Abstract
Receptor agonism remains poorly understood at the molecular and mechanistic level. In this study, we identified a fully human anti-Fas antibody that could efficiently trigger apoptosis and therefore function as a potent agonist. Protein engineering and crystallography were used to mechanistically understand the agonistic activity of the antibody. The crystal structure of the complex was determined at 1.9 Å resolution and provided insights into epitope recognition and comparisons with the natural ligand FasL (Fas ligand). When we affinity-matured the agonist antibody, we observed that, surprisingly, the higher-affinity antibodies demonstrated a significant reduction, rather than an increase, in agonist activity at the Fas receptor. We propose and experimentally demonstrate a model to explain this non-intuitive impact of affinity on agonist antibody signalling and explore the implications for the discovery of therapeutic agonists in general.
Keywords: agonist antibody, Fas, affinity maturation, phage display, scFv
Receptor agonism is a key step in the transmission of signals from the outside to the inside of a cell. Despite many years of study, the precise mechanisms underlying ligand-mediated agonism of cell surface receptors remain poorly understood. Indeed, if one takes the well-studied T-cell receptor as an example, a recent review1 highlights that three competing theories, namely receptor aggregation, conformational change and segregation of receptors from intracellular inhibitors, are all supported by experimental data and may all be partly responsible for signal transduction.
An important group of agonistic receptors is the tumour necrosis factor (TNF) receptor superfamily, members of which are known to form trimeric signalling complexes upon ligation with their cognate trimeric ligands.2 As many TNF receptor superfamily members have significant roles in the progression of human disease,3 a better understanding of their signalling mechanisms could enable the design of potent agonist and antagonist molecules as candidate drugs for clinical development. Monoclonal antibodies, with their high affinity, target selectivity and long serum half-lives, are well suited as receptor agonist drug molecules. Accordingly, there are agonistic antibodies in preclinical and clinical development against several TNF receptor family members, such as TNF-related apoptosis inducing ligand (TRAIL) receptor 1,4 TRAIL receptor 2,5 glucocorticoid-induced tumor necrosis factor receptor (GITR),6 CD1377 and CD40.8
The TNF receptor superfamily member Fas (CD95, APO-1, TNFRSF6) is a receptor, which participates in signal transduction to trigger cellular apoptosis after binding to Fas ligand (FasL).9 Because of its expression on many tumours and its ability to trigger apoptosis,10 Fas could be considered an interesting target for therapeutic intervention. However, the finding that an agonistic Fas antibody administered to mice caused severe liver damage and rapid death has led to the conclusion that Fas is not a valid therapeutic target.11 Despite the possible limitations of Fas in the therapeutic setting it can still be considered a relevant model system to explore the nature of agonistic antibody signalling in the TNF receptor superfamily. Agonistic antibodies, as opposed to the more frequently isolated antagonistic or neutralising antibodies, have rarely been studied from a structure–function perspective and little is known about the importance of epitope and affinity in the biological potency of such antibodies. Whereas neutralising antibodies are expected to exhibit a direct positive correlation between affinity and potency, agonist antibodies are potentially more subtle and difficult to understand and as a consequence it could be more challenging to enhance their potency.
In this study, phage-display technology was used to identify a human anti-Fas antibody, which could efficiently trigger apoptosis and therefore demonstrate potent in vitro tumour cell-killing activity. This model agonistic antibody was then used as a starting point for mutational and crystallographic studies to explore the binding interface and better understand the agonistic activity. This systematic analysis of an agonistic antibody interacting with its receptor, in particular the exploration of the relationship between affinity and potency, has led to some surprising conclusions about the nature of agonistic antibody signalling.
Results
Isolation of agonistic anti-Fas antibody E09 and comparison with other agonists
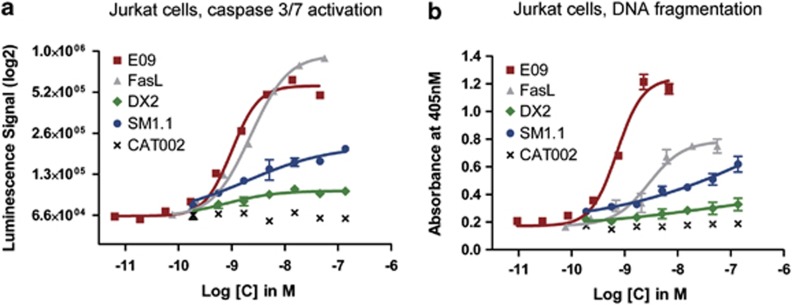
Antibodies to human Fas receptor were isolated by performing phage-display selections12 on the recombinant extracellular domain (ECD) of Fas. Antibodies specific for Fas ECD were detected by phage ELISA and a total of 264 unique scFv were sequenced. Of the 264 different scFv antibodies screened for agonism in a cell-viability assay, only one was identified as having anti-proliferative activity. This scFv, E09, was converted to human IgG1 antibody format for further characterisation. To confirm the agonistic activity towards the human Fas receptor, assays were performed on Jurkat cells to measure caspase 3/7 activation and DNA fragmentation, which, respectively, are early and late readouts for apoptosis. The E09 antibody was compared with the natural ligand FasL in recombinant form and two agonistic anti-Fas antibodies, the mouse monoclonal antibodies DX2 and SM1.1.10, 13 All agonists were able to induce caspase 3/7 activity and DNA fragmentation, as shown in Figure 1, but to differing extents. E09 was as potent as the natural ligand FasL at triggering caspase 3/7 activity and even more potent than FasL at inducing DNA fragmentation with an EC50 of 0.7 and 2.8 nM for E09 and FasL, respectively.
Figure 1.
Analysis of antibody E09 agonism of Fas by comparison to other agonists. Jurkat cells were incubated for 8 h with the indicated IgGs or FasL at different concentrations. CAT002 was an unrelated IgG negative control. (a) The turnover of the effector caspase substrate z-DEVD-aminoluciferin was measured in relation to the concentration of each agonist or control antibody. (b) The amount of oligonucleosomes present in the cytosol, an indicator of DNA fragmentation during apoptosis, was detected by an antibody-based assay and plotted versus the concentration. All experiments were performed in triplicate and error bars indicate the standard deviation between replicates
In a Jurkat cell-viability assay, two parameters could be determined. Efficiency was defined as the maximal cell killing (in percentage) that could be obtained and EC50 as the molar concentration of agonist required to obtain half-maximal killing. E09 demonstrated an efficiency of 80% (Table 1), which is slightly lower than FasL (94%) but had a significantly lower EC50 than the natural ligand (0.9 nM and 7 nM for E09 and FasL, respectively). The other agonist antibodies DX2 and SM1.1 showed reduced cell-killing efficiencies, of 16% and 26% respectively (Table 1). As the agonistic anti-Fas antibodies exhibited different apoptotic potencies, we explored the possible reasons.
