Abstract
The adiponutrin (PNPLA3) rs738409 polymorphism has been found to be associated with susceptibility to non-alcoholic fatty liver disease (NAFLD) in various cohorts. We further investigated the association of this polymorphism with non-alcoholic steatohepatitis (NASH) severity and with histological features of NAFLD. A total of 144 biopsy-proven NAFLD patients and 198 controls were genotyped for PNPLA3 gene polymorphism (rs738409 C>G). The biopsy specimens were histologically graded by a qualified pathologist. We observed an association of G allele with susceptibility to NAFLD in the pooled subjects (OR 2.34, 95% CI 1.69–3.24, p < 0.0001), and following stratification, in each of the three ethnic subgroups, namely Chinese, Indian and Malay (OR 1.94, 95% CI 1.12–3.37, p = 0.018; OR 3.51, 95% CI 1.69–7.26, p = 0.001 and OR 2.05, 95% CI 1.25–3.35, p = 0.005, respectively). The G allele is associated with susceptibility to NASH (OR 2.64, 95% CI 1.85–3.75, p < 0.0001), with NASH severity (OR 1.85, 95% CI 1.05–3.26, p = 0.035) and with presence of fibrosis (OR 1.95, 95% CI 1.17–3.26, p = 0.013) but not with simple steatosis nor with other histological parameters. Although the serum triglyceride level is significantly higher in NAFLD patients compared to controls, the G allele is associated with decreased level of triglycerides (p = 0.029) in the NAFLD patients. Overall, the rs738409 G allele is associated with severity of NASH and occurence of fibrosis in patients with NAFLD.
Introduction
Non-alcoholic fatty liver disease (NAFLD) is a common cause of chronic liver disease and has emerged as an important global health problem (Adams et al. 2005; Angulo 2002). The reported prevalence of NAFLD in the general population is as high as 35% (Cheung and Sanyal 2010). NAFLD is a spectrum ranging from simple steatosis (fat in the liver) to non-alcoholic steatohepatitis (NASH—fat with inflammation and/or fibrosis) and cirrhosis (Charlton 2004; Malaguarnera et al. 2009). NAFLD represents the hepatic component of the metabolic syndrome and is strongly associated with obesity and insulin resistance (Adams et al. 2009; Bertrand et al. 2011; Fatani et al. 2011; Vanni et al. 2010). Genetic factors have been shown to play a significant role in the pathogenesis of NAFLD (Alisi et al. 2011; Day 2010; Merriman et al. 2006). A familial aggregation study comparing overweight children with NAFLD, and overweight children without NAFLD reported that fatty liver was more significant in siblings of children with NAFLD (Schwimmer et al. 2009).
Adiponutrin encoded by PNPLA3 has been reported to have both lipolytic and lipogenic properties; however, the exact function of adiponutrin is unknown (Jenkins et al. 2004). A common variant of PNPLA3 gene (rs738409) has been reported to be associated with hepatic fat content in the Dallas Heart Study of different ancestries; Hispanics, European Americans and African-Americans. The Hispanics bear the highest propensity to develop NAFLD followed by the European Americans with African-Americans being the group with the lowest risk of NAFLD (Romeo et al. 2008). Since then, several other studies in different populations have been conducted (Cox et al. 2011; Hotta et al. 2010; Kantartzis et al. 2009; Lin et al. 2011; Romeo et al. 2010a, b; Sookoian et al. 2009; Valenti et al. 2010).
A diagnosis of fatty liver can be made clinically, based on radiological and laboratory findings. However, the spectrum of NAFLD, particularly the diagnosis of NASH can only be confirmed by liver biopsy (Cortez-Pinto and Camilo 2004; Serfaty and Lemoine 2008). While most of the studies have attempted to predict risk of NAFLD, it is more important to predict the risk of steatohepatitis as it is potentially progressive (Vuppalanchi and Chalasani 2009). Recent studies have therefore moved into looking at the association of PNPLA3 variant with disease severity (Hotta et al. 2010; Rotman et al. 2010; Sookoian et al. 2009).
The Malaysian population comprises of three major ethnic subgroups, namely the Malays, Chinese and Indians, each of which are presumably of different genetic pool, hence presenting a good opportunity to study the ethnic difference of NAFLD susceptibility with regard to PNPLA3 gene polymorphism. In the present study, we investigated the association of PNPLA3 rs738409 with severity of NASH and with histological features of NAFLD in the three major ethnic groups of Malaysian NAFLD patients.
Methods
Study population
The study involved a total of 144 consecutive biopsy-proven NAFLD patients and 198 controls without fatty liver based on histology or ultrasound and normal liver enzymes. Subjects were recruited from the University Malaya Medical Centre (UMMC). The study protocol was approved by the Medical Ethics Committee of UMMC. Informed consent was obtained from all study subjects. Participants with the following criteria were excluded from the study; current or previous alcohol consumption >10 g/day (Ruhl and Everhart 2005), hepatitis B or C infection, autoimmune hepatitis, were taking drugs that are known to cause steatosis, or Wilson’s disease. Cases were patients who were clinically diagnosed with NAFLD and confirmed by liver biopsy. The Brunt system was used for the grading and staging of the liver histology (Brunt et al. 2011; Kleiner et al. 2005). Patients with NAFLD were classified to three groups: simple steatosis, NASH without significant fibrosis (fibrosis score of <2) and NASH with significant fibrosis (fibrosis score of 2 or greater). The control group without fatty liver consists of those with normal BMI (<25 kg/m2), normal fasting plasma glucose (<110 mg/dl) and normal lipid profile.
