Abstract
The fungus Magnaporthe oryzae is a serious pathogen of rice and other grasses. Telomeric restriction fragments in Magnaporthe isolates that infect perennial ryegrass (prg) are hotspots for genomic rearrangement and undergo frequent, spontaneous alterations during fungal culture. The telomeres of rice-infecting isolates are very stable by comparison. Sequencing of chromosome ends from a number of prg-infecting isolates revealed two related non-LTR retrotransposons (M. oryzae Telomeric Retrotransposons or MoTeRs) inserted in the telomere repeats. This contrasts with rice pathogen telomeres that are uninterrupted by other sequences. Genetic evidence indicates that the MoTeR elements are responsible for the observed instability. MoTeRs represent a new family of telomere-targeted transposons whose members are found exclusively in fungi.
Keywords: genome instability, genome evolution, transposition
TELOMERES are the sequences that form the ends of linear chromosomes and are essential for maintaining the integrity of terminal DNA. In most organisms, the telomeres are composed of tandem arrays of short sequence motifs that are added on to the 3′ ends of chromosomes by telomerase—a specialized reverse transcriptase (Greider and Blackburn 1989; Yu et al. 1990). This prevents the loss of DNA that would normally occur as a result of conservative DNA replication. The telomeres are bound by numerous proteins that shelter the terminal sequences from degradation (Garvik et al. 1995; Vodenicharov and Wellinger 2006) and illegitimate recombination (Dubois et al. 2002). Certain Diptera lack telomerase and, instead, their chromosome termini are maintained by different types of repeats (Saiga and Edstrom 1985; Biessmann et al. 1998; Abad et al. 2004). The most striking example is in Drosophila whose telomeres are composed of arrays of non-LTR retrotransposons (Abad et al. 2004; George et al. 2006), which are capable of transposing to free DNA ends (Traverse and Pardue 1988; Biessmann et al. 1990). The extension of chromosome ends through the addition of transposon sequences not only solves the end-replication problem but also serves to establish a nucleoprotein complex that is essential for protecting the chromosome ends and maintaining telomere homoeostasis (Fanti et al. 1998; Perrini et al. 2004).
In many organisms, the telomeres are attached to a specific sequence that is duplicated (although often not perfectly) at many, and sometimes all, chromosome ends. Such sequences, which usually are TG-rich and often contain short tandem repeats, define a distinct distal subtelomere domain (Pryde et al. 1997). In addition to possessing distally located subtelomere domains, several organisms have proximal domains that contain genes and gene families that are also variously dispersed among different chromosome ends (Pryde et al. 1997). High levels of subtelomere polymorphism are found in many organisms, including vertebrates (Wilkie et al. 1991; Bassham et al. 1998; Baird et al. 2000; Mefford et al. 2001), insects (Biessmann et al. 1998; Anderson et al. 2008; Kern and Begun 2008), plants (Yang et al. 2005), and fungi (Naumov et al. 1995, 1996; Naumova et al. 1996; Cuomo et al. 2007; Kasuga et al. 2009). Indeed, genome-wide analyses of polymorphism often point to the subtelomere regions as being the most variable parts of the genome (Kellis et al. 2003; Winzeler et al. 2003; Cuomo et al. 2007; Kasuga et al. 2009), suggesting that they are “hotbeds” for the rapid evolution of genes and gene families (Mefford et al. 2001; Fan et al. 2008; Kern and Begun 2008).
The variable nature of telomere regions provides an adaptive advantage to a number of pathogens, such as the protists Plasmodium and Trypanosoma and the fungus Pneumocystis carinii. The subtelomeres of these organisms contain highly divergent gene families coding for surface proteins. Only one gene copy is expressed at any given time, and occasional switching of the expressed copy enables these pathogens to evade the host’s immune system. In Trypanosomes and Pneumocystis, switching involves ectopic recombination between a subtelomeric gene copy and another at a different chromosomal locus (Wada and Nakamura 1996; Horn and Cross 1997). Similarly, ectopic recombination plays a role in the evolution of the subtelomeric virulence factor genes in Plasmodium (Freitas-Junior et al. 2000).
In the plant pathogenic fungus Magnaporthe oryzae, there is also an association between chromosome ends and genes that control interactions with the host. As the causal agent of a devastating disease of rice, M. oryzae is best known as the “rice blast fungus.” However, the fungus exists as several different host-specialized forms that together cause diseases in >50 species of grasses (Ou 1985). The ability of Magnaporthe to infect a given host species—or, in some cases, a specific cultivar of a species—is controlled by “avirulence genes” that code for secreted proteins that are translocated in the host cytoplasm. To date, approximately half of the 20-plus avirulence genes that have been identified in M. oryzae map very close to telomeres (Farman 2007). Sequencing of subtelomere regions in a rice-infecting strain of M. oryzae revealed that they are hotspots for transposon insertions, many of which show evidence of having undergone ectopic recombination events (Rehmeyer et al. 2006). Presumably, avirulence genes tend to be enriched in these regions because the rearrangement and loss of DNA sequences that accompany such recombination promote avirulence gene (and, hence, pathogenic) diversity within M. oryzae populations.
Even though the chromosome ends of rice-pathogenic isolates show evidence of historical rearrangements, these events appear to be fairly infrequent because fungal isolates that are clonally related—as judged with repetitive DNA fingerprinting probes—tend to have very similar telomeric restriction fragment profiles. This is true even of isolates collected several years apart. In striking contrast, M. oryzae strains from perennial ryegrass (prg) often exhibit “absolute” telomere polymorphism—i.e., they share no telomeric fragments in common, even when they have little or no polymorphism at internal loci. Furthermore, analysis of clonal isolates derived from single spores revealed that telomeric fragments underwent frequent rearrangement during mitotic growth both in culture and in planta (Farman and Kim 2005).
The goal of the present study was to determine the molecular basis for the apparent difference in telomere stability between M. oryzae isolates that infect rice vs. those from prg. First, however, we performed a direct comparison of the rates of telomere change in a prg pathogen vs. a rice-infecting strain. Then, to determine if frequent telomere rearrangements in the prg pathogens can be attributed to differences in the structural organization of the chromosome ends, we cloned and characterized telomeres from three different prg-infecting isolates and, where possible, compared individual telomeres to their homologous counterparts in the rice-infecting strain. This led to the identification of non-LTR retrotransposons embedded in the telomere repeats of the prg pathogens. We present genetic evidence that the retrotransposon insertions promote instability of the telomeres that contain them.
Materials and Methods
Fungal strains/isolates
The fungal strains and other isolates that were used in the study are listed in the Supporting Information, Table S1. Stocks were reactivated by placing them on oatmeal agar (OA) and allowing them to grow under constant fluorescent illumination for 7 days. Spores were harvested by gently brushing sporulating colonies with a sterilized glass rod. They were then spread across a 4% water agar (WA) and placed at room temperature overnight. The spores were visualized under a dissecting microscope, and a single germinated spore was transferred to a fresh OA plate. To minimize telomeric diversity, all strains/isolates used for the study were single-spored before analysis.
Serial passaging of the fungus through plants
Single spore cultures of 70-15 and LpKY97-1A were grown on OA plates for 7 days. Conidia were collected by flooding the agar surface with 10 ml of a 0.1% gelatin solution and dislodging them with a bent glass rod. The spore suspensions then were filtered through two layers of cheesecloth and adjusted to a final density of 1 × 105 conidia/ml by using a hemacytometer. Two hundred microliters of each conidial suspension was spread on a 0.4% WA plate, and single conidia were collected as outlined above. Twenty monoconidial isolates were collected for each fungal strain, and these are termed generation zero (G0) cultures. The remainder of each conidial suspension was used to infect plants as follows: Rice cultivars 51583 (Oryza sativa) and perennial ryegrass (Lolium perenne) cultivar Linn were placed in separate plastic bags and sprayed with the conidial suspension of 70-15 and LpKY97-1A, respectively, using an artist’s airbrush. Inoculation bags were placed at room temperature in the dark for ∼18 hr and then transferred to a growth chamber with a 12-hr day/night cycle at 27° day and 21° night. The bags were opened slightly, and the humidity was allowed to equilibrate over a period of ∼4 hr. After equilibration, the plants were removed from the bags, and infections were allowed to proceed for 7 days. Leaves showing lesions were then clipped and placed in a moist chamber to promote sporulation. After 3–5 days, spores were picked off the lesions using a dry glass rod and resuspended in 2 ml of a 0.1% gelatin solution. The suspension was filtered through two layers of cheesecloth and adjusted to a final concentration of 1 × 105 conidia/ml. Two hundred microliters of the spore suspension was spread on 0.4% water agar and incubated overnight to allow germination to occur. Twenty “first-generation” (G1) single spores were then collected. The remainder of the spore suspension was used to inoculate a second set of plants. Inoculation and plant growth was performed as described above, and a second set of 20 single spores was collected. These are referred to as “second-generation” spores.
Crosses
Crosses were set up by placing 2-mm3 mycelial plugs ∼5 cm apart on OA plates. The plates were then incubated at room temperature under constant white-light illumination until the colonies had merged (∼10 days after inoculation). The plates were then transferred to an incubator set at 18° with constant illumination from a black light. After the perithecia had formed (∼21 days after inoculation), they were excised from the agar using a scalpel and transferred to 4% water agar. The perithecia were then cut open to release the eight-spored asci, which were spread across the agar surface using a drawn-out Pasteur pipette. The plates were incubated overnight at room temperature. Germinated asci were individually picked to oatmeal agar and incubated at room temperature until conidiospores had formed. Single spores were then isolated and grown to produce cultures representing individual meiotic products.
DNA extraction
Mycelium was grown at room temperature with shaking for ∼10 days. The mycelial ball was harvested with forceps, blotted dry, snap-frozen in liquid nitrogen, and then placed in a 96-deep-well plate. After freeze-drying for 24 hr, the mycelium was ground to a powder by shaking with steel beads in a 2000 GenoGrinder (Spex Certiprep, Metuchen, NJ). One milliliter of preheated (65°) lysis buffer (0.5 M NaCl; 1% SDS; 10 mM Tris–HCl, pH 7.5; 10 mM EDTA) was added to the ground mycelium and incubated at 65° for 30 min, after which 0.66 ml of phenol:chloroform:isoamyl alcohol (25:24:1) was added and incubated for another 30 min at 65°. The plate was then centrifuged for 30 min at 3000 × g to pellet the cell debris. Four hundred microliters of supernatant was transferred to a fresh 96-well plate, and 240 µl of isopropanol was added. The DNA was then pelleted by centrifugation at 3000 × g for 20 min, followed by two 70% ethanol washes. The DNA pellets were then dried and redissolved in TE containing RNase A (100 µg/ml). Finally, the DNA samples were quantified by fluorimetry.
Creation of random genomic mini-libraries enriched for telomeres
Polysaccharides were removed from genomic DNA preparations by using differential ethanol precipitation (Michaels et al. 1994). The genomic DNA samples (∼2 µg) were then blunt-ended using the End-It kit (Epicentre Technologies, Madison, WI) and ligated to EcoRV-digested pBluescript in a 10-µl volume. This step enriches for DNA sequences adjacent to natural DNA ends (i.e., telomeres) and regions flanking physically fragile sites. The ligase was killed by heat inactivation, 1 µl (20 units) of ApaI (New England Biolabs, Beverly, MA) was added, and digestion was allowed to proceed overnight at 25°. The ApaI was then killed by heat inactivation, the reaction mix was diluted 10-fold with 1× ligation enzyme buffer, and 0.1 µl T4 DNA ligase (NEB) was added. After ligation, circularized DNA molecules were rescued by transformation into Escherichia coli strain DH5α or EPI300. This resulted in libraries of clones whose inserts extend from a natural DNA end—either a telomere or a site of breakage—to the first ApaI site. Telomere-containing clones were then identified by colony hybridization using a (TTAGGG)200-300 probe (Farman 2011).
