Abstract
Objective
Familial hypercholesterolemia (FH) is an autosomal dominant inherited disorder caused by mutations in the low density lipoprotein receptor (LDLR) gene. FH is characterized by elevated plasma LDL cholesterol, premature atherosclerosis, and a high risk of premature myocardial infarction. In general, mutations within LDLR gene can cause five different classes of defects, namely: class I defect: no LDLR synthesis; class II defect: no LDLR transport; class III defect: no low density lipoprotein (LDL) to LDLR binding; class IV defect: no LDLR/LDL internalization; and class V defect: no LDLR recycling. One might expect that both the class of LDLR defect as well as the precise mutation influences the severity of hypercholesterolemia on one hand and the response on drug treatment on the other. To clarify this question we studied the effect of the LDLR mutation p.W556R in two heterozygote subjects.
Results
We found that two heterozygote FH patients with the LDLR mutation p.W556R causing a class II LDLR defect (transport defective LDLR) respond exceedingly well to the treatment with simvastatin 40 mg/ezetimibe 10 mg. There was a LDL cholesterol decrease of 55 and 64%, respectively. In contrast, two affected homozygote p.W556R FH patients, in the mean time undergoing LDL apheresis, had no response to statin but a 15% LDL cholesterol decrease on ezetimibe monotherapy.
Conclusions
The LDLR mutation p.W556R is a frequent and severe class II defect for FH. The affected homozygote FH patients have a total loss of the functional LDLR and—as expected—do not respond on statin therapy and require LDL apheresis. In contrast, heterozygote FH patients with the same LDLR defect respond exceedingly well to standard lipid-lowering therapy, illustrating that the knowledge of the primary LDLR defect enables us to foresee the expected drug effects.
Keywords: Familial hypercholesterolemia, Low density lipoprotein receptor (LDLR), Statin treatment, LDL apheresis
Introduction
The LDL receptor (LDLR) is an essential receptor for the uptake of low density lipoprotein (LDL) and accounts for the clearance of 70% of all plasma-circulating LDL [1]. LDLR/LDL complexes are internalized by endocytosis, mainly in hepatocytes and ligand dischargement in the acidic environment of the endosome enables the recycling of LDL receptors for another round of LDL binding [1–3].
The term “familial hypercholesterolemia” (FH) is generally used for LDLR deficiency which is inherited as an autosomal dominant trait. Homozygous LDLR deficiency is rare, with a frequency of 1 per million in the general population [4, 5]. It is characterized by severely elevated LDL cholesterol ( > 15.5 mmol/L; > 600 mg/dL). The clinical symptoms are xanthomas, thickened achilles tendons, carotis and coronary artery stenosis, and aortic valve stenosis that develop in the first decade of life, leading to premature death from stroke or myocardial infarction in childhood. The molecular basis of FH has been elucidated by the fundamental work of Goldstein and Brown and Rader et al. [6, 7]. They revealed that defects of the LDL receptor are caused by mutations within the LDL receptor gene. The LDLR is located on chromosome 19 [8]. Worldwide, more than 1,000 FH causing LDLR gene mutations ranging from single nucleotide substitutions to extended deletions had been identified in different ethnic groups [9–11].
According to the nature and location of the mutations within the LDLR gene, five different classes of FH-causing mutations have been defined. Class I mutations include null alleles with no detectable LDL receptor protein. Class II mutations produce transport-defective LDLR proteins that are either completely (class II a) or partially blocked (class II b or leaky LDLRs) in their transport from the endoplasmic reticulum to the Golgi apparatus due to impaired glycosylation [12]. Class III mutations encode LDL receptors with normal intracellular transport but defective LDL binding. Class IV mutations produce LDL receptors with normal transport and cell surface LDL binding but defective clustering in clathrin-coated pits for internalization. Finally, class V mutations produce recycling defective receptors that internalize normally, but are unable to release bound ligand within the acidic environment of the endosome, and thus do not recycle to the cell surface [12] (Fig. 1a).
Fig. 1.
