Abstract
Autophagy is well established as a starvation-induced process in yeast and mammalian cells and tissues. To elucidate the cellular mechanisms induced by starvation in fish, we characterized the induction of autophagy in cultured zebrafish cells under starvation conditions. As an autophagic marker protein, the microtubule-associated protein 1-light chain 3B protein (MAP1-LC3B) was cloned from the fish cells, and its expression and localization were characterized. In zebrafish embryonic (ZE) cells, posttranslational modifications produced two distinct forms of MAP1-LC3B, i.e., a cytosolic form and a membrane-bound form (types I and II, respectively). Immunofluorescence microscopy revealed fluorescently labeled autophagosomes in cells stably transfected with a green fluorescent protein (GFP)–MAP1-LC3B fusion protein and showed that this protein accumulated in punctate dots in a time-dependent manner in response to amino acid starvation. Starvation also induced the degradation of long-lived proteins. Treatment with 3-methyladenine and wortmannin, two class-III inhibitors of phosphoinositide 3-kinase (PI3K), repressed autophagy under starvation conditions, indicating that the PI3K class-III pathway regulates starvation-induced autophagy in fish.
Keywords: Amino acid starvation, Autophagy, Adaptation, Microtubule-associated protein 1-light chain 3, Zebrafish
Introduction
Many species of fishes undergo natural periods of starvation attributable to spawning migration activity or seasonal changes in food supply, and some can survive without feeding for several months or years (Lin et al. 1977; Moon and Johnston 1979). Numerous studies on the effects of starvation on fish tissues and muscle and hepatic enzymes have shown that the metabolic response to starvation includes the induction of proteolytic enzyme activities and energy consumption (Larsson and Lewander 1973; Lin et al. 1977; Moon and Johnston 1979; Patterson et al. 1974). Spawning migration of salmon, which is apparently supported by protein catabolism, is accompanied by the depletion of carbon and energy sources from muscle and liver and the stimulation of gluconeogenesis from amino acids (French et al. 1983). In carp, proteins and particular amino acids appear to be favored over carbohydrates as energy sources (Shimeno et al. 1990). Studies focusing on the autophagosomal–lysosomal pathway in rainbow trout recognized that this pathway is regulated during muscle wasting (Mommsen 2004; Salem et al. 2006). Thus, protein degradation by autophagy is an important adaptive mechanism in fish that allows them to survive nutritional starvation.
When autophagy is induced by nutrient starvation or other stresses or chemical agents, autophagosomes envelop cytoplasmic constituents, including organelles (Cuervo 2004). The double membrane-bound autophagosome subsequently fuses with lysosomes to become an autolysosome; within this structure, lysosomal hydrolytic enzymes degrade the cytoplasm-derived material.
The most important marker protein for detecting autophagic induction is microtubule-associated protein 1-light chain 3B protein (MAP1-LC3B) (Ichimura et al. 2000; Kirisako et al. 2000; Lang et al. 1998; Schlumpberger et al. 1997; Tanida et al. 1999). MAP1-LC3 exists in two modified forms: an 18-kDa cytoplasmic form that was originally identified as a MAP1-LC3 subunit (Kuznetsov and Gelfand 1987; Mann and Hammarback 1994) and a 16-kDa form that is associated with autophagosome membranes (Kabeya et al. 2000). The latter form is generated through a multistep posttranslational modification process that includes proteolytic cleavage of the C-terminal residue (Gly-120) from the newly synthesized MAP1-LC3 protein (Kabeya et al. 2000). In cultured cells and transgenic animals, autophagosomes can be fluorescently labeled in vivo by the transduction and expression of a chimeric protein consisting of green fluorescent protein (GFP) fused to MAP1-LC3B (Mizushima et al. 2004). The genes related to autophagy have been isolated in zebrafish, and the expression of GFP–MAP1-LC3B expression has been reported (He et al. 2009; He and Klionsky 2010; Makky et al. 2007). More recently, muscle protein degeneration in rainbow trout in vivo and in vitro was investigated in several autophagy-related genes, i.e., LC3B, gabarapl1, atg12l, and atg4b (Seiliez et al. 2010). Fasting fish for 14 days or serum depletion of trout myocytes strongly induced the expression of all studied genes. Insulin-like growth factor-1 (IGF1) induced FoxO3 phosphorylation in an in vitro primary culture of rainbow trout muscle cells but no effect on the expression of autophagy-related genes (Seiliez et al. 2010). Schiøtz et al. (2010) used electron microscopy to observe the induction of double membrane autophagosomes in cultured Atlantic salmon cells infected with infectious salmon anemia virus. Our preceding studies observed macroautophagy and chaperone-mediated autophagy with the expression of MAP1-LC3B and HSC70 expression as biomarkers in cells for the fish cultured cells derived from zebrafish and yellowtail under heat-shock conditions by Western blotting and immunocytochemistry (Yabu and Yamashita 2008; Yabu et al. 2011; Yamashita 2010). These previous studies indicate that MAP1-LC3 genes can be isolated and establish the fish models for autophagy and starvation studies. Thus, the GFP–MAP1-LC3B fusion protein can be used as an important biomarker to visualize fluorescent autophagosomes in fish cells in response to starvation.
In this study, we examined the autophagic response to starvation in fish using MAP1-LC3B as a marker for autophagosome formation. By searching the zebrafish Expressed Sequence Tag (EST) DNA database in GenBank, we identified three distinct zebrafish homologs (α, β, and γ; MAP1-LC3A, MAP1-LC3B, and MAP1-LC3C, respectively) of rat MAP1-LC3. We cloned these zebrafish proteins and also cloned MAP1-LC3B homologs from three other fish species: bluefin tuna (Thunnus orientalis), yellowtail (Seriola quinqueradiata), and Japanese flounder (Paralichthys olivaceus). We generated stable zebrafish embryonic (ZE) cell lines transfected with a GFP–MAP1-LC3B fusion gene construct and examined protein degradation by autophagy in these cells under conditions of nutritional stress. Treatment with inhibitors of phosphoinositide 3-kinase (PI3K) repressed starvation-induced autophagy, allowing us to identify the major autophagic pathway operating in cultured fish cells and tissues under starvation conditions.
Materials and Methods
Reagents
Wortmannin, 3-methyladenine (3-MA), mouse anti-tubulin monoclonal antibody (T6074), and mouse anti-bovine HSC70 IgG (BRM-22) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Cell culture reagents, ThermoScript RT-PCR System, and Xpress System synthetic oligonucleotides were obtained from Invitrogen (Carlsbad, CA, USA). A monoclonal antibody against enhanced GFP, the mammalian expression vector pEGFP-C1, a Smart Race cDNA Amplification Kit, Premix Taq ExTaq Version 2, and PrimeStarHS DNA polymerase were purchased from Takara Biomedical (Otsu, Japan). l-[U-14 C]Valine (7.2 MBq mM/mL), Hybond-P polyvinylidene fluoride (PVDF) membranes, an Enhanced Chemiluminescent (ECL) Western Blotting Detection Kit, and secondary antibodies (RPN2109) were acquired from GE Healthcare (Piscataway, NJ, USA). Rabbit anti-human MAP-LC3B polyclonal antibody, anti-GFP polyclonal antibody, and human recombinant human MAP-LC3B protein were purchased from Medical & Biological Laboratories (Nagoya, Japan). Goat anti-aldolase and rabbit anti-transferrin receptor polyclonal antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA).