Table 1. Summary of binding affinity, epitope competition and in vitro cell-killing data for Fas agonists.
|
Competition with FasL |
Competition with E09 IgG |
Jurkat cell killing |
|||||
|---|---|---|---|---|---|---|---|
| Ligand | Fas Kd, nM | IC50, nM | Max. Inh., % | IC50, nM | Max. Inh., % | EC50, nM | Efficiency, % |
| FasL | 14.0 | ND | ND | ND | ND | 7.1 | 94.0 |
| E09 | 17.0 | 1.4 | 100.0 | 0.9 | 100.0 | 0.9 | 80.0 |
| DX2 | 7.3 | 2.4 | 98.0 | 1.6 | 34.0 | 1.4 | 16.0 |
| SM1.1 | 5.2 | 5.5 | 86.0 | 1.8 | 100.0 | 4.0 | 26.0 |
The Kd values were determined by SPR analysis of binding to recombinant Fas. Each antibody was also tested for their ability to compete the Fas:FasL and the Fas:E09 interactions in biochemical competition assays to calculate an IC50 value (nM) and maximum inhibition value (%). Cell viability assays were performed on Jurkat cells using a titration of anti-Fas molecules to determine the maximal killing efficiency (%) and EC50 (nM)
Does the efficacy depend on the affinity or epitope?
The first obvious cause for a difference in agonism could be a difference in the affinity, as previously proposed.14 Therefore, we determined the dissociation constants of the complexes between the human Fas ECD and each agonist by surface plasmon resonance (SPR). The Kd values of all antibodies were in the range between 5 and 17 nM (Table 1) and therefore similar to each other and to human FasL (14 nM). This suggested that the difference in biological activity between the agonists was not simply due to different affinities for the receptor. As an alternative explanation, we considered whether the epitope might have a potential impact on the agonism and performed a competition assay with the different agonists. All antibodies totally competed the FasL:Fas interaction with similar IC50 values, suggesting that the antibodies could be functioning as ligand mimetics (Table 1). However, the three agonistic antibodies did not all compete with each other for a single binding epitope. Although antibody SM1.1 was able to completely inhibit the E09:Fas interaction, antibody DX2 showed only partial competition for Fas binding. These results imply that, although all of the antibodies compete with the natural ligand FasL, there is not a single binding site on the receptor, which is critical for agonism. It should be noted that epitope competition assays only offer an approximate mapping of binding epitopes rather than any precise level of molecular detail.
Molecular details revealed by the structure of Fas ECD in complex with E09 Fab
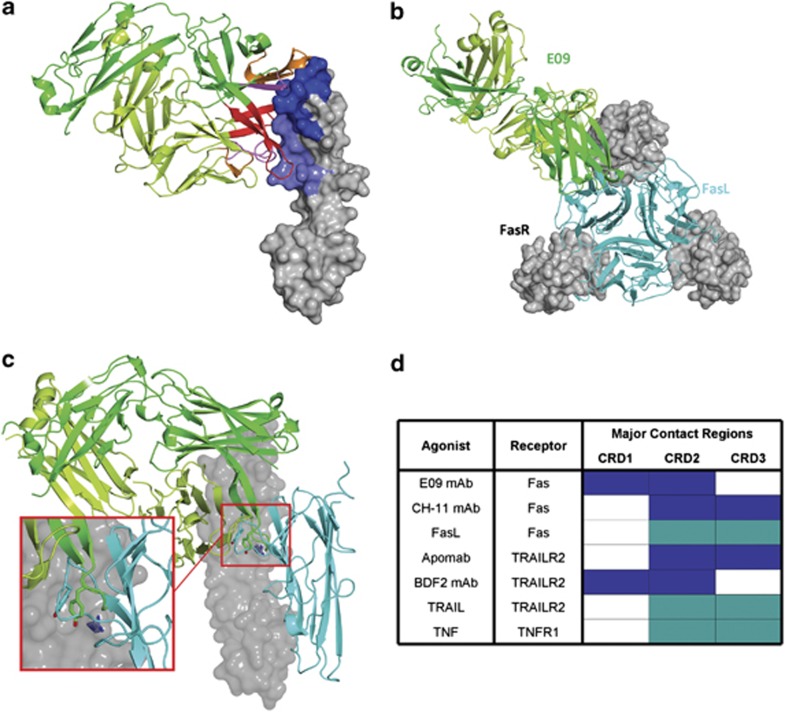
To gain further insight into the interaction of E09 with Fas, we determined the structure of the complex by X-ray crystallography of the Fab fragment of antibody E09 in complex with Fas ECD. The crystals of the complex diffracted to 1.9 Å resolution in the space group C2221 (Supplementary Table 1). The solved structure showed that Fab E09 bound to cysteine-rich domain (CRD) 1 and 2 from Fas receptor, burying 1030 Å2 of accessible surface on the receptor and 890 Å2 on the variable domain of both heavy and light chains (Figure 2a, Supplementary Figure 1 and Supplementary Table 2). The epitope on Fas was formed by a continuous surface patch spanning the outermost tip of the molecule on CRD1 (Fas_58-61) and reaching the last residue of CRD2 (Fas_86).
Figure 2.
Structural analysis of Fas agonist antibody E09. (a) The structure of Fas ECD in complex with E09 Fab. Fas ECD is represented with a surface rendering and the epitope on CRD1 and CRD2 coloured in darker blue and lighter blue, respectively. The E09 is shown as a cartoon with the heavy chain in darker green and the light chain in lighter green. The CRD1, 2 and 3 are coloured in light pink, dark pink and red, respectively. (b) Model of a trimeric Fas-FasL complex that shows the binding of E09 in green and FasL in cyan in top view. (c) A representation of Fas ECD in complex with E09 Fab in which the epitope overlap between FasL and E09 is visible. A single tyrosine in FasL (Y218) or E09 (VH_100d) makes contact with Fas_R86 (zoom). (d) Comparative analysis of broad epitopes on death receptors covered by natural ligands and agonistic antibodies
The antibody binding epitope on Fas overlaps only little with the predicted binding epitope for FasL, which is located on CRD2 and CRD3 of Fas.15 Of those residues, which are crucial for FasL binding,16 only two residues are found in the antibody epitope, namely Fas_81F and Fas_86R. Fas_86R was previously deemed essential for binding to FasL, whereas mutation of Fas_81F reduced binding.17 We modelled FasL and positioned it onto our structure of the complex according to the positions death ligands usually take when interacting with their cognate receptor (Figure 2b). In this model only one loop of FasL overlaps with a loop of the agonistic antibody (Figure 2c). This loop (the DE loop) harbours the highly conserved XYP motif important for the interaction between death ligands and their receptors.18 The tyrosine of this conserved motif (FasL Y218) overlaps with VH Y100d of the antibody E09 (Figure 2c) and was even predicted to interact directly with Fas receptor residue FasR86.18 Interestingly, in Fas:E09 Fas R86 of the receptor also interacts with the overlapping tyrosine residue VH Y100d and stacks with its guanidinium group on top of the aromatic side chain, thereby forming a cation-π-interaction (Figure 2c and Supplementary Figure 1). To understand whether VH Y100d really had a crucial role in the E09 bioactivity, we mutated this position to an arginine, which was shown to have a deleterious effect on apoptosis induction in the context of FasL.18 However, in the context of the E09 antibody this mutant did not reduce apoptosis induction (data not shown).