Hybridization probes
Probes were synthesized as follows: Telomere probe was generated in a template-less PCR reaction using the complementary primers TelomereF and TelomereR (Table S2) at a concentration of 200 nM. The PCR parameters were: 94° for 5 min, followed by 35 cycles of 94° for 30 sec; 50° for 30 sec; and 72° for 1 min. The final extension was at 72° for 2 min. The reaction products were separated by gel electrophoresis, and fragments of 1.5–2.0 kb were excised from the gel and purified using Qiaquik columns (Qiagen, Valencia, CA). The MoTeR1 and MoTeR2 probes were amplified with primer pairs MoTeR1001F and MoTeR1001R and MoTeR2001F and MoTeR2001R, respectively, using highly diluted plasmid clones as templates. All primer sequences are listed in Table S2. PCR was performed using ExTaq polymerase with the buffer and nucleotides supplied by the manufacturer (Takara, Shiga, Japan). Amplification products were purified by gel extraction and labeled with α32P dCTP (GE Healthcare, Piscataway, NJ) using the Prime-a-Gene Labeling System (Promega, Madison, WI).
Southern blotting and hybridization experiments
One microgram of genomic DNA was digested in a total reaction volume of 50 µl in the reaction buffer supplied by the enzyme manufacturer (NEB). Approximately 400 ng of the digested DNA was loaded onto a 0.7% agarose gel made up in 0.5 × TBE, and the gels were run for 24 or 48 hr in a 4° cold room. After staining with ethidium bromide and photographing, the gels were electroblotted for 2 hr at 12 V onto Pall Biodyne B-charged nylon membranes (Pall, East Hills, NY) using a GENIE electroblotter (Idea Scientific, Minneapolis, MN) and a power supply purchased from the manufacturer. After electroblotting, the immobilized DNA was denatured by soaking the membrane in 0.4 N NaOH for 10 min and then neutralized by washing with 2× SSC for 10 min. Finally, the membrane was covalently attached to the membrane using a crosslinker (Spectronics, Westbury, NY).
Prehybridization was conducted for 30 min at 65° in hybridization buffer minus probe (0.125 M NaHPO4, pH 6.2; 7% SDS; 1 mM EDTA). The probe was denatured with 0.1 vol 2 N NaOH for 8 min and then neutralized with 0.1 vol 1 M Tris–HCl, pH 7.4. The “prehybridization” solution was then decanted and replaced with 5 ml of fresh hybridization buffer. The probe was added, and hybridization was conducted for 24 hr at 65°. The blots were washed twice with 2× SSC for 30 min at 65°, followed by a high-stringency wash (0.1% SSC and 0.1% SDS) for 30 min at 65°. The membranes were then blotted dry, sandwiched in plastic wrap, and exposed to Storage Phosphor Screens for 3 days at room temperature. The screen was then scanned in a phosphorimager (GE Systems, Sunnyvale, CA).
Sequencing
Plasmids were sequenced using BigDye V3.0 chemistry (Applied Biosystems, Carlsbad, CA) and submitted to the University of Kentucky Advanced Genetic Technologies Center (UK-AGTC) for analysis by capillary electrophoresis. Cosmids were sheared using a Hydroshear (GeneMachines, San Carlos, CA). Fragments were size-selected using agarose gel electrophoresis, purified using the Qiagen gel extraction kit, and ligated to the CloneSmart LCamp vector (Lucigen, Middleton, WI). Colonies were picked to 384-well microtiter plates, grown overnight, and stored at −80°. Prior to freezing, the cultures were replicated into 2 ml of terrific broth in 96-deep-well microtiter plates. After overnight culture in a HiGro (GeneMachines), the cells were pelleted and submitted to the UK-AGTC for template preparation and sequencing using BigDye chemistry.
Sequences were assembled using Phred/Phrap (Ewing and Green 1998; Ewing et al. 1998) and visualized in Consed (Gordon et al. 1998). Repeated sequences were frequently collapsed into a single contig due to sequence identity. In these cases, restriction mapping and comparison of overlapping cosmid clones were used to reconstruct the organization of individual repeats within arrays. Relevant sequences have been deposited in the GenBank database under the accession nos. JQ747487–JQ7747492.
Phylogenetic analysis
Protein sequences were downloaded from the NCBI or translated from DNA sequence where protein sequence was unavailable. The protein (or translated DNA) sequences used in this study are listed in Table S3.
Reverse transcriptase domains:
The putative reverse transcriptase (RT) domains were identified using BLAST alignments, and the protein sequences were then trimmed to remove flanking regions and aligned using Kalign (Lassmann and Sonnhammer 2005). Where necessary, the multiple alignments were manually edited in Jalview version 2.4 (Waterhouse et al. 2009). Phylogenetic analysis using the neighbor-joining method (NJ) was performed with MEGA version 4 (Tamura et al. 2007). Bootstrapping (1000 replications) was used to evaluate the statistical support for the NJ tree. The Poisson correction method (PCM) was used to calculate evolutionary distance. The PCM calculates distance as = −(1 − p), where p is the proportion of sites that differ between two sequences.
Endonuclease domains:
All ambiguous positions were removed for each sequence pair. The evolutionary distances were computed using the Dayhoff matrix-based method (Schwartz and Dayhoff 1979). Analyses were conducted in MEGA5.
Results
Rates of telomere change in M. oryzae isolates from prg and rice
M. oryzae isolates collected from rice fields show much less isolate-to-isolate variation in telomeric restriction fragments than do isolates collected from diseased prg turf (Farman and Kim 2005; Farman 2007). This observation suggested that the telomeres of the prg pathogens are less stable. To test this hypothesis, we monitored changes in telomeric restriction fragment profiles after representative strains had been genetically purified via single-spore isolation. M. oryzae spores contain three haploid nuclei that are derived from a single progenitor nucleus, which means that any telomere rearrangements detected in the cultures must have arisen in the developing spore or subsequent to its isolation. Thus, single-sporing allowed us to compare the rate of telomeric fragment changes directly. Single-spore cultures of LpKY97-1A (from prg) and 70-15 (from rice) each were put through two successive rounds of plant infection on prg cultivar Linn and rice cultivar 51583, respectively. New single spores were recovered from the lesions that formed on the second set of diseased plants, and Southern hybridization analysis was used to compare the telomeric restriction fragment (TRF) profiles of 19 single-spore isolates with that of the original starting culture.
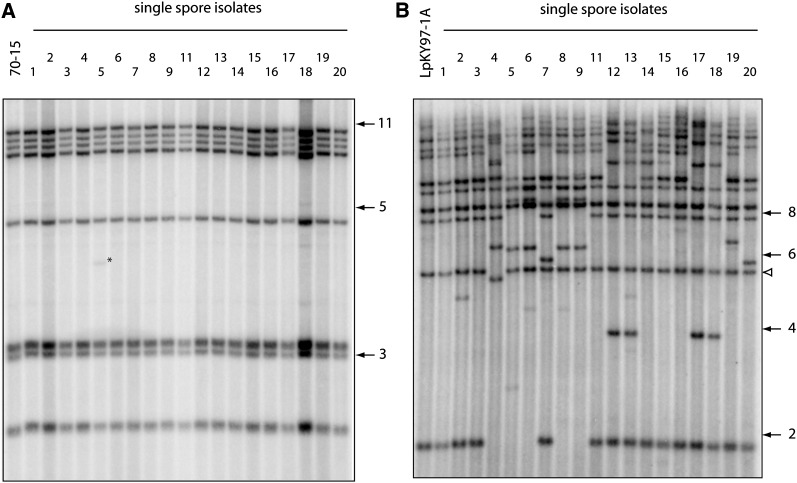
Note that the M. oryzae strains analyzed here have seven chromosomes and, therefore, should yield 14 hybridization signals. However, it is not uncommon for more than one telomeric fragment to migrate. In addition, very large fragments are sometimes sheared during DNA isolation, and smaller fragments sometimes run off the gel. For this reason, Southern blots rarely show a full telomere complement. The Southern hybridization data revealed that the telomeres of 70-15 were highly stable. After two disease cycles, only one single spore isolate (#5) exhibited a change in its telomere profile. This involved the appearance of a single novel band (Table S4), which, judging by its intensity, was not yet fixed in the culture (Figure 1A). By contrast, there were at least 20 independent, novel telomeres among the single-spore progeny from LpKY97-1A. Most of the single-spore cultures from LpKY97-1A exhibited extensive telomeric fragment variation (Figure 1B), with the estimated number of band changes in each isolate ranging from 1 to 8 (Table S4). Ten isolates showed changes in at least five telomeric fragments. Here, it is worth pointing out that the numbers of rearranged telomeres in the cultures derived from LpKY97-1A were underestimated because the use of a probe for an individual chromosome end revealed that some telomeres had been altered but the rearrangements were masked by the coincidental appearance of similarly sized bands (results not shown). Some newly formed telomeric fragments appeared to be present in multiple single-spore isolates. For example, fragments of ∼6.3 and ∼8.5 kb were present in isolates 5, 6, 8, and 9, while isolates 12, 13, 17, and 18 all possessed a novel 3.9-kb fragment (Figure 1B). The isolates within these two groups also had similar telomere profiles to one another. Conversely, a novel fragment of ∼14 kb is shared by isolates that have quite different profiles. Importantly, the observed changes cannot be explained simply by expansion and contraction of telomere repeats because most of the novel telomeric fragments hybridized with a similar intensity to the ones that did not change. A number of new fragments were exceptional in this regard because they showed very faint hybridization signals. These represent novel telomeric fragments that arose within the mycelium as it was being grown for DNA extraction. Interestingly, only three rearrangements were detected among 19 LpKY97-1A spore cultures sampled from the inoculum that was used for the first round of plant infection (results not shown).
Figure 1 .
Changes in telomeric restriction fragments following two cycles of plant infection. Spores were harvested from leaf lesions, and DNA was extracted from single-spore cultures. DNAs were digested with PstI, fractionated by electrophoresis, and electroblotted to membranes. The immobilized DNAs were then hybridized with a 32P-labeled telomere probe and, finally, exposed to phosphorimage screens. (A) Telomere changes in the rice pathogen 70-15. (B) Telomere changes in LpKY97-1A. The numbers on the right of each phosphorimage represent molecular sizes in kilobases. The white arrowhead marks LpKYTEL2, a telomeric fragment that undergoes rearrangement very rarely.
The above host-related difference in telomere stability appears to be a general phenomenon because analysis of single spores from plate-grown cultures of six additional rice-infecting strains revealed only seven telomere changes among 78 spore cultures (0.1 changes/telomere/single spore), while plate cultures of five prg-infecting isolates exhibited 24 rearrangements among 64 spores (0.47 changes/telomere/spore) (Table S5). Not all prg pathogens had unstable telomeres, however, because two isolates each yielded only one novel band among 8 and 10 spore cultures.
Telomere structure in M. oryzae isolates from rice and perennial ryegrass
It seemed likely that the difference in telomere stability between the two host-specific forms of M. oryzae is due to underlying differences in the organization of the telomeres or associated subtelomere regions. The 14 telomeres of strain 70-15 had been sequenced in a prior study (Rehmeyer et al. 2006). In this strain, the chromosome ends consisted of an average of 26 copies of the repeating unit, TTAGGG. Some very short TTAGGG repeats were also found at internal locations, but the nontelomeric sites never had more than two tandem units. To characterize the telomeres in strains from prg, we used a telomere probe to screen chromosome end-enriched, plasmid mini-libraries. This resulted in the recovery of four telomeres from strain LpKY97-1A, five from strain FH, and three from strain CHW. Two of the clones contained telomeres with nothing unusual about them. The insert into one of them, LpKYTEL1, consisted of telomere repeats attached to rDNA sequences (Figure 2A) and, therefore, represents a homolog of telomere 3 (Rehmeyer et al. 2006). The other, CHTEL9, had telomere repeats attached to a sequence that lacked matches to the genome sequence of strain 70-15 and, as such, could not be assigned a specific telomere designation (Figure 2J).