Fish Samples
Cultured bluefin tuna (body weight 39 kg) were obtained from an aquaculture farm on Kyushu Island. Yellowtail (body weight 3 kg) were purchased at a local market. Japanese flounder (body weight 0.3 kg) were reared on artificial feed for 1 year at the National Research Institute of Fisheries Science.
Cell Culture
ZE cells were cultured in Leibovitz’s L-15 medium (Gibco, Grand Island, NY, USA) containing 2% fetal calf serum (FCS, JRH Biosciences, Lenexa, KS, USA) at 28.5°C in the presence of 5-U/mL penicillin and 50-μg/mL streptomycin. For amino acid starvation, the cells were cultured in Hank’s balanced salt solution (HBBS) containing 10-mM HEPES (pH 7.6) without amino acids or FCS for 0–12 h at 28.5°C.
Western Blotting
Proteins were extracted using a modification of a previously described method (Yabu et al. 2009). Briefly, each 100 mg of zebrafish tissue, such as, brain, heart, kidney, intestine, hepatopancreas, skeletal muscle, and spleen, as well as embryos and ZE cells (1 × 107/mL), was homogenized in 1 mL of cold 20-mM Tris-HCl buffer (pH 7.5) containing 250-mM sucrose, 10-mM MgCl2, and 1× protease inhibitor cocktail (Roche Molecular Biochemicals, Mannheim, Germany) by passage through a 27 G needle. After centrifugation of the homogenate at 1,300 × g for 15 min at 4°C, the 0.1-mL supernatant was recovered and solubilized in 0.05-mL lysis buffer [150-mM Tris-HCl buffer (pH 6.8), 6% SDS (w/v), 15% 2-mercaptoehathol (v/v), and 30% glycerol (v/v)]. The samples were separated by SDS-PAGE on 20% or 15% polyacrylamide gels and then electroblotted onto PVDF membranes as described previously (Yabu et al. 2008, 2009). The blotted membranes were blocked with 10% skim milk in Tris-based saline (TBS) containing 25-mM Tris-HCl buffer (pH 7.5), 150-mM sodium chloride, and 0.05% Tween20 (w/v) for 1 h. After washing the membrane with TBST, diluted TBST containing 10% skim milk was added and incubated overnight at 4°C. Proteins were detected by Western blotting with primary antibodies against MAP1-LC3B (Medical & Biological Laboratories), GFP (Medical & Biological Laboratories), aldolase (Santa Cruz Biotechnology), and transferrin receptor (Santa Cruz Biotechnology); a horseradish peroxidase-conjugated secondary antibody (GE Healthcare); and an ECL Western Blotting Detection kit (GE Healthcare) according to the manufacturer’s instructions. Protein concentration was determined using a Bio-Rad Protein Assay Kit (Bio-Rad, Hercules, CA, USA).
Subcellular Fractionation
Subcellular fractionation was performed using a modification of a previously described method (Yabu et al. 2008). Cultured cells (1 × 107/mL) were homogenized in 20-mM Tris-HCl buffer (pH 7.5) containing 1-mM EDTA, 1-mM EGTA, 10-mM KCl, and 1× protease inhibitor cocktail (Roche Molecular Biochemicals) by passage through a 27 G needle. Samples were centrifuged at 1,300 × g for 15 min at 4°C to remove nuclei and unbroken cells, and the resulting supernatant fractions were centrifuged at 100,000 × g for 1 h at 4°C. The resulting secondary supernatant fractions were used as a cytosolic fraction. The secondary pellet was resuspended in the same buffer by homogenization and sonication and then centrifuged at 12,000 × g for 1 h at 4°C. The resulting tertiary pellet was used as a crude microsomal fraction. After determination of the protein concentrations in the cytosolic and crude microsomal fractions, both fractions were stored at −80°C until used for Western blotting.
Cloning of Zebrafish MAP1-LC3 Genes
The rat MAP1-LC3 amino acid sequence (GenBank accession number U05784) was used to search the National Center for Biotechnology Information (NCBI) GenBank zebrafish EST database for homologous ESTs. The resulting ESTs were assembled into three contigs by the Pileup module of the GCG software package (Genetics Computer Group Inc., Madison, WI, USA). Three pairs of primers were designed based on the contig sequences.
Total RNA was extracted from ZE cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. For first-strand synthesis of cDNA, 5 μg of RNA was used in a 20-μL reaction mixture utilizing the ThermoScript RT-PCR System (Invitrogen) according to the manufacturer’s instructions. Resulting reverse transcription (RT) products were diluted 1:10 and stored at −20°C for later use, and synthetic primers were used in polymerase chain reactions (PCRs) to amplify MAP1-LC3A (540-bp fragment) [5′-GCCCA CGCGT CCGAA GTTGA CCGAC AATTC-3′ (upstream) and 5′-GTGAT GATAG GGACA GAGCT AAATA ATCGG-3′ (downstream)]; MAP1-LC3B (469-bp fragment) [5′-GCAGA TCTGC CTAAA ACAAA CGTGT TTAAT-3′ (upstream) and 5′-TAGGT TGTAT CATTG TCCAT GAGTG AAAG-3′ (downstream)]; and MAP1-LC3C (480-bp fragment) [5′-GAATTA GTGCG CAGGT CAGAG GTGCA GCTC-3′ (upstream) and 5′-CAAAG ACTTA AATAA TGCAT CAGTT CAGTT-3′ (downstream)]. PCR was carried out in 40-μL reaction mixtures using Premix Taq (Takara Biomedical) and reaction conditions consisting of denaturation at 96°C for 2 min; 25 cycles of denaturation at 96°C for 20 s, annealing at 65°C for 20 s, polymerization at 72°C for 30 s; and extension at 72°C for 7 min. The PCR products were subcloned into the pGEM-T Easy vector (Promega, Madison, WI, USA). The cloned nucleotide sequences were determined by DNA sequence analysis using a BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA).