Clinical and laboratory assessments
Body mass index (BMI), waist circumference and blood pressure were determined according to standard protocol. Blood was taken for determination of hemoglobin A1c (HbA1c), total cholesterol, triglycerides, high-density lipoprotein cholesterol (HDL), low-density lipoprotein cholesterol (LDL), alanine transferase (ALT), aspartate aminotransferase (AST), and gamma glutamyl transpeptidase (GGT) level in all subjects.
Genotyping
Blood samples obtained from the subjects were centrifuged at 1,500 rpm for 10 min. The buffy coat layer were separated and transferred into 1.5-mL centrifuge tubes. Genomic DNA was extracted from the concentrated lymphocytes of the buffy coat using the QiAamp DNA Mini Kit (Qiagen, Hilden, German). PNPLA3 rs738409 I148M variant was genotyped using a predesigned Taqman SNP genotyping assay (Applied Biosystems, Foster City, CA, USA) on a Step One Real-Time PCR instrument (Applied Biosystems). The total reaction volume for each well was 10 μl containing 5 μl universal mastermix (Applied Biosystems), 0.5 μl assay mix, 3.5 μl distilled water and 1 μl genomic DNA. The plate was set up at 95°C holding stage for 20 s, 45 cycles of 95°C denaturation for 3 s and 60°C annealing for 20 s and ran on a fast reaction (approximately 40 min for each run). Negative controls were introduced for every run to ensure the genotyping quality.
Statistical
All values were presented as mean ± standard deviation for continuous data and as percentages for categorical data. All variables were tested for normality using Kolmogorov–Smirnov test. For normally distributed variables (age, BMI, waist circumference, ALT, HDL cholesterol, LDL cholesterol, total cholesterol, triglycerides, and systolic blood pressure), Student’s t test was performed to determine the associations between the NASH and simple steatosis. For skewed variables—HbA1c, AST, GGT, diastolic blood pressure, steatosis grade, lobular inflammation, ballooning, and fibrosis—Mann–Whitney U test was performed. Hardy–Weinberg equilibrium (HWE) was checked for all the groups using a goodness-of fit χ 2 test with one degree of freedom. Association of allele was performed using the binary logistic regression. Multiple logistic regression was performed to adjusting for age, gender and ethnicity. Analysis of variance (ANOVA) and Kruskal–Wallis test were conducted for comparison of means between three groups for normally and non-normally distributed variables, respectively.
Univariate analysis of association of genotypes and histological ordinal variables was assessed using the Jonckheere–Terpstra test. Ordinal regression is performed for multivariate analysis of histological ordinal variables. To correct for testing for multiple histological parameters, the false discovery rate (FDR) was calculated using Benjamini–Hochberg procedure (Benjamini and Hochberg 1995). For easier interpretation of the results, binary logistic regression was used to obtain the odds ratio. Analyses were performed using SPSS 16.0 (Chicago IL) and Microsoft Excel, and a p value <0.05 was considered statistically significant.
We estimated that a sample size of 135 cases and controls would provide 80% power at an α of 0.05 with the following assumptions: the allele frequency ranges from 0.24 to 0.45, the baseline risk for the Malaysian population was 0.17, and the minimum detectable odds ratio was 2.0.
Results
The demographic and clinical data of the NAFLD and control groups are shown in Table 1. The 144 patients consisted of 54 Chinese, 31 Indians, and 59 Malays. Out of the 198 controls, 54 were Chinese, 64 Indians and 80 Malays. Ethnicity of the subjects was confirmed by affirmations of absence of mixed marriages for at least three generations.
Table 1.
Demographic and clinical data of the subjects
| Characteristics | n (%) or mean ± SD | |
|---|---|---|
| Control (n = 198) | NAFLD (n = 144) | |
| Gender | ||
| Males | 85 (43) | 77 (53) |
| Females | 113 (57) | 67 (47) |
| Ethnicity | ||
| Malays | 80 (41) | 59 (41) |
| Chinese | 54 (27) | 54 (38) |
| Indians | 64 (32) | 31 (21) |
| Age (years) | 53.1 ± 11.5 | 51.2 ± 12.0 |
| BMI (kg/m2) | 22.7 ± 2.6 | 28.7 ± 4.4 |
| HbA1c (%) | 5.7 ± 0.8 | 6.6 ± 1.7 |
| HDL cholesterol (mg/dl) | 49.5 ± 12.9 | 48.5 ± 12.7 |
| LDL cholesterol (mg/dl) | 89.5 ± 22.4 | 117.1 ± 40.0 |
| Total cholesterol (mg/dl) | 176.4 ± 26.9 | 196.7 ± 44.0 |
| Triglycerides (mg/dl) | 118.4 ± 32.3 | 155.0 ± 62.7 |
| AST (IU/L) | 21.8 ± 9.5 | 42.9 ± 25.4 |
| ALT (IU/L) | 36.0 ± 16.6 | 83.0 ± 48.5 |
| GGT (IU/L) | 44.0 ± 25.4 | 111.6 ± 115.5 |
Data are expressed in mean ± SD for continuous data and percentage for categorical data
ALT alanine transferase, AST aspartate aminotransferase, BMI body mass index, GGT gamma glutamyl transpeptidase, HbA1c hemoglobin A1c, HDL high-density lipoprotein, LDL low-density lipoprotein, NAFLD non-alcoholic fatty liver disease
Table 2 shows the demographic and clinical data of the patients with simple steatosis and with NASH. The BMI, HbA1c, waist circumference, triglycerides, systolic and diastolic blood pressure were significantly higher in the NASH group (p < 0.05) as compared to the simple steatosis group.