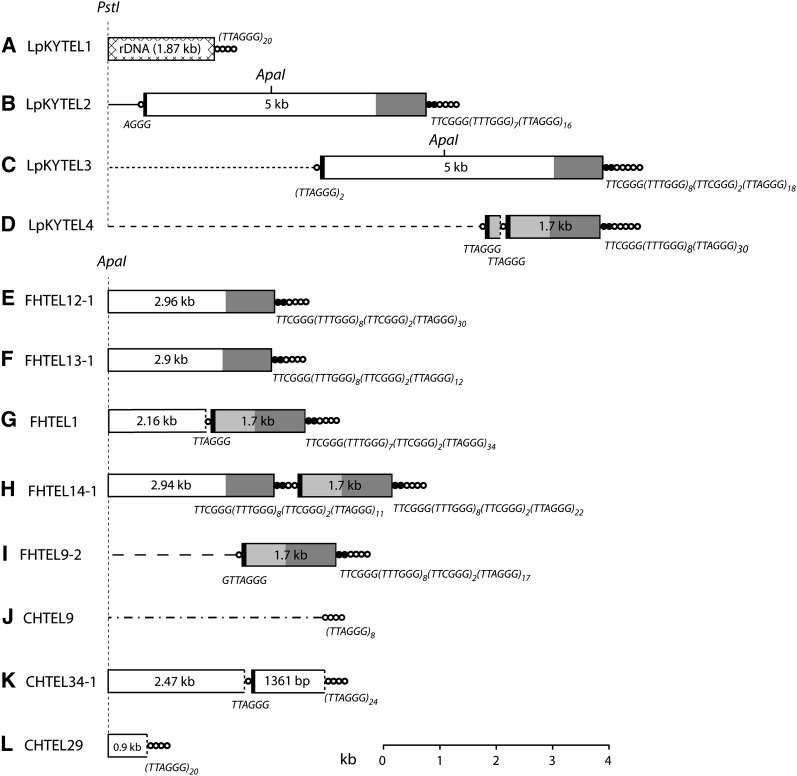
Figure 2 .
Organization of telomeric DNA in three prg-infecting strains of M. oryzae. The three strains analyzed were LpKY97-1A, FH, and CHW. Telomeres were cloned either as PstI to blunt end or ApaI to blunt end restriction fragments, and their organizations were determined by restriction mapping and sequencing. Panels A\x{2013}K show the organization of 11 cloned telomeric fragments. Telomeric sequence ([TTAGGG]n) is represented by concatenated circles and the variant telomere tract (consensus sequence: [TTTGGG]8[TTCGGG]2) is depicted by concatenated, closed circles. Repeats inserted in the telomere array are shown as boxes, and regions within the boxes having the same shading represent sequences that are identical (or nearly identical). The numbers within the boxes represent the length of each element. Truncated repeats are indicated by using dotted lines to mark the truncation boundary. Chromosome unique DNA is illustrated with a line, and the different line styles indicate that they have different sequences. The sequences of short telomere tracts (and telomere-like tracts) at the junctions of adjacent insertions are listed below the relevant junctions. Positions of PstI and ApaI restriction sites are shown. Boxes and lines are drawn to scale with the exception of the circles that represent the telomeres and telomere-like sequences.
Analysis of the remaining clones revealed novel repetitive elements embedded within the telomere repeat tract. Clone LpKYTEL2 contained an ∼5-kb sequence at the proximal end of the telomere tract immediately after a partial telomere repeat (AGGG) (Figure 2B). Interestingly, at the distal end of the inserted DNA there was a variant telomere sequence, TTCGGG(TTTGGG)7, followed by canonical (TTAGGG) telomere repeats that extended out to the boundary of the clone insert, which likely corresponds to the actual chromosome end. The chromosome-unique sequence adjacent to the telomere matched a region near telomere 5 of 70-15, except that the LpKY97-1A chromosome end had an ∼50-kb truncation relative to the 70-15 homolog.
Clone FHTEL14 contained a 1.7-kb sequence arranged in tandem with a copy of the 5-kb element, with the latter sequence terminating at an ApaI site due to the cloning process (Figure 2H). LpKYTEL4 also contained the 1.7-kb sequence in tandem with a drastically truncated version of the 5-kb element, which retained only 262 bp from the proximal end (Figure 2D). Interestingly, the boundaries of the full-length 1.7-kb sequences were identical to those in the 5-kb repeat, with the two elements sharing >800 bp of sequence at their distal termini (including the variant telomere repeat) and 77 bp at their proximal ends. However, the variant repeats were slightly different from the ones in the LpKYTEL2 clone: FHTEL14 contained TTCGGG(TTTGGG)8(TTCGGG)2 while LpKYTEL4 had TTCGGG(TTTGGG)8. The remaining clones contained various combinations of 5- and 1.7-kb elements arranged singly or in tandem and in full-length and truncated forms (Figure 2, C, E–G, I, K, and L). The truncated elements lacked the variant repeats at their distal boundaries. Instead, they were directly capped with a canonical telomere when they were the most distal element in the array (i.e., at the chromosome end). When they were in the middle of an array, they were separated from their distal neighbor by short TTAGGG tracts of varying lengths (Figure 2, K and L).
Comparison of homologous telomeres in M. oryzae strains FH and 70-15
To gain additional insights into the organization of repeats within the Magnaporthe telomeres and to determine if there is additional structural variation in the subterminal regions that could contribute to telomere instability, we screened a cosmid library of FH genomic DNA with probes derived from the 5- and 1.7-kb elements. Restriction mapping and end-sequencing of the clones that were recovered identified four different chromosome ends. A single clone representing each end (two clones for the rDNA end) was sequenced to 5× coverage and primer walking was used to fill key gaps. The organizations of the FH chromosome ends were then compared with their homologous counterparts in the rice pathogen, 70-15.
Telomere 3:
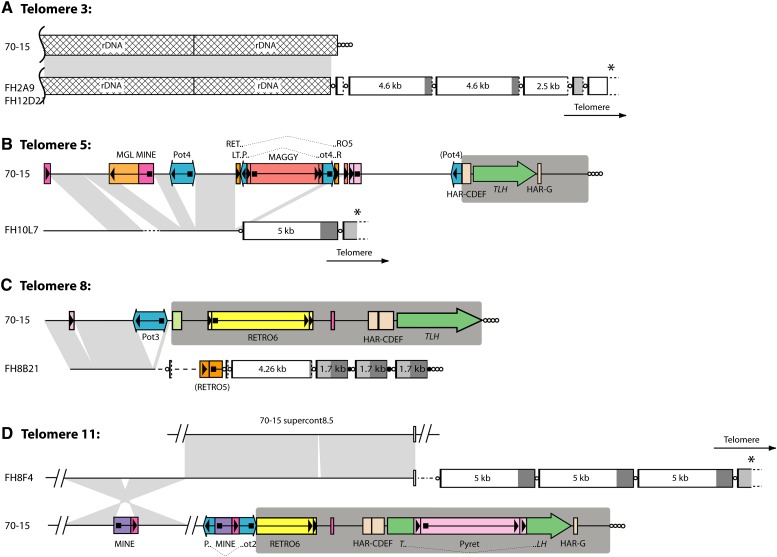
In 70-15, telomere 3 caps the rDNA array and is attached to sequences in the untranscribed spacer region between the 18S and 26S rRNA genes at a position ∼1.25 kb downstream of a 26S rRNA gene copy (Figure 3A). The corresponding telomere of FH also contained rDNA sequences, but the telomere began only 500 nucleotides downstream of one of the 26S rRNA genes. Unlike the “rDNA telomere” in LpKY97-1A, telomere 3 in FH cosmids 2A9 and 12D21 contained four 5′-truncated copies of the 5-kb element (∼0.25, 4.6, 4.6, and 2.5 kb long), a 5′-truncated 1.7-kb element (∼700 bp), and a fifth copy of the 5-kb element that was truncated by the cloning process.
Figure 3 .
Comparison of homologous chromosome ends in strains 70-15 and FH. Panels A\x{2013}D show alignments of four pairs of chromosome ends. Telomere and telomere-like sequences are represented with circles, and the telomere-embedded repeats are depicted as boxes as described in the legend to Figure 2. The TLH regions in the 70-15 subtelomeres are highlighted with dark gray background shading. For cosmid clones that did not contain the terminal telomere repeat array, the direction of the telomere is indicated. Transposon insertions and other repeats in the subtelomere regions are represented by colored boxes with arrows or arrowheads indicating their orientations. Features that are described in the text are labeled. Regions of alignment between the 70-15 chromosome and the FH homolog are connected by light gray shading. Sequences linked to telomere 11 of FH align with two sequences that are well separated in the 70-15 genome, and so two alignments are shown. Sequences that were present near the FH telomeres but absent from the 70-15 genome are drawn as dotted lines, with different line styles representing different sequences. The star-shaped area of shading between telomere 11 homologs represents an inversion that contains a transposon insertion, which in turn caused a deletion in the recipient chromosome. All features are drawn to scale with the exception of the circles that represent the telomeres and telomere-like sequences.
Telomere 5:
This telomere had already been captured in the LpKYTEL2 clone described above. However, while strain LpKY97-1A was found to be missing ∼50 kb of subterminal sequence present in 70-15, the FH “allele” contained in cosmid FH10L7 was missing only ∼17.5 kb. Even so, this deletion encompassed the entire subtelomere region present in 70-15, as well as ∼9 kb of proximal sequence (Figure 3B). Alignment of the homologous regions showed that the FH chromosome end lacks six transposon insertions that are present in 70-15. Conversely, it contains a short stretch of sequence that was missing in 70-15 (dotted line in Figure 3B). The FH telomere contains a full-length copy of the 5-kb element embedded in the telomere repeats followed by a copy of the 1.7-kb repeat which, again, was truncated by the cloning process.
Telomere 8:
Cosmid clone FH8B21 contains sequence that aligns uniquely with the chromosome-unique region located near telomere 8 of 70-15. Comparison of the homologous ends revealed that FH again lacks transposon copies that were present in the 70-15 chromosome (Figure 3C). The region of homology ended just 209 bp proximal to the subtelomere border in 70-15. Beyond that divergence point, the FH chromosome contained ∼2.6 kb of sequence that is not present in the 70-15 genome, followed by a short stretch of sequence from the RETRO5 retrotransposon (Farman 2002; Dean et al. 2005). The FH telomere contained a truncated 5-kb element and three tandem copies of the 1.7-kb repeat with a short TTAGGG tract separating each element (Figure 3C). The most distal 1.7-kb repeat was followed by 24 copies of the canonical telomere repeat sequence, which was adjoined to the cloning site in the cosmid vector. On the basis of their terminal position within the insert DNA, these repeats likely correspond to the actual chromosome end.
Telomere 11:
Cosmid FH8F4 contains sequence that is homologous to terminal sequence near 70-15 telomere 11 (Figure 3D). Within the region of alignment, the 70-15 chromosome contains a MINE retrotransposon (Fudal et al. 2005) that is not present in the FH homolog. The alignment between FH8F4 and the telomere 11-associated sequence ended abruptly when the FH sequence transitioned to match a 70-15 genomic sequence that is not associated with a telomere in 70-15. This similarity stopped at a short repetitive sequence beyond which the FH chromosome contains a sequence that is not present at all in the 70-15 genome. The telomere was adjoined to this novel sequence and contained three variant copies of the 5-kb element. The most distal copy was joined to a truncated 1.7-kb element which, again, was truncated by the cloning process. Interestingly, this chromosome end was homologous to the one in LpKYTEL4 that contained the 5-kb element and a truncated copy in tandem. However, in LpKY97-1A, the telomere contained just one intact copy of the 5-kb element (see Figure 2D).