Isolation of bluefin tuna, yellowtail, and Japanese flounder MAP1-LC3B cDNAs Poly(A)+ mRNA was extracted from T. orientalis, S. quinqueradiata, and P. olivaceus skeletal muscle, and single-strand cDNA was synthesized according to a method described previously (Yabu et al. 2001). To obtain the partial nucleotide sequences of these cDNAs, RT-PCR was performed using primers (sense primer 5′-ATGCC TTCGG AAAAG ACATT YAARC A-3′ and anti-sense primer 5′-CCAAA TGTCT CCTGG GAAGC RTANA CCAT-3′) based on the cDNA sequences of zebrafish and rat MAP1-LC3B. PCR was carried out in 40-μL reaction mixtures using Premix Taq and reaction conditions consisting of denaturation at 96°C for 2 min; 40 cycles of denaturation at 94°C for 20 s, annealing at 55°C for 20 s, polymerization at 72°C for 30 s; and extension at 72°C for 7 min. The PCR products were subcloned into a pGEM-T Easy vector. DNA sequence analysis confirmed that the nucleotide sequences of the RT-PCR products were homologous to that of the rat MAP1-LC3B gene. They were then used with a Smart Race cDNA Amplification kit (Takara Biomedical) to create MAP1-LC3B cDNA clones for T. orientalis, S. quinqueradiata, and P. olivaceus. The PCR products were subcloned into a pGEM-T Easy vector, and their nucleotide sequences were determined by DNA sequence analysis.
DNA sequences were deposited in the DNA Databank of Japan (DDBJ) database under the accession numbers AB264293 (zebrafish MAP1-LC3A), AB264294 (zebrafish MAP1-LC3B), AB264295 (zebrafish MAP1-LC3C), AB461861 (T. orientalis MAP1-LC3B), AB461862 (S. quinqueradiata MAP1-LC3B), and AB461863 (P. olivaceus MAP1-LC3B). The amino acid sequences for rat MAP1-LC3 and human MAP1-LC3A, MAP1-LC3B, and MAP1-LC3C were obtained from the NCBI protein database and had the accession numbers NM_022867, AF276658, AF087871, and AF276659, respectively.
Plasmid Construction
The zebrafish MAP1-LC3B cDNA was amplified by PCR using a sense primer containing an EcoRI site (5′-GAATT CTATG CCTTC GGAAA AGACA TTTAA-3′) and an anti-sense primer containing a BamHI site (5′-GGATC CTTAC TGAAA TCCAA ATGTC TCCTG-3′). The resulting product was subcloned into pGEM-T Easy using the TA-Cloning method (Promega), and the cloned nucleotide sequence was confirmed by sequence analysis. The MAP1-LC3B coding sequence was then subcloned into the EcoRI–BamHI site of the multicloning site of vector pEGFP-C1 (Takara). The resulting construct, designated pGFP–MAP1-LC3B, encodes a chimeric protein (designated GFP–MAP1-LC3B) consisting of enhanced GFP fused to MAP1-LC3B at the MAP1-LC3B N-terminus.
Quantification of Autophagy
Induction of autophagy in pGFP–MAP1-LC3B-transfected ZE cells was measured by determining the percentage of GFP–MAP1-LC3B-positive cells that contained GFP–MAP1-LC3B punctate dots by counting under a fluorescence microscope (R400, Edge Scientific Instruments, Santa Monica, CA, USA) as described previously (Furuya et al. 2005; Pattingre et al. 2005). For each experiment, triplicate samples containing at least 200 cells each were analyzed for each condition.
Analysis of Protein Degradation
To label intracellular proteins, ZE cells were incubated for 48 h at 28.5°C with 370-MBq/mL l-[14 C]valine in Leibovitz’s L-15 medium containing 2% FCS, 5-U/mL penicillin, and 50-μg/mL streptomycin. Unincorporated l-[14 C]valine was removed by three rinses with phosphate-buffered saline. To stimulate autophagy, the labeled cells were then incubated in HBBS containing 10-mM valine, which does not interfere with autophagy in most cell types (Bauvy et al. 2009). When required, 3-MA, a potent inhibitor of the formation of autophagic vacuoles (Pattingre et al. 2003), was added during the “chase” period. After 6 h of incubation to allow the degradation of short-lived proteins, the medium was replaced with fresh medium. After 6 h, the medium was removed to a new tube, and trichloroacetic acid (TCA) was added to a final concentration of 10%. The medium/TCA mixture was centrifuged at 1,000 × g for 10 min at 4°C, and the acid-soluble radioactivity in the supernatant fraction was measured by liquid scintillation counting. The cells were washed twice with cold 10% TCA and dissolved in 200 μl of Soluene 350. The acid-precipitable cellular radioactivity in the pellet fraction was then measured by liquid scintillation counting. The rate of degradation of long-lived proteins was calculated as the ratio of the acid-soluble radioactivity in the medium to that of the acid-precipitable fraction in the cells.
Generation of Stably Transfected ZE Cells
ZE cells were cultured in Leibovitz’s L-15 medium (Invitrogen) containing 5% FCS, 5-U/mL penicillin, and 50-μg/mL streptomycin. To obtain stable transfectants, 5 × 106 ZE cells were transfected with pGFP–MAP1-LC3B vector DNA (5 μg) using FuGene 6 Transfection Reagent (Roche Molecular Biochemicals) according to the manufacturer’s instructions. Transfectants were selected in the presence of 0.5-mg/mL geneticin (Invitrogen). For amino acid starvation, cells were cultured in HBBS containing 10-mM HEPES (pH 7.6) without any amino acids or FCS.
Fish Culture
Zebrafish were maintained at 28.5°C on a 14-h light/10-h dark cycle as described previously (Westerfield 1995). The zebrafish used in this study were at least 6 months old and had a mean body mass of 350 ± 17 mg.
Statistical Analysis
The results are expressed as the mean ± standard deviation. Differences among groups were analyzed using one-way analysis of variance (ANOVA) followed by Bonferroni’s post-hoc t-test. Comparisons between two experimental groups were based on two-tailed t-tests. P < 0.01 was deemed statistically significant.
Results
Molecular Cloning of MAP1-LC3 from Fish
A GenBank EST database search for zebrafish homologs of rat MAP1-LC3 yielded ESTs with lengths of 540, 469, and 530 bases. Subcloning and sequencing of the cDNA clones corresponding to these ESTs yielded three open reading frames encoding putative polypeptides of 121, 122, and 128 amino acids corresponding to three genetically distinct isoforms of MAP1-LC3, i.e., MAP1-LC3A (DDBJ accession no. AB264293), MAP1-LC3B (AB264294), and MAP1-LC3C (AB264295), respectively. The three zebrafish proteins exhibited a high degree of sequence identity to rat MAP1-LC3B and to the human MAP1-LC3 isoforms MAP1-LC3B (Fig. 1), MAP1-LC3A, and MAP1-LC3C. The predicted amino acid sequences of zebrafish MAP1-LC3A, MAP1-LC3B, and MAP1-LC3C were 96%, 93%, and 72% identical to human MAP1-LC3A, MAP1-LC3B, and MAP1-LC3C, respectively.
