Abstract
Telomeres protect chromosome ends from being repaired as double-strand breaks (DSBs). Just as DSB repair is suppressed at telomeres, de novo telomere addition is suppressed at the site of DSBs. To identify factors responsible for this suppression, we developed an assay to monitor de novo telomere formation in Drosophila, an organism in which telomeres can be established on chromosome ends with essentially any sequence. Germline expression of the I-SceI endonuclease resulted in precise telomere formation at its cut site with high efficiency. Using this assay, we quantified the frequency of telomere formation in different genetic backgrounds with known or possible defects in DNA damage repair. We showed that disruption of DSB repair factors (Rad51 or DNA ligase IV) or DSB sensing factors (ATRIP or MDC1) resulted in more efficient telomere formation. Interestingly, partial disruption of factors that normally regulate telomere protection (ATM or NBS) also led to higher frequencies of telomere formation, suggesting that these proteins have opposing roles in telomere maintenance vs. establishment. In the ku70 mutant background, telomere establishment was preceded by excessive degradation of DSB ends, which were stabilized upon telomere formation. Most strikingly, the removal of ATRIP caused a dramatic increase in telomeric retrotransposon attachment to broken ends. Our study identifies several pathways thatsuppress telomere addition at DSBs, paving the way for future mechanistic studies.
TELOMERES are nucleoprotein structures that serve two vital functions. First, they overcome chromosome end shortening by regulating the addition of new sequences. Second, they prevent chromosome ends from being recognized as double-strand breaks (DSBs). Failure in this second function (the capping function) results in fusions between telomere ends and subsequently in genomic instability. Just as DSB repair is suppressed at the telomere, de novo telomere establishment is suppressed at the site of DSBs. Telomere healing, the addition of telomere functions at a DSB, stabilizes the broken chromosome but leads to loss of heterozygosity due to aneuploidy. Understanding the process of de novo telomere formation will shed light on the cellular mechanisms that normally prevent aneuploidy formation for genome maintenance. In particular, primordial germ cells might lack the apoptotic response elicited by the presence of persistent DSBs possibly due to their quiescent state during early development (Hanyu-Nakamura et al. 2004; Renault et al. 2004; Sano et al. 2005). In these cells, unrepaired breaks might be allowed to persist long enough to acquire telomeres increasing the potential of forming aneuploid cells. Telomere healing can result in human diseases (Flint et al. 1994; Varley et al. 2000; Bonaglia et al. 2011).
In most eukaryotic organisms studied, telomere elongation is carried out by the telomerase enzyme. In these systems, telomere formation on DSBs depends on telomerase function to add short repeats to broken ends. However, it remains unknown how the telomerase function was initially recruited to DSBs with little or no homology to natural telomeres.
In the fruit fly Drosophila, chromosome ends are maintained by the addition of telomere specific retrotransposons (reviewed in Mason et al. 2008; Pardue and Debaryshe 2008). However, these transposons have been shown to be neither necessary nor sufficient for establishing the protective cap at telomeres. Instead flies establish telomere identity via an epigenetic mechanism without a specific sequence requirement (reviewed in Rong 2008). Drosophila cells are remarkably tolerant of terminal deficiencies (TDs), which are terminally deleted chromosomes stabilized by the establishment of a new telomere. Some of these neotelomeres have no retrotransposon sequence but are capped by the same capping complexes that protect natural telomeres and can be maintained for generations (Beissmann and Mason 1988; Levis 1989; Beissmann et al. 1992; Donaldson and Karpen 1997; Mason et al. 1997; Ahmad and Golic 1998; Gao et al. 2010). Therefore, de novo telomeres can form on essentially any DNA sequence without the requirement of the addition of natural telomeric DNA in Drosophila. Drosophila might thus represent the best system to study the early steps required for telomere formation on DSBs.
The genetic requirements for TD formation have been studied previously in Drosophila. In the developing soma, the loss of a telomere induces a significant DNA damage response and subsequent apoptosis. Yet some cells acquire telomere function on the broken chromosome and proliferate to constitute up to 20% of the cells found in the adult structures (Ahmad and Golic 1999; Titen and Golic 2008). How these cells are able to bypass checkpoints and apoptosis is not clear. It was recently discovered that checkpoint response mediated by p53 and Chk2 limits the proliferation of cells that suffered the loss of a telomere. In the male mitotic germline, loss of a telomere also elicits an initial DNA damage response (Kurzhals et al. 2011) but TD chromosomes can be readily recovered as progeny. In the female germline that carries mutations in the mutator 2 (mu2) gene, which encodes the fly homolog of the MDC1 checkpoint protein (Dronamraju and Mason 2009), X-ray irradiation results in a dramatic increase in the recovery of TD-harboring progeny (Mason et al. 1984). Elegant genetic experiments led to the suggestion that DSBs persist in mu2 mutant oocytes, allowing de novo telomere formation upon fertilization (Mason et al. 1997).
During the generation and/or propagation of flies with TDs, occasional addition of telomere-specific retrotransposons can occur and was estimated to have a rate of ∼1% per generation (Beissmann et al. 1992). Several mutant backgrounds have been reported to elevate this frequency of transposon addition: heterozygosity of Irbp, which encodes Ku70 in Drosophila, ku80 or Su(var)205, which encodes HP1 (Savitsky et al. 2002; Melnikova et al. 2005). The dominant Tel mutation results in longer arrays of telomeric transposon on natural telomeres (Siriaco et al. 2002).
Previous studies of de novo telomere formation suffered from one or both limitations: relatively low frequencies of TD formation and lack of control over the precise location at which telomeres would form. In this study, we developed a new assay that allows highly efficient de novo telomere formation at a specific site generated by the rare cutting I-SceI endonuclease. Using this assay, we quantified the frequency of telomere formation and transposon attachment at DSBs in wild-type and mutant backgrounds. This has allowed us to conduct the largest study of the genetic requirements for telomere establishment at DNA breaks in the Drosophila germline.
Materials and Methods
Fly stocks and genetics
Construction of the D4A line was described in Gao et al. (2010). All mutants were on the third chromosome with appropriate balancers except for Lig411, which is on the X and maintained as a homozygous stock (Wei and Rong 2007). The tefuZIII-5190 allele, a mutation disrupting the function of the ATM checkpoint kinase, was discovered by Rickmyre et al. (2007). The tefuf3 allele is a white-eyed derivative of the tefustg allele described by Bi et al. (2004) and Gong et al. (2005). Briefly, the duplicated tefu locus in tefustg was subjected to I-CreI–induced reduction as described (Rong et al. 2002). White-eyed derivative lines were tested for their ability to complement lethality caused by other tefu mutations. The tefuf3 allele failed to complement, suggesting that it is a tefu loss-of-function mutation. The nbs2K allele was described in Gao et al. (2009) and the Irbp7B2 and IrbpEX8 alleles in Johnson-Schlitz et al. (2007). All other stocks were obtained from the Bloomington Stock Center and described in FlyBase (flybase.net). Crossing schemes are provided in supporting information.
Scoring method for the TD assay
Progeny that inherited the uncut chromosome were identified by the presence of Sco. Progeny inheriting the D4A chromosome were all Sco + and further categorized as “w + KrIF” if they retained the “irregular facet” eye phenotype and reporter eye color. Progeny scored as w KrIF retained the irregular facet eye phenotype but lost white expression. Progeny were scored as having a terminal deficiency “w + TD” if they had mottled eye color and normal-sized eyes (Kr +). Progeny were scored as “w TD” if reporter eye color was lost but normal eye shape regained. Transposon addition was scored as restoration of wild-type eye color and normal eye size.