In a comparative analysis of known epitopes of agonistic antibodies in the death receptor family we found that, similar to FasL, the other natural ligands interact mostly with CRD2 and CRD3 of the death receptors (Figure 2d). In addition, the agonistic anti-Fas CH-11 IgM antibody and anti-TRAIL receptor 2 IgG antibody Apomab recognise this region of the death receptors,14, 19 whereas anti-DR5 BDF2 and our antibody anti-Fas E09 rather bind to CRD1 and CRD2.20 There was generally a tendency for agonistic antibodies to bind to the outside of the trimeric receptor, as opposed to the natural ligands, which bound inside the trimer. From all these observations we can infer that E09's binding mode does not correlate well with the natural ligand recognition site on Fas, and that there is likely to be a different mechanism involved.
Affinity maturation of anti-Fas E09 antibody
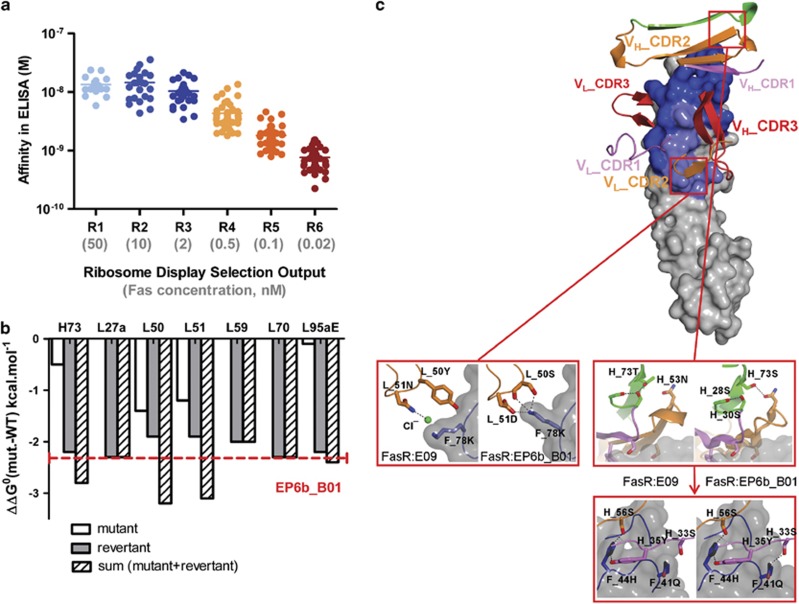
To understand the agonistic mechanism, we attempted to improve the biological activity of E09 by increasing the affinity for Fas. Six rounds of ribosome-display selections were performed on a library of randomly mutated E09 variants and the 44 variants with the highest affinity from each round were identified using an affinity-based screen.21 These scFv were purified and then re-analysed with the same affinity assay to demonstrate the effectiveness of the evolution process in enriching for higher-affinity variants (Figure 3a).
Figure 3.
Directed evolution of Fas antibody E09. (a) Analysis of the 44 highest affinity E09 variant antibodies from the six selection rounds, R1-R6. Surrogate affinity was determined using a six-point antigen titration in an ELISA-based assay. (b) Individual energy contribution of the seven mutations in the top clone EP6b_B01 to affinity improvement. Free energy of binding compared with the parent (ΔΔG0 (mut-WT)) of single-point mutants and revertants in the EP6b_B01 sequence context are shown in white and grey, respectively. Theoretical additive values of the point mutant and the revertant are shown in dashed black. Energy calculations were based on BIAcore affinity measurement of scFvs for recombinant Fas. (c) Structural basis for affinity improvement in EP6b_B01. The lower panels show close-up views from the squares in the upper panel. The left part of each panel shows the zoom from the structure FasR:E09 complex, the right part the zoom from FasR:EP6b_B01. Hydrogen bonds are indicated as black dashed lines
Four variants with a range of affinities for Fas were converted to human IgG1 format for more precise affinity determination by SPR. The sequences of these variants are shown in Supplementary Figure 2. The highest affinity variant EP6b_B01 had a Kd for monomeric human Fas of 350 pM and was improved 49-fold compared with the parent antibody E09 (Table 2). The affinity improvement of EP6b_B01 was due to a faster on-rate (3.4-fold) and slower off-rate (14-fold), compared with E09.
Table 2. Fas binding kinetics of E09 and variant antibodies.
| IgG name | Optimisation method | kon, M−1 s−1 | koff, s−1 | Kd, nM | Kd gain vs parent |
|---|---|---|---|---|---|
| E09 | (Parent antibody) | 1.9E+05 | 3.2E-03 | 17 | 1 |
| EP12r_E01 | Directed Evolution | 1.8E+05 | 1.8E-03 | 10 | 2 |
| EP4b_E03 | Directed Evolution | 2.2E+05 | 6.4E-04 | 2.9 | 6 |
| EP5b_E05 | Directed Evolution | 3.4E+05 | 3.1E-04 | 0.91 | 19 |
| EP6b_B01 | Directed Evolution | 6.4E+05 | 2.3E-04 | 0.35 | 49 |
| E09_Y58W | Point Mutation | 1.7E+05 | 1.7E-02 | 98 | 0 |
| E09_Y52aR | Point Mutation | 1.7E+05 | 8.2E-03 | 47 | 0 |
| E09_R52bS | Point Mutation | 1.5E+05 | 3.5E-04 | 2.4 | 7 |
Values for kon, koff and Kd were calculated following SPR analysis of binding to recombinant Fas. The Kd gain of each variant relative to the parent antibody E09 was also calculated
The sequence of EP6b_B01 contained seven amino-acid substitutions from E09. Six of them were present in over 40% of variants in the affinity matured population at selection round six, and were thus classified as hot-spot mutations (Supplementary Figure 3). To quantify the energetic contribution of the mutations, each was individually added to the E09 backbone or removed from EP6b_B01. Energetic analysis was performed as before,21 following affinity measurements (Supplementary Table 3). Three of the seven mutations, VH T73S, VL Y50S and VL N51D, were shown to have a clear beneficial effect on affinity (Figure 3b). The other single mutations had no impact on affinity despite the fact that three of them were defined as hotspots and were strongly selected during the ribosome-display process.
The increased affinity of EP6b_B01 towards Fas ECD was analysed by structural means. The complex of Fas ECD with Fab EP6b_B01 crystallised in the space group P3121 and the structure was solved at 2.1 Å resolution. EP6b_B01 covered the same epitope on Fas as its parent antibody E09. Analysis of the interface revealed that the number of hydrogen bonds increased by a factor of two and electrostatic interactions were introduced. The energetically relevant VL S50 and VL D51 indeed appeared to be key residues in these extended interactions (Figure 3c, lower left panel). Vernier residue VH 73 contributes to the increased affinity by a rearrangement of the CDRs, thereby allowing VH S33 to form an additional hydrogen bond with Fas Q41 (Figure 3c, lower right panel).