Fig. 1.
Amino acid sequence alignments of microtubule-associated protein 1-light chain 3B (MAP1-LC3B) from zebrafish, bluefin tuna, yellowtail, Japanese flounder, rat, and human. Amino acid identity is indicated by dots, and possible cleavage sites are indicated by the arrowhead
We also isolated MAP1-LC3B cDNA clones from bluefin tuna, yellowtail, and Japanese flounder. Conceptual translation of the open reading frames for these clones yielded putative MAP1-LC3B polypeptides containing 122, 122, and 122 amino acids, respectively (Fig. 1). These predicted amino acid sequences of these polypeptides were highly homologous to those of rat and zebrafish MAP1-LC3B.
MAP1-LC3B Translocates from the Cytosol into Microsomes During Amino Acid Starvation of ZE Cells
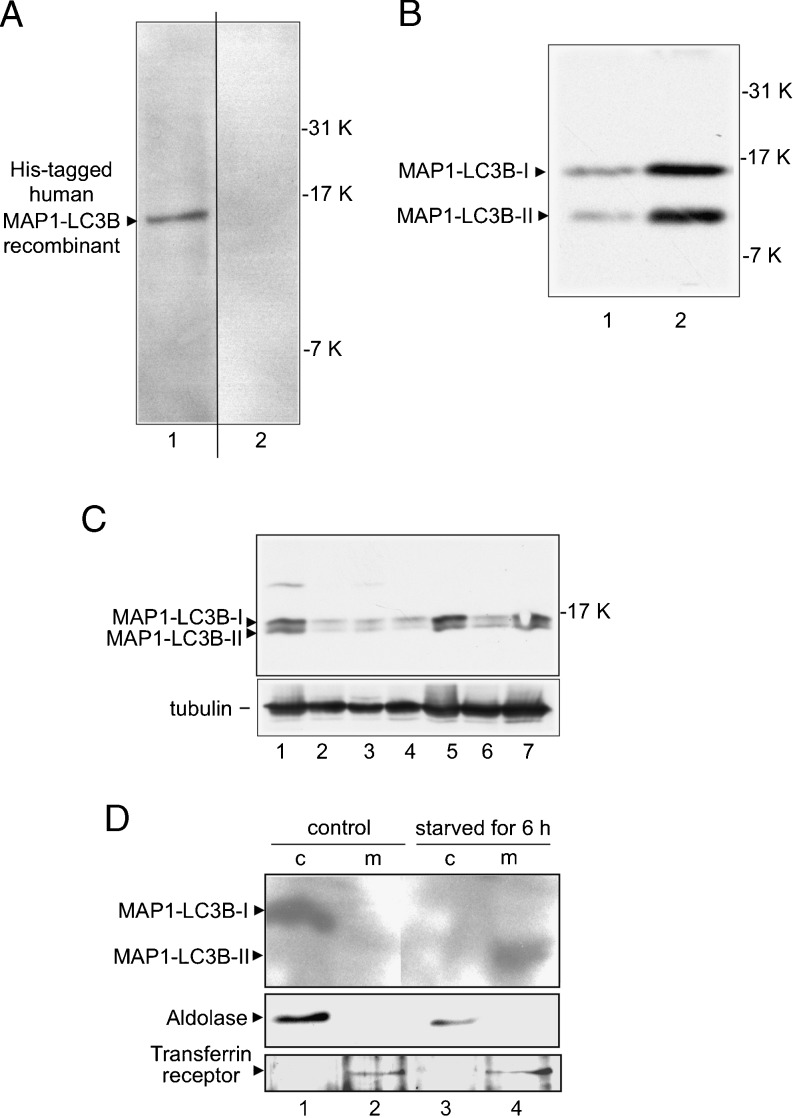
We used Western blot analysis to examine anti-human MAP1-LC3B polyclonal antibody immunoreactivity against zebrafish MAP1-LC3B protein. The anti-human MAP1-LC3B antibody cross-reacted with the human MAP1-LC3B recombinant protein (Fig. 2a, lane 1), and absorption of the anti-human MAP1-LC3B polyclonal antibody with human MAP1-LC3B recombinant protein abolished immunoreactivity in the ZE cell lysate (Fig. 2a, lane 2). Zebrafish embryos at the 1-somite-stage (Fig. 2b) and brain, heart, kidney, intestine, hepatopancreas, skeletal muscle, and spleen were examined to investigate zebrafish MAP1-LC3B protein expression in ZE cells (Fig. 2c). Rat MAP1-LC3B has been well established to undergo posttranslational modifications in vivo (Kabeya et al. 2000) that result in two modified forms: an 18-kDa cytoplasmic form (MAP1-LC3B-I) and a 16-kDa form that is associated with the autophagosome membrane (MAP1-LC3B-II). In our Western blot analysis, zebrafish MAP1-LC3B was detected as 14.5- and 12-kDa bands in ZE cells, zebrafish embryos, and all tissues (Fig. 2b, c).
Fig. 2.
Detection of MAP1-LC3B proteins in zebrafish. a MAP1-LC3B was analyzed by Western blotting using an anti-human MAP1-LC3B antibody. His-tagged human MAP1-LC3B recombinant protein (10 ng) was analyzed with an anti-human MAP1-LC3B antibody (lane 1), and lysate (20-μg protein) from zebrafish embryonic (ZE) cells was analyzed with an anti-human MAP1-LC3B antibody preparation, pre-incubated by absorption of the cross-reactive antibodies with human MAP1-LC3B recombinant protein (lane 2). b Lysates of ZE cells (lane 1) and zebrafish embryos at the 1-somite stage (lane 2) were analyzed for MAP1-LC3B expression by Western blotting using an anti-human MAP1-LC3B antibody. c Lysates of zebrafish tissues, brain (lane 1), heart (lane 2), kidney (lane 3), intestine (lane 4), hepatopancreas (lane 5), skeletal muscle (lane 6), and spleen (lane 7) were analyzed for MAP1-LC3B expression by Western blotting using an anti-human MAP1-LC3B and anti-tubulin antibodies. d Translocation of MAP1-LC3B from the ZE cytosol into microsomes during amino acid starvation. Supernatant (lanes 1 and 3) and pellet (lanes 2 and 4) fractions from ultracentrifugation (100,000 × g) of homogenates of normal ZE cells (lanes 1 and 2) and ZE cells starved of amino acids for 6 h (lanes 3 and 4) were analyzed by Western blotting using antibodies against MAP1-LC3B, aldolase (a cytosolic marker), and transferrin receptor (a membrane protein marker)
Subcellular localization of endogenous MAP1-LC3B in ZE cells was examined by ultracentrifugal fractionation and Western blot analysis using antibodies against MAP1-LC3B, transferrin receptor (a cell membrane marker), and aldolase (a cytosolic marker). MAP1-LC3B was detected primarily in the cytosolic fraction (Fig. 2d, lane 1) under normal conditions and in the crude microsomal fraction under starvation conditions (Fig. 2d, lane 4). The results indicate that MAP1-LC3B translocates from the cytosol into the membrane during amino acid starvation of ZE cells.