Calculations and statistics
The mean frequency for each progeny class was calculated as “progeny class/total D4A progeny” for each individual germline. Total TD frequency was calculated as the number of Kr + progeny over total D4A-inheriting progeny for each germline. Means were then compared to that from wild type with either Student’s T test if both data sets followed a normal distribution or Mann–Whitney for nonnormal distributions. Fisher’s exact test was used to test for significance in alteration in the frequency of retrotransposon addition. Each cross was classified as either “producing” or “not producing” attachment events regardless of the actual number of attachment events recovered.
Molecular analyses
All primers are listed in supporting information, Table S1. Southern blots were performed using the AlkPhos labeling and detection kit from GE Biosciences (RPN3680). DNA was cut with EcoRV, ClaI, or PvuI individually. Enzymes were purchased from NEB. Inverse PCR were done as in flypush.imgen.bcm.tmc.edu/pscreen/GDP_iPCRProtocol_051611.pdf. For cloning ends of TD, genomic DNA was cut with EcoRV. For cloning of transposon attachment, genomic DNA was cut with HhaI.
Results
De novo telomere formation induced at a site-specific DSB
We generated a P-element construct called P[Iw], which contains a mini-white reporter gene with an I-SceI recognition site immediately upstream of the promoter (Rong and Golic 2003; Figure 1A). The reporter gene gives rise to eye pigmentation in an otherwise white-null background. The D4A insertion of this construct is located at the tip of chromosome 2R ∼200 kb from its natural telomere. Located within the 200-kb distal sequence is the Krupple (Kr) locus. Using meiotic recombination in the female germline, we introduced the dominant irregular facets allele (KrIF) onto the D4A chromosome. Flies with one copy of KrIF have small and rough eyes (Figure 1B).
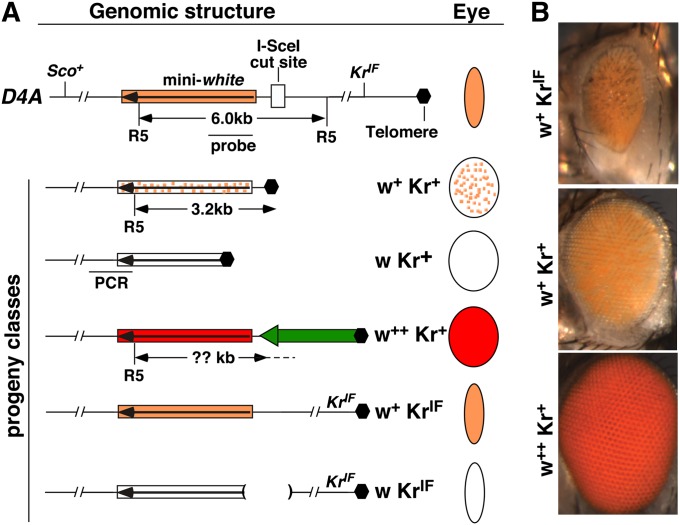
Figure 1 .
The terminal deficiency (TD) assay. (A) Genomic structure of the starting D4A chromosome is shown at the top, which has the wild-type Sco + gene and the dominant KrIF mutation. The structures of the D4A region in different progeny are shown below. The box with a long arrow depicts the mini-white marker gene, which is color coded in accordance with the different eye colors and patterns that it produces in different progeny. Schematic representation of the different eye colors and shapes are given in the “Eye” column. Different progeny classes are also defined by their phenotypes, which are given to the left of the eye diagrams. In the w + Kr + class, placement of white at the telomere resulted in a mottled pattern of eye pigmentation. In the w Kr + class, the white gene is 5′ truncated losing its expression. In the w + + Kr + class, a transposon (green block arrow) attached 5′ to white leading to its higher expression. In the w KrIF class, the deletion (parentheses) was generated during NHEJ repair, abolishing white expression. The positions of the cut site for EcoRV (R5) are given along with the location of the probe and the predicted size of the fragment, except in the case of w + + Kr + as the R5 location on the transposon is not known (question mark). The position of the PCR fragment used to test the presence of D4A sequence is shown for the w Kr + class of progeny. (B) Eye pictures of some of the progeny recovered with phenotypes given to the left.
We used an I-SceI transgene under the control of the regulatory elements from the armadillo locus (arm-I–SceI; Gong et al. 2005) to constitutively produce the enzyme. Cutting at the D4A insertion can result in a terminally deleted chromosome 2R with loss of the distal 200-kb fragment. This TD restores normal eye structure due to the removal of the dominant KrIF mutation and places the white marker at the new telomere, resulting in a variegated eye pigmentation pattern (Figure 1B). We named this terminal deletion of 2R as D4ATD. Animals homozygous for D4ATD are inviable, due to the loss of essential genes at the distal fragment. Nevertheless, D4ATD lines can be maintained as heterozygotes for many generations.
In addition to using phenotypic criteria to confirm the recovery of potential D4ATD chromosomes, i.e., loss of KrIF, loss of viability in homozygotes, and change in pigmentation pattern, we verified the genomic structure of the terminal deletion (Figure 2) by molecular analyses. First, in the Southern blot analysis shown in Figure 2A, the EcoRV enzyme cuts once within the D4A insertion and once distal to the insertion site resulting in a 6.0-kb fragment from the original D4A line. The size of this fragment is reduced to 3.2 kb in animals with D4ATD, consistent with it being a terminal fragment starting from the restriction cut within D4A and ending around the I-SceI cut site. Similar size changes were observed using two other restriction digestions (data not shown). We also measured the size of this terminal fragment in D4ATD flies for consecutive generations after TD formation and observed that its size decreases by ∼100 bp per generation (Figure 2B), a rate similar to the one previously estimated for TDs generated in Drosophila (Beissmann and Mason 1988; Levis 1989). Therefore, we posit that this decrease can be entirely attributed to sequence loss due to incomplete end replication. Second, we used an inverse PCR design to clone the very end of D4ATD (Figure 2C). In this scheme, the chromosome end in D4ATD was ligated to the blunt end generated by EcoRV within the D4A construct. The junction of this ligation was PCR amplified and sequenced. In all cases (n = 70), we observed sequences centromere–proximal to the I-SceI cut site ligated to the remnant of the EcoRV site, identifying the last nucleotide of the chromosome end (Figure 2C).
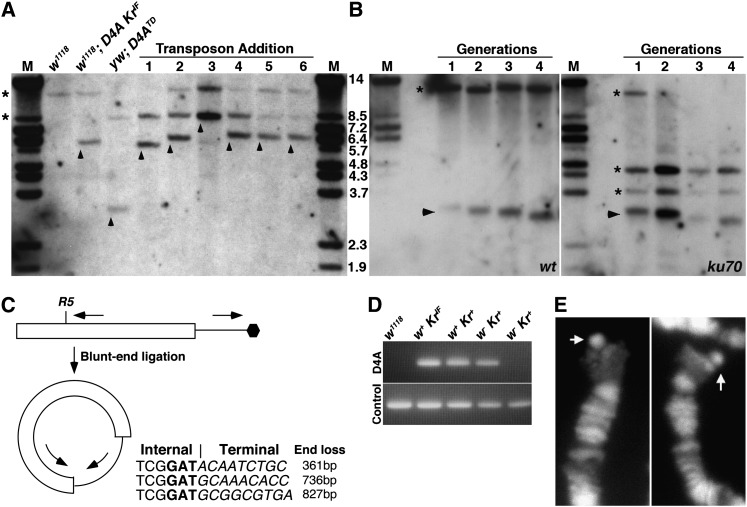
Figure 2 .