Linking affinity and biological activity of anti-Fas antibodies
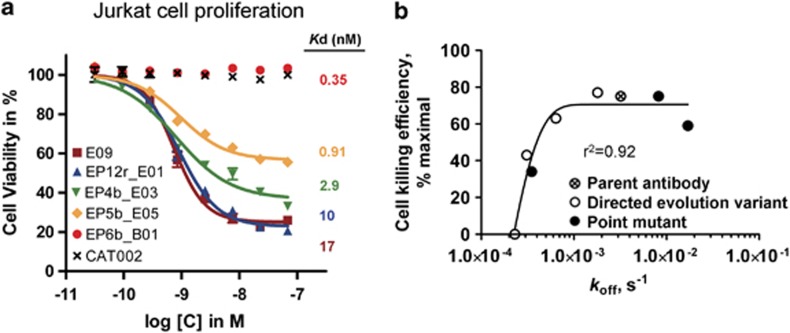
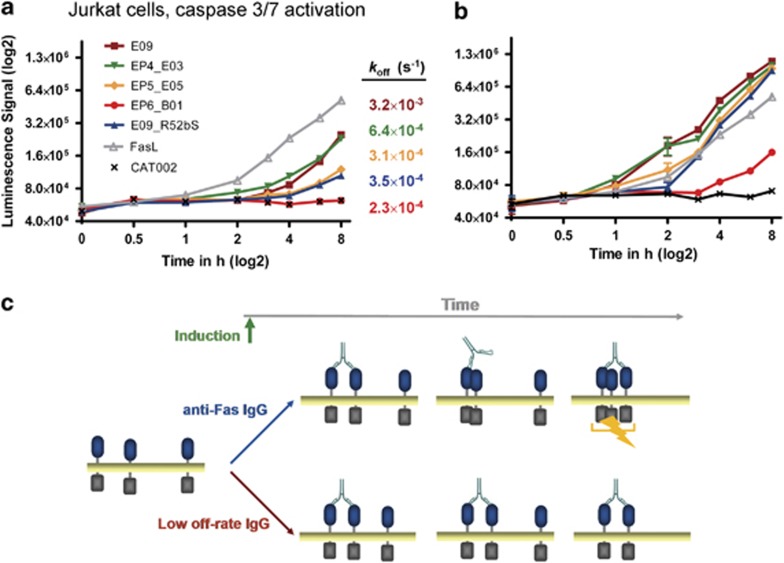
Four selected IgG variants exhibiting a range of affinities for Fas receptor were then characterised for their apoptotic activity. The in vitro viability assay using Jurkat cells demonstrated a surprising negative correlation between Fas affinity and cell-killing efficiency (Figure 4a and Supplementary Table 4). For instance, E09 and the intermediate affinity-optimised variant EP5b_E05 showed efficiencies of 75% and 43% respectively. Most significantly, the highest affinity antibody EP6b_B01 did not show activity at all.
Figure 4.
E09 variants with higher affinity are less potent agonists. (a) Dose-dependent killing of Jurkat cells by E09 parent antibody, affinity optimised variants, or negative control CAT002. Dose response curves were plotted from cell-viability data following treatment with each of the antibodies. The cell-viability assay was performed in quadruplicate and error bars indicate the S.D. between replicates. The affinity of each antibody for Fas is also labelled next to the relevant curve. (b) The rate of koff (s−1) for E09 and variant antibodies, as determined by SPR, was plotted against the Jurkat cell-killing efficiency. The parent antibody E09 is indicated by a circle containing a cross, the directed evolution variants are shown as unfilled circles and point mutants are shown as filled circles
To confirm that this result was not just an artefact of the ribosome display directed evolution process, rational point mutants of E09 were generated to modulate the affinity for Fas. By randomising six contact residues in E09 and screening the variants, antibodies with improved affinity could be isolated (Supplementary Figure 4), primarily by targeting VH Y52a and VH R52b. Mutations at those positions were not seen during the ribosome-display process and, when tested for agonism, these point mutants also exhibited an inverse correlation between affinity and biological activity (Supplementary Table 4).
In all cases, the increase of affinity for the matured antibodies, whether derived by directed evolution or rational point mutagenesis, was due to a decrease in the off rate (Table 2). Furthermore, the off rates of the E09 antibody and variants, when plotted against cell-killing efficiency (Figure 4b), showed a good correlation (r2 value of 0.92).
Kinetic analysis of Fas agonism
As the E09 antibody and its variants were assumed to all bind a similar epitope on Fas, based on their similar sequences and also the elucidation of near identical epitopes in the E09:Fas and EP6b_B01:Fas crystal structures, the main difference between the variants was their altered binding kinetics. To explore whether the different binding kinetics could be affecting agonism, a time-course experiment was performed with FasL, E09 and the variant antibodies, measuring the rate of caspase 3/7 activation in Jurkat cells. FasL initiated caspase 3/7 activity in Jurkat cells more rapidly than any of the five tested IgGs (Figure 5a). The antibodies with faster off rates, such as E09 or EP4b_E03, were able to trigger the cascade more rapidly than antibodies EP5b_E05, E09_R52bS and EP6b_B01, which demonstrated slower off rates. Such results were in accordance with the cell-killing efficiency data (Figure 4).
Figure 5.
Kinetic explanation of Fas agonist activity. Jurkat cells were incubated with the selected antibodies or FasL for the indicated time points. Agonistic activity was monitored by caspase substrate z-DEVD cleavage. (a) Experiment performed without cross-linking the antibodies, showing the rate of caspase induction over time. Also labelled next to the relevant curve is the rate of koff (s−1) for each antibody, as determined by SPR. (b) The same experiment was performed following cross-linking of the antibodies with protein A. These experiments were performed in triplicate and error bars indicate the S.D. between replicates. (c) Schematic illustration of the difference in agonism between an antibody with relatively fast dissociation from Fas (above) and an antibody with relatively slow dissociation (below). The antibody with the faster rate of koff (above) is better able to recruit new Fas monomers and create an active signalling complex
On the basis of the previous observations that an increased avidity of anti-death receptor antibodies improved the bioactivity tremendously,20, 22 we investigated the effect of cross-linking on a selection of our previously characterised IgGs. We used the pentavalent recombinant protein A to cross-link the antibodies via their Fc domains and evaluated the rate at which the Fas agonists could trigger caspase activation. The trimeric recombinant FasL was again used as a positive control for a rapid induction of apoptosis.
With protein A cross-linking, caspase activation was accelerated even beyond FasL, for the majority of agonist antibodies (Figure 5b). Interestingly, cross-linking enhanced the speed of caspase activation of all antibodies independently of their Fas affinities or off rates, resulting in very similar kinetic profiles for all variants, except EP6b_B01. In addition, the cell-killing efficiency and EC50 of the IgGs improved upon cross-linking (Supplementary Figure 5). For example, the efficiency and EC50 of E09 IgG improved from 69.4% to 88.5% and from 0.83 nM to 0.10 nM, respectively.
On the basis of our findings, we propose a model in which partial dissociation of a bivalent antibody from Fas is crucial for receptor activation. In this hypothesis, dissociation of one Fab arm must occur in order to recruit more Fas monomers to the receptor cluster and for agonism to be triggered (Figure 5c). Antibodies with a very slow dissociation rate would therefore be predicted to ‘lock' two Fas monomers into a non-signalling complex.
Discussion
The TNF receptor superfamily is a rich source of therapeutic targets for agonistic monoclonal antibody approaches and yet there is little knowledge of the molecular details of agonism or strategies for improving agonist potency. The work described here used antibody-mediated Fas receptor agonism, resulting in cellular apoptosis, as a model system to explore these questions.