One of the repeats within the Magnaporthe telomeres is a non-LTR retrotransposon; the other is a derivative element
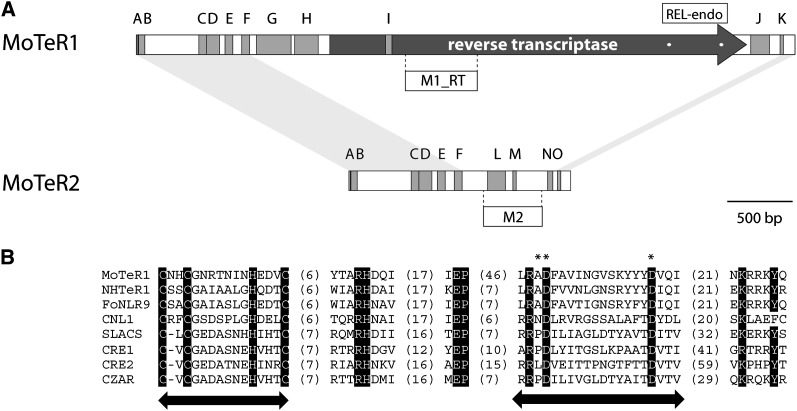
Translation of the nucleotide sequence of the 5-kb element revealed a large open reading frame (ORF) that is predicted to code for a 1070-amino-acid protein with a RT domain (Pfam ID: PF00078.18) (Figure 4A). A BLAST search of the GenBank database produced top matches to hypothetical proteins in the fungi Nectria hematococca, Fusarium oxysporum, and Cryptococcus neoformans. Further investigation revealed that the Nectria proteins also are encoded by repeated elements embedded in telomeres; the Fusarium proteins are encoded by the FoNLR9 non-LTR retroelement previously described by Novikova and et al. (2009); and the Cryptococcus proteins are encoded by the telomere-associated non-LTR retroelement Cnl1 (Goodwin and Poulter 2001). There was also significant similarity to reverse transcriptases encoded by the SLACS non-LTR retrotransposons found in Trypanosoma brucei gambiense (Aksoy et al. 1990) and Leishmania braziliensis (Peacock et al. 2007) and to the CRE1 and CRE2 retrotransposons in Crithidia fasciculate (Gabriel et al. 1990; Teng et al. 1995). On the basis of these findings, we conclude that the elements in the Magnaporthe telomeres are non-LTR retrotransposons, or MoTeRs. The 5-kb element is designated MoTeR1 and the 1.7-kb element, MoTeR2. Likewise, we have named the related Nectria element, NhTeR1.
Figure 4 .
Organization of MoTER1 and MoTER2. (A) Overall structures showing positions of relevant features. Both elements are drawn to scale. Coding regions are depicted by a dark-gray arrow. Blocks of tandem repeats are shown as medium-gray boxes. The sequences and copy numbers of the individual repeat units are listed in Table S6. Light-gray shading connecting the termini of MoTER1 and MoTER2 shows regions of significant sequence identity between the two elements. The positions of the REL-endo domains and the probes used in this study are shown. (B) Alignment of the MoTeR1 REL-endo domain with those found in other retrotransposons. The names of the retroelements are listed on the left. Arrows indicate the positions of the CCHC and REL domains. Asterisks indicate the characteristic AD..D residues. Values in parentheses indicate the numbers of amino acid residues between the highly conserved motifs.
Identification of the reverse transcriptase gene in MoTeR1 established that the 5′ terminus of the element is the one that occurs proximal to the chromosome tip and is the end that contains the variant telomere repeat. It is also the end that is missing from truncated elements. The terminus of MoTeR2 is identical to that of MoTeR1, with 860 bp from the 5′ end and 77 bp from the 3′ end being common to the two elements (Figure 4A). MoTeR2 does not encode its own reverse transcriptase but contains a single ORF that codes for a predicted protein of 280 amino acids. This protein lacked similarity to sequences in the databases. The orientation of MoTeR2 can be inferred through its alignment with MoTeR1. It, too, was invariably inserted with its 5′ terminus pointed toward the chromosome tips.
Both MoTeR1 and MoTeR2 contain numerous short tandem repeat (STR) motifs, mostly concentrated in their 5′ ends, upstream of where the RT ORF starts in MoTeR1. Eleven different repeating units were identified in MoTeR1 and 9 in MoTeR2. Six of the STRs are present in both elements and reside in the region of 5′ shared sequence (Figure 4A). The sequences and locations of the individual repeat units are listed in Table S6.
The SLACS and CRE elements have strict insertion-site preferences, with both elements transposing into specific nucleotide positions in spliced leader sequence genes (Aksoy et al. 1990; Gabriel et al. 1990; Teng et al. 1995). Integration involves an endonuclease activity possessed by the RT protein (Yang et al. 1999; McClure et al. 2002). Motivated by the close similarity to the SLACS and CRE RT proteins, and the apparent site specificity of the MoTeR elements, we searched for a putative endonuclease domain in the MoTeR1 RT protein sequence. The C-terminal regions of the RT proteins of SLACS, CRE1, and CRE2, as well as other elements, have restriction endonuclease-like “PD-(D/E)” domains, with the consensus organization C[X1-3]C[X7-8]H[X3-4]C[X9-10]RHD/N..X19-33..E..X9-21..R/KPD..X12-14..D/E) (Yang et al. 1999). A good match to this motif was identified in the C-terminal region of the predicted MoTeR1 RT, which has the sequence C[X2]C[X6]H[X3]C[X9]RHD..X20..E..X49..RAD..X12..E (Figure 4B). Similar motifs were found in the proteins encoded by the Nectria, Cryptococcus, and Fusarium elements.
Multiple alignments of four full-length copies of MoTeR1 found in the genome of strain FH revealed that the copies present in cosmid FH8F4 had significance sequence differences relative to the others (Figure S1). In particular, 77 bp from positions 874 to 950 in the consensus element were replaced by an unrelated 38-bp sequence. In addition, the “H” tandem repeat array was one repeat unit shorter than the consensus. The sequence replacement was also present in the MoTeR in clone FHTEL13-1 but the “H” array in the latter element was full length (results not shown). In addition to the major sequence differences, a few single nucleotide polymorphisms (SNPs) and short indels were also detected. Alignment of sequences from four MoTeR2 copies revealed only SNPs and short indels (Figure S2).
Phylogenetic analysis of a representative set of reverse transcriptase protein sequences placed the MoTeR RT in a group that contains proteins encoded by NhTeR1, Cnl1, FoNLR9, SLACS, and CRE1/2 (Figure S3A). These elements were in a sister group to Giardia Genie (GilM and GilT) elements, which insert into a telomere-associated tandem repeat but are not telomeric per se (Arkhipova and Morrison 2001; Burke et al. 2002). The MoTeR RT was only distantly related to other retrotransposons that insert specifically into (or onto) telomeres (Figure S3A, telomeric elements highlighted in dark gray). The MoTeRs were more distantly related to other retrotransposons that have been identified in Magnaporthe (light gray shading). A phylogenetic analysis based on the restriction-enzyme like endonuclease (REL-endo) domains in reverse transcriptases from select site-specific elements produced the same tree architecture as that obtained with the RT sequences (Figure S3B).
Analysis of MoTeR 5′ and 3′ insertion junctions
Non-LTR retrotransposons often produce target-site duplications (TSDs) upon integration. To determine if there are TSDs flanking the MoTeR insertions, we examined in more detail a number of 5′ and 3′ insertion junctions for several full-length and truncated MoTeRs, some of which were inserted at distal positions and others of which were embedded in the middle of arrays. As shown in Figure S4, regardless of whether the elements were full length, the 3′ insertion junctions always occurred in the same register within the telomere repeats, with the three cytosines of the telomere repeat adjoining the four adenines at the 3′ end of the MoTeRs (AAAA.CCCTAA or AAGA.CCCTAA in the case of insertion #2 in LpKYTEL4; Figure S4D). For full-length elements containing at least a portion of the variant telomere motif, the variant repeats were always inserted in precise register with the telomere (i.e., CCCTAA.CCCGAA or CCCTAA.CCCAAA). In contrast, 5′-truncated elements were found in several different registers within the telomere repeats. In most cases there was a stretch of microhomology between the boundary of the truncated element and the flanking telomere sequence, which precluded precise determination of the insertion site. However, analysis of several truncated MoTeR 5′ junctions that lacked homology revealed a significant predominance of the CCCTAA, truncated MoTeR register (Table S7).
We also amplified several individual (unpaired) MoTeR-MoTeR junctions from a number of different strains. This revealed significant variation in the number of telomere repeats separating adjacent elements, ranging from half a repeat to 28. In addition, some junctions appeared to contain more than one variant repeat array. For example, LpKY97-1A exhibited a number of junctions with noncontiguous CCCAAA motifs (Table S8).
Because the variant repeats at the MoTeR 5′ ends were in the same register as the telomere repeats, full-length MoTeR insertions automatically exhibited what appear to be TSDs with the sequence CCCTAA (see Figure S4). However, because the telomere is repetitive to start off with, it is not possible to say if these are true TSDs resulting from staggered cleavage of the target site. Even if they are TSDs, their sequences and lengths are unclear because, in most instances, the ends of the MoTeRs and the start of the telomere could not be precisely defined. Exceptions to this rule were the 5′ truncated MoTeR (tMoTeR) junctions generated via nonhomologous end joining (Table S7). Surprisingly, the majority of these junctions also exhibited a structure consistent with TSD (i.e., CCCTAA-tMoTeR-CCCTAA), although there were a number of other junctions with alternative registers.
Genomic distribution of MoTeR elements
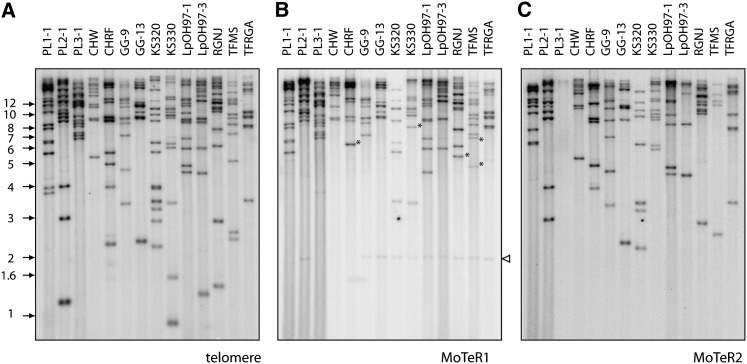
To determine if the MoTeRs also occur at internal genomic locations, DNA samples from several M. oryzae isolates from prg were digested with PstI, which lacks sites within either MoTeR1 or MoTeR2. The digested DNAs were fractionated by electrophoresis and electroblotted to nylon membranes. The blots were hybridized sequentially with probes for the telomere repeat, MoTeR1, and MoTeR2. Comparison of the hybridization patterns for the three probes revealed that MoTeR1 and MoTeR2 sequences usually occurred on fragments that hybridized with the telomere probe (Figure 5). There were only five obvious exceptions where fragments that hybridized strongly to the MoTeR1 probe did not hybridize to the telomere probe. These were in isolates CHRF, KS330, RGNJ, and TFMS and are highlighted with asterisks in Figure 5. Cloning of the exceptional fragment from RGNJ revealed an insertion of the MGLR retrotransposon in MoTeR1. This insertion introduced a PstI site distal to the probe’s target sequence, thereby obscuring a probable linkage between the MoTeR and the telomere. Another faint, nontelomeric, MoTeR1-hybridizing fragment was observed in several isolates (indicated with a white arrowhead in Figure 5B). Cloning of this fragment revealed that it corresponds to a highly mutated, internal fragment of MoTeR1 situated at a nontelomeric location. Neither the MoTeR1 nor the MoTeR2 probe hybridized to 70-15 DNA (results not shown).
Figure 5 .