Molecular analyses of events from the TD assay. (A) Southern blot analyses of terminal deletions from various genetic backgrounds. Genomic DNA was digested with EcoRV. “M” represents marker lanes with marker sizes in kilobases. Genotypes were shown at the top for the first three lanes. Lines 1–6 are from events with transposon attachment (w + + Kr + class in Figure 1A) with the germline genotype as followed: 1, wt; 2, mu2; 3, tefu; 4, mus304; 5, Irbp; and 6, spnA. Bands from the endogenous white gene are denoted with asterisks. Note that both the w1118 and yw1 backgrounds were used in the experiments. Bands from mini white at D4A are labeled with arrowheads. (B) Southern blot analyses on TD flies of different generations from either wild-type (wt) or Irbp backgrounds. Bands from endogenous white are labeled with asterisks. The D4A band shows a steady decrease in sizes in both backgrounds. The extra white bands in the Irbp background originated from the w1 allele. (C) Design of inverse PCR to clone the very end of D4ATD. The small arrows depict the positions of the PCR primers. The hexagon represents the end of the chromosome. An EcoRV digestion followed by blunt-end ligation created a circular piece of DNA allowing productive PCR reactions. Partial sequences from three clones are given with bold-typed “GAT” separating the internal sequences from the terminal sequences (in italics). GAT represents half of the GATATC cut site for R5. The extent of end loss was estimated for these three lines starting from the position at which I-SceI cut. (D) PCR results from assaying the presence of D4A sequences in different progeny with phenetypes given at the top. Note that for one of the w Kr + events, the control PCR was positive but not for the D4A PCR. (E) Polytene telomeres from progeny recovered from the tefu germline in which the distal KrIF piece of DNA transposed to the tip of chromosome 3L. The flies were heterozygous for the transposition so that an extra bit of chromosomal material (arrow) is present on only one of the two homologous telomeres.
In addition to progeny with terminally deleted 2R chromosomes, we recovered several other classes of offspring from parents with both D4A and arm-I–SceI (Figure 1A). First, some progeny inherited an intact chromosome 2R, as evidenced by the presence of KrIF, with the overall genomic structure at D4A, including the white marker preserved. These w + KrIF progeny could have arisen from DSB repair or I-SceI failing to cut. Second, DSB repair by either imprecise end joining preceded by a large nucleolytic degradation of the ends or by gene conversion with the homologous chromosome would result in the loss of white expression and give rise to w KrIF progeny. Third, among progeny with a potential TD (Kr +), we observed two classes of rare progeny in addition to the more common ones with mottled eye color. These two classes are described in more details below.
The first class of Kr + offspring had uniformly pigmented eyes but darker than the original D4A pigmentation (w + + Kr + in Figure 1A). We suspect that in these progeny the D4ATD chromosome had acquired a telomeric retrotransposon, similar to previously reported cases of transposon attachment to broken chromosomes (Traverse and Pardue 1988; Levis 1989; Beissmann et al. 1992). The increased white expression is likely driven by the promoter located at the 3′-UTR of the HeT-A or TARHE elements (Danilevskaya et al. 1997; Kahn et al. 2000; George et al. 2006; Frydrychova et al. 2007), which would have been positioned to the immediate upstream of white in D4ATD (Figure 1A). Southern blot analyses of these lines showed that the terminal restriction fragment increased by various sizes depending on the restriction enzyme used (Figure 2A), consistent with the attachment of a DNA fragment to the broken end. We also used inversed PCR to gain sequence information for the attached fragment. In three cases in which we recovered a PCR product, the attached sequence is homologous to 3′-UTR of HeT-A in the database, suggesting that at least in those cases a HeT-A element has attached to the new end (Table S1).
The second class of rare Kr + progeny has white eyes (w Kr +). As shown below, the majority if not all of them are D4ATD accompanied by loss of white sequences essential for its expression. We classified this class of progeny as “w D4ATD.” Since we recovered the presumed D4A chromosome by following the Scutoid (Sco) dominant marker on the non-D4A homolog, another mechanism that can give rise to progeny phenotypically as w D4ATD (w Kr + and non-Sco) is a mitotic crossing over between homologous chromosomes at the site of D4A during gene conversion (GC) repair of the I-SceI–induced DSB. These interhomolog GC events are believed to be rare since it requires the removal of the entire P element (∼6 kb) to expose DSB-flanking homology for GC. In addition, mitotic GC associated with crossing over is also very rare, estimated to be <0.7% of the interhomolog GC events in one instance (Rong and Golic 2003). Nevertheless, w Kr + arising from interhomolog GC can be distinguished from w D4ATD events by the loss of the entire D4A insertion and the preservation of homozygous viability due to the presence of the 200-kb segment distal to the D4A insertion site.
Efficient TD formation in the wild-type germline
To measure the efficiency of TD formation in the germline, we crossed individual flies that carried both D4A and the arm-I–SceI transgene to white-eyed mates and scored all the progeny from this cross. Data are reported in Figure 3 and Table 1. First, we were interested in whether the efficiency of TD formation differs between the male and the female germline. For this test, we used individual males or females in which D4A was paternally supplied while arm-I–SceI was maternally inherited. The siblings of these flies included flies with only the D4A insertion, yet they often displayed eyes mosaic for white and KrIF, indicating that the D4A chromosome was cut and repaired even though the arm-I–SceI transgene was not present. This suggests that embryos from females carrying arm-I–SceI have a significant maternal deposition of I-SceI. Therefore, in flies with both D4A and a maternal arm-I–SceI, cutting could occur during early development before zygotic expression. We calculated the frequency of TD for each individual germline as the instances of TD events over the total progeny that inherited the D4A chromosome. The mean frequency of TD formation for the 49 female germlines examined was 0.63. Of the 56 male germlines examined, the mean frequency of TD formation was 0.49. The two frequencies are statistically different (P = 0.01), suggesting that TD formation is slightly more efficient in the female germline. Since the cutting and generation of TDs are a mitotic event, it is highly unlikely that the lack of meiotic recombination in males can explain the differences.
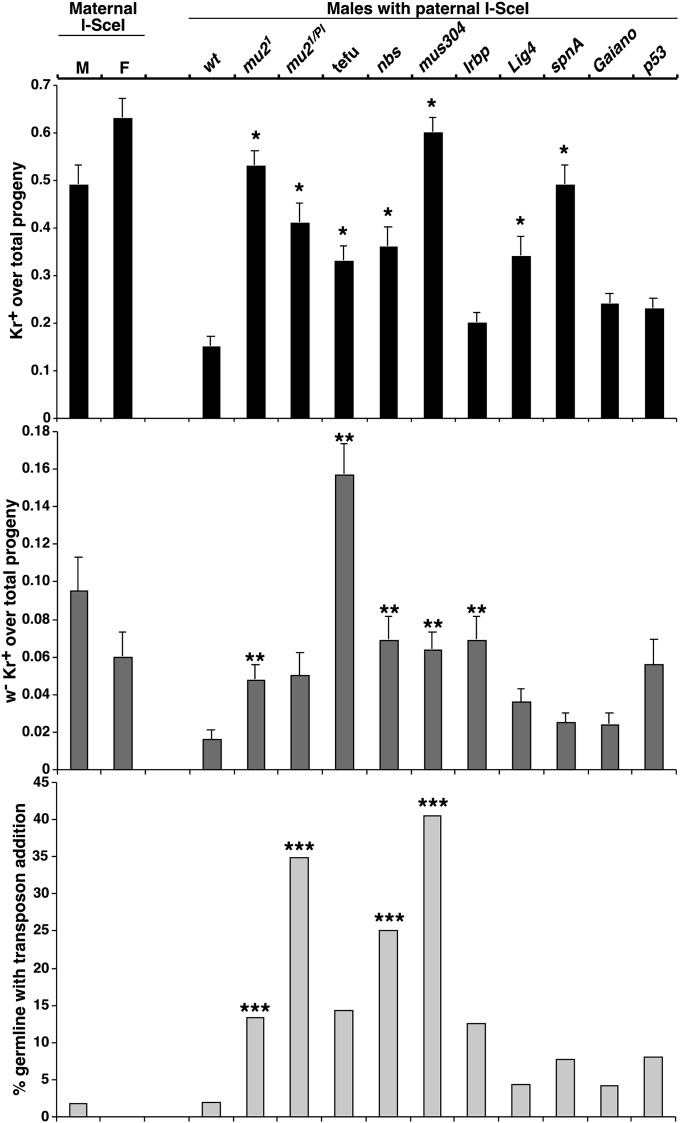
Figure 3 .