Table 2.
Demographic and clinical data of the NAFLD patients
| Characteristics | n (%) or mean ± SD | ||
|---|---|---|---|
| Simple steatosis (n = 33) | NASH (n = 111) | p value | |
| Gender, n (%) | 0.461 | ||
| Males | 20 (61) | 57 (51) | |
| Females | 13 (39) | 54 (49) | |
| Age (years) | 50.7 ± 11.8 | 51.2 ± 12.1 | 0.82 |
| BMI (kg/m2) | 26.7 ± 3.9 | 29.2 ± 4.4 | 0.003 |
| HbA1c (%)* | 6.1 ± 1.3 | 6.7 ± 1.7 | 0.021 |
| Waist circumference (cm) | 89.2 ± 11.2 | 95.2 ± 10.4 | 0.005 |
| HDL cholesterol (mg/dl) | 50.2 ± 15.1 | 48.0 ± 11.9 | 0.391 |
| LDL cholesterol (mg/dl) | 114.7 ± 42.8 | 117.8 ± 39.3 | 0.698 |
| Total cholesterol (mg/dl) | 191.6 ± 44.9 | 198.2 ± 43.8 | 0.448 |
| Triglycerides (mg/dl) | 124.6 ± 42.0 | 164.1 ± 65.1 | 0.001 |
| AST (IU/L)* | 37.6 ± 21.3 | 44.5 ± 26.3 | 0.139 |
| ALT (IU/L) | 71.9 ± 50.0 | 86.4 ± 47.8 | 0.134 |
| GGT (IU/L)* | 99.4 ± 106.6 | 115.3 ± 118.2 | 0.132 |
| Systolic blood pressure (mmHg) | 125.2 ± 13.0 | 134.2 ± 14.1 | 0.001 |
| Diastolic blood pressure (mmHg)* | 78.2 ± 9.1 | 83.8 ± 9.7 | 0.003 |
ALT alanine transferase, AST aspartate aminotransferase, BMI body mass index, GGT gamma glutamyl transpeptidase, HbA1c hemoglobin A1c, HDL high-density lipoprotein, LDL low-density lipoprotein, NAFLD non-alcoholic fatty liver disease, NASH non-alcoholic steatohepatitis
* p values obtained using Mann–Whitney U test, all other comparisons used independent t test
Genotypes and allele frequencies of PNPLA3 rs738409 polymorphism
The association tests of PNPLA3 rs738409 for the different NAFLD spectrum are presented in Table 3. The frequency of the risk allele G is significantly higher in the NASH (48%) and in the NAFLD (45%) when each is compared to the control group (24%). The genotype distribution was in Hardy–Weinberg equilibrium for all the patients and controls of each ethnic subgroup.
Table 3.
Association tests of PNPLA3 rs738409 in different NAFLD spectrum
| NAFLD spectrum | G allele frequency | Unadjusted | Adjusted for age, gender and ethnicity | ||
|---|---|---|---|---|---|
| p value | OR (CI) | p value | OR (CI) | ||
| Control as reference | |||||
| NAFLD vs. control | |||||
| Overall | 0.45 vs. 0.24 | <0.0001 | 2.34 (1.69–3.24) | <0.0001 | 2.23 (1.60–3.11) |
| Malays | 0.42 vs. 0.24 | 0.005 | 2.05 (1.25–3.35) | 0.007 | 2.00 (1.21–3.21) |
| Chinese | 0.49 vs. 0.32 | 0.018 | 1.94 (1.12–3.37) | 0.02 | 1.93 (1.11–3.36) |
| Indians | 0.44 vs. 0.18 | 0.001 | 3.51 (1.69–7.26) | 0.001 | 3.50 (1.67–7.33) |
| Simple steatosis vs. control | 0.35 vs. 0.24 | 0.083 | 1.61 (0.94–2.75) | 0.272 | 1.37 (0.78–2.42) |
| All NASH vs. control | 0.48 vs. 0.24 | <0.0001 | 2.64 (1.85–3.75) | <0.0001 | 2.58 (1.80–3.69) |
| NASHa vs. control | 0.39 vs. 0.24 | 0.011 | 1.96 (1.17–3.29) | 0.023 | 1.86 (1.09–3.18) |
| NASHb vs. control | 0.52 vs. 0.24 | <0.0001 | 3.02 (2.02–4.50) | <0.0001 | 2.99 (1.99–4.50) |
| Simple steatosis as reference | |||||
| All NASH vs. simple steatosis | 0.48 vs. 0.35 | 0.083 | 1.63 (0.94–2.81) | – | – |
| NASHa vs. simple steatosis | 0.39 vs. 0.35 | 0.6 | 1.20 (0.61–2.38) | – | – |
| NASHb vs. simple steatosis | 0.52 vs. 0.35 | 0.035 | 1.85 (1.05–3.26) | – | – |
| NASH a as reference | |||||
| NASHb vs. NASHa | 0.52 vs 0.39 | 0.088 | 1.62 (0.93–2.81) | – | – |
CI confident interval, OR odds ratio, NAFLD non-alcoholic fatty liver disease, NASH non-alcoholic steatohepatitis, NASH a non-alcoholic steatohepatitis without significant fibrosis, NASH b non-alcoholic steatohepatitis with significant fibrosis
There was a significant association between the risk allele G and NAFLD in the pooled subjects (OR 2.34, 95% CI 1.69–3.24, p < 0.0001) as well as after stratification into the three ethnic subgroups, (OR 1.94, 95% CI 1.12–3.37, p = 0.018; OR 3.51, 95% CI 1.69–7.26, p = 0.001 and OR 2.05, 95% CI 1.25–3.35, p = 0.005, for Chinese, Indian and Malay, respectively). Following adjustment for age and gender (include adjustment for ethnicity for pooled subjects), the association between the risk allele G and NAFLD in the pooled subjects and after stratification into the three ethnic subgroups remained (OR 2.23, 95% CI 1.60–3.11, p < 0.0001; OR 1.93, 95% CI 1.11–3.36, p = 0.02; OR 3.50, 95% CI 1.67–7.33, p = 0.001 and OR 2.00, 95% CI 1.21–3.21, p = 0.007, for pooled, Chinese, Indian and Malay, respectively) (Table 3).