Genomic distribution of MoTER elements. PstI-digested DNA samples (∼200 ng/lane) were fractionated by electrophoresis and electroblotted to membranes. The immobilized DNAs were then hybridized with a MoTER1 probe (A). After exposure to a phosphorimage screen, the probe was stripped off and the DNAs were reprobed with a MoTER2-specific fragment (B). Finally, the DNA samples were hybridized with the telomere probe (C). Asterisks highlight MoTeR-containing fragments that did not hybridize to the telomere probe. Isolate names are listed above the relevant lanes, and molecular sizes are shown on the left. The white arrowhead marks a highly degenerate internal MoTeR sequence.
Several of the previously described telomere clones revealed the presence of both MoTeR1 and MoTeR2 inserted in a single telomere. The hybridization data showed that this is a common occurrence, as evidenced by many cases in which both MoTeRs hybridized to a single telomeric fragment (Figure 5). There were also numerous examples of telomeric fragments that contained only MoTeR1 or MoTeR2 in single or multiple copies. Finally, there were a number of telomeres that appeared to lack MoTeR elements altogether (Table S9), although we cannot rule out the possibility that some of these telomeres contain MoTeR arrays with other (PstI site-containing) transposons embedded within them.
The copy numbers of the MoTeR elements in the various prg pathogen genomes were estimated by counting the fragments that hybridized to each probe, taking into account the relative hybridization intensities of each fragment. We also used densitometric scanning of the Southern blot lanes to provide an independent assessment. The two sets of estimates are presented in Table S10. On the basis of densitometric scanning, the copy number of MoTeR1 ranged from ∼10 to 27 with an average of 15.5, while MoTeR2 copy number ranged from 0 to ∼23 (average = 11.3). Similar values were obtained by counting bands, although in general this method tended to underestimate the values due to the conservative interpretation of intense hybridization signals. Table S10 also provides estimates of the number of telomeric fragments. In most cases, our estimates were close to the expected number of 14.
Analysis of the cosmid sequences for FH revealed one severely truncated MoTeR copy at an internal location ∼2 kb from the telomere. This vestige was truncated within the T-rich repeat and consisted of only 129 bp from the 3′ terminus. Despite its internal disposition, this vestigial copy still had a short telomere tract at the 3′ insertion boundary. Surprisingly, analysis of the 70-15 genome sequence also revealed four greatly truncated MoTeR copies at internal locations. Two copies were closely linked to one another and were positioned ∼35 kb away from telomere 1, while the others were ∼100 kb and 3 Mb away from telomeres 12 and 2, respectively.
Mitotic instability is restricted to telomeres that contain MoTeR insertions
The MoTeR insertions represent a major structural difference between the telomeres of LpKY97-1A and those of 70-15, which led us to hypothesize that they were the underlying cause of the observed telomere instability. However, an alternative possibility is that instability is due to an unrelated factor in the prg pathogens. For example, they might possess an inefficient telomerase gene or lack a key telosome protein, either of which could result in the telomeres experiencing frequent rearrangements. To distinguish between these two scenarios, we crossed the MoTeR-containing, “unstable telomere” strain FH (a prg pathogen) with 2539, a strain whose telomeres largely lack MoTeRs and are comparatively stable. Thus, if FH contains a major factor conferring telomere instability, this trait should segregate in a simple Mendelian fashion.
Progeny were collected and genetically purified by single-spore isolation, and each isolate was then used to initiate a series of subcultures that were continued either for 10 generations on small (60-mm) petri plates or for 5 generations on standard ones (85 mm). DNA was extracted from the original (G0) isolates, as well as from the 5th (G5) or 10th generation (G10) cultures, and instability was assessed by comparing the telomere profiles of the G5/G10 DNA samples with those of the starting cultures (G0), as well as with the original parental isolates.
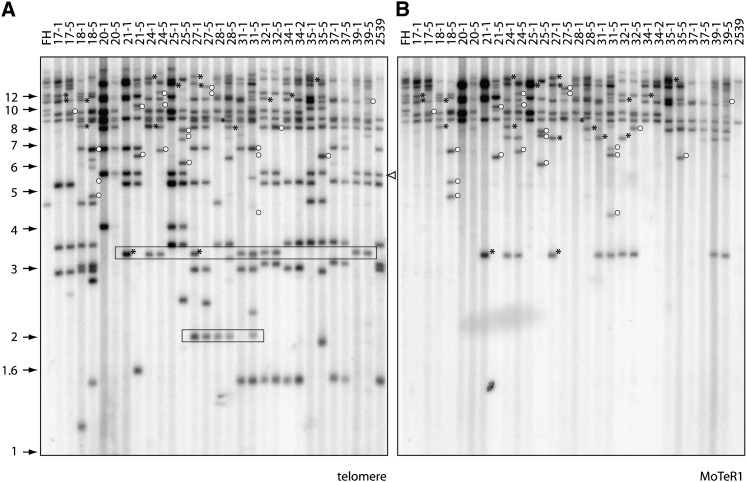
M. oryzae is a haploid fungus, and, therefore, if telomere instability is due to a recessive mutation in a single gene, one would expect to see the trait segregate 1:1 among the progeny. In actuality, numerous telomere changes were observed in all 29 of the progeny isolates examined (Figure 6A). This effectively ruled out the possibility that instability is due to a simple Mendelian factor (or indeed multiple recessive genes). Instead, the data were consistent with the presence of multiple dominant factors, each of which functions independently of the others. Significantly, Southern hybridization showed that all of the progeny inherited several copies of MoTeR1 (Figure 6B) and MoTeR2 (data not shown), a finding consistent with the notion that the MoTeR elements are responsible for the observed instability.
Figure 6 .
Segregation of telomere instability in a genetic cross. DNA was extracted from single-spore cultures of each progeny isolate (G1). These isolates were subcultured for four additional generations, new single-spore cultures (G5) were established, and DNA was extracted from the resulting cultures. The genomic DNA samples were digested with PstI, fractionated by electrophoresis, and electroblotted to a membrane. The membrane was probed sequentially with 32P-labeled telomere and MoTeR1 fragments. The left and right lanes contain the DNAs from the parental strains. DNA samples from the G1 and G5 subcultures of an individual progeny isolate were loaded in adjacent lanes to allow comparison of their telomeric restriction fragment profiles. Molecular sizes are shown on the left. The white arrowhead marks 2539TEL10, a MoTeR2-containing telomere that was highly stable. (A) Phosphorimage of the TTAGGG-probed gel. Boxes outline fragments that segregated among the progeny but that were not present in either parent. (B) The same membrane used in A was stripped and reprobed with a fragment from the MoTeR1 RT. Asterisks mark hybridizing fragments that were present in the G0 culture but absent in the G5. Open circles show fragments that appeared in the G5 cultures.
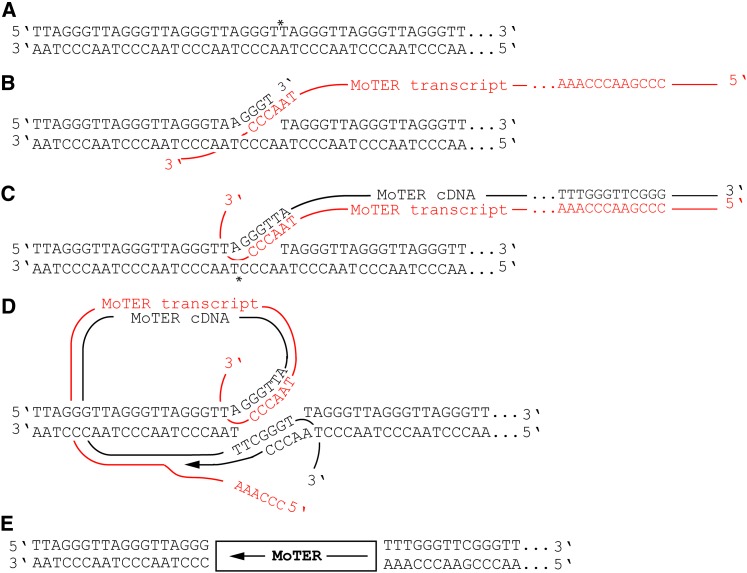
Figure 7 .
Proposed transposition mechanism for the MoTeR elements. (A) Nicking of the telomere repeat top strand by the MoTeR RT. The nick site is identified with an asterisk (note that the specific nick position is not known). (B) Annealing between the 3′ end on the top strand of the nicked telomeric DNA and the CCCTAA sequences predicted to occur at, or near, the 3′ end of a MoTeR transcript. (C) First-strand synthesis of MoTeR cDNA and nicking of the bottom telomeric DNA strand. The model shows a 3′ overhang with four nucleotides, but the length and type of overhang may be different (note that a 5′ overhang would produce a target-site deletion, as opposed to a duplication). (D) Annealing between the 3′ end released by nicking the bottom strand and the 3′ end of the newly synthesized MoTeR cDNA, followed by second-strand synthesis with concomitant RNA strand displacement. (E) Ligation of remaining nicks would result in a MoTeR element inserted precisely in the telomere repeats.
Further evidence in support of this hypothesis came from the analysis of instability at individual telomeres: Eleven of the 14 telomeres in 2539 lack MoTeR insertions. Owing to good electrophoretic resolution, it was straightforward to monitor the segregation of these MoTeR-less telomeres. Ten were perfectly stable during extended vegetative growth of the progeny isolates that had inherited them; i.e., if the telomere was visible in the G0 culture of a given progeny isolate, it was also present in the G5/G10 culture (Figure 6A). In total, we monitored stability at 136 telomeric loci lacking MoTeR insertions and only 2 showed evidence of rearrangement (Figure 6A). In both cases, it was 2539 Tel-3 (the rDNA telomere) that was affected.
It was not possible to score the segregation and stability for most of the telomeres inherited from FH or for the MoTeR-containing telomeres from 2539. First, their large size prevented sufficient resolution of the different telomeric fragments. More importantly, however, so many of the novel telomere fragments were large that, in most cases, it was not possible to tell if a progeny had inherited a particular telomere from FH or if it contained a newly rearranged telomere of a similar size. Therefore, to monitor stability of MoTeR-containing telomeres, we rehybridized the Southern blot shown in Figure 6A with MoTeR1 and MoTeR2 probes and looked for individual MoTeR-containing telomeres (either inherited from FH or newly formed) that were present in the G0 culture but missing in the G5/10. This identified 155 score-able telomeric fragments containing MoTeR insertions, 41 (over one quarter) of which were rearranged during subculture (representative results for MoTeR1 are shown in Figure 6B). It is important to note that all of these rearrangements were independent events.
Not all of the MoTeR-containing telomeres were unstable, however. For example, 2539 Tel-10 contains a tandem array of two MoTeR2 insertions, but no rearrangements of this telomere were detected among the progeny isolates (arrowed in Figure 6A). We were unable to monitor changes at the two other MoTeR-containing telomeres contributed by 2539 (2539 Tel-13 and 2539 Tel-14) because similarly sized telomeric fragments inherited from FH obscured them. However, analysis of just six progeny from a different cross (2539 × Guy11, a strain that lacks telomeric MoTeRs altogether) revealed mitotic rearrangements in both of these telomeres, with frequencies of 0.25 (one of four progeny that inherited 2539 Tel-13) and 0.6 (three of five with 2539 Tel-14). As before, the non-MoTeR telomeres and 2539 Tel-10 were mitotically stable in the progeny from the 2539 × Guy11 cross (results not shown).