Frequency plots from the TD assay. The numerical data and statistical analyses were given in Table 1. The genotypes are given at the top of the charts. For experiments with a maternal contribution of I-SceI, both male (M) and female (F) parents were used. The top chart depicts total TD frequencies in different genetic backgrounds. *P value <0.0001 when compared to the wild-type sample. The middle chart depicts the frequencies of the appearance of TD progeny that had loss part of the white gene due to end degradation. **P value ≤0.001. The bottom chart depicts the level of transposonattachment to DSBs in different backgrounds. ***P value ≤0.05.
Table 1 . Frequencies of progeny classes.
| Genotype | Na | Total progeny D4A/homologb | w + Kr + plus w Kr +c | Pd | w Kr +c | Pe | Transposon additionf | Pg |
|---|---|---|---|---|---|---|---|---|
| +/ + femaleh | 49 | 1125/1528 | 0.63 ± 0.04 | 0.06 ± 0.013 | 0 | |||
| +/ + maleh | 56 | 1977/2570 | 0.49 ± 0.04 | 0.01 | 0.095 ± 0.018 | 0.25 | 1 | 0.5 |
| Male | ||||||||
| +/ + | 49 | 1948/2475 | 0.15 ± 0.02 | 0.016 ± 0.005 | 1 | |||
| mu21 | 45 | 2043/2387 | 0.53 ± 0.03 | <0.0001 | 0.048 ± 0.008 | 0.0013 | 6 | 0.05 |
| mu21/Pl00250 | 23 | 1197/1451 | 0.41 ± 0.04 | <0.0001 | 0.050 ± 0.012 | 0.0036 | 8 | 0.0003 |
| atm5190/F3 | 49 | 1885/2056 | 0.33 ± 0.03 | <0.0001 | 0.157 ± 0.016 | <0.0001 | 7 | 0.06 |
| nbs2k | 24 | 1100/1286 | 0.36 ± 0.04 | <0.0001 | 0.069 ± 0.012 | <0.0001 | 6 | 0.004 |
| mus304D1/D3 | 42 | 2169/2465 | 0.60 ± 0.03 | <0.0001 | 0.064 ± 0.009 | <0.0001 | 17 | 0.0002 |
| ku707b2/ex8 | 48 | 1773/2304 | 0.20 ± 0.02 | 0.0605 | 0.069 ± 0.012 | <0.0001 | 6 | 0.05 |
| lig411 | 45 | 1459/1896 | 0.34 ± 0.04 | <0.0001 | 0.036 ± 0.007 | 0.0093 | 2 | 0.5 |
| spnA1/Dfi | 39 | 1995/2164 | 0.49 ± 0.04 | <0.0001 | 0.025 ± 0.005 | 0.0284 | 3 | 0.283 |
| Gaiano | 48 | 2035/2094 | 0.24 ± 0.02 | 0.0023 | 0.024 ± 0.006 | 0.3935 | 2 | 0.5 |
| p535A-14 | 50 | 2268/2633 | 0.23 ± 0.02 | 0.0072 | 0.056 ± 0.013 | 0.0043 | 4 | 0.36 |
Number of crosses.
Total progeny counted that inherited either D4A or its homologous chromosome.
Mean frequencies of TD progeny calculated as progeny class/total progeny inheriting the D4A chromosome.
Derived from Student’s T tests.
Derived from Mann–Whitney tests.
Number of germline that produced at least one progeny with sequence addition.
Derived from Fisher’s exact tests.
Both males and females were tested under the condition in which maternal I-SceI was present. When I-SceI was solely supplied zygotically, only males were tested.
Df = Df(3R)X3F.
Our goal was to use this assay to examine the effect of different genetic backgrounds on TD formation. We chose to examine the male germline due to female sterility associated with some of the mutants to be examined. In all subsequent crosses, D4A was maternally supplied while arm-I–SceI was paternally inherited. This was to delay I-SceI expression and was done for two reasons: to reduce the chances of jackpot events and to reduce the frequency of TD formation in wild-type flies, which would allow for an increased resolution of potential effects in mutant backgrounds. Under these conditions, the TD frequency for wild-type male germline was reduced from 0.49 to 0.15.
In a wild-type background, only one germline of the 49 examined had TD progeny with an attached transposon. This event resulted in homozygous lethality and its genomic structure was confirmed by Southern blot analysis (Figure 2A). Of the 49 males examined, 13 had white-eyed progeny that had lost KrIF (w Kr + in Figure 1A), with a mean frequency of 0.016. We implemented two tests to determine the frequency of w D4ATD of these progeny. First, we examined individual progeny from each of the 13 males by PCR for the presence of a centromere–proximal region in the D4A construct (Figure 2D). Seven of the 13 examined still contained part of D4A. Second, a TD would lead to recessive lethality that cannot be complemented by an existing TD. We examined 10 of these 13 events and discovered that this was true for all 10 independent w Kr + events, which indicates that they were also truncations that have lost the distal piece. These results indicate that at least 7 of 13 (54%) and possibly 100% (10/10) of the w Kr + progeny actually had a terminal deletion. The 54% is likely an underestimate as some of the white loss could have been so extensive that the entire D4A construct was lost before establishment of the new telomere, leading to a negative outcome in the PCR test.
Drosophila MDC1 inhibits TD formation
We chose to examine the effect of removing the function of Mu2, the Drosophila homolog of the MDC1 checkpoint protein (Dronamraju and Mason 2009). Previously, Mason et al. 1997 showed that irradiation of oocytes from adult females resulted in an increased incidence of TDs in a mu2 background but no effect was seen when sperm was irradiated. Here we tested the effect of loss of Mu2 in the male mitotic germline. The results show that the mu21 mutant background resulted in a dramatic increase in the proportion of progeny with TDs, with a shift of frequency from 0.15 in wild type to 0.53 in mu2. This increase was reproduced (0.41 for TD recovery) using a trans-heterozygous combination of mu2 [mu21/Pl00252 (mu21/Pl)], confirming that the effect was due to the loss of Mu2 functions. There was also a significant increase in the proportion of w Kr + progeny, from 0.016 in wild type to 0.048 in mu21 and to 0.050 in mu21/Pl. We examined 19 independent w Kr + progeny and found that 13 of them retained some D4A sequences. This ratio (13/19) is similar to that from wild-type males (7/13, P = 0.5). We conclude that there is a significant increase in TD progeny that had lost white expression from the mu2 germline. In addition, we observed a higher frequency for transposon attachment events in mu21 (6 of 45 germlines) than in wild type (1 of 48) although not statistically significant (P = 0.0518). A more dramatic increase was observed for mu21/Pl (8/23, P = 0.0003). Therefore, loss of Mu2 results in more efficient telomere formation and transposon addition at DSBs in the mitotic compartment of the male germline.
