Abstract
The vacuolar-type ATPase (V-ATPase) is a proton pump composed of two sectors, the cytoplasmic V1 sector that catalyzes ATP hydrolysis and the transmembrane Vo sector responsible for proton translocation. The transmembrane Vo complex directs the complex to different membranes, but also has been proposed to have roles independent of the V1 sector. However, the roles of the V1 sector have not been well characterized. In the nematode Caenorhabditis elegans there are two V1 B-subunit genes; one of them, vha-12, is on the X chromosome, whereas spe-5 is on an autosome. vha-12 is broadly expressed in adults, and homozygotes for a weak allele in vha-12 are viable but are uncoordinated due to decreased neurotransmission. Analysis of a null mutation demonstrates that vha-12 is not required for oogenesis or spermatogenesis in the adult germ line, but it is required maternally for early embryonic development. Zygotic expression begins during embryonic morphogenesis, and homozygous null mutants arrest at the twofold stage. These mutant embryos exhibit a defect in the clearance of apoptotic cell corpses in vha-12 null mutants. These observations indicate that the V1 sector, in addition to the Vo sector, is required in exocytic and endocytic pathways.
Keywords: vacuolar proton-translocating ATPase (V-ATPase), synapse, neurotransmission, apoptotic corpse
ONE role of vacuolar-type proton ATPases (V-ATPase) is to acidify organelles in secretory and endocytic pathways (Mellman et al. 1986; Futai et al. 2000; Forgac 2007). The V-ATPase is composed of two substructures: a catalytic domain of eight different subunits called the V1 sector and a membrane-anchored set of subunits called the Vo sector (Figure 1A). Both sectors are required for acidification of cellular organelles; however, genetic analysis suggests that the Vo domain may have an additional role in promoting membrane fusion (Peters et al. 2001; Hiesinger et al. 2005; Liégeois et al. 2006; Sun-Wada et al. 2006; Peri and Nüsslein-Volhard 2008; Di Giovanni et al. 2010; Williamson et al. 2010; Strasser et al. 2011). To distinguish the roles of acidification and potential additional roles of the V-ATPase, it is particularly important to identify the functions of the catalytic V1 sector.
Figure 1 .
Molecular analysis of the vha-12 locus. (A) C. elegans V-ATPase. The C. elegans genome contains homologs of each of the 14 unique vertebrate subunits of the V-ATPase (S. K. Lee et al. 2010) (http://www.wormbase.org, release WS224, April 3, 2011). The V1 sector is shaded orange, and the Vo sector is shaded blue. Four subunits have multiple isoforms; the V1 subunits B and H have two isoforms, the Vo a subunit has four isoforms, and the Vo c subunit has three isoforms. (B) vha-12 cloning. (Top) Genetic map: mapping and rescue of the n2915 and mg41 mutations. The n2915 mutation was mapped between lin-18 and dpy-23 loci. (Middle) Physical map: cosmid C04E10 rescued the n2915 locomotion defect and contains the coding region for 10 genes. A 5-kb PCR amplification of F20B6.2 was sufficient to completely rescue vha-12(n2915) mutants. (Bottom) Sequencing the corresponding region in vha-12(n2915) adults and in arrested vha-12(mg41) mutant embryos identified the nucleotide changes corresponding to nonsense (stop) and missense (stars) mutations.
The differential expression and localization of specialized V1 sector subunits suggest that the proton pump could be adopted for different purposes (Toei et al. 2010). For example, in mammals different B isoforms of the V1 sector are localized to either the cell surface or internal membranes. The B1 isoform is localized to the surface of specialized kidney, inner ear, and vas deferens epithelial cells where it pumps protons into the extracellular space, whereas the B2 subunit is typically associated with endosomes and pumps protons into the lumen of organelles (Brown et al. 2009). Consistent with the epithelial localization of B1 subunits, mutations in the B1 surface isoform are linked to renal tubule acidosis and deafness in humans (Karet et al. 1999; Stover et al. 2002; Hahn et al. 2003; Vargas-Poussou et al. 2006; Gil et al. 2007; Fuster et al. 2008).