Observation of Autophagosomes in Zebrafish Cells
To visualize autophagosomes in cultured zebrafish cells in vivo, the zebrafish MAP1-LC3B cDNA was cloned into the eukaryotic expression vector pEGFP-C1 to create a plasmid encoding a MAP1-LC3B fusion protein that contained an N-terminal enhanced GFP tag (GFP–MAP1-LC3B). When ZE cells were stably transfected with this plasmid, the level of overexpressed GFP–MAP1-LC3B was approximately 120 times higher than that of endogenous MAP1-LC3B (Fig. 3c, lane 1).
Fig. 3.
Amino-acid-starvation-induced autophagy in ZE cells expressing a fluorescent autophagosome marker. ZE cells were stably transfected with the plasmid encoding GFP–MAP1-LC3B fusion protein. a, b Immunofluorescence images of cells fixed after incubation for 6 h at 28.5°C in Leibovitz’s L-15 medium containing 2% fetal calf serum (FCS) (a) or in Hanks’ buffered salt solution (HBBS) (b). Bar = 10 μm. Representative cells containing GFP–MAP1-LC3B dots (i.e., autophagosomal structures) can be seen in panel b. c The transfected cells were treated with HBSS for 0 (lane 1), 1 (lane 2), 2 (lane 3), 3 (lane 4), 4 (lane 5), and 6 h (lane 6). The cell lysates were analyzed by Western blotting with anti-human MAP1-LC3B and anti-tubulin antibodies. Arrowheads indicate stained signals for MAP1-LC3B. Asterisk indicates the band of the degraded GFP–MAP1-LC3B products. Tubulin was detected as the internal control. d Time-dependent induction of autophagy in amino acid-starved cells was quantified by counting punctate structures under a fluorescence microscope as described in the “Materials and Methods” section. e Supernatant (lanes 1 and 3) and pellet (lanes 2 and 4) fractions from ultracentrifugation (100,000 × g) of normal transfectant homogenates (lanes 1 and 2) and transfectant starved of amino acids for 6 h (lanes 3 and 4) were analyzed by Western blotting using antibodies against GFP, aldolase (a cytosolic marker), and transferrin receptor (a membrane protein marker). GFP–MAP1-LC3B-I, GFP–MAP1-LC3B-II, aldolase, and the transferrin receptor are indicated by arrowheads. Each value shown represents the mean of three independent experiments. Error bars represent one standard deviation. * P < 0.01 vs. control
In rats, the amount of MAP1-LC3B is highly correlated with the number of autophagosomes, and MAP1-LC3B localizes in the autophagosome membrane in a punctate pattern that increases in number and intensity of dots when the cells are starved of amino acids (Kabeya et al. 2000). A similar pattern appears for the Atg8 protein in yeast (Kirisako et al. 2000). Therefore, ZE cells stably expressing GFP–MAP1-LC3B were examined to determine whether the fusion protein, like endogenous MAP1-LC3B, would function as a specific marker for autophagic membranes during starvation. Indeed, amino acid starvation induced the time-dependent appearance of many ring-shaped GFP–MAP1-LC3B structures in the ZE cells (Fig. 3d), confirming that GFP–MAP1-LC3B functions as an autophagosome marker even when it is overexpressed.
Subcellular localization of exogenous GFP–MAP1-LC3B in the ZE transfectant was confirmed by ultracentrifugal fractionation and Western blot analysis using antibodies against GFP, transferrin receptor (a cell membrane marker), and aldolase (a cytosolic marker). GFP–MAP1-LC3B-I was detected primarily in the cytosolic fraction (Fig. 3e, lane 1) under normal growing conditions and in the crude microsomal fraction under starved conditions (Fig. 3e, lane 4). These results indicate that MAP1-LC3B translocates from the cytosol into the crude microsomal fraction, including lysosomes and endoplasmic reticulum, during amino acid starvation of the ZE transfectant.
In rats, the induction of autophagy by amino acid starvation causes the cytosolic MAP1-LC3B-I to be converted to MAP1-LC3B-II, which is associated with autophagosomes and conjugated to phosphatidylethanolamine (Kabeya et al. 2004). Forms I and II of MAP1-LC3B can be distinguished by SDS-PAGE analysis. Using our stable MAP1-LC3B-transfected ZE cell line, we found that MAP1-LC3B is also present in two forms in ZE cells (Fig. 2). The first form (MAP1-LC3B-I) is converted to the second form (MAP1-LC3B-II) in this cell line after 6 h of starvation (Fig. 3c, lane 2), suggesting that this cell line is a suitable model for investigations of autophagic activity.
Inhibition of Starvation-Induced Autophagy by Class-III PI3K Signaling
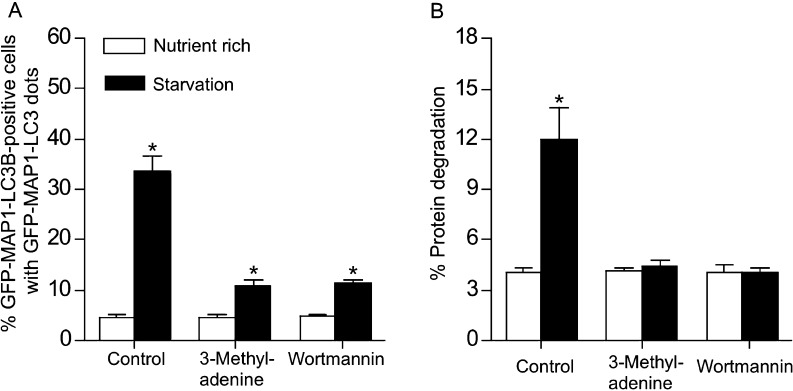
We next examined the effect of inhibiting class-III PI3K on amino-acid-starvation-induced autophagy in our stably transfected ZE cell line. Treatment of the cells with either of the class-III PI3K inhibitor 3-MA or wortmannin repressed autophagic activity (Fig. 4a). By measuring the degradation of long-lived proteins in the stably transfected ZE cells, we found that starvation was associated with significant proteolysis of long-lived cellular proteins in untreated control cells, but not in 3-MA- or wortmannin-treated cells (Fig. 4b). Together, these results suggest that PI3K inhibitors block autophagy at the stage prior to the association of MAP1-LC3B with the pre-autophagosomal membrane.
Fig. 4.