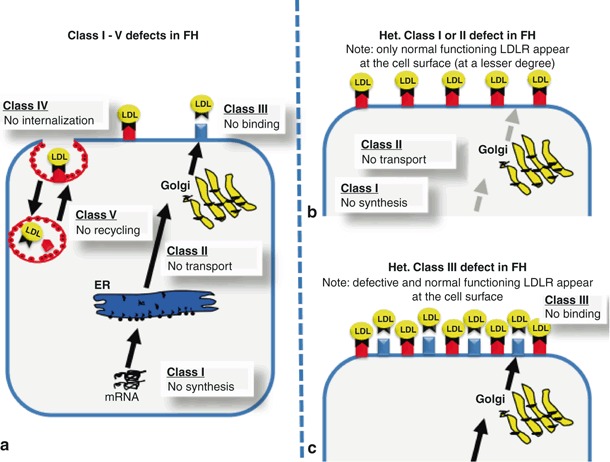
(a) Illustrates the five different classes for defects of LDL receptor function and the consequences of the different defects on the cell surface for class I and II defects (b) or class III defects (c). Class I mutations include null alleles with no detectable LDL receptor protein. Class II mutations produce transport-defective LDLR proteins that are either completely (class II a) or partially blocked (class II b or leaky LDLRs) in their transport from the endoplasmic reticulum to the Golgi apparatus due to impaired glycosylation. Class III mutations encode LDL receptors with normal intracellular transport but defective LDL binding. Class IV mutations produce LDL receptors with normal transport and cell surface LDL binding but defective clustering in clathrin-coated pits for internalization. Class V mutations produce recycling defective receptors that internalize normally, but are unable to release bound ligand within the acidic environment of the endosome, and thus do not recycle to the cell surface
Methods
Lipid profiles
Lipid analysis was performed from plasma drawn under fasting conditions (fasting period of at least 12 h) without any lipid-lowering medication and was repeated after 6 weeks treatment with simvastatin 40 mg and ezetimibe 10 mg as a single tablet given once daily. Total cholesterol and triglycerides were measured enzymatically (Roche Diagnostics, Mannheim, Germany). High-density lipoprotein (HDL) was measured after the precipitation of apoB-containing lipoproteins. LDL was calculated using the Friedewald equation [13]. ApoE phenotyping was performed by isoelectric focusing and immunofixation as described previously [14].
Mutation scanning of the LDLR and apoB-100 genes
Genomic DNA was isolated from whole blood by standard procedures. Oligonucleotides and PCR reaction conditions for the amplification of the promoter region and 18 exons of the LDLR and the apoB gene sequence encoding the carboxyterminal modulator element of apoB-100 (residues 3448–3561) were exactly as previously described [15, 16]. Denaturing gradient gel electrophoresis (DGGE) screening for mutations in the LDLR and apoB genes were performed in a Protean II electrophoresis system (Bio-Rad, Munich, Germany). PCR products of LDLR exons with aberrant DGGE patterns were sequenced in forward and reverse directions with the BigDyeTM terminator cycle sequencing ready reaction kit (Applied Biosystems, Warrington, UK) on an ABI 3730 DNA sequencer.
Results
The index family is a Turkish family with two identical male twins, who are homozygote for the p.W556R LDLR mutation. Their parents are both heterozygote for the p.W556R LDLR defect. The ApoE phenotype was 3/3 and by this normal in all patients. At the age of 2 years, the twins showed multiple tuberoeruptive xanthomas over the extensor surface of the elbows, knees, subcutaneous nodules in the extensor tendons of the hands and xanthelasma palpebrarum. The parents have none of these signs. Lipid studies were carried out in all family members. The two boys had extremely high levels of total cholesterol and LDL cholesterol above 25.8 mmol/L ( > 1000 mg/dL). By this the twins meet the diagnostic criteria for homozygous FH which was confirmed by identifying the primary LDLR defect [17]. The boys were initially treated with statins, however, there was no measureable decrease in LDL cholesterol ( < 5%). Interestingly, the treatment with ezetimibe resulted in a decrease in LDL cholesterol of roughly 15% which is remarkable considering the total lack of any functioning LDLR. The twins were clinically stable and underwent weekly LDL apheresis for more than 10 years . In this report, we would like to focus mainly on the heterozygote parents of these two boys. Both subjects were free of any signs for coronary artery disease (CAD); however, there was a strong family history of CAD. The male subject had untreated a total cholesterol of 9.8 mmol/L ( = 379 mg/dL) and LDL cholesterol of 7.7 mmol/L ( = 298 mg/dL), whereas the female subject with the identical genetic defect had initially a total cholesterol of 7.6 mmol/L ( = 296 mg/dL) and LDL cholesterol of 5.9 mmol/L ( = 203 mg/dL). After 6 weeks treatment with simvastatin 40 mg in combination with ezetimibe 10 mg once daily, the LDL levels dropped by 55 and 64% respectively. The medication was well tolerated. The total lipid profile prior to and under lipid-lowering treatment is summarized in Table 1.