Loss of ATRIP increases transposon attachment to broken chromosomes
Another important checkpoint pathway is controlled by the ATR kinase and its interacting partner ATRIP, which is encoded by the mus304 gene in Drosophila. Loss of Mus304 resulted in a large increase in the frequency of TD recovery to 0.60 in our assay, the highest among all mutants tested. This increase was accompanied by a proportional increase in the number of w Kr + progeny recovered (0.064), most of which retained D4A sequences (11/16) and hence represented w D4ATD events. The most striking effect of removing ATRIP function was the increase of transposon attachment events. We recovered such progeny from 17 of the 42 germlines examined, which represents an ∼20-fold increase (P < 0.0001). Four independent events were examined by Southern blot analysis and were found to contain sequence additions similar to addition events seen in other backgrounds (Figure 2A and data not shown). Therefore, loss of ATRIP in Drosophila enhances de novo telomere formation and transposon attachment to DSBs.
The Gaiano genetic background does not affect transposon attachment to broken ends
We suspected that the Tel mutation, originated from the Gaiano stock, would have an effect on the efficiency of transposon attachment to DSBs, since its presence leads to a higher level of telomeric retrotransposon repeats than seen in regular lab stocks (Siriaco et al. 2002). In our de novo telomere assay, we observed an increase of TD formation frequency (from 0.15 to 0.24) in Tel homozygous germline (P = 0.0023). However, only 3 of 48 Tel germlines produced transposon attachment events and 2.4% of the progeny were w Kr +, both numbers were not significantly different from those obtained from wild-type males.
Reduced ATM or Nbs function promotes TD formation
We previously showed that the ATM checkpoint kinase, which is encoded by the tefu gene in Drosophila, and the Mre11–Rad50–Nbs (MRN) complex are essential for preventing telomere fusion and that they function in the same pathway in telomere maintenance (Bi et al. 2004, 2005). We are interested in whether they serve a function in telomere establishment. We characterized the hypomorphic nbs2K allele that supports male fertility (Gao et al. 2009) and recently characterized a hypomorphic allele of tefu (tefu5190) with similar phenotypes (P. Morciano, G. Gao, and Y. S. Rong, unpublished results). Using these viable alleles, we were able to examine the functions of these genes using our de novo telomere assay. We found a similar and significant increase in the frequency of TD progeny for both mutants, 0.33 for tefu and 0.36 for nbs. Remarkably, the frequency for the w Kr + progeny in tefu (0.157) was almost ninefold higher than the wild-type level of 0.016, while nbs had a less dramatic but still significant increase to 0.069. From both germlines, we observed similar proportions of w Kr + progeny that retained D4A sequences using the PCR test (22/30 for tefu; 9/16 for nbs). In addition, none of the nine w Kr + lines that were tested from tefu complemented an existing TD. Therefore, D4ATD progeny that suffered white loss were recovered more frequently from both tefu and nbs background. There does not seem to be a significant increase in the number of transposon attachment events from the tefu germline (7/49 germlines, P = 0.0592), but a significant one from the nbs germline (6/24 germlines, P = 0.0043).
There was an interesting class of progeny recovered from both the tefu and nbs germlines, which might have resulted from the low level of telomere fusion observed for both hypomorphic alleles (Gao et al. 2009; P. Morciano, G. Gao, and Y. S. Rong, unpublished results). The homolog of the D4A and KrIF-containing chromosome has the Sco dominant marker. Normally, Sco and KrIF segregate from each other due to the lack of meiotic recombination in Drosophila males. We recovered multiple Sco KrIF offspring from both germlines (14/49 for tefu, 2/24 for nbs). Further analysis showed that KrIF was no longer mapped to chromosome 2. Further mapping of three independent events from the tefu germline localized two of them to chromosome 3 and one to the X. In light of the fact that these KrIF transposition events were recovered only from mutant males known to have telomere decapping defects (i.e., tefu and nbs), we hypothesize that they were the result of a telomere fusion between the natural 2R telomere with a telomere from a nonhomologous chromosome. Upon I-SceI–induced cutting at D4A, the distal KrIF fragment, now physically attached to a nonhomologous chromosome, segregated with the Sco-marked chromosome 2 to the same gamete. Cytological analyses of polytene chromosomes heterozygous for these KrIF transposition events clearly identified the transposed material attached to an existing chromosome end (Figure 2E). We cannot formally rule out that the KrIF transposition occurred as a result of a ligation between the telomere of a nonhomologous chromosome (3L in the case shown in Figure 2E) to the distal end of the DSB generated at D4A.
Inhibition of NHEJ promotes end degradation prior to telomere formation
The DSB generated by I-SceI cutting could be repaired by precise end joining using the overhangs of the DSB, which would recreate a cut site for I-SceI. Since I-SceI is continuously expressed throughout development in our assay, cutting and repair can occur repeatedly until either the end becomes a telomere or the cut site sequence is altered due to other modes of DSB repair such as imprecise nonhomologous end joining (NHEJ). We wanted to examine the effect on TD formation when NHEJ is affected in mutant backgrounds that eliminate either DNA ligase IV (Lig4), which functions primarily in NHEJ, or Ku70, which functions prominently in both NHEJ and telomere maintenance.
Loss of Lig4 leads to a significant increase in frequency of TD progeny to 0.34 and an increase in frequency of w D4ATD progeny (from 0.016 to 0.036, P = 0.0093, with seven of nine progeny tested retaining D4A sequences). This moderate increase in white loss is likely due to excessive DSB degradation before telomere formation, which would be consistent with our previous results from studying the effect of Lig4 on DSB repair (Wei and Rong 2007). Only 2 of the 46 germlines examined had transposon attachment events, which is not significantly different from the wild-type level.
In contrast to removing Lig4, loss of Ku70, which is encoded by the Irbp gene in Drosophila, did not lead to an increase in TD recovery (Table 1). Transposon attachment events were seen in 6 of the 48 Irbp germlines, which is more frequent than wild type but not statistically significant (P = 0.05), indicating that DSB ends without the protection of Ku70 are not more susceptible to transposon attachment. In contrast to these small effects that we observed so far, there was a significant increase in the frequency of w Kr + progeny (Table 1). This is remarkable considering that previous cases of elevated recovery of w Kr + progeny were all accompanied by elevated recovery of total TD progeny (w + Kr + plus w Kr +). Most of these w Kr + progeny from Irbp males (16/23 tested) retained D4A sequences. In all eight cases but one, w Kr + progeny failed to complement an existing TD, consistent with their having a TD. The one exceptional w Kr + line was homozygous viable, indicating its being a non-TD event. We suggest that this event arose from a mitotic crossing over between Sco and KrIF, occurring at the site of the D4A insertion.
The high rate of white loss in TD events from Irbp germline could be due to excessive end processing before or after telomere establishment at D4A. As Irbp mutants in other organisms suffer extensive telomere processing (reviewed in Riha et al. 2006), it might be expected that the telomere at D4ATD, although functionally capped, suffered excessive degradation in the absence of Ku70. To investigate this possibility, we monitor the rate of sequence loss at the end of D4ATD over four generations in the Irbp mutant background. By Southern blot analyses of the size of the terminal restriction fragment, we did not observe an elevated loss of end sequences in Irbp (Figure 2B), suggesting that telomeres without Ku70 do not suffer attrition beyond the normal level caused by incomplete end replication (Figure 2B). Therefore, the high incidence of white loss in progeny from the Irbp germline was likely due to excessive DSB degradation before telomere formation.
In summary, inhibiting NHEJ can promote telomere formation, which is sometimes preceded by excessive degradation of the less protected ends.