However, a rigid B-subunit specialization does not seem to be common in all animals. The mouse genome also encodes two B subunits. But unlike humans, mutation of the apical B1 subunit in mice does not result in deafness or kidney dysfunction (Dou et al. 2003; Finberg et al. 2005). The B2 subunit, normally localized to endosomes, can partially compensate for the loss of B1 subunits (Păunescu et al. 2007), suggesting that B-subunit specialization is not universal. Only one B-subunit gene is encoded in the Drosophila genome, suggesting that there is no specialization of subunits (Du et al. 2006). The fly VHA55 protein is localized on internal membranes as well as on the apical surface on the kidney-like Malpighian tubules, consistent with acidification of organelles and proton extrusion (Du et al. 2006). In contrast to B1 mutations in mice, mutations in vha55 are larval lethal (Davies et al. 1996). Thus, mutations in B-subunit genes in mice and flies exhibit two phenotypic extremes: they are nearly wild type or lethal.
The genome of the nematode Caenorhabditis elegans encodes two B-subunit genes, called vha-12 and spe-5, as well as multiple genes for other V-ATPase subunits (Figure 1A). To investigate whether these B-subunit genes have specialized functions, we and the authors of the accompanying article in this issue (Gleason et al. 2012) have cloned and characterized mutations in each of these loci. We isolated a null allele and a missense allele in the vha-12 locus, which is on the X chromosome. The missense allele allowed us to examine the effects of impaired V1 V-ATPase in adult cells. The VHA-12 B subunit is required for acidification of synaptic vesicles and the release of normal levels of neurotransmitter from adult neurons. VHA-12 is also required maternally for early embryogenesis and is required zygotically during morphogenesis. We also show a previously unrecognized role of the catalytic B subunit in the clearance of apoptotic cell corpses in embryos. vha-12 is thus required broadly, whereas spe-5 is required only during spermiogenesis (Gleason et al. 2012). Together these articles suggest that B-subunit diversity in C. elegans may have arisen to escape X chromosome inactivation rather than to provide functional diversity for the V-ATPase proton pump.
Materials and Methods
Strains and alleles
C. elegans strains were cultured using standard methods. The wild type is N2 Bristol (Brenner 1974).
Reference strains:
MT7907 vha-12(n2915sd) X is a viable mutation isolated in an EMS screen for jerky uncoordinated mutants and outcrossed to N2 four times. GR1029 lon-2(e678) let-44(mg41) X; mnDp31 is a recessive X-linked embryonic lethal mutation. Briefly, lon-2 males were mutagenized with EMS and crossed to unc-20(e112) X animals. Coordinated cross-progeny were individually screened for the absence of the lon-2(e678) X-linked marker in the second filial generation. The lethal mutation mg41 was outcrossed seven times to N2 and balanced using the free duplication, mnDp31. CB189 unc-32(e189) III is a mutation in subunit a of the Vo sector (Pujol et al. 2001).
Rescuing strains:
MT7907 vha-12(n2915sd) X was rescued by microinjection of DNA from single genes contained in the rescuing cosmid C04E10. The vha-12 gene was amplified from genomic DNA using oligonucleotides oRW025 and oRW026 to generate a 5-kb PCR fragment (see below). Transformants were identified by co-injection of 20 ng/μl plasmid pEK1[lin-15(+)] into EG7341 vha-12(n2915) lin-15(n765ts) X (Clark et al. 1994). Two of two lin-15(+) lines were rescued for the Vha-12 Unc phenotype {EG2371 vha-12(n2915sd) lin-15(n765ts) X; oxEx397[vha-12(+); lin-15(+)]}. The 5-kb rescuing PCR fragment was subcloned to generate plasmid pRW05. pRW05 was microinjected into MT7907 vha-12(n2915sd) to generate EG3250 vha-12(n2915sd); oxEx552 [vha-12(+) pRW05 (2 ng/μl); Pmyo-3::GFP (10 ng/μl); Bluescript (76 ng/μl)] for quantitative analyses of rescue. Embryonic lethality of vha-12(mg41) was rescued by injecting GR1029 with the vha-12(+) pRW05 plasmid to produce EG3249 lon-2(e678) vha-12(mg41) X; oxEx551[vha-12(+) pRW05 (2 ng/μl); Pmyo-3::GFP (10 ng/μl); Bluescript (76 ng/μl)]. To perform mosaic analysis we generated a strain that contained a rescuing vha-12(+) gene array that expresses GFP under the control of an early developmental promoter. The vha-12(+) rescuing plasmid was injected into GR1029 to generate strain EG7188 lon-2(e678) vha-12(mg41); oxEx1703[vha-12(+) pRW05 (0.5 ng/μl); pCFJ420 Peft-3::GFP::HIS-58 (histone H2B) (10 ng/μl); Punc-122::GFP (60 ng/μl) coelomocyte marker; Promega DNA ladder (32.5 ng/μl)].