Table 1.
Lipid levels before and after 6 weeks of treatment in two heterozygote patients with the p.W556R LDL receptor mutation
| Patients | Sex | Age (years) | TC (mmol/L) | TG (mmol/L) | LDL-C (mmol/L) | HDL-C (mmol/L) | LDLR (p.W556R) |
|---|---|---|---|---|---|---|---|
| No lipid-lowering therapy | M | 36 | 9.8 | 1.7 | 7.7 | 1.2 | Heteroz |
| Simvastatin 40 mg/ezetimibe 10 mg | 5.2 | 1.1 | 3.5 | 1.1 | |||
| No lipid-lowering therapy | F | 30 | 7.6 | 1.3 | 5.2 | 1.8 | Heteroz |
| Simvastatin 40 mg/ezetimibe 10 mg | 3.5 | 0.6 | 1.9 | 1.4 |
Age refers to years of age at blood sampling. Lipid values are given in millimoles/litre
F female; M male; TC total cholesterol; TG triglycerides; LDL-C LDL cholesterol; HDL-C HDL cholesterol; LDLR LDL receptor
Discussion
We identified the molecular basis of LDLR deficiency in some of the most severely affected patients with the highest LDL cholesterol levels reported so far in humans. The disease was found to be due to a p.W556R mutation of the LDL receptor resulting in class II defect with total loss of function [18]. Sozen et al. [19] revealed this mutation to be rather frequent in the Turkish population . Most recently we reported the kinetic consequences of this mutation by studying the in vivo kinetics in the heterozygote father of our twins. The heterozygote p.W556R subject had both a slow-fractional catabolic rate (FCR) for LDL Apo B-100 (FCR in FHMarburg: 0.19/day versus FCR in controls: 0.35 ± 0.1/day) and a high LDL production rate (PR; FHMarburg: 13.0 mg/kg/day versus PR in controls: 10.3 ± 1.9 mg/kg/day) resulting in an increase in LDL cholesterol [20].
In FH, two thirds of all known mutations are located in the epidermal growth factor-like (EGF) domain of the LDLR. Among these, one third affects the YWTD repeats of the LDL receptor and alter conserved scaffolding residues of the b -propeller that disrupt its structure. These results in inefficient LDL receptor transport to the cell surface—a so-called class II defect of the LDLR as those in the patients described here [21]. There are several interesting findings we would like to share from our studies in the heterozygote FHMarburg (p.W556R) patients:
The LDL cholesterol levels of the female subject is remarkably lower compared with those of her (in terms of identical LDLR defect) male counterpart (LDL in the female subject: 5.9 mmol/L ( = 203 mg/dL) versus LDL in the male subject: 7.7 mmol/L ( = 298 mg/dL). This finding is of special interest since both subjects have the identical genetic defect leading to FH; however, simply the gender seems to influence the LDL cholesterol levels for up to one third of their individual LDL cholesterol levels. Since this finding is seen in other FH subjects as well, we assume that understanding the gender-specific mechanisms might open a new and promising approach to lower LDL cholesterol.
The LDL cholesterol-lowering potency of simvastatin 40 mg/ezetimibe 10 mg was impressive in both subjects. However, despite the identical mutation the effect of simvastatin 40 mg/ezetimibe 10 mg was greater in the female subject compared with the male subject (64 versus 55%). This finding is in contrast to the data reported recently by Abramson et al. [22]. Abramson et al. analyzed the data of more than 22,000 hyperlipidemic patients from 27 studies. They found that men treated with statin and ezetimibe experienced significantly greater changes in LDL cholesterol (p = 0.0066) compared with women. This underlines the heterogeneity in hyperlipidemic patients and the need of appropriate knowledge of the primary defects.