The biochemical and structural comparison of epitopes between Fas antibody E09 and the natural FasL, as well as two commercially available antibody agonists, did not identify a single ‘key' binding site on the receptor, which could initiate a signalling cascade. The minimal predicted overlap between the E09 epitope and that of FasL (Figure 2), suggested that E09 was not functioning as a direct ligand mimetic. Indeed the orientation of E09 binding was totally different, with the antibody binding to the outside of the receptor complex and FasL binding to the inside, when the receptor is viewed in the expected trimeric configuration (Figure 2). That both E09 and FasL appeared to interact via a tyrosine side chain with an arginine at residue 86 of Fas receptor did initially suggest that this could be a key interaction in the signalling process. However, this tyrosine in the E09 VH CDR3 loop, could be mutated to several other side chains without any major impact on potency (data not shown), indicating that it was not a critical residue for signal transduction.
Affinity maturation of E09 was performed by directed evolution and also by the rational approach of side chain replacement to isolate antibodies with different affinities for Fas. This series of antibodies, when tested for agonistic activity, showed a surprising inverse correlation between affinity and agonism. We believe this to be the first demonstration of an increase in antibody affinity leading to a decrease in biological potency, in contrast to the many published examples of positive correlations between the two attributes. Although the initial observation was a correlation between higher affinity and reduced receptor agonism, further analysis showed a clearer correlation between a slower rate of dissociation and reduced agonism (Figure 4b). Effectively, the antibodies with the slowest dissociation rates were impaired in both their maximal level of receptor agonism and the time required to initiate signalling. As the E09 antibody was also capable of antagonising the Fas:FasL interaction (Table 1), we could also explore the correlation between affinity and antagonism using this series of antibodies. We would predict, based on the many other antagonistic antibodies studied to date, that we would see a direct correlation between affinity and antagonism.
The accumulated evidence led us to hypothesise a model for agonism of receptors in the TNF receptor superfamily, which takes into account that these receptors are normally brought into an active trimeric configuration by their naturally trimeric ligands. The model predicts that bivalent agonists, such as IgG antibodies, must be able to bind to Fas receptor and then partially dissociate in order to free one Fab arm to recruit further Fas monomers and form an active signalling complex (Figure 5c). Antibodies with too slow a dissociation rate would, in effect, become locked in a non-signalling, dimeric state. Supporting this hypothesis are the previous observations that the Fab arms of an antibody are capable of considerable movement around the flexible hinge region23 and also that many receptors, including Fas,24, 25 are believed to move freely within the plasma membrane in order to form clusters for optimal signalling.26
To test our hypothesis we used antibody cross-linking to demonstrate that the impairment caused by slow dissociation could be overcome in most cases by increasing the valency of the agonist antibodies to avoid the non-signalling, dimeric complexes (Figure 5 and Supplementary Figure 4). We show that cross-linked IgG and the trimeric FasL were more effective agonists than the standard bivalent IgG molecule (Figure 5). Others have also shown that the higher valency IgM format of the Fas agonist CH-1127 leads to very high potency. This observation about valency has led some to pursue higher order multimers, such as Mega-FasL,28 as potential therapeutic agonists, notwithstanding the safety concerns.
Although many others have observed that higher valency Fas agonists are more potent at triggering apoptosis,20, 22, 27, 28, 29 and our data is in agreement with those studies, this is the first detailed analysis of the relationship of affinity to potency. By using antibodies of a single lineage, which only differ by a few amino acids, we have maintained a constant epitope on Fas while varying the antibody affinity. We have shown that lower-affinity Fas antibodies can have higher potency, a theory that has in fact been put forward in a previous publication29 and may have much in common with the ‘serial triggering' hypothesis for peptide-MHC activation of T-cell receptors.30
The main implication of our finding is that bivalent agonists of receptors in the TNF receptor superfamily could have a potency ceiling, which is not surmountable by the conventional approach of affinity maturation. In fact, as shown here, affinity maturation would tend to impair the antibody potency. Alternative engineering strategies would be required to overcome this limitation. One example of such engineering would be to use bispecific antibody technology to engineer a bivalent molecule in which one arm contained a high-affinity binding site to anchor the antibody onto one Fas molecule and the other arm contained a low affinity binding site to efficiently recruit further Fas monomers. Additional engineering of the IgG hinge region to maximise the Fab arm flexibility could further enhance agonism of such a molecule.
In summary we describe the first detailed structural and kinetic analysis of a related lineage of agonist molecules to the Fas receptor, which has enabled us to better understand the nature of bivalent agonist signalling in the TNF receptor superfamily. We propose a model to explain the surprising inverse correlation between antibody affinity and potency and use the implications of the model to make suggestions, which are relevant to future drug discovery efforts on targets within the TNF receptor superfamily.
Materials and Methods
Cell lines and reagents
Tumour cell lines HeLa (cervix epitheloid carcinoma) and Jurkat (T-cell leukaemia) were obtained from ATCC (Manassas, VA, USA) and were maintained in DMEM medium (Gibco, Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (Invitrogen) and 1% non-essential amino acids (Invitrogen) or RPMI medium 1640 with GlutaMAX (Invitrogen) supplemented with 10% fetal bovine serum, respectively. Commercial mouse anti-human Fas antibodies ZB4 and CH-11 are from Upstate (Millipore, Billerica, MA, USA) and SM1.1 from Chemicon (Millipore). DX2 antibody and human Fas ligand (hFasL) were purchased from R&D Systems (Minneapolis, MN, USA). Human Fas receptor in fusion with an Fc fragment and hFasL are from R&D Systems. The monomeric format of the Fas ECD for Biacore and crystallisation studies was obtained from Peprotech (Rocky Hill, NJ, USA).
Antibody isolation
Phage-display technology was used to isolate scFv antibodies to the recombinant ECD of Fas receptor. A large non-immunised human scFv phage-display library containing up to 1011 binding members12 was used for antibody isolation as described previously.31 Briefly, human Fas receptor ECD in fusion with an Fc fragment was immobilised overnight at 4 °C onto MaxiSorp plates (Nunc, ThermoFisher, Waltham, MA, USA) at 10 μg/ml in PBS before presentation to the antibody library. Soluble selections were performed as described previously32 using biotinylated Fas receptor ECD/Fc protein and magnetic streptavidin beads (Dynal, Invitrogen) for capturing the antigen-antibody complex.
Three to four rounds of selection were performed after a deselection step using a large excess of the irrelevant human IgG1 antibody CAT002 to minimise the enrichment of anti-Fc antibodies. Specificity to Fas receptor ECD was confirmed by phage-scFv ELISAs as described in Vaughan et al.,31 using CAT002 as irrelevant control.
Expression of scFv and IgG1 antibodies
Escherichia coli periplasmic extracts were prepared in 96-well plate as previously described.31 ScFv expression was induced overnight with 20 μM final of IPTG and bacterial pellets were resuspended the following day in cold TES buffer before centrifugation to clarify the extract from cellular debris. ScFv were purified from bacterial periplasmic extract by immobilised metal affinity chromatography and gel filtration to buffer exchange into PBS as described in Ruter et al.8 Protein concentrations were determined by bicinchoninic acid assay (Sigma-Aldrich, St. Louis, MO, USA). For IgG expression, the VH and VL chains of selected antibodies were cloned into human IgG1 expression vectors as described in Persic et al.33 except that an oriP fragment was included in the vectors to facilitate use with human embryonic kidney Epstein–Barr virus-encoded nuclear antigen-293 (HEK EBNA-293) cells and to allow episomal replication. Co-transfection of the heavy chain vector pEU15.1 and lambda light chain vector pEU4.4 into HEK EBNA-293 allowed whole IgG to be expressed and purified by protein A affinity chromatography (GE Healthcare, Little Chalfont, UK). Human IgG1 concentration was determined spectrophotometrically using an extinction coefficient based on the amino-acid sequence of the antibody.