PNPLA3 rs738409 polymorphism and liver histology
The NAFLD patients were then divided into simple steatosis and NASH group following which, association analysis was performed (Table 3). We showed that the G allele is associated with NASH (OR 2.64, 95% CI 1.85–3.75, p < 0.0001) but not with simple steatosis. We further stratified the NASH patients into two groups: NASH without significant fibrosis (fibrosis score <2) and NASH with significant fibrosis (fibrosis score ≥2) and went on to compare these two groups of patients with that of the simple steatosis group. Results revealed that there is a significant association in G allele frequency between the NASH group with significant fibrosis and that of the simple steatosis (OR 1.85, 95% CI 1.05–3.26, p = 0.035), and a suggestive association between the NASH group without significant fibrosis and the simple steatosis group.
We investigated the association of PNPLA3 rs738409 with the histological features of NAFLD: steatosis, lobular inflammation, hepatocellular ballooning and fibrosis. We showed that there is significant association between the GG genotype with fibrosis stage (p = 0.038) but not with the other histological features (patients with genotype GG compared to CC, had mean score 2.03 vs. 1.52) (Table 4). We further investigated the magnitude of the effect of the risk allele G with the histological features. Patients bearing at least a single risk allele G of rs738409 were more likely to have a fibrosis score ≥2 (OR 1.95, 95% CI 1.17–3.26, p = 0.013). The association remains significant after the adjustment of other histological features (p = 0.012). However, we did not observe any association between the risk allele and steatosis, lobular inflammation and hepatocellular ballooning (Table 5).
Table 4.
Comparison of various clinical and histological parameters between the PNPLA3 rs738409 genotypes among NAFLD patients
| Characteristics | NAFLD, n = 144 (mean ± SD) | |||
|---|---|---|---|---|
| CC (n = 48) | CG (n = 63) | GG (n = 33) | p value | |
| Age (years) | 50.9 ± 12.2 | 52.1 ± 12.0 | 49.5 ± 12.1 | 0.585 |
| BMI (kg/m2) | 28.8 ± 4.4 | 28.5 ± 4.7 | 28.7 ± 3.8 | 0.936 |
| HbA1c (%)* | 6.5 ± 1.8 | 6.6 ± 1.6 | 6.6 ± 1.6 | 0.759 |
| Waist circumference (cm) | 94.3 ± 10.2 | 93.9 ± 12.0 | 92.9 ± 9.8 | 0.838 |
| HDL cholesterol (mg/dl) | 49.4 ± 13.91 | 47.7 ± 13.6 | 48.7 ± 8.6 | 0.768 |
| LDL cholesterol (mg/dl) | 117.9 ± 37.1 | 119.9 ± 40.2 | 110.7 ± 44.1 | 0.558 |
| Total cholesterol (mg/dl) | 203.9 ± 41.0 | 195.0 ± 44.3 | 189.6 ± 47.3 | 0.327 |
| Triglycerides (mg/dl) | 170.3 ± 81.8 | 155.0 ± 51.5 | 132.8 ± 41.8 | 0.029 |
| AST (IU/L)* | 43.8 ± 25.1 | 40.7 ± 23.2 | 45.9 ± 29.8 | 0.858 |
| ALT (IU/L) | 86.1 ± 50.3 | 79.2 ± 40.8 | 86.0 ± 59.2 | 0.705 |
| GGT (IU/L)* | 121.0 ± 117.7 | 106.4 ± 114.9 | 107.9 ± 116.2 | 0.276 |
| Systolic BP (mmHg) | 130.4 ± 13.3 | 133.4 ± 13.9 | 132.3 ± 16.6 | 0.548 |
| Diastolic BP (mmHg)* | 81.5 ± 10.2 | 83.4 ± 8.8 | 82.2 ± 11.3 | 0.477 |
| Steatosis grade* | 1.8 ± 0.8 | 1.9 ± 0.8 | 1.7 ± 0.8 | 0.434 |
| Lobular inflammation* | 1.2 ± 0.6 | 1.3 ± 0.6 | 1.4 ± 0.6 | 0.472 |
| Ballooning* | 1.1 ± 0.6 | 1.2 ± 0.6 | 1.3 ± 0.6 | 0.396 |
| Fibrosis* | 1.5 ± 1.0 | 1.7 ± 1.0 | 2.0 ± 0.9 | 0.038 |
ALT alanine transferase, AST aspartate aminotransferase, BMI body mass index, GGT gamma glutamyl transpeptidase, HbA1c hemoglobin A1c, HDL high-density lipoprotein, LDL low-density lipoprotein, NAFLD non-alcoholic fatty liver disease
* p values obtained using Kruskal–Wallis test, all other comparisons used ANOVA
Table 5.