The rather strong LDL-lowering effect of simvastatin 40 mg/ezetimibe 10 mg in both heterozygote subjects is remarkable, even more so since the homozygote children showed none (statin) to only moderate (ezetimibe) LDL-lowering effects. A more than 60% LDL cholesterol-lowering effect, as seen in the female FH subject, has been reported by Goldberg et al. [23] in a lipid-lowering trial for the highest simvastatin 80 mg/ezetimibe 10 mg doses ( - 61%) whereas simvastatin 40 mg/ezetimibe 10 mg provides a decrease in LDL cholesterol of − 55%.
How it comes that the same genetic defect is totally disastrous in its homozygote state and easy to be treated in its heterozygote state? Most likely, that the class of defect is crucial and accountable for its treatability. Once—like in our case with the p.W556R mutation as a class II defect—we have a defect resulting in the lack of LDLR transport. In the homozygote state we see no LDLR at all on the cell surface, making this defect to be one of the worst LDLR mutations known so far. However, in the heterozygote state we see only fully functional LDLR’s on the cell surface, since the LDLR defect is not coming through the LDLR pathway (see Fig. 1b). Once the LDLR system is stimulated by statin treatment we expect to see more (and only fully functional) LDLR on the cell surface, resulting in a clear LDL decrease as seen in both of our patients. In contrast, a class III or class IV LDLR defect would produce (maybe still partially functional) LDLR which show up on the cell surface. In the homozygote state, these defects (depending on the location of the mutation) increase LDL cholesterol more or less severely. However, in its heterozygote state we will see—different from class I or II defects—always a mixture of functional and non functional LDLR on the cell surface. This mixture of functional and non functional LDLR will increase under statin therapy. Different from heterozygote LDLR class I or II mutations, the non functional LDLR class III or IV will compete with functional LDLR on the cell surface and by this interact with an appropriate LDL-lowering effect (see Fig. 1c). Of course, our study is highly limited by the exceedingly small number of patients involved. However, these subjects were well defined and our findings might explain why some heterozygote FH patients respond extremely well to statin therapy whereas others fail. For example, it is known that FH patients with xanthomas have significantly higher LDL cholesterol levels than those without and are less likely to achieve LDL cholesterol target levels under lipid-lowering therapy [24]. Differences between the classes of LDLR mutations had been described by Miltiadous et al. [25]. However, different from our observations, they found only moderate decrease of LDL cholesterol (− 34 ± 9%) in 28 patients with a class II LDLR defect (G1646A, C858A) with statin treatment (atorvastatin 20 mg) compared with 21 patients with a class V LDLR mutation (G1775A) with a higher decrease in LDL cholesterol (− 49 ± 9%).
In the near future, thanks to the worldwide efforts and upcoming screening procedures, we should be able to predict the potential and benefit of any given lipid-lowering drug and the need of LDL apheresis, once we know the precise genetic defects leading to hypercholesterolemia and its disastrous consequences [26–30]. Till then, we should be careful in future study designs of lipid-lowering drugs, in which certain genetic backgrounds could easily over- or underestimate the treatment benefits of the drugs under trial.
Acknowledgment
This work was supported by grants of the Prof. Dr. Reinfried Pohl-Stiftungsprofessur for Preventive Cardiology at the Philipps-University Marburg to Juergen R. Schaefer.
This article is part of a supplement sponsored by an unrestricted educational grant from B. Braun and Fresenius Medical Care.
Conflict of interest
Juergen R. Schaefer serves as a scientific advisor for MSD and ESSEX pharma. He received research grants and/or lecture fees by MSD, ESSEX, Bayer Healthcare, Takeda Pharma, Genzyme, B. Braun Melsungen. None of the other authors declare any potential conflict of interest.