Kinetic and affinity measurement by Biacore analysis
The affinity and kinetic parameters of anti-Fas antibodies for the human Fas receptor were determined by SPR using a BIAcore 2000 instrument set up at 25 °C. For scFv analysis, protein G' (Sigma-Aldrich) immobilised on a CM5 sensor chip by the amine coupling method was used to capture 300–400 RUs of Fc-tagged recombinant human Fas. Serial dilution of scFv from 200 nM to 6.25 nM in HBS-EP buffer (Biacore, Uppsala, Sweden) with 0.1% BSA were injected at 30 μl/min for 2 min. Dissociation was analysed for a short time to minimise avidity effect of potential dimeric scFv. IgG analysis was performed by first capturing ∼1000 RUs of the antibody on the protein G' – CM5 sensor chip and running a serial dilution from 400 to 12.5 nM of monomeric human Fas ECD (Peprotech) as analyte for 1 min association followed by a 10-min dissociation phase. Curve fitting was done on the BIAevaluation 3.2 RC1 software (Biacore) using a 1 : 1 Langmuir binding model.
Biochemical competition assays
Biochemical ligand and epitope competition assays were performed by immobilising anti-Fas E09 human IgG1 or FasL onto MaxiSorp plates (Nunc) at 5 or 2.5 μg/ml, respectively. After blocking, serial dilutions of anti-Fas IgG or irrelevant CAT002 were added to the coated plates in competition with biotinylated human Fas receptor Fc fusion protein at a final concentration of 0.5 μg/ml or 0.75 μg/ml for E09 and FasL competition assays, respectively. Biotinylated Fas receptor bound to the plate was detected using europium-labelled streptavidin (Perkin Elmer, Waltham, MA, USA) by measuring fluorescence at 340 nm excitation and 615 nm emission.
DNA fragmentation assay
Jurkat cells were seeded onto 96-well tissue culture plates the day before the assay at a density of 2 × 103 cells per well in 90 μl media and grown overnight at 37 °C and 5%. CO2. After adding 10 μl of anti-Fas IgG or FasL at different concentrations, cells were further incubated for 8 h before determining the level of DNA fragmentation using the Cell Death Detection ELISAPLUS (Roche, Indianapolis, IN, USA) according to the manufacturer's instructions. Briefly, cells were pelleted and lysed for 30 min at room temperature. Lysates were centrifuged and 20 μl of supernatant transferred to a streptavidin coated plate before adding 80 μl of immunoreagents for the sandwich ELISA containing a biotinylated anti-histone and a peroxidase-conjugated anti-DNA antibodies. Plates were incubated for 2 h at room temperature before adding the peroxidase detection reagents and reading on a Wallac (Perkin Elmer) 1420 workstation at 405 nm.
Caspase assay
Jurkat cells were seeded onto 96-well tissue culture plates the day before the assay at a density of 1 × 104 cells per well in 90 μl media and grown overnight at 37 °C and 5%. CO2. For titration experiments, cells were incubated for 8 h after addition of 10 μl of IgG or FasL at different concentrations before adding the caspase assay reagent.
Kinetic activation experiments were performed at a concentration of 5 nM final of IgG and FasL or 1 nM of IgM CH-11 in order to maintain the molarity of Fas receptor binding molecules. For cross-linking experiment, IgGs at 50 nM were incubated overnight at 4 °C with 12.5 nM of recombinant protein A (Pierce, ThermoFisher) to pre-form the complex before adding 10 μl to the cells. Activity assay was performed using a Caspase-Glo 3/7 assay kit (Promega, Madison, WI, USA) according to the manufacturer's instructions and plates were incubated for 1 h at room temperature before reading on a luminometer.
Cell-viability assay
Tumour cells were seeded in culture medium onto 96-well tissue culture plates at a density of 3 × 104 cells/well or 2 × 104 cells/well in 100 μl for HeLa and Jurkat cell lines, respectively. Periplasmic extract of anti-Fas scFv were added to HeLa cells in combination with a sub-lethal dose of cycloheximide (Sigma-Aldrich) at 6.6 μg/ml final as sensitising agent. Titration experiments were performed by adding 15 μl of a serial dilution of FasL or IgG in PBS plus 35 μl of medium supplemented with penicillin and streptomycin to Jurkat cells. Cyclohemixide sensitisation was not required for that cell line to obtain Fas-mediated cell killing. For cross-linking experiment, IgG-Protein A complexes were pre-formed by incubating 0.1 mg/ml of antibody with 170 nM of Protein A (4 : 1 molar ratio) before the serial dilution. After 16–18 h incubation at 37 °C and 5% CO2, Alamar Blue (Serotec, Oxford, UK) was added in an amount equal to 10% of the culture volume. The plates were returned to the incubator for an additional 7 h at 37 °C and viability assessed by measuring fluorescence on a Wallac 1420 workstation at 560 nm excitation and 590 nm emission. Data were analysed by using PRISM software (GraphPad, La Jolla, CA, USA).
Ribosome-display affinity maturation
The gene encoding the E09 anti-Fas scFv was used as the template for constructing the randomised library. Error-prone PCR was performed as described21 using Taq polymerase in the presence of 7 mM MgCl2 and 0.3 mM MnCl2 to introduce an average of 4.5 base changes per scFv gene. Ribosome-display selections were performed in a soluble format as previously described34 using human Fas-Fc protein and capturing the antigen-scFv complexes, thanks to Protein G-coated magnetic beads (Dynal). Stringent selections for higher-affinity scFv variants were applied according to previously published principles,32 whereby the concentration of antigen was reduced over successive rounds of selection from 50 nM in round 1 down to 20pM in round 6.
Affinity maturation screening assays
Screening assays were essentially performed as described.21 Primary screening was performed by immobilising supernatant-leaked scFv onto high binding COSTAR (Corning, NY, USA) plates. Binding of human Fas receptor Fc fusion protein incubated in skimmed-milk blocked plates at 0.35 ng/ml were detected using 0.5 μg/ml of europium-labelled anti-human IgG (Perkin Elmer). Fluorescence signals detected at 340 nm excitation and 615 nm emission were normalised by the cell-culture density at 600 nm to take into account difference in cell growth. Confirmatory assay was based on a surrogate affinity screen using the same principle as the primary assay. Purified scFvs were immobilised on COSTAR plate and incubated for 2 h with a serial dilution of human Fas receptor Fc fusion protein before detection with the europium-labelled anti-human IgG. Surrogate affinity of the scFvs for Fas receptor was calculated from the equilibrium binding titrations using PRISM software (GraphPad).