Association of risk allele G with histological features in NAFLD patients
| Histology | Univariate p valuea | Multivariate p valueb (FDR q valueg) | OR (95% CI) |
|---|---|---|---|
| Steatosis >33 vs. <33% | 0.91 | 0.892 (0.446) | 0.88 (0.52–1.47)c |
| Lobular inflammation ≥2 foci vs. <2 foci | 0.237 | 0.202 (0.135) | 1.13 (0.64–1.99)d |
| Hepatocellular ballooning ≥1 vs. <1 | 0.176 | 0.168 (0.168) | 1.07 (0.61–1.87)e |
| Fibrosis ≥2 vs. <2 | 0.013 | 0.012 (0.024) | 1.95 (1.17–3.26)f |
OR odds ratio, CI confident interval
aJonckheere–Terpstra test
bOrdinal regression
c,d,e,fMultivariate logistic regression
gFalse discovery rate, q < 0.05 is significant
Analysis of various clinical parameters with PNPLA3 rs738409 genotypes
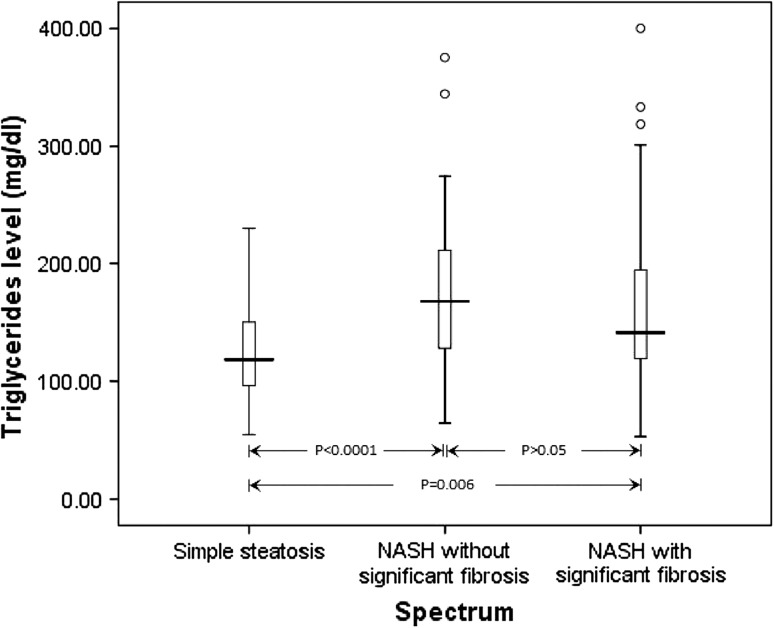
Next, we compared the clinical parameters between the PNPLA3 rs738409 genotypes among patients with NAFLD (Table 4). Patients with GG genotype have a significantly lower level of plasma triglycerides compared to those with the CC genotype (p = 0.029). We then compared the levels of triglyceride between the three groups: simple steatosis, NASH without significant fibrosis and NASH with significant fibrosis (Fig. 1). We observed a significant difference between the three groups (p = 0.002). The triglycerides level is significantly higher in the NASH group with (p = 0.006) and without significant fibrosis (p < 0.0001) compared to those with simple steatosis. The triglyceride level is lower in the NASH group with significant fibrosis compared to the NASH group without significant fibrosis, but this did not achieve statistical significance.
Fig. 1.
The comparison of plasma triglycerides level in the NAFLD spectrum. The box plot showed the extreme values, 25th, 50th (median) and 75th percentiles and outliers
Discussion
In the present study, we confirm the association of PNPLA3 rs738409 G allele with susceptibility to NAFLD as has been shown in a recent genome-wide association study (Romeo et al. 2008). More importantly, we showed a positive association of the risk allele G with risk of NASH and with fibrosis stage but not with other histological parameters. Thus, the G allele is not only associated with occurence of NASH, but also with presence of fibrosis.