Homologous recombination repair limits TD formation
We previously showed that recombinational repair of DSB are highly active in the mitotic germline (Rong and Golic 2003; Wei and Rong 2007). In particular, gene conversion with the sister chromatid might be the most significant repair pathway employed to repair DSBs induced by I-SceI. As this mode of repair also recreates a functional cut site, we were interested in studying the effect of removing homologous recombination on telomere establishment. We used mutations in the spnA gene, which encodes the Drosophila homolog of the Rad51 recombinase. Removal of Rad51 function resulted in a significant increase in the frequency of total TD progeny to 0.49 as well as a significant increase in the frequency of w Kr + progeny to 0.02. There was no significant increase in the number of transposon attachment events associated with Rad51 removal (4/39).
Removal of p53-mediated cell death minimally affects TD formation
Recently, it was discovered that reduction of p53 function results in a dramatic increase in somatic cell survival after telomere loss due to the breakage of a nonessential dicentric chromosome (Kurzhals et al. 2011). We wanted to see whether a similar situation is found in the mitotic germline using our assay. Removal of p53 activity resulted in a modest increase in TD formation (from 0.15 to 0.23). There was an increase in the frequency of w Kr + progeny to 0.056. Finally, there does seem to be an increase in transposon addition at TD ends (Table 1).
Discussion
Reconciliation with Muller’s inability to recover terminal deficiencies
In this study we induced de novo telomere formation on an endonuclease-induced DSB. Remarkably, as high as 63% of the progeny on average had acquired a new telomere at the DSB site under continuous I-SceI production through development starting from the earliest stages of embryonic divisions (Figure 3 and Table 1). This high rate of TD recovery in the germline is in startling contrast to Muller’s inability to recover terminally deleted chromosomes in Drosophila that had led to the very concept of “telomere” (Muller 1940; Muller and Herkowitz 1954).
We offer four lines of explanation for reconciliation. First, Muller used X-ray irradiation as the DSB inducing agent, whereas we used a site-specific endonuclease. Breaks generated by irradiation might need to be processed differently or more extensively than ends from a nuclease digestion to become suitable substrates for telomere formation. Second, the DSB at D4A is relatively close to an existing telomere. It is possible that there is a higher concentration of capping proteins surrounding the telomeres in the nucleus, making it more likely for a DSB end to be capped as a telomere. This has been previously suggested by Mason et al. (2008). Third, I-SceI induces one DSB per diploid genome in our system, whereas the number of breaks induced by X-ray was difficult to control and some cells might have more than one. Cells respond differently to DSB dosage. Yeast cells in the G1 phase respond differently to one vs. four DSBs induced by a nuclease (Zierhut and Diffley 2008). This different response might lead to inefficient telomere formation when a cell encounters more than one break. Although the above three factors might contribute to the decreased likelihood of recovering TDs in the Drosophila germline, they remain a priori assumptions to explain Muller’s results since TDs can be nevertheless recovered in the female germline by X-ray irradiation (Mason et al. 1997).
We offer our fourth proposition as a key difference between our experiments and those of Muller’s: Muller induced DSBs to male germ cells in advanced stages of spermatogenesis, yet the DSB induced in our assays are limited to the mitotic compartment of the male germline. We recently discovered that paternal telomeres that have lost the protection of the K81 capping protein engage in highly efficient telomere fusion before the first zygotic division (Gao et al. 2011). This result implies that first, de novo telomere establishment is highly inefficient on decondensed sperm DNA even in the presence of abundant maternal deposition of capping components (our unpublished data); and second, DNA repair, particularly end joining, is highly active during the early embryonic cycles. In Muller’s experiments, the DSBs generated in the male germline likely persist until after fertilization (Muller 1940), making it unlikely to be capped during the first zygotic division. On the contrary, the DSBs in our assay can be generated throughout development as I-SceI is continuously and ubiquitously expressed. In addition, simple rejoining of an I-SceI–induced DSB or intersister GC repair would recreate a functional cut site allowing a second round of cutting. Therefore, the DSB at D4A had multiple opportunities to acquire a telomere during all stages of development. Evidence supporting our last proposition already exists. In light of the increase in TD recovery from irradiated mu2 females, Mason et al. (1997) postulated that DNA lesions induced in mu2 oocytes persist through oogenesis followed by telomere establishment on DSBs in the early embryo. A similar increase was not observed when sperm instead of oocytes were irradiated, suggesting DNA lesions on sperm chromatin are poor substrates for telomere formation. In addition, neotelomere formation on ends of broken dicentric chromosomes can be readily recovered when induced in the mitotic male germline (Ahmad and Golic 1998; Titen and Golic 2010).
We also have evidence suggesting that de novo telomere formation can occur very early in the somatic lineages. This was derived from scoring flies that inherited both D4A and a maternal I-SceI gene. In our assay, TD formation leads to the loss of the dominant KrIF mutation, restoring the eye to its normal size. In addition, TD formation leads to a variegated eye pigmentation pattern (Figure 1B). We often recovered flies that had variegated eyes with sizes that are fully normal. These eyes likely developed from a cell with D4ATD formed at early stages of development.
Defective checkpoints allow telomere formation on persisting DSBs
The first set of mutants that increase TD recovery in the germline have defects in DNA checkpoint functions: mu2 and mus304. From germlines in both backgrounds, we recorded increases of TD formation that are among the highest in all mutants tested. It is possible that cells able to establish telomeres on DSBs had a survival advantage over cells with persisting DSBs so that cells with TD are selected for in our assay. If this were true and if many cells in mu2 or mus304 were unable to establish telomeres at D4A and later died, we would have observed preferential recovery of the uncut homolog and impaired male fertility due to germ cell loss. We observed neither for any of the mutants that we have tested, suggesting that apoptosis is not a normal response in these cells defective for damage response or repair. We support the previous proposition by Dronamraju and Mason (2009) that defective checkpoints lead to persistence of DSBs allowing more time for telomere establishment.
Our hypomorphic tefu and nbs mutations enhanced TD recovery similarly but to a lesser extent than mu2 and mus304 null mutations. It is possible that the underlying mechanism, i.e., persisting DSBs due to defective checkpoints, is common for both groups of mutants. However, we and others have used null mutations and showed that the checkpoint functions of ATM and, to some degrees, MRN are less prominent in Drosophila than the ones controlled by ATR and ATRIP (Bi et al. 2005; Oikemus et al. 2006). We thus speculate that other functions of ATM and MRN might help inhibit de novo telomere formation. In particular, ATM and MRN are essential for end tethering during DSB repair (Kaye et al. 2004; Lobachev et al. 2004; Callen et al. 2007; Lee et al. 2008). We imagine that the two ends of the DSB induced at D4A are allowed to separate in great distance in tefu or nbs cells, which would impede repair, giving more time for telomere formation. In addition, the MRN complex is important for both HR and NHEJ repair of DSBs (reviewed in Haber 2000). Our nbs mutation might affect telomere formation via its function in DSB repair as our results suggest that inhibiting DSB repair facilitates telomere formation (see below).
Defective DSB repair channels DSBs to telomere addition
Two modes of repair of the DSB at D4A will recreate the I-SceI cut site: precise end joining and recombination with the sister chromatid, making the chromosome susceptible to a second round of cutting. Therefore, any events leading to the disruption of the cut site would be favored in the presence of continuous I-SceI expression. De novo telomere formation represents one of these events. Consistently, when HR was impaired by the spnA mutation or end joining by Lig4, TD recovery rate increased. Loss of SpnA has a larger effect than loss of Lig4, which is consistent with our previous observation that intersister HR is the predominant pathway for the repair of I-SceI–induced DSBs in the male germline (Rong and Golic 2003; Wei and Rong 2007). Therefore, channeling of DSBs is likely the cause for elevated TD recovery in repair-defective germline.