Effects of 3-methyladenine (3-MA) and wortmannin on amino-acid-starvation-induced autophagy in ZE cells stably transfected with GFP–MAP1-LC3B. a The class III phosphatidylinositol 3-kinase inhibitors (i.e., 3-MA and wortmannin) repressed amino-acid-starvation-induced autophagy. After ZE cells were treated with 3-MA (1 mM) or wortmannin (100 nM) for 1 h, they were amino-acid-starved by incubation in HBBS for 6 h at 28.5°C, fixed and visualized by fluorescence microscopy. b Effects of 3-MA and wortmannin on amino-acid-withdrawal-induced degradation of long-lived proteins in ZE cells. ZE cells were radiolabeled with 370-MBq/mL l-[U-14 C]valine for 48 h at 28.5°C, incubated in the presence of 3-MA (1 mM), wortmannin (100 nM), or no inhibitor for 1 h, and then amino-acid-starved by incubation in HBBS for 6 h at 28.5°C. The cell medium and cell lysates were precipitated with trichloroacetic acid, and the ratio of acid-soluble radioactivity in the medium to the acid-precipitable radioactivity in the cell lysate was determined by liquid scintillation counting and used as the rate of long-lived protein degradation as described in the “Materials and Methods” section. Each value represents the mean of three independent experiments. Error bars represent one standard deviation. Asterisks denote a statistical difference between nutrient rich and starvation conditions (P < 0.01)
Conversion of MAP1-LC3B-I to MAP1-LC3B-II During Starvation In Vivo
Starvation of zebrafish for 24 and 48 h caused their mean body weight to decrease from 347 ± 42 mg to 336 ± 3 mg and 327 ± 7 mg, respectively. Body weight began to decrease within 24 h of starvation and peaked within 48 h of starvation (Fig. 5a). Western blot analysis of endogenous MAP1-LC3B in the skeletal muscle and hepatopancreas showed that endogenous MAP1-LC3B-I is converted to MAP1-LC3B-II during starvation (Fig. 5b, c). The high level of MAP1-LC3B-II was observed in skeletal muscle after 36 h of starvation. MAP1-LC3B-II was detected in hepatopancreas under feeding condition and within 36 h of starvation and disappeared after 48 h of starvation. Thus, MAP1-LC3B-II is induced by starvation, and MAP1-LC3B-I is converted to MAP1-LC3B-II primarily by Atg4 as a result of activation of autophagy at an early stage after starvation.
Fig. 5.
Conversion of MAP1-LC3B-I to MAP1-LC3B-II in zebrafish skeletal muscle and hepatopancreas during starvation. Change in whole body weight following starvation (a). Error bars represent one standard deviation. * P < 0.01 vs. fish before starvation (n = 7). The skeletal muscle (b) and hepatopancreas (c) of the fed fish (lane 1) and the starved fish for 24 (lane 2), 36 (lane 3), or 48 h (lane 4) were subjected to Western blot analysis with anti-human MAP1-LC3B and anti-tubulin antibodies. Arrows indicate the MAP1-LC3B-I and MAP1-LC3B-II bands
Discussion
In this study, we successfully cloned an autophagic marker protein, MAP1-LC3B, from zebrafish and other fish species and used this marker protein to characterize the autophagic response in fish models at the cellular and organismal levels in vivo.
We cloned three genetically distinct isoforms of MAP1-LC3 from zebrafish (MAP1-LC3A, MAP1-LC3B, and MAP1-LC3C), and we also cloned MAP1-LC3B homologs from three other fish species. Amino acid sequence analysis showed that the fish and mammalian MAP1-LC3B proteins have common structural features, such as a putative cleavage site at the C-terminal glycine residue for the proteins encoded by the cloned sequences (Fig. 1). Western blotting and immunofluorescence studies in zebrafish cells showed that MAP1-LC3B is expressed in zebrafish and activated in response to amino acid withdrawal; it also acts as an autophagosome marker in ZE cells, thus behaving similarly to known mammalian MAP1-LC3B proteins in its response to starvation (He et al. 2003; Kabeya et al. 2000). Subcellular fractionation studies showed that MAP1-LC3B localized in the cytosolic fraction in one form (MAP1-LC3B-I) and in the crude microsomal fraction in another form (MAP1-LC3B-II) during starvation, indicating that MAP1-LC3B-II can be used as a biomarker for autophagic activation in ZE cells (Figs. 2, 3, and 4). We also found that amino acid withdrawal induces the degradation of long-lived proteins in ZE cells because treatment of the cells with 3-MA or wortmannin repressed autophagic activity, indicating that the PI3K class-III pathway regulates starvation-induced autophagy in fish. The MAP1-LC3B-II form was also detected in zebrafish muscle and hepatopancreas after 24-h starvation in vivo (Fig. 5). In flounder muscle, the MAP1-LC3B-II form was detected after starvation for 10 days, and HSC70, aldolase, and cathepsin L were induced in the lysosomal fraction, indicating that both macroautophagy and chaperone-mediated autophagy were induced under severe starvation conditions (Yabu et al., unpublished data). Recently, we have detected a stronger signal for MAP1–LC3 active form II and enhanced macroautophagy signaling through PI3 and TOR kinases in heat-shocked yellowtail cells (Yabu et al. 2011). We also detected MAP1–LC3 active form II, autophagic activation, and extensive lysosomal proteolytic degradation in the muscles of bluefin tuna, yellowtail, and mackerel in “burnt” meat, which lacks a bright red meat color and has a more watery, softer texture, under the oxidative stress conditions caused by selenium deficiency and hypoxia when fish are caught during the spawning period in summer (Yamashita 2010; Yamashita et al. 2010). Therefore, fish possess the autophagic pathways that serve as an adaptive response to starvation, and the hepatopancreas and muscle are the principal organs responsible for recycling cytoplasmic components to provide nutrients to other tissues during starvation, as suggested in mammalian models (Mizushima et al. 2004).
Previous mammalian studies have shown that autophagy is induced in most organs in response to nutrient starvation, although its regulation varies among the organs (Mizushima et al. 2004). However, autophagy remains constant, increases slightly (Mortimore et al. 1983), or decreases during starvation depending on the circumstances (de Waal et al. 1986). In studies using transgenic mice systemically expressing a GFP–MAP1-LC3 fusion protein, nutrient starvation induced autophagy in most tissues (Mizushima et al. 2004), and in some tissues, including thymic epithelial cells, lens epithelial cells, podocytes, and some exocrine gland cells, autophagy occurred even in the absence of starvation, suggesting that autophagy is regulated differently in different organs and might not be restricted to the starvation response (Mizushima et al. 2004). In mice with Pompe disease, a genetic deficiency of glycogen-degrading lysosomal glucosidase, skeletal and cardiac muscles are functionally deficient in autophagy because of impaired autophagosomal–lysosomal fusion (Mizushima et al. 2004). As a result, autophagic substrates, including potentially toxic, aggregation-prone, ubiquitinated proteins, accumulate in Pompe myofibrils and may cause profound muscle damage. Thus, autophagy mediates lysosomal protein degradation of cytoplasmic components, including the most long-lived proteins and organelles.
