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 2007;26:4078–4088. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri F, Zippo A, Krepelova A, Cherubini A, Rocchigiani M, Oliviero S. Myc regulates the transcription of PRC2 to control the expression of developmental genes in embryonic stem cells. Mol Cell Biol. 2011;32:840–851. doi: 10.1128/MCB.06148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzan F, Pellegatta S, Poliani PL, Pisati F, Caldera V, Menghi F, Kapetis D, Marras C, Schiffer D, Finocchiaro G. Enhancer of Zeste 2 (EZH2) is up-regulated in malignant gliomas and in glioma stem-like cells. Neuropathol Appl Neurobiol. 2011;37:381–394. doi: 10.1111/j.1365-2990.2010.01132.x. [DOI] [PubMed] [Google Scholar]
- Palacios D, Mozzetta C, Consalvi S, Caretti G, Saccone V, Proserpio V, Marquez VE, Valente S, Mai A, Forcales SV, Sartorelli V, Puri PL. TNF/p38alpha/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell. 2010;7:455–469. doi: 10.1016/j.stem.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12:136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen MT, Helin K. Histone demethylases in development and disease. Trends Cell Biol. 2010;20:662–671. doi: 10.1016/j.tcb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JD, Sansom SN, Smith J, Dobenecker MW, Tarakhovsky A, Livesey FJ. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci U S A. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: postponing commitment. Curr Opin Cell Biol. 2008;20:201–207. doi: 10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Polycomb repression under the skin. Cell. 2009;136:992–994. doi: 10.1016/j.cell.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Prezioso C, Orlando V. Polycomb proteins in mammalian cell differentiation and plasticity. FEBS Lett. 2011;585:2067–2077. doi: 10.1016/j.febslet.2011.04.062. [DOI] [PubMed] [Google Scholar]
- Rao RC, Tchedre KT, Malik MT, Coleman N, Fang Y, Marquez VE, Chen DF. Dynamic patterns of histone lysine methylation in the developing retina. Invest Ophthalmol Vis Sci. 2010;51:6784–6792. doi: 10.1167/iovs.09-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Poth EM, Zhu H, Blackshaw S. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev. 2011;6:32. doi: 10.1186/1749-8104-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richie ER, Schumacher A, Angel JM, Holloway M, Rinchik EM, Magnuson T. The Polycomb-group gene eed regulates thymocyte differentiation and suppresses the development of carcinogen-induced T-cell lymphomas. Oncogene. 2002;21:299–306. doi: 10.1038/sj.onc.1205051. [DOI] [PubMed] [Google Scholar]
- Richly H, Aloia L, Di Croce L. Roles of the Polycomb group proteins in stem cells and cancer. Cell Death Dis. 2011;2:e204. doi: 10.1038/cddis.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riising EM, Boggio R, Chiocca S, Helin K, Pasini D. The polycomb repressive complex 2 is a potential target of SUMO modifications. PloS One. 2008;3:e2704. doi: 10.1371/journal.pone.0002704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawarkar R, Paro R. Interpretation of developmental signaling at chromatin: the Polycomb perspective. Dev Cell. 2010;19:651–661. doi: 10.1016/j.devcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, Bunker RD, Wirth U, Bouwmeester T, Bauer A, Ly- Hartig N, Zhao K, Chan H, Gu J, Gut H, Fischle W, Muller J, Thoma NH. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jager N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Fruhwald MC, Roggendorf W, Kramm C, Durken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- Seenundun S, Rampalli S, Liu QC, Aziz A, Palii C, Hong S, Blais A, Brand M, Ge K, Dilworth FJ. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 2010;29:1401–1411. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher F, Rossler R, Brouwer N, Balasubramaniyan V, Boddeke E, Copray S. Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells. 2008;26:2875–2883. doi: 10.1634/stemcells.2008-0121. [DOI] [PubMed] [Google Scholar]
- Shi B, Liang J, Yang X, Wang Y, Zhao Y, Wu H, Sun L, Zhang Y, Chen Y, Li R, Zhang Y, Hong M, Shang Y. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol. 2007;27:5105–5119. doi: 10.1128/MCB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, Ellis J, Lipshitz HD, Cordes SP. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell. 2009;138:885–897. doi: 10.1016/j.cell.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Singh K, Dilworth FJ. Redirecting traffic in the nucleus. Dev Cell. 2011;21:390–392. doi: 10.1016/j.devcel.2011.08.025. [DOI] [PubMed] [Google Scholar]
- Snowden AW, Gregory PD, Case CC, Pabo CO. Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr Biol. 2002;12:2159–2166. doi: 10.1016/s0960-9822(02)01391-x. [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- Stojic L, Jasencakova Z, Prezioso C, Stutzer A, Bodega B, Pasini D, Klingberg R, Mozzetta C, Margueron R, Puri PL, Schwarzer D, Helin K, Fischle W, Orlando V. Chromatin regulated interchange between polycomb repressive complex 2 (PRC2)-Ezh2 and PRC2-Ezh1 complexes controls myogenin activation in skeletal muscle cells. Epigenetics Chromatin. 2011;4:16. doi: 10.1186/1756-8935-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, Viale A, Reinberg D, Wulfing C, Tarakhovsky A. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121:425–436. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 2010;7:288–298. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Testa G. The time of timing: how Polycomb proteins regulate neurogenesis. Bioessays. 2011;33:519–528. doi: 10.1002/bies.201100021. [DOI] [PubMed] [Google Scholar]
- Turner BM. Defining an epigenetic code. Nat Cell Biology. 2007;9:2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Wang J, Kumar RM, Biggs VJ, Lee H, Chen Y, Kagey MH, Young RA, Abate-Shen C. The Msx1 Homeoprotein Recruits Polycomb to the Nuclear Periphery during Development. Dev Cell. 2011;21:575–588. doi: 10.1016/j.devcel.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Jin Q, Lee JE, Su IH, Ge K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc Natl Acad Sci U S A. 2010;107:7317–7322. doi: 10.1073/pnas.1000031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B, Yang CC, Yang JY, Lin CY, Lai CC, Hung MC. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol. 2011;13:87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, Zhang J, Gajjar A, Dyer MA, Mullighan CG, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Ellison DW, Baker SJ. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyngaarden LA, Delgado-Olguin P, Su IH, Bruneau BG, Hopyan S. Ezh2 regulates anteroposterior axis specification and proximodistal axis elongation in the developing limb. Development. 2011;138:3759–3767. doi: 10.1242/dev.063180. [DOI] [PubMed] [Google Scholar]
- Yuzyuk T, Fakhouri TH, Kiefer J, Mango SE. The polycomb complex protein mes-2/E(z) promotes the transition from developmental plasticity to differentiation in C. elegans embryos. Dev Cell. 2009;16:699–710. doi: 10.1016/j.devcel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Chen S, Huang H. Phosphorylation of EZH2 by CDK1 and CDK2: a possible regulatory mechanism of transmission of the H3K27me3 epigenetic mark through cell divisions. Cell Cycle. 2011;10:579–583. doi: 10.4161/cc.10.4.14722. [DOI] [PMC free article] [PubMed] [Google Scholar]