End degradation during telomere addition
In our assay, scoring progeny that inherited a TD but have lost white sequences helps illustrate the extent of nucleolytic degradation of chromosome ends before and after de novo telomere formation. We surmise that a longer half-life of DSB in checkpoint mutants would result in more extensive degradation. Consistently, mutants with suspected defects in checkpoint functions (mu2, mus304, tefu, and nbs) all led to increased white loss in TD progeny. However, in tefu and, to a lesser extent, nbs mutants, this increase is disproportionally larger than the increase in total TD recovery. For example, close to half of TD progeny from tefu suffered a loss of white expression (0.157/0.33 = 48%). These results suggest that ATM and NBS normally inhibit end degradation at telomere ends.
Contrary to mutants that prolong the presence of DSBs, we consider that mutants defective in individual DSB repair pathways are unlikely to cause extensive end degradation before telomere formation. As we and others have shown, different repair pathways compete for the available DSBs (Johnson-Schlitz et al. 2007; Wei and Rong 2007) so that when one is defective, DSBs are efficiently repaired by others. This suggests that defects in a single repair pathway is unlikely to prolong the presence of DSBs. Consistently, the increases in TD recovery in spnA and Lig4 germline were not accompanied by significantly elevated levels of white loss.
The Irbp mutant is interesting in that it behaved differently from any other mutants in our assay, causing a dramatic increase of white-loss TD events but without a significant increase in the overall TD recovery. We deduced several points concerning Ku70’s function from these results. First, loss of Ku70 does not impact precise end joining to a degree similar to the Lig4 mutation. Second, imprecise NHEJ is infrequently used for DSB repair at D4A in the male germline so that its disruption by the loss of Ku70 does not lead to significant channeling of DSBs for telomere formation. Third, the excessive end degradation in Irbp germ cells is likely specific to telomeric ends and occurs after the commitment of the DSB to a telomeric fate but before the establishment of a functional telomere. This last point was based on our observation that events with white loss followed by successful NHEJ (scored as w KrIF progeny, Figure 1A) were not recovered at a higher frequency in the mutant background, a result similar to ones from a previous study (Johnson-Schlitz et al. 2007). We also showed that once a functional telomere has been established at D4A, loss of Ku70 has no effect on the rate of end attrition. We speculate that once a DSB is committed to a telomeric fate, the binding of Ku70 prevents excessive nucleotytic attrition before the functional establishment of a protective cap. Intriguingly, Ku70 seems to have no role in either fate determination of DSB ends or cap establishment on ends.
Transposon addition to broken ends
Remarkably, we observed an ∼20-fold increase in new telomere formation accompanied by a transposon attachment event in mus304 germ cells. This frequency is likely to be an underestimate due to the fact that transposon attachment to the D4A end that has lost part of white could not be identified in our assay.
In yeast Saccharomyces ceravisiae, the Mec1/ATR kinase, and presumably its partner ATRIP, prevents accumulation of the Cdc13 protein at DSBs (Zhang and Durocher 2010). Cdc13 is a member of the Cdc13–Stn1–Ten1 (CST) telomeric complex essential for telomere protection and the recruitment of telomerase activities to telomeres (reviewed in Giraud-Panis et al. 2010). Interestingly, the Drosophila Verrocchio (Ver) protein was recently identified as an essential capping protein and shares limited homology with Stn1 proteins from other organisms (Raffa et al. 2010). This suggests that a similar CST complex might exist in Drosophila. We speculate that Drosophila CST might accumulate at DSBs in the absence of Mus304/ATRIP, leading to more efficient recruitment of the transposon machinery, similar to CST’s role in telomerase recruitment in the other systems.
Ku70 heterozygosity has been shown to lead to elevated rates of transposon attachment in the female germline. This increase happens over a few successive generations (Melnikova et al. 2005). We did not observe evidence of rampant transposon attachment to D4ATD from Southern blot analyses on TDs that have been kept in the Irbp background for several generations (Figure 2A). However, our crossing scheme only allowed TDs to be present in the mutant germline from males.
Concluding remark
We are not surprised by the fact that almost all mutations tested in our study lead to increases in the recovery of events associated with de novo telomere formation. It is consistent with the idea that telomere formation might be a backup mechanism to all modes of damage repair and response in germ cells. Although our candidate approach in identifying factors essential for telomere establishment is far from comprehensive, a picture has emerged in which factors responsible for the recruitment and execution of damage repair and response activities are also responsible for inhibiting telomere formation. In the absence of these activities, the DNA and chromatin structures at the ends might be sufficient for the recruitment of the protective cap. If this were true, only defects in the protective cap itself would have a negative effect on telomere establishment on DSBs. We did not test this hypothesis due to the lack of hypomorphic mutations in capping components.
Supplementary Material
Acknowledgments
We thank members of the Rong lab for comments on the manuscript and Patrizia Morciano for sharing unpublished results. This work was supported by the intramural research program of the Center for Cancer Research at the National Cancer Institute.
Footnotes
Communicating editor: T. Wu
Literature Cited
- Ahmad K., Golic K. G., 1998. The transmission of fragmented chromosomes in Drosophila melanogaster. Genetics 148: 775–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad K., Golic K. G., 1999. Telomere loss in somatic cells of Drosophila causes cell cycle arrest and apoptosis. Genetics 151: 1041–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissmann H., Mason J. M., 1988. Progressive loss of DNA sequences from terminal chromosome deficiencies in Drosophila melanogaster. EMBO J. 7: 1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissmann H., Champion L. E., O’Hair M., Ikenaga K., Kasravi B., et al. , 1992. Frequent transpositions of Drosophila melanogaster HeT-A transposable elements to receding chromosome ends. EMBO J. 11: 4459–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Wei S. C., Rong Y. S., 2004. Telomere protection without a telomerase; the role of TEFU and Mre11 in Drosophila telomere maintenance. Curr. Biol. 14: 1348–1353 [DOI] [PubMed] [Google Scholar]
- Bi X., Srikanta D., Fanti L., Pimpinelli S., Badugu R., et al. , 2005. Drosophila TEFU and ATR checkpoint kinases control partially redundant pathways for telomere maintenance. Proc. Natl. Acad. Sci. USA 102: 15167–15172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaglia M. C., Giorda R., Beri S., De Agostini C., Novara F., et al. , 2011. Molecular mechanisms generating and stabilizing terminal 22q13 deletions in 44 subjects with Phelan/McDermid syndrome. PLoS Genet. 7(7): e1002173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E., Jankovic N. M., Difilippantonio S., Daniel J. A., Chen H. T., et al. , 2007. TEFU prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell 130: 63–75 [DOI] [PubMed] [Google Scholar]
- Danilevskaya O. N., Arkhipova I. R., Traverse K. L., Pardue M. L., 1997. Promoting in tandem: the promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell 88: 647–655 [DOI] [PubMed] [Google Scholar]
- Donaldson K. M., Karpen G. H., 1997. Trans-suppression of terminal deficiency-associated position effect variegatin in a Drosophila minichromosome. Genetics 145: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dronamraju R., Mason J. M., 2009. Recognition of double strand breaks by a mutator protein (MU2) in Drosophila melanogaster. PLoS Genet. 5(5): e1000473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J., Craddock C. F., Villegas A., Bently D. P., Williams H. J., et al. , 1994. Healing of broken human chromosomes by the addition of telomeric repeats. Am. J. Hum. Genet. 55: 505–512 [PMC free article] [PubMed] [Google Scholar]