The risk allele G was found to be significantly associated with susceptibility to NAFLD after stratification into the three major ethnic subgroups, the Chinese, Indian and Malay. The Malaysian Indians migrated to Malaysia from Southern India in the 1800s (Periasamy 2007). Studies have suggested that Indians from India were proto-Asian origin with West Eurasian admixture, hence giving them the genetic affinity towards both Asian and European (Bamshad et al. 2001; Jorde and Wooding 2004). Compared to the other ethnic groups, the Malaysian Indians appear to have a high prevalence of NAFLD as is so with the Caucasians. The Malaysian Chinese on the other hand, are of the Hans descendance from Southern China (Hock 2007) and it is noted that the results of the Chinese subgroup in our study is consistent with the finding in the Taiwanese Hans (Lin et al. 2011) as proven by positive associations in both populations. Our results are consistent with the results of the previous studies of different populations including Hispanics (Romeo et al. 2008), Argentinian (Sookoian et al. 2009), Germans (Kantartzis et al. 2009), Italian (Romeo et al. 2010a, b; Valenti et al. 2010), Japanese (Hotta et al. 2010), Taiwanese (Lin et al. 2011), and African-Americans (Cox et al. 2011). The present study thus confirms that the rs738409 plays a significant role in susceptibility to NAFLD.
We also successfully replicated the finding of an association of the PNPLA3 variant with NASH in our population as was observed in several studies (Hotta et al. 2010; Rotman et al. 2010; Sookoian et al. 2009). There are more patients with NASH in our study population compared to those with simple steatosis, which is probably reflective of the patients who are seen at the UMMC, which is a tertiary referral center.
A novel finding in this study is that the G allele is associated with the severity of NASH. Most of the NAFLD biopsy-proven studies to date only looked at two spectrum of NAFLD: simple steatosis and NASH (Hotta et al. 2010; Rotman et al. 2010; Sookoian et al. 2009). Predicting steatohepatitis is more important as NASH can potentially progress and treatment should be considered at an early stage of NASH. Compared to those with simple steatosis, the G allele is significantly higher in patients with NASH with significant fibrosis but not in NASH patients without significant fibrosis. Furthermore, the G allele frequency in NASH with significant fibrosis is higher than that of NASH without significant fibrosis. We also revealed that the G allele is associated with a higher fibrosis score but not with the other histological features of NAFLD. The association of rs738409 G allele with higher fibrosis score was also observed in other studies (Hotta et al. 2010; Rotman et al. 2010; Valenti et al. 2010). One Asian study found that the G allele is associated with fibrosis score but not with steatosis (Hotta et al. 2010). Our results were in concordance with that of Hotta et al. (2010), and in addition, there was no association with lobular inflammation and hepatocellular ballooning.
There have been conflicting reports of the correlation between presence of G allele and serum transaminase levels (Hotta et al. 2010; Kotronen et al. 2009; Lin et al. 2011; Romeo et al. 2008, 2010a, b; Sookoian et al. 2009; Valenti et al. 2010). The rs738409 G allele was shown to be associated with increased levels of both serum AST and ALT in the Argentinian (Sookoian et al. 2009), Italian (Romeo et al. 2010a, b), Japanese (Hotta et al. 2010), and the Taiwanese (Lin et al. 2011). In the Hispanics (Romeo et al. 2008) and in an Italian population (Valenti et al. 2010), the G allele was found to be related with increased serum ALT only. The G allele was found to be associated with increased serum AST level only in the Finnish population (Kotronen et al. 2009). However, the serum level of AST/ALT was found not to be significantly associated with the G allele in the African-Americans (Cox et al. 2011; Romeo et al. 2008), European Americans (Romeo et al. 2008), Germans (Kantartzis et al. 2009), and in the present study. The differences in these findings are probably due to the differences in the recruited subjects in the various studies, ranging from adults with NAFLD (Hotta et al. 2010; Kotronen et al. 2009; Romeo et al. 2008; Sookoian et al. 2009; Valenti et al. 2010), either obese or diabetic adults (Kantartzis et al. 2009), obese adults (Romeo et al. 2010b), and obese children (Lin et al. 2011; Romeo et al. 2010a).
Two studies reported a decreased level of plasma triglycerides in the NAFLD subjects harboring the G allele (Hotta et al. 2010; Speliotes et al. 2010). This finding is in agreement with our study, in which we found decreased level of plasma triglycerides amongst the NAFLD patients with GG genotype despite a significantly higher level triglyceride in the NAFLD patients as compared to controls, suggesting that genetic susceptibility of G allele plays a much more important role than triglyceride levels in the pathogenesis of NAFLD. In the present study, we further analyzed the triglyceride levels in the NAFLD spectrum. We found that the triglyceride levels are higher in NASH without significant fibrosis compared to NASH with significant fibrosis, suggesting that the triglyceride level subsides along with the degree of hepatocellular injury (Jou et al. 2008).
There are several limitations and strengths to this study. Due to ethical considerations, no liver biopsies were performed for the controls leading to a possibility of misclassification of controls. However, this likelihood was reduced by using the stated selection criteria for the controls. The distribution of BMI, HbA1c, triglycerides, and liver enzymes in controls reflects this selection criteria. The causal pathway between rs738409 and NASH has been postulated to be mediated through triglycerides. As this study conditions on BMI, the case–control association of rs738409 and NASH is vulnerable to confounding bias between the pathway of triglycerides and NASH. Thus, residual confounding from collider stratification bias due to conditioning of a downstream covariate might explain away some of the association between rs738409 and NASH. However, the associations between rs738409 and the different grades of NAFLD among the cases are not likely to be confounded and provide valid estimates. A major strength of this study was the ability to compare the association between rs738409 and NAFLD among three Asian ethnic groups. This study suggests that ethnicity modifies the relationship between rs738409 and NAFLD. It appears that the role of rs738409 on NAFLD is amplified among Indians compared to Chinese and Malays.