References
- 1.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 2.Cummings RD, Kornfeld S, Schneider WJ, Hobgood KK, Tolleshaug H, Brown MS, Goldstein JL. Biosynthesis of N- and O-linked oligosaccharides of the low density lipoprotein receptor. J Biol Chem. 1983;258:15261–15273. [PubMed] [Google Scholar]
- 3.Davis CG, Van Driel IR, Russell DW, Brown MS, Goldstein JL. The low density lipoprotein receptor. Identification of amino acids in cytoplasmic domain required for rapid endocytosis. J Biol Chem. 1987;262:4075–4082. [PubMed] [Google Scholar]
- 4.Hobbs HH, Russell DW, Brown MS, Goldstein JL. The LDL receptor locus in familial hypercholesterolemia: mutational analysis of a membrane protein. Annu Rev Genet. 1990;24:133–170. doi: 10.1146/annurev.ge.24.120190.001025. [DOI] [PubMed] [Google Scholar]
- 5.Austin MA, Hutter CM, Zimmern RL, Humphries SE. Familial hypercholesterolemia and coronary heart disease: a HuGE association review. Am J Epidemiol. 2004;160:421–429. doi: 10.1093/aje/kwh237. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein JL, Brown MS (1989) Familial Hypercholesterolemiahypercholesterolemia. In: Scriver CR, Beaudet AL, Sly WS, Valle D, (eds) The metabolic basis of inherited disease. McGraw-Hill, New York pp 1215–1250
- 7.Rader DJ, Cohen J, Hobbs HH. Monogenic hypercholesterolemia: new insights in pathogenesis and treatment. J Clin Invest. 2003;111:1795–1803. doi: 10.1172/JCI18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sudhof TC, Goldstein JL, Brown MS, Russell DW. The LDL receptor gene: a mosaic of exons shared with different proteins. Science. 1985;228:815–822. doi: 10.1126/science.2988123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villeger L, Abifadel M, Allard D, Rabes JP, Thiart R, Kotze MJ. The UMD-LDLR database: additions to the software and 490 new entries to the database. Hum Mutat. 2002;20:81–87. doi: 10.1002/humu.10102. [DOI] [PubMed] [Google Scholar]
- 10.Leigh SE, Foster AH, Whittall RA, Hubbart CS, Humphries SE. Update and analysis of the university college london low density lipoprotein receptor familial hypercholesterolemia database. Ann Hum Genet. 2008;72:485–498. doi: 10.1111/j.1469-1809.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- 11.Varret M, Abifadel M, Rabès JP, Boileau C. Genetic heterogeneity of autosomal dominant hypercholesterolemia. Clin Genet. 2008;73(1):1–13. doi: 10.1111/j.1399-0004.2007.00915.x. [DOI] [PubMed] [Google Scholar]
- 12.Hobbs HH, Brown MS, Goldstein JL. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum Mutat. 1992;1:445–466. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- 13.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low density lipoprotein cholesterol without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 14.Hackler R, Schaefer JR, Motzny S, Brand S, Kleine TO, Kaffarnik H, Steinmetz A. Rapid determination of apolipoprotein E phenotypes from whole plasma by automated isoelectric focusing using PhastSystem and immunofixation. J Lipid Res. 1994;35:153–158. [PubMed] [Google Scholar]
- 15.Nissen H, Guldberg P, Hansen AB, Petersen NE, Horder M. Clinically applicable mutation screening in familial hypercholesterolemia. Hum Mutat. 1996;8:168–177. doi: 10.1002/(SICI)1098-1004(1996)8:2<168::AID-HUMU9>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 16.Soufi M, Sattler AM, Maerz W, Starke A, Herzum M, Maisch B, Schaefer JR. A new but frequent mutation of apoB-100-apoB His3543Tyr. Atherosclerosis. 2004;174:11–16. doi: 10.1016/j.atherosclerosis.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 17.Zschocke J, Schaefer JR. Homozygous familial hypercholesterolaemia in identical twins. Lancet. 2003;361(9369):1641. doi: 10.1016/S0140-6736(03)13303-X. [DOI] [PubMed] [Google Scholar]