Point mutagenesis
Single-point mutants of E09 scFv were generated by saturation mutagenesis targeting the following six contact residues (numbered according to Kabat35): VH52 Ala, VH52a Tyr, VH52b Arg, VH55 Gly, VH56 Ser or VH58 Tyr. Site-directed mutagenesis has been performed using a classic molecular biology approach based on PCR with the following mutagenic primers:
>E09_NSS_A52_fwd
5′-GGGTTGGCAGTATCNSSTATAGGGGGAATAGCAATAGTGG-3′
>E09_NSS_Y52a_ fwd
5′-GGGTTGGCAGTATCGCCNSSAGGGGGAATAGCAATAGTGG-3′
>E09_NSS_R52b_ fwd
5′-GGGTTGGCAGTATCGCCTATNSSGGGAATAGCAATAGTGG-3′
>E09_NSS_G55_ fwd
5′-GGGGAATAGCAATAGTNSSAGCACCTACTATAATCCGTCCCTCAAG-3′
>E09_NSS_S56_ fwd
5′-GGGGAATAGCAATAGTGGGNSSACCTACTATAATCCGTCCCTCAAG-3′
>E09_NSS_Y58_ fwd
5′-GGGGAATAGCAATAGTGGGAGCACCNSSTATAATCCGTCCCTCAAG-3′
After cloning in the pCANTAB6 vector,31 ligations were transformed in E. coli TG1 strain. Mutants were identified by sequencing, and a total of 32 clones were selected for scFv purification and affinity characterisation.
Production of E09 and EP6b_B01 Fabs for crystallisation
Anti-Fas E09 human IgG1 was transiently expressed in EBNA-293 and purified by protein A chromatography using MabSelect SuRe (GE Healthcare). Purified IgG was digested by papain (Sigma-Aldrich) at a ratio of 1 mg enzyme per 100 mg of antibody for 5 h at room temperature in 30 nM DL-cysteine buffer. Digestion was stopped by adding iodoacetamide at a final concentration of 50 mM. Fab fragment was then purified on MabSelect SuRe column in a non-binding mode followed by a gel-filtration step on Superdex 75 (GE Healthcare) in 50 mM acetate pH 5.5 buffer containing 100 mM NaCl. At the end of the process, 1.45 ml of purified Fab at 5.48 mg/ml was obtained. Binding activity of the E09 Fab fragment was shown to be similar to the human IgG1 by Biacore analysis using a CM5-Protein G' chip to capture human Fas receptor –Fc fusion protein and running a serial dilution of the Fab fragment as analyte. The E09 variant EP6b_B01 was produced as a Fab fused with a C-terminal 10 × His tag in CHO cells. The Fab was purified by immobilised metal affinity chromatography using HisTrap (GE Healthcare) and by size exclusion chromatography in PBS
Crystallisation and structure determination
Excess Fab was mixed with the ECD of human Fas and incubated on ice. The excess protein was separated from the complex by gel filtration using a Superdex 200 10/300 GL (GE Healthcare) in 50 mM MES/NaOH, pH 6.5, 100 mM NaCl. The complexes were concentrated to 11 mg/ml and crystallised by sitting drop vapour diffusion at 4 °C. The E09-Fas complex crystallised in 100 mM Tris/HOAc, pH 8.5, 5 mM CdCl2, 20% PEG4000, whereas the EP6b_B01-Fas complex crystallised in 100 mM HEPES/NaOH, pH 7.5, 10% isopropanol, 20% PEG4000. For data collection the crystals were cryo-protected in a mother liquor solution containing additional 20% ethylene glycol and prefrozen in liquid propane (E09) or nitrogen (EP6b_B01). Diffraction data of the E09-Fas and EP6b_B01-Fas complexes were collected on a Pilatus detector at the Swiss Light Source (Villigen, Switzerland) using a wavelength of 0.9000 and 1.0000 Å, respectively. Data were processed using XDS36 and the phases solved by molecular replacement using Phaser v1.3 (E09) and v2.1.4 (EP6b_B01).37 For the phase solution of the first Fab:Fas structure (E09) an iterative process involving the four domains of the antibody HC19 (1GIG) was applied.38 The obtained solution was refined with Phenix before the molecular replacement was finished by searching the Fas receptor using a polyserine model of the DR5 structure 2H9G.20
The model was automatically built with ARP/wARP v739 and rebuilt in Coot v0.3.3.40 Refinement of both complexes was carried out using Phenix v1.6.4 and Coot v0.6.1.41 Data collection and refinement statistics of the two structures are listed in Supplementary Table 1.
The atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession number 3TJE for the E09:Fas complex and 3THM for the EP6b_B01 complex.
Acknowledgments
We thank the DNA Chemistry, Tissue Culture and High-Throughput Expression teams at MedImmune for their invaluable support of this work. We also thank Jon Large for his assistance with the artwork.
Glossary
- TNF
tumour necrosis factor
- TRAIL
TNF-related apoptosis-inducing ligand
- GITR
glucocorticoid-induced tumor necrosis factor receptor
- FasL
Fas ligand
- ECD
extracellular domain
- CRD
cysteine-rich domain
- CDR
complementarity-determining region
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by S Nagata
Supplementary Material
References
- van der Merwe PA, Dushek O. Mechanisms for T cell receptor triggering. Nat Rev Immunol. 2011;11:47–55. doi: 10.1038/nri2887. [DOI] [PubMed] [Google Scholar]
- Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- Tansey MG, Szymkowski DE. The TNF superfamily in 2009: new pathways, new indications, and new drugs. Drug Discov Today. 2009;14:1082–1088. doi: 10.1016/j.drudis.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Pukac L, Kanakaraj P, Humphreys R, Alderson R, Bloom M, Sung C, et al. HGS-ETR1, a fully human TRAIL-receptor 1 monoclonal antibody, induces cell death in multiple tumour types in vitro and in vivo. Br J Cancer. 2005;92:1430–1441. doi: 10.1038/sj.bjc.6602487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CL, Main S, Newton P, Chodorge M, Cadwallader K, Humphreys R, et al. Human monomeric antibody fragments to TRAIL-R1 and TRAIL-R2 that display potent in vitro agonism. MAbs. 2009;1:552–562. doi: 10.4161/mabs.1.6.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AD, Schaer DA, Liu C, Li Y, Hirschhorn-Cymmerman D, Kim SC, et al. Agonist anti-GITR monoclonal antibody induces melanoma tumor immunity in mice by altering regulatory T cell stability and intra-tumor accumulation. PLoS One. 2010;5:e10436. doi: 10.1371/journal.pone.0010436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascierto PA, Simeone E, Sznol M, Fu YX, Melero I. Clinical experiences with anti-CD137 and anti-PD1 therapeutic antibodies. Semin Oncol. 2010;37:508–516. doi: 10.1053/j.seminoncol.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Ruter J, Antonia SJ, Burris HA, Huhn RD, Vonderheide RH. Immune modulation with weekly dosing of an agonist CD40 antibody in a phase I study of patients with advanced solid tumors. Cancer Biol Ther. 2010;10:983–993. doi: 10.4161/cbt.10.10.13251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T, Takahashi T, Golstein P, Nagata S. Molecular cloning and expression of the Fas ligand, a novel member of the tumor necrosis factor family. Cell. 1993;75:1169–1178. doi: 10.1016/0092-8674(93)90326-l. [DOI] [PubMed] [Google Scholar]