Discussion
DNA fingerprinting studies have shown that M. oryzae isolates from prg exhibit extreme variation in their telomere profiles, even when there is little or no polymorphism at internal DNA loci (Farman and Kim 2005). In contrast, rice-infecting isolates that have similar “internal DNA” profiles to one another tend also to have highly similar telomere banding patterns (Farman 2007). In the present study, we found that the telomeres of a number of prg pathogens underwent frequent, spontaneous rearrangement during growth in planta, while those from rice-infecting strains were very stable by comparison. Thus, we conclude that the differences in telomere variation among isolates within the two host-specialized populations is a reflection of inherent differences in telomere stability. In isolate LpKY97-1A, the rate of telomeric fragment change was quite extraordinary. After only two disease cycles, some single spores experienced rearrangement of at least half of the parental telomere fragments. Therefore, it is reasonable to suppose that as few as four or five disease cycles could be sufficient for all traces of a parental telomere profile to be erased. A paucity of rearrangements among spores in the initial inoculum suggested that most changes occurred during growth in planta. This was surprising because the fungal growth within the plant was very limited (average colony diameter: <0.5 cm) compared with the growth on the initial plate (colony diameter: 8.5 cm). Consequently, the fungus ought to have undergone more nuclear divisions prior to inoculum production than it would have done in the plant. This suggests that telomere instability is promoted either by growth in planta or by the process of sporulation.
In several instances, specific newly formed telomere fragments were present in more than one single-spore isolate. Although we cannot rule out the possibility that these were identical, independent rearrangements, the general similarity in telomere profiles indicates that these spores inherited telomere variants descending from events that occurred quite early in the experiment, possibly in sectors of the colony used to produce the spores for infection cycle 1.
Only certain telomeres in LpKY97-1A were hypervariable; others appeared to be quite resistant to change. LpKY97TEL-2 is an example of a stable telomere, with the rearrangement in single spore isolate #4 (Figure 1B) being one of only two such alterations detected in >150 single spores analyzed to date. Recalcitrance to change does not appear to persist over the long term, however, because no telomere restriction fragments appeared to be even remotely conserved among field isolates (Farman and Kim 2005). Indeed, as reported above, the LpKY97-1A and FH clones that contain this telomere (LpKY97TEL-2 and cosmid FH10L7) were polymorphic.
A previous sequencing study showed that the telomeres of 70-15 consist of an average of 26 TTAGGG repeats and are devoid of MoTeR sequences. Sequences of telomeres from other rice pathogens (Gao et al. 2002; M. L. Farman, unpublished data) as well as Southern hybridization data indicate that this telomere organization is typical of the rice-infecting population as a whole. Strain 70-15 did, however, have four highly truncated MoTeR vestiges in its genome, and the presence of short TTAGGG motifs at the 3′ insertion junctions of these relics suggests that they are inserted in former telomeres that have since become internalized.
All of the prg-infecting isolates that we have analyzed to date possess MoTeRs in their telomeres (Figure 6; J. H. Starnes, O. S. Novikova, and M. L. Farman, unpublished data), and, although Southern hybridization revealed very few nontelomeric copies, genome sequencing revealed numerous extremely short 3′ terminus relics at internal locations. It seems likely that the MoTeRs are responsible for the observed instability because sequencing of several kilobases of subterminal DNA in strain FH revealed no other features that would be expected to promote rearrangements. In fact, with the exception of the rDNA units and the MoTeRs, the subterminal regions of the FH chromosomes appear to be largely devoid of repeated sequences (Figure 3). The most compelling evidence for MoTeR involvement in instability came from a genetic analysis, which gave no indication that other loci were at play. Moreover, when MoTeR-containing and MoTeR-less telomeres were recombined into the same genetic background, only the telomeres with MoTeR insertions were rearranged during subculture. The rDNA telomere from 2539 was a notable exception. However, telomeres linked to the rDNA array have proven to be highly unstable even when they lack MoTeR insertions (as an example, see the bottom band in Figure 1B), so it was not surprising to detect rearrangements at this particular chromosome end. The lack of changes in the majority of MoTeR-less telomeres was significant from another perspective because it indicated that de novo transposition events are not very frequent, at least onto telomeres that lack MoTeR insertions. Nevertheless, we have identified de novo MoTeR insertions in telomeres that formerly lacked them (“naked” telomeres), which indicates that naked telomeres are receptive to transposition and, furthermore, that the MoTeRs are mobile (O. S. Novikova, J. H. Starnes, and M. L. Farman, unpublished results).
Interestingly, not all MoTeR-containing telomeres underwent rearrangement. As discussed above, the highly stable telomere represented by clone LpKYTEL-2 contains a single MoTeR1 insertion. Likewise, 2539TEL-10 contains two MoTeR2s in tandem, yet it also remained unaltered during vegetative growth. A single truncation of telomere 10 was observed among 26 progeny from the 2539 × Guy11 cross although, in this case, it is possible that the rearrangement occurred during meiosis. MoTeR2 insertions do, nevertheless, promote mitotic rearrangements, as evidenced by a highly unstable telomere in LpKY97-1A that contains only tandem insertions of MoTeR2 (O. S. Novikova, J. H. Starnes, and M. L. Farman, unpublished results). We speculate that the mutability of individual MoTeR arrays is determined by the specific length of the internal telomere tract that is created when a MoTeR inserts, with short arrays being recalcitrant to change. This could explain why some of the MoTeR-containing strains show infrequent telomere rearrangement (Table S4).
Aside from the transposition events, it is not yet known by what mechanism the MoTeRs promote rearrangements, although it seems likely that the initiating event is some kind of double-strand break. Possibly, the tracts of interstitial telomere sequence created when MoTeRs insert cause replication fork stalling during replication, resulting in subsequent strand breakage, as has been shown for interstitial telomeres in yeast (Ivessa et al. 2002). An even more attractive idea, however, is that overexpression or hyperactivity of the MoTeR RT protein leads to indiscriminate scission of telomeric DNA without attendant reverse transcription. Finally, perhaps the MoTeR sequences themselves—specifically the variant telomeres or the extensive internal homopolymeric tracts—promote breakage. Repair of exposed ends could then proceed by de novo telomere formation, by break-induced replication using MoTeR arrays or internal sequences as templates, or by nonhomologous end-joining.
MoTeRs are site-specific retrotransposons that target telomere repeats
Here we show that the vast majority of intact MoTeRs are located on telomeric restriction fragments, and characterization of one exceptional fragment suggested that it most likely was telomeric, too. Also, every insertion-site junction characterized to date contained short tracts of telomere repeat sequence immediately flanking the MoTeR border. There are two possible reasons for such telomere specificity. One is that the MoTeRs have the same transposition strategy as the TRAS and SART elements in Bombyx mori that insert into telomere repeats following target-site cleavage by element-encoded endonucleases (Okazaki et al. 1995; Takahashi et al. 1997). Alternatively, the MoTeRs may add on to chromosome ends when the terminal TTAGGG tract is compromised, a strategy that would be functionally similar to that of the HeT-A and TART transposons in Drosophila and to what has been proposed for the Penelope-like elements found in a number of organisms (Gladyshev and Arkhipova 2007). Current data favor the operation of a TRAS/SART-like insertion mechanism. First, the organization of MoTeR arrays is consistent with repeated insertions into telomere repeats and maintenance of an extended terminal telomere tract. If, instead, the elements were to insert onto compromised chromosome ends, the observed structural organization would necessitate that the terminal tract is first degraded and then replenished after each MoTeR insertion. Although we cannot rule out such occurrences, this is a less economical explanation of the data. A second reason to suspect insertion into telomeres is that the predicted MoTeR1 RT protein shares amino acid residues found in the REL-endo domains of RTs encoded by site-specific retroelements that recognize and cleave their genomic targets. The MoTeRs encode proteins with a slight variant of the PD-(D/E) motif, namely AD-D, which happens to be present in the HincII restriction enzyme (Menon et al. 2010) and several newly identified type II endonucleases (Kosinski et al. 2005). The presence of a REL-endo domain suggests that MoTeR insertion also initiates through the recognition and cleavage of a specific sequence—in this case, telomere repeats. Here, it is important to note that, while the MoTeR features suggest that they primarily cleave DNA before inserting, we cannot exclude the possibility that they are capable also of adding on to compromised chromosome ends, as has been shown for endonuclease-deficient LINE-1 elements (Morrish et al. 2007).
MoTeR1 appears to code for everything that would be required for transposition and is, therefore, likely an autonomous element. On the other hand, MoTeR2 apparently was derived from MoTeR1 through the substitution of internal sequences, which resulted in the loss of the RT gene. Consequently, MoTeR2 is unlikely to be capable of transposition by itself and probably requires the RT protein supplied by MoTeR1. The conservation of sequence at the 5′ and 3′ ends of the two MoTeR elements is intriguing and suggests that these regions are important for transposition. Most likely, the short stretch of sequence that is common to the 3′ ends is important for binding to the RT protein. In this regard, MoTeR2 resembles SINE elements. The similarity stops there, however, because MoTeR2 lacks other features characteristic of SINEs, namely poly(A) tails and putative RNA polymerase III binding sites (results not shown).
Both elements contain numerous blocks of tandem and interspersed repeats. In fact, such sequences compose at least 50% of MoTeR2. Interestingly, two major blocks of tandem repeats end just upstream of the predicted start site of the reverse transcriptase gene in MoTeR1 and the start of the predicted protein encoded by MoTeR2. This organization is strikingly reminiscent of the promoter regions of telomere-linked helicase genes present in the subtelomere regions of rice pathogenic isolates of M. oryzae (Rehmeyer et al. 2006) and in the fungus Aspergillus nidulans (Clutterbuck and Farman 2007). Possibly, tandem repeats are important for the expression or regulation of telomere-linked genes.
MoTeR elements are exquisitely adapted for insertion into telomeres
Many non-LTR retrotransposons exhibit variable-length poly(A) tracts at their 3′ insertion junctions (Biessmann et al. 1992; Higashiyama et al. 1997; Moran and Gilbert 2002; Fujiwara et al. 2005), which indicates that the elements are mobilized via a polyadenylated RNA intermediate and, furthermore, that reverse transcription initiates within the poly(A) sequence (Luan et al. 1993; Symer et al. 2002). MoTeR elements represent examples of non-LTR retrotransposons that lack poly(A) sequences at their insertion junctions. Although MoTeR insertions have four adenine residues at their 3′ borders (5′..CGCGAATTAAAA 3′), which could be construed as a poly(A) tail, the strict conservation of the length and position of this sequence argues against post-transcriptional addition and is more consistent with its presence in the primary transcript. A similar situation exists with the R2 elements in B. mori. R2 insertion boundaries also consist of four adenine residues joined directly to the 28S rRNA target sequence (Luan et al. 1993). Originally, the A4 motif was thought be derived from a poly(A) tail (Luan and Eickbush 1995). However, subsequent in vitro transposition studies showed that transcripts extending into the downstream 28S rRNA sequences faithfully recreated the insertion junctions seen in vivo (Luan and Eickbush 1996), while transcripts ending with the four adenines did not (Luan and Eickbush 1995). Accordingly, it was proposed that R2 utilizes read-through transcripts for transposition in vivo (Luan and Eickbush 1996). Considering the structural similarity between the R2 and MoTeR 3′ junctions and the fact that intact MoTeRs always insert in precise register with the telomere repeats, it seems likely that MoTeR transposition also utilizes read-through transcripts that would allow accurate positioning at the target site (see Figure 7). As such, the MoTeRs appear to be better adapted for telomeric insertion than TRAS or SART whose poly(A) tails have essentially no complementarity with the Bombyx telomere sequence.
The most striking feature of the MoTeRs, however, is the presence of variant telomere repeats at their 5′ ends. We propose that these serve a number of functions related to the MoTeRs’ telomeric existence. First, because the variant repeats always occur in precise register with the flanking telomere sequences, it appears that they could be involved in the process of 5′ end insertion via homology-mediated end-joining (see Figure 7). Presumably, 3′ end insertion is initiated by a nick (Figure 7A), and annealing of the MoTeR transcript to the free 3′ end would allow DNA synthesis to proceed via target-primed reverse transcription (Figure 7B). Once reverse transcription proceeds through the repeats, the newly synthesized DNA strand would have near its 3′ end a region of homology to the bottom strand of the target site (Figure 7C). Annealing of homologous sequences would allow the second MoTeR strand to be completed via target-primed DNA synthesis (Figure 7D). The differences in variant repeat length shown in Table S8 could be explained by annealing at different positions within the variant array. Similarly, discontinuous arrays could arise through MoTeR insertions into existing variant repeats. The availability of significant homology to facilitate 5′-end insertion could explain why there are so many full-length MoTeRs in the M. oryzae telomeres, while the telomere-specific and telomere-associated elements found in other organisms (which lack variant repeats) are almost always truncated to varying degrees (Higashiyama et al. 1997; Arkhipova and Morrison 2001).