In summary, we showed an association between rs738409 of the PNPLA3 gene and the risk of NAFLD in the pooled subjects and after stratification into the three ethnic subgroups. The rs738409 G allele was significantly associated with progression to NASH and fibrosis.
Acknowledgments
We would like to thank the patients and staffs of UMMC for their participation in this study and Batool Sadat Haerian for her technical support. This study was supported by University Malaya Research Grant RG069/09HTM and RG364/11HTM, and by the HIRMOHE Grant E000024-20001.
Conflict of interest
All authors do not have any conflict of interest to disclose.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Shamsul Mohd Zain, Phone: +603-79673195, FAX: +603-79674791, Email: soulz712@gmail.com.
Zahurin Mohamed, Email: zahurin@ummc.edu.my.
References
- Adams L, Angulo P, Lindor K. Nonalcoholic fatty liver disease. CMAJ. 2005;172:899–905. doi: 10.1503/cmaj.045232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams LA, Waters OR, Knuiman MW, Elliott RR, Olynyk JK. NAFLD as a risk factor for the development of diabetes and the metabolic syndrome: an eleven-year follow-up study. Am J Gastroenterol. 2009;104:861–867. doi: 10.1038/ajg.2009.67. [DOI] [PubMed] [Google Scholar]
- Alisi A, Cianfarani S, Manco M, Agostoni C, Nobili V (2011) Non-alcoholic fatty liver disease and metabolic syndrome in adolescents: pathogenetic role of genetic background and intrauterine environment. Ann Med (in press) [DOI] [PubMed]
- Angulo P. Nonalcoholic fatty liver disease. N Engl J Med. 2002;346:1221–1231. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- Bamshad M, Kivisild T, Watkins WS, Dixon ME, Ricker CE, Rao BB, Naidu JM, Prasad BV, Reddy PG, Rasanayagam A, Papiha SS, Villems R, Redd AJ, Hammer MF, Nguyen SV, Carroll ML, Batzer MA, Jorde LB. Genetic evidence on the origins of Indian caste populations. Genome Res. 2001;11:994–1004. doi: 10.1101/gr.GR-1733RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57:12
- Bertrand JB, Lefevre H, Prevot S, Perlemuter G. Nonalcoholic fatty liver disease in a severely obese adolescent. An arguable liver biopsy. Arch Pediatr. 2011;18:62–65. doi: 10.1016/j.arcped.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Brunt EM, Kleiner DE, Wilson LA, Belt P, Neuschwander-Tetri BA. Nonalcoholic fatty liver disease (NAFLD) activity score and the histopathologic diagnosis in NAFLD: distinct clinicopathologic meanings. Hepatology. 2011;53:810–820. doi: 10.1002/hep.24127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton M. Nonalcoholic fatty liver disease: a review of current understanding and future impact. Clin Gastroenterol Hepatol. 2004;2:1048–1058. doi: 10.1016/S1542-3565(04)00440-9. [DOI] [PubMed] [Google Scholar]
- Cheung O, Sanyal AJ. Recent advances in nonalcoholic fatty liver disease. Curr Opin Gastroenterol. 2010;26:202–208. doi: 10.1097/MOG.0b013e328337b0c4. [DOI] [PubMed] [Google Scholar]
- Cortez-Pinto H, Camilo ME. Non-alcoholic fatty liver disease/non-alcoholic steatohepatitis (NAFLD/NASH): diagnosis and clinical course. Best Pract Res Clin Gastroenterol. 2004;18:1089–1104. doi: 10.1016/j.bpg.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Cox AJ, Wing MR, Carr JJ, Hightower RC, Smith SC, Xu J, Wagenknecht LE, Bowden DW, Freedman BI. Association of PNPLA3 SNP rs738409 with liver density in African Americans with type 2 diabetes mellitus. Diabetes Metab. 2011;37(5):452–455. doi: 10.1016/j.diabet.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CP. Genetic and environmental susceptibility to non-alcoholic fatty liver disease. Dig Dis. 2010;28:255–260. doi: 10.1159/000282098. [DOI] [PubMed] [Google Scholar]
- Fatani S, Itua I, Clark P, Wong C, Naderali EK. The effects of diet-induced obesity on hepatocyte insulin signaling pathways and induction of non-alcoholic liver damage. Int J Gen Med. 2011;4:211–219. doi: 10.2147/IJGM.S17376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock SS. Chinese migration. The population of Malaysia. Singapore: ISEAS Publishing; 2007. [Google Scholar]
- Hotta K, Yoneda M, Hyogo H, Ochi H, Mizusawa S, Ueno T, Chayama K, Nakajima A, Nakao K, Sekine A. Association of the rs738409 polymorphism in PNPLA3 with liver damage and the development of nonalcoholic fatty liver disease. BMC Med Genet. 2010;11:172. doi: 10.1186/1471-2350-11-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004;279:48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- Jorde LB, Wooding SP. Genetic variation, classification and ‘race’. Nat Genet. 2004;36:S28–S33. doi: 10.1038/ng1435. [DOI] [PubMed] [Google Scholar]