- Frydrychova R. C., Biessmann H., Konev A. Y., Golubovsky M. D., Johnson J., et al. , 2007. Transcriptional activity of the telomeric retrotransposon HeT-A in Drosophila melanogaster is stimulated as a consequence of subterminal deficiencies at homologous and nonhomologous telomeres. Mol. Cell. Biol. 27: 4991–5001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Bi X., Chen J., Srikanta D., Rong Y. S., 2009. Mre11-Rad50-Nbs complex is required to cap telomeres during Drosophila embryogenesis. Proc. Natl. Acad. Sci. USA 106: 10728–10733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Walser J. C., Beaucher M. L., Morciano P., Wesolowska N., et al. , 2010. HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. EMBO J. 29: 819–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Cheng Y., Wesolowska N., Rong Y. S., 2011. Paternal imprint essential for the inheritance of telomere identity in Drosophila. Proc. Natl. Acad. Sci. USA 108: 4932–4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J. A., DeBaryshe P. G., Traverse K. L., Celniker S. E., Pardue M. L., 2006. Genomic organization of the Drosophila telomere retrotransposable elements. Genome Res. 16: 1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud-Panis M. J., Teixeira M. T., Geli V., Gilson E., 2010. CST meets shelterin to keep telomeres in check. Mol. Cell 39: 665–676 [DOI] [PubMed] [Google Scholar]
- Gong M., Bi X., Rong Y. S., 2005. Targeted mutagenesis of Drosophila tefu and mre11 genes. Drosoph. Inf. Serv. 88: 79–83 [Google Scholar]
- Haber J. E., 2000. Partners and pathways repairing a double-strand break. Trends Genet. 16: 259–264 [DOI] [PubMed] [Google Scholar]
- Hanyu-Nakamura K., Kobayashi S., Nakamura A., 2004. Germ-cell autonomous Wunen2 is required for germline development in Drosophila embryos. Development 131: 4545–4553 [DOI] [PubMed] [Google Scholar]
- Johnson-Schlitz D. M., Flores C., Engels W. R., 2007. Multiple-pathway analysis of double-strand break repair mutations in Drosophila. PLoS Genet. 3: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn T., Savitsky M., Georgiev P., 2000. Attachment of HeT-A sequences to chromosomal termini in Drosophila melanogaster may occur by different mechanisms. Mol. Cell. Biol. 20: 7634–7642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J. A., Melo J. A., Cheung S. K., Vaze M. B., Haber J. E., et al. , 2004. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr. Biol. 14: 2096–2106 [DOI] [PubMed] [Google Scholar]
- Kurzhals R. L., Titen S. W., Xie H. B., Golic K. G., 2011. Chk2 and p53 are haploinsufficient with dependent and independent functions to eliminate cells after telomere loss. PLoS Genet. 7: e1002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Zhang Y., Lee S. E., 2008. Saccharomyces cerevisiae TEFU orthologue suppresses break-induced chromosome translocations. Nature 454: 543–546 [DOI] [PubMed] [Google Scholar]
- Levis R., 1989. Viable deletions of a telomere from a Drosophila chromosome. Cell 58: 791–801 [DOI] [PubMed] [Google Scholar]
- Lobachev K., Vitriol E., Stemple J., Resnick M. A., Bloom K., 2004. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the MRX repair complex. Curr. Biol. 14: 2107–2112 [DOI] [PubMed] [Google Scholar]
- Mason J. M., Champion L. E., Hook G., 1997. Germ-line effects of a mutator, mu2, in Drosophila melanogaster. Genetics 146: 1381–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason J. M., Strobe E., Green M. M., 1984. mu-2: mutator gene in Drosophila that potentiates the induction of terminal deficiencies. Proc. Natl. Acad. Sci. USA 81: 6090–6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason J. M., Frydrychova R. C., Biessmann H., 2008. Drosophila telomeres: an exception providing new insights. Bioessays 30: 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova L., Biessmann H., Georgiev P., 2005. The Ku protein complex is involved in length regulation of Drosophila telomeres. Genetics 170: 221–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J. 1940. An analysis of the process of structural change in chromosomes of Drosophila. J. Genet. 40: 1–66 [Google Scholar]
- Muller H. J., Herkowitz I. H., 1954. Concerning the healing of chromosome ends produced by breakage in Drosophila melanogaster. Am. Nat. 88: 177–208 [Google Scholar]
- Oikemus S. R., Queiroz-Machado J., Lai K., McGinnis N., Sunkel C., et al. , 2006. Epigenetic telomere protection by Drosophila DNA damage response pathway. PLoS Genet. 2: e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue M. L., DeBaryshe P. G., 2008. Drosophila telomeres: a variation on the telomerase theme. Fly (Austin) 2: 101–110 [DOI] [PubMed] [Google Scholar]
- Raffa G. D., Raimondo D., Sorino C., Cugusi S., Cenci G., et al. , 2010. Verrocchio, a Drosophila OB fold-containing protein, is a component of the terminin telomere-capping complex. Genes Dev. 24: 1596–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault A. D., Sigal Y. J., Morris A. J., Lehmann R., 2004. Soma-germline competition for lipid phosphate uptake regulates germ cell migration and survival. Science 305: 1963–1966 [DOI] [PubMed] [Google Scholar]
- Rickmyre J. L., DasGupta S., Liang-Yee Ooi D., Keel J., Lee E., et al. , 2007. The Drosophila homolog of MCPH1, a human microcephaly gene, is required for genomic stability in the early embryo. J. Cell Sci. 120: 3565–3577 [DOI] [PubMed] [Google Scholar]
- Riha K., Heacock M. L., Shippen D. E., 2006. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu. Rev. Genet. 40: 237–277 [DOI] [PubMed] [Google Scholar]
- Rong R. S., Golic K. G., 2003. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics 165: 1831–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y. S., 2008. Telomere capping in Drosophila: dealing with chromosome ends that most resemble DNA breaks. Chromosoma 117: 235–242 [DOI] [PubMed] [Google Scholar]
- Rong Y. S., Titen S. W., Xie H. B., Golic M. M., Bastiani M., et al. , 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 16: 1568–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky M., Kravchuk O., Melnikova L., Georgiev P., 2002. Heterochromatin protein 1a is involved in control of telomere elongation in Drosophila melanogaster. Mol. Cell. Biol. 22: 3204–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H., Renault A. D., Lehmann R., 2005. Control of lateral migration and germ call elimination by the Drosophila melanogaster lipid phosphate phosphatases Wunen and Wunen 2. J. Cell Biol. 171: 675–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriaco G. M., Cenci G., Haoudi A., Champion L. E., Zhou C., et al. , 2002. Telomere elongation (Tel), a new mutation in Drosophila melanogaster that produces long telomeres. Genetics 160: 235–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titen S. W., Golic K. G., 2008. Telomere loss provokes multiple pathways to apoptosis and produces genomic instability in Drosophila melanogaster. Genetics 180: 1821–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titen S. W., Golic K. G., 2010. Healing of euchromatic chromosome breaks by efficient de novo telomere addition in Drosophila melanogaster. Genetics 184: 309–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverse K. L., Pardue M. L., 1988. A spontaneously opened ring chromosome of Drosophila melanogaster has acquired HeT DNA sequences at both new telomeres. Proc. Natl. Acad. Sci. USA 85: 8116–8120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley H., Di S., Scherer S. W., Royle N. J., 2000. Characterization of terminal deletions at 7q32 and 22q13.3 healed by de novo telomere addition. Am. J. Hum. Genet. 67: 610–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D. S., Rong Y. S., 2007. A genetic screen for DNA double-strand repair mutations in Drosophila. Genetics 177: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Durocher D., 2010. De novo telomere formation is suppressed by the Mec1-dependent inhibition of Cdc13 accumulation at DNA breaks. Genes Dev. 24: 502–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C., Diffley J. F., 2008. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 27: 1875–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.