A number of 5′-truncated MoTeRs were identified by querying several genome sequences. Most of these elements were inserted at positions having short stretches of homology to their truncated 5′ borders. The frequency distribution of overlap sizes was inconsistent with random nucleotide matches, leading us to conclude that integration of truncated 5′ ends is microhomology-driven, as has been reported for truncated LINE-1 elements (Zingler et al. 2005). There were also a number of truncated elements inserted at locations lacking homology to the 5′ borders. Surprisingly, the majority of such insertions shared the same telomeric register (CCCTAA-tMoTeR). Possibly, this reflects a template jump from an incomplete MoTeR transcript onto the 3′ overhang of the target site (Bibillo and Eickbush 2004). Alternatively, it may result from direct ligation between a blunt-ended target site and the DNA moiety of a DNA/RNA hybrid.
The variant telomere sequence might perform other functions beneficial to the MoTeRs’ existence within telomeres. First, should the 5′ end of the MoTeRs fail to become incorporated into the existing telomere, the variant tract might act as a seed for de novo telomere addition by telomerase, thereby slowing or preventing erosion of MoTeR sequences. Finally, the variant repeats could also affect telomere function in ways that would be beneficial to MoTeR propagation. For example, they might influence the protein complement of the telomere or its chromatin configuration, thereby serving the dual purposes of promoting MoTeR transcription and making the telomeres more receptive to subsequent MoTeR insertions.
MoTeR elements represent a new class of telomere-targeted retrotransposons unique to fungi
The MoTeR RT was most similar to hypothetical proteins encoded by the C. neoformans Cnl1 element, N. hematococca NhTeR1, and F. oxysporum FoNLR9. The original report of Cnl1 stated that the element is telomere-associated and that it inserts preferentially into other Cnl1 copies (Goodwin and Poulter 2001). However, a careful examination of the published data, as well as genome sequence information, revealed that nearly all Cnl1 insertions are sandwiched between telomere repeats (data not shown). Therefore, it is clear that Cnl1 inserts into telomeres and not into other Cnl1 copies, as previously reported. Interestingly, however, only 6 of the ∼40 Cnl1 copies in C. neoformans strain B-3501A are currently in telomeres; the remainder are at internal locations. Likewise, some copies of NhTeR1 were present at internal locations (data not shown).
FoNLR9 was originally identified as a CRE-like element in the F. oxysporum genome. At the same time, a related element (FvNLR4) was identified in Fusarium verticillioides (FvNLR4) (Novikova et al. 2009). Although no MoTeR matches were identified by BLAST, a tBLASTn search of the F. verticillioides genome revealed that FvNLR4 codes for a predicted protein that shares 46% identity with the MoTeR1 RT. A survey of FoNLR9 and FvNLR4 sequences within the respective genomes revealed that neither contained intact copies of FoNLR9 or FoNLR4 (all copies were missing their 5′ ends), and none of the elements were telomeric. However, several 3′ termini were identified, and all of them all possessed short tracts of telomere sequence at their insertion junctions (data not shown). This suggests that the FoNLR4 and FoNLR9 are also telomere-targeted transposons and, furthermore, that the telomeric sequences at their 3′ insertion junctions represent former telomeres.
The fungal elements were only distantly related to the three other groups of telomeric transposons. Instead, their nearest phylogenetic neighbors were CRE1/2, SLACS, and CZAR, which, although site-specific in nature, insert at nontelomeric rDNA loci. Intriguingly, all of the major groups of telomeric transposons described to date are most closely related to nontelomeric elements. For example, TART and TAHRE are related to elements in the Jockey clade (Figure S3A; Malik et al. 1999; Abad et al. 2004), whose members mostly lack target-site specificity and insert at random chromosomal locations. Likewise, the recently described Penelope-like elements, which are proposed to add onto chromosome ends (Gladyshev and Arkhipova 2007), belong to a distinct clade that contains the non-site-specific, nontelomeric Penelope transposons Poseidon and Neptune (Evgen’ev et al. 1997; Volff et al. 2001). Although TRAS and SART are quite different from the fungal elements from a phylogenetic standpoint, they share the property of being most closely related to elements that insert into 28S rDNA repeats. This raises the question as to whether these close relationships between telomeric transposons and rDNA transposons are more than coincidental. Finally, on the basis of the phylogenetic distribution of the four classes of telomeric retroelements, it appears that telomere specificity has arisen independently a number of times.
It should be noted that a number of other telomere-associated retrotransposons have been reported in the literature. However, while these elements tend to be found near telomeres, they are not truly telomeric. For example, the Ty5 retrotransposon in Saccharomyces cerevisiae inserts into regions of telomeric heterochromatin but not at telomeres per se (Zou et al. 1996). GilM and GilT from Giardia lamblia were first described as telomeric elements (Arkhipova and Morrison 2001), but a subsequent study showed them to be site-specific transposons that insert into a subtelomeric tandem repeat (Burke et al. 2002). Finally, the Zepp element is found in association with Chlorella telomeres (Higashiyama et al. 1997; Noutoshi et al. 1998) but actually inserts into other Zepp copies, and it is not yet known if the element is truly telomere-specific.
In conclusion, we have shown that telomere hypervariability in M. oryzae isolates from perennial ryegrass is caused by the presence of MoTeRs within the telomere repeats. Although telomeric transposons have been identified in a number of other organisms, this is the first example where their presence causes the chromosome ends to be extremely unstable. It is intriguing that there is no obvious fitness cost to having telomeres that undergo continual rearrangement. This is evident because Magnaporthe isolates from perennial ryegrass are aggressive pathogens that cause devastating losses of perennial ryegrass turf (Vincelli 1999). This raises the question as to whether the MoTeR-induced telomere dynamics provide an immediate adaptive advantage to the pathogen. In Drosophila, the expansion and contraction of HeT-A/TART arrays can affect the expression of neighboring genes through alteration of the chromatin environment (Golubovsky et al. 2001). If MoTeR sequences also have the capacity to affect chromatin structure, then MoTeR-induced rearrangements at chromosome ends potentially could lead to terminally located genes being turned on and off in a stochastic fashion. Functionally, such a system would resemble the telomere-based mechanisms that switch expression of surface proteins in Plasmodium (Scherf et al. 1998), Trypanosomes (Horn and Cross 1997), and Pneumocystis (Wada and Nakamura 1996), which allow these pathogens to evade the host’s immune system. For this reason, it will be interesting to determine if MoTeR activity affects the expression of neighboring genes and if these genes have roles in host interactions and pathogen survival or spread.
Supplementary Material
Acknowledgments
The authors thank Pete Mirabito for helpful comments on the manuscript. This is research publication no. 12-12-026 from the University of Kentucky Experiment Station and is published with the permission of the director. This research was supported by National Science Foundation grants MCB-0653930 and MCB-0135462 to M.L.F.
Footnotes
Communicating editor: T. Wu
Literature Cited
- Abad J. P., De Pablos B., Osoegawa K., De Jong P. J., Martin-Gallardo A., et al. , 2004. Genomic analysis of Drosophila melanogaster telomeres: full-length copies of HeT-A and TART elements at telomeres. Mol. Biol. Evol. 21: 1613–1619 [DOI] [PubMed] [Google Scholar]
- Aksoy S., Williams S., Chang S., Richards F. F., 1990. SLACS retrotransposon from Trypanosoma brucei gambiense is similar to mammalian LINEs. Nucleic Acids Res. 18: 785–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. A., Song Y. S., Langley C. H., 2008. Molecular population genetics of Drosophila subtelomeric DNA. Genetics 178: 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkhipova I. R., Morrison H. G., 2001. Three retrotransposon families in the genome of Giardia lamblia: two telomeric, one dead. Proc. Natl. Acad. Sci. USA 98: 14497–14502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird D. M., Coleman J., Rosser Z. H., Royle N. J., 2000. High levels of sequence polymorphism and linkage disequilibrium at the telomere of 12q: implications for telomere biology and human evolution. Am. J. Hum. Genet. 66: 235–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham S., Beam A., Shampay J., 1998. Telomere variation in Xenopus laevis. Mol. Cell. Biol. 18: 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibillo A., Eickbush T. H., 2004. End-to-end template jumping by the reverse transcriptase encoded by the R2 retrotransposon. J. Biol. Chem. 279: 14945–14953 [DOI] [PubMed] [Google Scholar]
- Biessmann H., Mason J. M., Ferry K., D’Hulst M., Valgeirsdottir K., et al. , 1990. Addition of telomere-associated HeT DNA sequences “heals” broken chromosome ends in Drosophila. Cell 61: 663–673 [DOI] [PubMed] [Google Scholar]
- Biessmann H., Champion L. E., O’Hair M., Ikenaga K., Kasravi B., et al. , 1992. Frequent transpositions of Drosophla melanogaster HeT-A elements to receding chromosome ends. EMBO J. 11: 4459–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann H., Kobeski F., Walter M. F., Kasravi A., Roth C. W., 1998. DNA organization and length polymorphism at the 2L telomeric region of Anopheles gambiae. Insect Mol. Biol. 7: 83–93 [DOI] [PubMed] [Google Scholar]
- Burke W. D., Malik H. S., Rich S. M., Eickbush T. H., 2002. Ancient lineages of non-LTR retrotransposons in the primitive eukaryote, Giardia lamblia. Mol. Biol. Evol. 19: 619–630 [DOI] [PubMed] [Google Scholar]
- Clutterbuck A. J., Farman M., 2007. Aspergillus nidulans linkage map and genome sequence: closing gaps and adding telomeres, pp. 57–73 in The Aspergilli: Genomics, Medical Aspects, Biotechnology, and Research Methods, edited by G. H. Goldman and S. A. Osmani. CRC Press, Taylor & Francis Group, Boca Raton, FL [Google Scholar]
- Cuomo C. A., Guldener U., Xu J. R., Trail F., Turgeon B. G., et al. , 2007. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317: 1400–1402 [DOI] [PubMed] [Google Scholar]
- Dean R. A., Talbot N. J., Ebbole D. J., Farman M. L., Mitchell T. K., et al. , 2005. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434: 980–986 [DOI] [PubMed] [Google Scholar]
- Dubois M. L., Haimberger Z. W., McIntosh M. W., Gottschling D. E., 2002. A quantitative assay for telomere protection in Saccharomyces cerevisiae. Genetics 161: 995–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evgen’ev M. B., Zelentsova H., Shostak N., Kozitsina M., Barskyi V., et al. , 1997. Penelope, a new family of transposable elements and its possible role in hybrid dysgenesis in Drosophila virilis. Proc. Natl. Acad. Sci. USA 94: 196–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B., Green P., 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194 [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M. C., Green P., 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Fan C., Zhang Y., Rounsley S., Long M., Wing R. A., 2008. The subtelomere of Oryza sativa chromosome 3 short arm as a hot bed of new gene origination in rice. Mol. Plant 1: 839–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanti L., Giovinazzo G., Berloco M., Pimpinelli S., 1998. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell 2: 527–538 [DOI] [PubMed] [Google Scholar]
- Farman M. L., 2002. Pyricularia grisea isolates causing gray leaf spot on perennial ryegrass (Lolium perenne) in the United States: relationship to P. grisea isolates from other host plants. Phytopathology 92: 245–254 [DOI] [PubMed] [Google Scholar]