Important and interesting phenomena relating to autophagy and starvation have also been characterized in various fish species. Metabolic suppression in response to oxygen or water deficiency or to cold is thought to be a survival strategy that, in effect, slows down biological time and fuel utilization (Hochachka and Somero 2002). In Pacific salmon, studies on the qualitative pattern of fuel selection during spawning migration have shown that fats are the preferred fuel during early stages of migration without feeding, but as fats are depleted, protein catabolism gradually becomes the dominant carbon and energy source (Mommsen et al. 1980). The proteins fueling starvation metabolism come primarily from the gut and white muscle tissue, but not all proteins are targets for protein degradation; myofibrillar proteins are preferentially protected, whereas glycolytic enzymes and muscle sarcoplasmic enzymes, for example, aldolase, enolase, and creatine kinase, are selectively utilized. In contrast, muscle and liver glycogen reserves are conserved until the end of the migration, when they are thought to be used to power the energetically expensive swimming behaviors associated with spawning. In response to migratory starvation, lysosomal proteolytic enzymes, such as cathepsins A, B, H, and L, are extensively induced in the muscle and liver of salmon (Mommsen et al. 1980; Yamashita and Konagaya 1990a, b, 1991a, b). In particular, fast-twitch muscles respond dramatically to spawning migration. Enhanced proteolytic activities have also been linked to starvation and physiological stressors in milkfish (Shiau et al. 2001). Starvation of these fish reduced the amount of lipid and protein in white muscle, and as starvation continued, the levels of free histidine, taurine, and glycine also fell (Shiau et al. 2001). After 40 days of starvation, the histidine concentration in white muscle had decreased by 46%. Brief starvation induces the accelerated release of amino acids from skeletal muscle and is associated with an increase in gluconeogenesis (Hochachka and Somero 2002). In addition, prolonged starvation is characterized by a decrease in gluconeogenesis, largely because the release of substrate amino acids from skeletal muscle is reduced (Hochachka and Somero 2002).
Several autophagy-related genes, including MAP1-LC3B, have been isolated from fish cells (He and Klionsky 2010; He et al. 2009; Makky et al. 2007; Schiøtz et al. 2010; Seiliez et al. 2010; Yabu and Yamashita 2008). Transgenic zebrafish expressing GFP–MAP1-LC3B fusion protein have been generated, and autophagic responses were assayed in embryos (He and Klionsky 2010; He et al. 2009). Schiøtz et al. (2010) showed that infectious salmon anemia virus induces double membrane autophagosomes in the Atlantic salmon cells. They used MAP1-LC3B fused to GFP to show the redistribution of fluorescence into the typical punctate pattern seen in cells undergoing autophagy. Additionally, they found no changes in the expression of the autophagy-related genes, such as atg3, atg5, atg6, atg7, atg8, atg10, and atg12 in relation to the first phase of viral infection. Seiliez et al. (2010) revealed that several autophagy-related genes are induced in rainbow trout muscle after 14 days of starvation in vivo and in serum-depleted myocytes in vitro. IGF1 induces FoxO3 phosphorylation of in myocytes in vitro, but has no effect on the gene expression (Seiliez et al. 2010). These studies suggest that both macroautophagy and chaperone-mediated autophagy play a major biological role in promoting catabolism during starvation in fish.
We used MAP1-LC3B and a GFP fusion protein to systematically monitor the autophagic response to starvation in zebrafish cells and tissues. This analysis provided some novel and fundamental insights into fish biology. This system can be utilized in investigations of other issues concerning autophagy in fish, such as the regulation of autophagy by food supply. It can be applied during spawning migration and by developmental stage or to study the effects of diseases and toxic contaminants on autophagy. This system can also be used to examine the specific roles of apoptosis, protein degradation, and substrate specificity in autophagy in fish.
Acknowledgments
This work was supported in part by grants from the Japan Science and Technology Corporation and the Japan Society for the Promotion of Science and the Ministry of Agriculture, Forestry and Fisheries of Japan.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Bauvy C, Meijer AJ, Codogno P. Chapter 4: assaying of autophagic protein degradation. Meth Enzymol. 2009;452:47–61. doi: 10.1016/S0076-6879(08)03604-5. [DOI] [PubMed] [Google Scholar]
- Cuervo AM. Autophagy: many paths to the same end. Mol Cell Biochem. 2004;263:55–72. doi: 10.1023/B:MCBI.0000041848.57020.57. [DOI] [PubMed] [Google Scholar]
- De Waal EJ, Vreeling-Sindelarova H, Schellens JP, Houtkooper JM, James J. Quantitative changes in the lysosomal vacuolar system of rat hepatocytes during short-term starvation. A morphometric analysis with special reference to macro- and microautophagy. Cell Tissue Res. 1986;243:641–648. doi: 10.1007/BF00218073. [DOI] [PubMed] [Google Scholar]
- French CJ, Hochachka PW, Mommsen TP. Metabolic organization of liver during spawning migration of sockeye salmon. Am J Physiol. 1983;245:R380–R827. doi: 10.1152/ajpregu.1983.245.6.R827. [DOI] [PubMed] [Google Scholar]
- Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Analyzing autophagy in zebrafish. Autophagy. 2010;6:642–644. doi: 10.4161/auto.6.5.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Dang Y, Dai F, Guo Z, Wu J, She X, Pei Y, Chen Y, Ling W, Wu C, Zhao S, Liu JOYuL. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem. 2003;278:29278–29287. doi: 10.1074/jbc.M303800200. [DOI] [PubMed] [Google Scholar]
- He C, Bartholomew CR, Zhou W, Klionsky DJ. Assaying autophagic activity in transgenic GFP-LC3 and GFP-Gabarap zebrafish embryos. Autophagy. 2009;5:520–526. doi: 10.4161/auto.5.4.7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochachka PW, Somero GN. Biochemical adaptation: mechanism and process in physiological evolution. New York: Oxford University Press; 2002. [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov SA, Gelfand VI. 18 kDa microtubule-associated protein: identification as a new light chain (LC-3) of microtubule-associated protein 1 (MAP-1) FEBS Lett. 1987;212:145–148. doi: 10.1016/0014-5793(87)81574-0. [DOI] [PubMed] [Google Scholar]