- Jou J, Choi SS, Diehl AM. Mechanisms of disease progression in nonalcoholic fatty liver disease. Semin Liver Dis. 2008;28:370–379. doi: 10.1055/s-0028-1091981. [DOI] [PubMed] [Google Scholar]
- Kantartzis K, Peter A, Machicao F, Machann J, Wagner S, Konigsrainer I, Konigsrainer A, Schick F, Fritsche A, Haring HU, Stefan N. Dissociation between fatty liver and insulin resistance in humans carrying a variant of the patatin-like phospholipase 3 gene. Diabetes. 2009;58:2616–2623. doi: 10.2337/db09-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A, Yeh M, McCullough AJ, Sanyal AJ. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- Kotronen A, Johansson LE, Johansson LM, Roos C, Westerbacka J, Hamsten A, Bergholm R, Arkkila P, Arola J, Kiviluoto T, Fisher RM, Ehrenborg E, Orho-Melander M, Ridderstrale M, Groop L, Yki-Jarvinen H. A common variant in PNPLA3, which encodes adiponutrin, is associated with liver fat content in humans. Diabetologia. 2009;52:1056–1060. doi: 10.1007/s00125-009-1285-z. [DOI] [PubMed] [Google Scholar]
- Lin YC, Chang PF, Hu FC, Yang WS, Chang MH, Ni YH. A common variant in the PNPLA3 gene is a risk factor for non-alcoholic fatty liver disease in obese Taiwanese children. J Pediatr. 2011;158:740–744. doi: 10.1016/j.jpeds.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Malaguarnera M, Di Rosa M, Nicoletti F, Malaguarnera L. Molecular mechanisms involved in NAFLD progression. J Mol Med (Berl) 2009;87:679–695. doi: 10.1007/s00109-009-0464-1. [DOI] [PubMed] [Google Scholar]
- Merriman RB, Aouizerat BE, Bass NM. Genetic influences in nonalcoholic fatty liver disease. J Clin Gastroenterol. 2006;40:S30–S33. doi: 10.1097/01.mcg.0000168643.16074.19. [DOI] [PubMed] [Google Scholar]
- Periasamy M (2007) Indian migration into Malaya and Singapore during the British period. Biblioasia, vol 3. National Library Board Singapore, Singapore
- Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo S, Sentinelli F, Cambuli VM, Incani M, Congiu T, Matta V, Pilia S, Huang-Doran I, Cossu E, Loche S, Baroni MG. The 148M allele of the PNPLA3 gene is associated with indices of liver damage early in life. J Hepatol. 2010;53:335–338. doi: 10.1016/j.jhep.2010.02.034. [DOI] [PubMed] [Google Scholar]
- Romeo S, Sentinelli F, Dash S, Yeo GS, Savage DB, Leonetti F, Capoccia D, Incani M, Maglio C, Iacovino M, O’Rahilly S, Baroni MG. Morbid obesity exposes the association between PNPLA3 I148M (rs738409) and indices of hepatic injury in individuals of European descent. Int J Obes (Lond) 2010;34:190–194. doi: 10.1038/ijo.2009.216. [DOI] [PubMed] [Google Scholar]
- Rotman Y, Koh C, Zmuda JM, Kleiner DE, Liang TJ. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology. 2010;52:894–903. doi: 10.1002/hep.23759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl CE, Everhart JE. Joint effects of body weight and alcohol on elevated serum alanine aminotransferase in the United States population. Clin Gastroenterol Hepatol. 2005;3:1260–1268. doi: 10.1016/S1542-3565(05)00743-3. [DOI] [PubMed] [Google Scholar]
- Schwimmer JB, Celedon MA, Lavine JE, Salem R, Campbell N, Schork NJ, Shiehmorteza M, Yokoo T, Chavez A, Middleton MS, Sirlin CB. Heritability of nonalcoholic fatty liver disease. Gastroenterology. 2009;136:1585–1592. doi: 10.1053/j.gastro.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serfaty L, Lemoine M. Definition and natural history of metabolic steatosis: clinical aspects of NAFLD, NASH and cirrhosis. Diabetes Metab. 2008;34:634–637. doi: 10.1016/S1262-3636(08)74597-X. [DOI] [PubMed] [Google Scholar]
- Sookoian S, Castano GO, Burgueno AL, Gianotti TF, Rosselli MS, Pirola CJ. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res. 2009;50:2111–2116. doi: 10.1194/jlr.P900013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Butler JL, Palmer CD, Voight BF, Hirschhorn JN. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 2010;52:904–912. doi: 10.1002/hep.23768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenti L, Al-Serri A, Daly AK, Galmozzi E, Rametta R, Dongiovanni P, Nobili V, Mozzi E, Roviaro G, Vanni E, Bugianesi E, Maggioni M, Fracanzani AL, Fargion S, Day CP. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51:1209–1217. doi: 10.1002/hep.23622. [DOI] [PubMed] [Google Scholar]
- Vanni E, Bugianesi E, Kotronen A, De Minicis S, Yki-Jarvinen H, Svegliati-Baroni G. From the metabolic syndrome to NAFLD or vice versa? Dig Liver Dis. 2010;42:320–330. doi: 10.1016/j.dld.2010.01.016. [DOI] [PubMed] [Google Scholar]
- Vuppalanchi R, Chalasani N. Nonalcoholic fatty liver disease and nonalcoholic steatohepatitis: selected practical issues in their evaluation and management. Hepatology. 2009;49:306–317. doi: 10.1002/hep.22603. [DOI] [PMC free article] [PubMed] [Google Scholar]