- Farman M. L., 2007. Telomeres in the rice blast fungus: the world of the end as we know it. FEMS Microbiol. Lett. 273: 125–132 [DOI] [PubMed] [Google Scholar]
- Farman M. L., 2011. Targeted cloning of fungal telomeres. Meth. Mol. Biol. 722: 11–31 [DOI] [PubMed] [Google Scholar]
- Farman M. L., Kim Y.-S., 2005. Telomere hypervariability in Magnaporthe oryzae. Mol. Plant Pathol. 6: 287–298. [DOI] [PubMed] [Google Scholar]
- Freitas-Junior L. H., Bottius E., Pirrit L. A., Deitsch K. W., Scheidig C., et al. , 2000. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature 407: 1018–1022 [DOI] [PubMed] [Google Scholar]
- Fudal I., Böhnert H. U., Tharreau D., Lebrun M. H., 2005. Transposition of MINE, a composite retrotransposon, in the avirulence gene ACE1 of the rice blast fungus Magnaporthe grisea. Fungal Genet. Biol. 42: 761–772 [DOI] [PubMed] [Google Scholar]
- Fujiwara H., Osanai M., Matsumoto T., Kojima K. K., 2005. Telomere-specific non-LTR retrotransposons and telomere maintenance in the silkworm, Bombyx mori.. Chromosome Res. 13: 455–467 [DOI] [PubMed] [Google Scholar]
- Gabriel A., Yen T. J., Schwartz D. C., Smith C. L., Boeke J. D., et al. , 1990. A rapidly rearranging retrotransposon within the miniexon gene locus of Crithidia fasciculata. Mol. Cell. Biol. 10: 615–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Khang C. H., Park S.-Y., Lee Y.-H., Kang S. K., 2002. Evolution and organization of a highly dynamic, subtelomeric helicase gene family in the rice blast fungus Magnaporthe grisea. Genetics 162: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B., Carson M., Hartwell L., 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15: 6128–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J. A., Debaryshe P. G., Traverse K. L., Celniker S. E., Pardue M.-L., 2006. Genomic organization of the Drosophila telomere retrotransposable elements. Genome Res. 16: 1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev E. A., Arkhipova I. R., 2007. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc. Natl. Acad. Sci. USA 104: 9352–9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovsky M. D., Konev A. Y., Walter M. F., Biessmann H., Mason J. M., 2001. Terminal retrotransposons activate a subtelomeric white transgene at the 2L telomere in Drosophila. Genetics 158: 1111–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin T. J., Poulter R. T., 2001. The diversity of retrotransposons in the yeast Cryptococcus neoformans. Yeast 18: 865–880 [DOI] [PubMed] [Google Scholar]
- Gordon D., Abajian C., Green P., 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8: 195–202 [DOI] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H., 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337: 331–337 [DOI] [PubMed] [Google Scholar]
- Higashiyama T., Noutoshi Y., Fujie M., Yamada T., 1997. Zepp, a LINE-like retrotransposon accumulated in the Chlorella telomeric region. EMBO J. 16: 3715–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D., Cross G. A., 1997. Analysis of Trypanosoma brucei vsg expression site switching in vitro. Mol. Biochem. Parasitol. 84: 189–201 [DOI] [PubMed] [Google Scholar]
- Ivessa A. S., Zhou J. Q., Schulz V. P., Monson E. K., Zakian V. A., 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 16: 1383–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga T., Mannhaupt G., Glass N. L., 2009. Relationship between phylogenetic distribution and genomic features in Neurospora crassa. PLoS ONE 4: e5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E., 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254 [DOI] [PubMed] [Google Scholar]
- Kern A. D., Begun D. J., 2008. Recurrent deletion and gene presence/absence polymorphism: telomere dynamics dominate evolution at the tip of 3L in Drosophila melanogaster and D. simulans. Genetics 179: 1021–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski J., Feder M., Bujnicki J. M., 2005. The PD-(D/E)XK superfamily revisited: identification of new members among proteins involved in DNA metabolism and functional predictions for domains of (hitherto) unknown function. BMC Bioinformatics 6: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T., Sonnhammer E. L., 2005. Kalign: an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan D. D., Eickbush T. H., 1995. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol. Cell. Biol. 15: 3882–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan D. D., Eickbush T. H., 1996. Downstream 28S gene sequences on the RNA template affect the choice of primer and the accuracy of initiation by the R2 reverse transcriptase. Mol. Cell. Biol. 16: 4726–4734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan D. D., Korman M. H., Jakubczak J. L., Eickbush T. H., 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72: 595–605 [DOI] [PubMed] [Google Scholar]
- Malik H. S., Burke W. D., Eickbush T. H., 1999. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 16: 793–805 [DOI] [PubMed] [Google Scholar]
- McClure M. A., Donaldson E., Corro S., 2002. Potential multiple endonuclease functions and a ribonuclease H domain in retroposon genomes. Virology 296: 147–158 [DOI] [PubMed] [Google Scholar]
- Mefford H. C., Linardopoulou E., Coil D., Van Den Engh G., Trask B. J., 2001. Comparative sequencing of a multicopy subtelomeric region containing olfactory receptor genes reveals multiple interactions between non-homologous chromosomes. Hum. Mol. Genet. 10: 2363–2372 [DOI] [PubMed] [Google Scholar]
- Menon S. K., Eilers B. J., Young M. J., Lawrence C. M., 2010. The crystal structure of D212 from Sulfobolus Spindle-Shaped Virus Ragged Hills reveals a new member of the PD-(D/E)XK nuclease superfamily. J. Virol. 84: 5890–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D., John M. J., Amasino R. M., 1994. Removal of polysaccharides from plant DNA by ethanol precipitation. BioTechniques 17: 274–276 [PubMed] [Google Scholar]
- Moran J. V., Gilbert N., 2002. Mammalian LINE-1 retrotransposons and related elements, pp. 836–869 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz. American Society for Microbiology, Washington, DC [Google Scholar]
- Morrish T. A., Garcia-Perez J. L., Stamato T. D., Taccioli G. E., Sekiguchi J., et al. , 2007. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature 446: 208–212 [DOI] [PubMed] [Google Scholar]
- Naumov G. I., Naumova E. S., Korhola M. P., 1995. Chromosomal polymorphism of MEL genes in some populations of Saccharomyces cerevisiae. FEMS Microbiol. Lett. 127: 41–45 [DOI] [PubMed] [Google Scholar]
- Naumov G. I., Naumova E. S., Sancho E. D., Korhola M. P., 1996. Polymeric SUC genes in natural populations of Saccharomyces cerevisiae. FEMS Microbiol. Lett. 135: 31–35 [DOI] [PubMed] [Google Scholar]
- Naumova E. S., Turakainen H., Naumov G. I., Korhola M., 1996. Superfamily of alpha-galactosidase MEL genes of the Saccharomyces sensu stricto species complex. Mol. Gen. Genet. 253: 111–117 [DOI] [PubMed] [Google Scholar]
- Noutoshi Y., Arai R., Fujie M., Yamada T., 1998. Structure of the Chlorella Zepp retrotransposon: nested Zepp clusters in the genome. Mol. Gen. Genet. 259: 256–263 [DOI] [PubMed] [Google Scholar]
- Novikova O., Fet V., Blinov A., 2009. Non-LTR retrotransposons in fungi. Funct. Integr. Genomics 9: 27–42 [DOI] [PubMed] [Google Scholar]
- Okazaki S., Ishikawa H., Fujiwara H., 1995. Structural analysis of TRAS1, a novel family of telomeric repeat-associated retrotransposons in the silkworm, Bombyx mori. Mol. Cell. Biol. 15: 4545–4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S. H., 1985. Rice Diseases. Commonwealth Agricultural Bureaux, Slough, UK [Google Scholar]
- Peacock C. S., Seeger K., Harris D., Murphy L., Ruiz J. C., et al. , 2007. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 39: 839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrini B., Piacentini L., Fanti L., Altieri F., Chichiarelli S., et al. , 2004. HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol. Cell 15: 467–476 [DOI] [PubMed] [Google Scholar]
- Pryde F. E., Gorham H. C., Louis E. J., 1997. Chromosome ends: All the same under their caps? Curr. Opin. Genet. Dev. 7: 822–828 [DOI] [PubMed] [Google Scholar]
- Rehmeyer C., Li W., Kusaba M., Kim Y.-S., Brown D., et al. , 2006. Organization of chromosome ends in the rice blast fungus Magnaporthe oryzae. Nucleic Acids Res. 34: 4685–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiga H., Edstrom J. E., 1985. Long tandem arrays of complex repeat units in Chironomus telomeres. EMBO J. 4: 799–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A., Hernandez-Rivas R., Buffet P., Bottius E., Benatar C., et al. , 1998. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 17: 5418–5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R., Dayhoff M., 1979. Matrices for detecting distant relationships, pp. 353–358 in Atlas of Protein Sequences, edited by Dayhoff M. National Biomedical Research Foundation, Silver Spring, MD [Google Scholar]
- Symer D. E., Connelly C., Szak S. T., Caputo E. M., Cost G. J., et al. , 2002. Human L1 retrotransposition is associated with genetic instability in vivo. Cell 110: 327–338 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Okazaki S., Fujiwara H., 1997. A new family of site-specific retrotransposons, SART1, is inserted into telomeric repeats of the silkworm, Bombyx mori. Nucleic Acids Res. 25: 1578–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S., 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Teng S. C., Wang S. X., Gabriel A., 1995. A new non-LTR retrotransposon provides evidence for multiple distinct site-specific elements in Crithidia fasciculata miniexon arrays. Nucleic Acids Res. 23: 2929–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverse K. L., Pardue M.-L., 1988. A spontaneously opened ring chromosome of Drosophila melanogaster has acquired HeT DNA sequences at both telomeres. Proc. Natl. Acad. Sci. USA 85: 8116–8120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincelli P., 1999. Gray leaf spot: an emerging disease of perennial ryegrass. Turfgrass Trends 7: 1–8 [DOI] [PubMed] [Google Scholar]
- Vodenicharov M. D., Wellinger R. J., 2006. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol. Cell 24: 127–137 [DOI] [PubMed] [Google Scholar]
- Volff J. N., Hornung U., Schartl M., 2001. Fish retroposons related to the Penelope element of Drosophila virilis define a new group of retrotransposable elements. Mol. Genet. Genomics 265: 711–720 [DOI] [PubMed] [Google Scholar]
- Wada M., Nakamura Y., 1996. Antigenic variation by telomeric recombination of major-surface-glycoprotein genes of Pneumocystis carinii. J. Eukaryot. Microbiol. 43: 8S. [DOI] [PubMed] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J., 2009. Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie A. O., Higgs D. R., Rack K. A., Buckle V. J., Spurr N. K., et al. , 1991. Stable length polymorphism of up to 260 kb at the tip of the short arm of human chromosome 16. Cell 64: 595–606 [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., Castillo-Davis C. I., Oshiro G., Liang D., Richards D. R., et al. , 2003. Genetic diversity in yeast assessed with whole-genome oligonucleotide arrays. Genetics 163: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Malik H. S., Eickbush T. H., 1999. Identification of the endonuclease domain encoded by R2 and other site-specific non-long terminal repeat retrotransposable elements. Proc. Natl. Acad. Sci. USA 96: 7847–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T. J., Yu Y., Chang S. B., De Jong H., Oh C. S., et al. , 2005. Toward closing rice telomere gaps: mapping and sequence characterization of rice subtelomere regions. Theor. Appl. Genet. 111: 467–478 [DOI] [PubMed] [Google Scholar]
- Yu G. L., Bradley J. D., Attardi L. D., Blackburn E. H., 1990. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature 344: 126–132 [DOI] [PubMed] [Google Scholar]
- Zingler N., Willhoeft U., Brose H.-P., Schoder V., Jahns T., et al. , 2005. Analysis of 5′ junctions of human LINE-1 and Alu retrotransposons suggest and alternative model for 5′-end attachment requiring microhomology-mediated end-joining. Genome Res. 15: 780–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S., Ke N., Kim J. M., Voytas D. F., 1996. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev. 10: 634–645 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.