- Trauth BC, Klas C, Peters AM, Matzku S, Moller P, Falk W, et al. Monoclonal antibody-mediated tumor regression by induction of apoptosis. Science. 1989;245:301–305. doi: 10.1126/science.2787530. [DOI] [PubMed] [Google Scholar]
- Ogasawara J, Watanabe-Fukunaga R, Adachi M, Matsuzawa A, Kasugai T, Kitamura Y, et al. Lethal effect of the anti-Fas antibody in mice. Nature. 1993;364:806–809. doi: 10.1038/364806a0. [DOI] [PubMed] [Google Scholar]
- Lloyd C, Lowe D, Edwards B, Welsh F, Dilks T, Hardman C, et al. Modelling the human immune response: performance of a 1011 human antibody repertoire against a broad panel of therapeutically relevant antigens. Protein Eng Des Sel. 2009;22:159–168. doi: 10.1093/protein/gzn058. [DOI] [PubMed] [Google Scholar]
- Cifone MG, De Maria R, Roncaioli P, Rippo MR, Azuma M, Lanier LL, et al. Apoptotic signaling through CD95 (Fas/Apo-1) activates an acidic sphingomyelinase. J Exp Med. 1994;180:1547–1552. doi: 10.1084/jem.180.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadeel B, Thorpe CJ, Yonehara S, Chiodi F. Anti-Fas IgG1 antibodies recognizing the same epitope of Fas/APO-1 mediate different biological effects in vitro. Int Immunol. 1997;9:201–209. doi: 10.1093/intimm/9.2.201. [DOI] [PubMed] [Google Scholar]
- Bajorath J. Identification of the ligand binding site in Fas (CD95) and analysis of Fas-ligand interactions. Proteins. 1999;35:475–482. [PubMed] [Google Scholar]
- Starling GC, Bajorath J, Emswiler J, Ledbetter JA, Aruffo A, Kiener PA. Identification of amino acid residues important for ligand binding to Fas. J Exp Med. 1997;185:1487–1492. doi: 10.1084/jem.185.8.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starling GC, Kiener PA, Aruffo A, Bajorath J. Analysis of the ligand binding site in Fas (CD95) by site-directed mutagenesis and comparison with TNFR and CD40. Biochemistry. 1998;37:3723–3726. doi: 10.1021/bi972959d. [DOI] [PubMed] [Google Scholar]
- Schneider P, Bodmer JL, Holler N, Mattmann C, Scuderi P, Terskikh A, et al. Characterization of Fas (Apo-1, CD95)-Fas ligand interaction. J Biol Chem. 1997;272:18827–18833. doi: 10.1074/jbc.272.30.18827. [DOI] [PubMed] [Google Scholar]
- Adams C, Totpal K, Lawrence D, Marsters S, Pitti R, Yee S, et al. Structural and functional analysis of the interaction between the agonistic monoclonal antibody Apomab and the proapoptotic receptor DR5. Cell Death Differ. 2008;15:751–761. doi: 10.1038/sj.cdd.4402306. [DOI] [PubMed] [Google Scholar]
- Li B, Russell SJ, Compaan DM, Totpal K, Marsters SA, Ashkenazi A, et al. Activation of the proapoptotic death receptor DR5 by oligomeric peptide and antibody agonists. J Mol Biol. 2006;361:522–536. doi: 10.1016/j.jmb.2006.06.042. [DOI] [PubMed] [Google Scholar]
- Chodorge M, Fourage L, Ravot G, Jermutus L, Minter R. In vitro DNA recombination by L-Shuffling during ribosome display affinity maturation of an anti-Fas antibody increases the population of improved variants. Protein Eng Des Sel. 2008;21:343–351. doi: 10.1093/protein/gzn013. [DOI] [PubMed] [Google Scholar]
- Dhein J, Daniel PT, Trauth BC, Oehm A, Moller P, Krammer PH. Induction of apoptosis by monoclonal antibody anti-APO-1 class switch variants is dependent on cross-linking of APO-1 cell surface antigens. J Immunol. 1992;149:3166–3173. [PubMed] [Google Scholar]
- Sandin S, Ofverstedt LG, Wikstrom AC, Wrange O, Skoglund U. Structure and flexibility of individual immunoglobulin G molecules in solution. Structure. 2004;12:409–415. doi: 10.1016/j.str.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Scott FL, Stec B, Pop C, Dobaczewska MK, Lee JJ, Monosov E, et al. The Fas-FADD death domain complex structure unravels signalling by receptor clustering. Nature. 2009;457:1019–1022. doi: 10.1038/nature07606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho KL, Harrington HA. Bistability in apoptosis by receptor clustering. PLoS Comput Biol. 2010;6:e1000956. doi: 10.1371/journal.pcbi.1000956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie P, Goetz JG, Dennis JW, Nabi IR. Lattices, rafts, and scaffolds: domain regulation of receptor signaling at the plasma membrane. J Cell Biol. 2009;185:381–385. doi: 10.1083/jcb.200811059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonehara S, Ishii A, Yonehara M. A cell-killing monoclonal antibody (anti-Fas) to a cell surface antigen co-downregulated with the receptor of tumor necrosis factor. J Exp Med. 1989;169:1747–1756. doi: 10.1084/jem.169.5.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisele G, Roth P, Hasenbach K, Aulwurm S, Wolpert F, Tabatabai G, et al. APO010, a synthetic hexameric CD95 ligand, induces human glioma cell death in vitro and in vivo. Neuro Oncol. 2011;13:155–164. doi: 10.1093/neuonc/noq176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komada Y, Inaba H, Li QS, Azuma E, Zhou YW, Yamamoto H, et al. Epitopes and functional responses defined by a panel of anti-Fas (CD95) monoclonal antibodies. Hybridoma. 1999;18:391–398. doi: 10.1089/hyb.1999.18.391. [DOI] [PubMed] [Google Scholar]
- Valitutti S, Muller S, Cella M, Padovan E, Lanzavecchia A. Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature. 1995;375:148–151. doi: 10.1038/375148a0. [DOI] [PubMed] [Google Scholar]
- Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, et al. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol. 1996;14:309–314. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- Hawkins RE, Russell SJ, Winter G. Selection of phage antibodies by binding affinity. Mimicking affinity maturation. J Mol Biol. 1992;226:889–896. doi: 10.1016/0022-2836(92)90639-2. [DOI] [PubMed] [Google Scholar]
- Persic L, Roberts A, Wilton J, Cattaneo A, Bradbury A, Hoogenboom HR. An integrated vector system for the eukaryotic expression of antibodies or their fragments after selection from phage display libraries. Gene. 1997;187:9–18. doi: 10.1016/s0378-1119(96)00628-2. [DOI] [PubMed] [Google Scholar]
- Hanes J, Jermutus L, Pluckthun A. Selecting and evolving functional proteins in vitro by ribosome display. Methods Enzymol. 2000;328:404–430. doi: 10.1016/s0076-6879(00)28409-7. [DOI] [PubMed] [Google Scholar]
- Kabat EA.Sequences of Proteins of Immunological Interest, 5th edn. NIH publication no. 91-3242US Department of Health and Human Services, Public Health Service, National Institutes of Health: Bethesda, MD; 1991 [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizebard T, Daniels R, Kahn R, Golinelli-Pimpaneau B, Skehel JJ, Knossow M. Refined three-dimensional structure of the Fab fragment of a murine IgGl,lambda antibody. Acta Crystallogr D Biol Crystallogr. 1994;50:768–777. doi: 10.1107/S0907444994001903. [DOI] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.