- Lang T, Schaeffeler E, Bernreuther D, Bredschneider M, Wolf DH, Thumm M. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 1998;17:3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A, Lewander K. Metabolic effects of starvation in the eel, Anguilla anguilla L. Comp Biochem Physiol. 1973;44:367–374. doi: 10.1016/0300-9629(73)90489-1. [DOI] [PubMed] [Google Scholar]
- Lin H, Romsos DR, Tack PI, Leveille GA. Effects of fasting and feeding various diets on hepatic lipogenic enzyme activities in coho salmon (Oncorhynchus kisutch (Walbaum) J Nutr. 1977;107:1477–1483. doi: 10.1093/jn/107.8.1477. [DOI] [PubMed] [Google Scholar]
- Makky K, Tekiela J, Mayer AN. Target of rapamycin (TOR) signaling controls epithelial morphogenesis in the vertebrate intestine. Dev Biol. 2007;303:501–513. doi: 10.1016/j.ydbio.2006.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann SS, Hammarback JA. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J Biol Chem. 1994;269:11492–11497. [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mommsen TP. Salmon spawning migration and muscle protein metabolism: the August Krogh principle at work. Comp Biochem Physiol B Biochem Mol Biol. 2004;139:383–400. doi: 10.1016/j.cbpc.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Mommsen TP, French CJ, Hochachka PW. Sites and patterns of protein and amino acid utilization during the spawning migration of salmon. Can J Zool. 1980;58:1785–1799. doi: 10.1139/z80-246. [DOI] [Google Scholar]
- Moon TW, Johnston IA. Starvation and enzyme activities in the skeletal muscles and liver of a teleost fish (Pleuronectes platessa) Biochem Soc Trans. 1979;7:1045–1046. doi: 10.1042/bst0071045. [DOI] [PubMed] [Google Scholar]
- Mortimore GE, Hutson NJ, Surmacz CA. Quantitative correlation between proteolysis and macro- and microautophagy in mouse hepatocytes during starvation and refeeding. Proc Natl Acad Sci USA. 1983;80:2179–2183. doi: 10.1073/pnas.80.8.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson S, Johnston IA, Goldspink G. The effect of starvation on the chemical composition of red and white muscles in the plaice (Pleuronectes platessa) Experientia. 1974;30:892–894. doi: 10.1007/BF01938344. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Bauvy C, Codogno P. Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. J Biol Chem. 2003;278:16667–16674. doi: 10.1074/jbc.M210998200. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Salem M, Kenney PB, Rexroad CE, 3rd, Yao J. Microarray gene expression analysis in atrophying rainbow trout muscle: a unique nonmammalian muscle degradation model. Physiol Gen. 2006;28:33–45. doi: 10.1152/physiolgenomics.00114.2006. [DOI] [PubMed] [Google Scholar]
- Schiøtz BL, Roos N, Rishovd AL, Gjoen T. Formation of autophagosomes and redistribution of LC3 upon in vitro infection with infectious salmon anemia virus. Virus Res. 2010;151:104–107. doi: 10.1016/j.virusres.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Schlumpberger M, Schaeffeler E, Straub M, Bredschneider M, Wolf DH, Thumm M. AUT1, a gene essential for autophagocytosis in the yeast Saccharomyces cerevisiae. J Bacteriol. 1997;179:1068–1076. doi: 10.1128/jb.179.4.1068-1076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiliez I, Gutierrez J, Salmeron C, Skiba-Cassy S, Chauvin C, Dias K, Kaushik S, Tesseraud S, Panserat S. An in vivo and in vitro assessment of autophagy-related gene expression in muscle of rainbow trout (Oncorhynchus mykiss) Comp Biochem Physiol B Biochem Mol Biol. 2010;157:258–266. doi: 10.1016/j.cbpb.2010.06.011. [DOI] [PubMed] [Google Scholar]
- Shiau CY, Pong YP, Chiou TK, Tin YY. Effect of starvation on free histidine and amino acids in white muscle of milkfish Chanos chanos. Comp Biochem Physiol B Biochem Mol Biol. 2001;128:501–506. doi: 10.1016/S1096-4959(00)00350-X. [DOI] [PubMed] [Google Scholar]
- Shimeno S, Kheyyali D, Takeda M. Metabolic adaptation to prolonged starvation in carp. Nippon Suisan Gakkaishi. 1990;56:35–41. doi: 10.2331/suisan.56.35. [DOI] [Google Scholar]
- Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E. Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. Eugene: University of Oregon Press; 1995. [Google Scholar]
- Yabu T, Yamashita M. Observation of stress-induced autophagy in fish cells. Bull Fish Res Agen. 2008;26:23–28. [Google Scholar]
- Yabu T, Kishi S, Okazaki T, Yamashita M. Characterization of zebrafish caspase-3 and induction of apoptosis through ceramide generation in fish fathead minnow tailbud cells and zebrafish embryo. Biochem J. 2001;360:39–47. doi: 10.1042/0264-6021:3600039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabu T, Imamura S, Yamashita M, Okazaki T. Identification of Mg2+-dependent neutral sphingomyelinase 1 as a mediator of heat stress-induced ceramide generation and apoptosis. J Biol Chem. 2008;283:29971–29982. doi: 10.1074/jbc.M805402200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabu T, Shimuzu A, Yamashita M. A novel mitochondrial sphingomyelinase in zebrafish cells. J Biol Chem. 2009;284:20349–20363. doi: 10.1074/jbc.M109.004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabu T, Imamura S, Mohammed MS, Touhata K, Minami T, Terayama M, Yamashita M. Differential gene expression of HSC70/HSP70 in yellowtail cells in response to chaperone-mediated autophagy. FEBS J. 2011;278:673–685. doi: 10.1111/j.1742-4658.2010.07989.x. [DOI] [PubMed] [Google Scholar]
- Yamashita M. Stress responses of fish during catching process. In: Konno K, Ochiai Y, Fukuda Y, editors. Quality control of tuna meat by optimization of fishing and handling. Tokyo: Koseisha-Koseikaku; 2010. pp. 81–94. [Google Scholar]
- Yamashita M, Konagaya S. The elevation of catheptic activity in muscle and liver of ayu Plecoglossus altivelis during maturation. Nippon Suisan Gakkaishi. 1990;56:1157. [Google Scholar]
- Yamashita M, Konagaya S. High activities of cathepsins B, D, H, and L in the white muscle of chum salmon in spawning migration. Comp Biochem Physiol. 1990;B95:149–152. doi: 10.1016/0305-0491(90)90262-r. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Konagaya S. Hydrolytic action of salmon cathepsins B and L to muscle structural proteins in respect of muscle softening. Nippon Suisan Gakkaishi. 1991;57:1917–1922. doi: 10.2331/suisan.57.1917. [DOI] [Google Scholar]
- Yamashita M, Konagaya S. Proteolysis of muscle proteins in the extensively softened muscle of chum salmon caught during spawning migration. Nippon Suisan Gakkaishi. 1991;57:2163. doi: 10.2331/suisan.57.1917. [DOI] [Google Scholar]
- Yamashita Y, Yabu T, Yamashita M. Discovery of the strong antioxidant selenoneine in tuna and selenium redox metabolism. World J Biol Chem. 2010;1:144–150. doi: 10.4331/wjbc.v1.i5.144. [DOI] [PMC free article] [PubMed] [Google Scholar]