- 18.Soufi M, Zschocke J, Quak E, Hofmann G, Maisch B, Schaefer JR. First description of homozygous familial hypercholesterolemia (FH) in twins. Atherosclerosis. 1999;Suppl. 2(147):34. [Google Scholar]
- 19.Sozen MM, Whittall R, Oner C, Tokatli A, Kalkanoglu HS, Dursun A. The molecular basis of familial hypercholesterolaemia in Turkish patients. Atherosclerosis. 2005;180:63–71. doi: 10.1016/j.atherosclerosis.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 20.Soufi M, Kurt B, Schweer H, Sattler AM, Klaus G, Zschocke J, Schaefer JR. Genetics and kinetics of familial hypercholesterolemia, with the special focus on FH-(Marburg) p.W556R. Atheroscler Suppl. 2009;10(5):5–11. doi: 10.1016/S1567-5688(09)71802-1. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt HH, Tietge UJ, Buettner J, Barg-Hock H, Offner G, Schweitzer S. Liver transplantation in a subject with familial hypercholesterolemia carrying the homozygous p.W577R LDL-receptor gene mutation. Clin Transplant. 2008;22:180–184. doi: 10.1111/j.1399-0012.2007.00764.x. [DOI] [PubMed] [Google Scholar]
- 22.Abramson BL, Benlian P, Hanson ME, Lin J, Shah A, Tershakovec AM. Response by sex to statin plus ezetimibe or statin monotherapy: a pooled analysis of 22,231 hyperlipidemic patients. Lipids Health Dis. 2011;10:146. doi: 10.1186/1476-511X-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldberg AC, Sapre A, Liu J, Capece R, Mitchel YB. Ezetimibe Study Group. Efficacy and safety of ezetimibe coadministered with simvastatin in patients with primary hypercholesterolemia: a randomized, double-blind, placebo-controlled trial. Mayo Clin Proc. 2004;79(5):620–629. doi: 10.4065/79.5.620. [DOI] [PubMed] [Google Scholar]
- 24.Heath KE, Gudnason V, Humphries SE, Seed M. The type of mutation in the low density lipoprotein receptor gene influences the cholesterol-lowering response of the HMG-CoA reductase inhibitor simvastatin in patients with heterozygous familial hypercholesterolaemia. Atherosclerosis. 1999;143(1):41–54. doi: 10.1016/S0021-9150(98)00274-3. [DOI] [PubMed] [Google Scholar]
- 25.Miltiadous G, Xenophontos S, Bairaktari E, Ganotakis M, Cariolou M, Elisaf M. Genetic and environmental factors affecting the response to statin therapy in patients with molecularly defined familial hypercholesterolaemia. Pharmacogenet Genomics. 2005;15(4):219–225. doi: 10.1097/01213011-200504000-00005. [DOI] [PubMed] [Google Scholar]
- 26.Graham CA, McClean E, Ward AJ, Beattie ED, Martin S, O’Kane M. Mutation screening and genotype:phenotype correlation in familial hypercholesterolaemia. Atherosclerosis. 1999;147:309–316. doi: 10.1016/S0021-9150(99)00201-4. [DOI] [PubMed] [Google Scholar]
- 27.Vergopoulos A, Knoblauch H, Schuster H. DNA testing for familial hypercholesterolemia: improving disease recognition and patient care. Am J Pharmacogenomics. 2002;2(4):253–262. doi: 10.2165/00129785-200202040-00005. [DOI] [PubMed] [Google Scholar]
- 28.Guardamagna O, Restagno G, Rolfo E, Pederiva C, Martini S, Abello F. The type of LDLR gene mutation predicts cardiovascular risk in children with familial hypercholesterolemia. J Pediatr. 2009;155:199–204. doi: 10.1016/j.jpeds.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 29.Ned Renée M, Sijbrands Eric JG (2011) Cascade screening for familial hypercholesterolemia (FH) (Internet). Version 11. PLoS currents: evidence on genomic tests. 2011: PMC3102597 [DOI] [PMC free article] [PubMed]
- 30.Setia N, Verma IC, Khan B, Arora A. Premature coronary artery disease and familial hypercholesterolemia: need for early diagnosis and cascade screening in the Indian population. Cardiol Res Pract. 2012;2012:658526. doi: 10.1155/2012/658526. [DOI] [PMC free article] [PubMed] [Google Scholar]