Transcriptional reporter:
The transcriptional reporter strain was EG2410 lin-15(n765ts) X; oxEx192 [Pvha-12::NLS::GFP; lin-15(+)].
Synaptic marker strains:
Synaptic varicosities in a subset of motor neurons can be visualized in transgenic animals expressing the worm homolog of the synaptic vesicle protein synaptobrevin SNB-1 fused at its C terminus to GFP (Jorgensen et al. 1995). The “wild-type” synaptic marker strain was MT8247 lin-15(n765ts) nIs52[Punc-25::SNB-1::GFP]; lin-15(+)] X. EG1956 vha-12(n2915sd) nIs52lin-15(n765ts) X was the “vha-12” synaptic marker strain.
Axon commissure reporter strains:
The unc-47 promoter drives GFP expression in GABA neurons (McIntire et al. 1997). Axon branching was analyzed by expressing cytoplasmic GFP in the GABA neurons, using the unc-47 promoter in the wild-type strain, EG1306 lin-15(n765ts) oxIs12[Punc-47::GFP; lin-15(+)] X, and the vha-12 strain, EG1961 vha-12(n2915sd) lin-15(n765ts) oxIs12 X.
pHluorin strains:
Vesicle pH reporter constructs were constructed similarly to the synaptic marker reporter except GFP was substituted with the pH-sensitive version of GFP, superecliptic pHluorin (Miesenböck et al. 1998; Sankaranarayanan et al. 2000). The X-ray–integrated line was outcrossed five times to wild-type N2 animals to produce the wild-type pHluorin strain EG3440 oxIs155 [Punc-25::snb-1::superecliptic pHluorin; lin-15(+)] IV; lin-15(n765ts) X. EG3478 oxIs155 IV; vha-12(n2915sd) lin-15(n765ts) X was the vha-12 pHluorin strain.
Cell death strains:
The caspase-defective strain was EG6313 ced-3(n717) IV; lon-2(e678) let-44(mg41) X; mnDp31.
Molecular biology
Sequencing:
Sequencing templates were prepared from the PCR-amplified vha-12 gene from whole worm lysates. To prepare the lysates a single adult hermaphrodite or 5–10 embryos were added to 7.5 μl of distilled water in a thin-walled PCR tube. Two microliters of 5× GC Phusion PCR buffer (Thermo Fisher Scientific) and 0.5 μl of 20 mg/ml proteinase K (New England Biolabs, Ipswich, MA) were incubated at 65° for 1 hr and at 95° for 30 min. PCR was performed with high-fidelity polymerase (Phusion; Thermo Fisher Scientific). PCR products were gel purified (Zymo Research, Irvine, CA) and cloned into a vector and sequenced. To collect vha-12(mg41) embryos lacking the free duplication (mnDp31), a cohort of 20 embryos laid by vha-12(n2915)/lon-2(e678) vha-12(mg41) parents were transferred to a fresh plate. Embryos that did not hatch overnight were collected for vha-12(mg41) sequencing.
Minimal rescuing fragment:
vha-12(+) was generated using the DNA polymerase cocktail Expand (Roche), wild-type genomic DNA, and oligonucleotides oRW025 and oRW026. The PCR product was purified by gel extraction (QIAGEN, Valencia, CA) before injection.
Transcriptional reporter construct:
Pvha-12::NLS::GFP was generated by subcloning the PCR product amplified with oligonucleotides oRW075 and oRW077 into pPD95-67 (http://www.addgene.org/Fire_Lab); thus 3 kb of sequence upstream of the vha-12 start codon was placed 5′ to the nuclear signal sequence contained in pPD95-67.
pHluorin construct:
Superecliptic pHluorin is a pH-sensitive version of the green fluorescent protein; at low pH the fluorescence of ecliptic versions of pHluorin is quenched (Miesenböck et al. 1998). Superecliptic contains additional mutations that shift the pKa from 7.07 to 7.18 and was described by Sankaranarayanan et al. (2000). Superecliptic pHluorin was amplified using Pfu DNA polymerase and oligonucleotides oRW109 and oRW110 and subcloned into plasmid pJL35 (Punc-47::SNB-1:GFP) to make the Punc-47::SNB-1::superecliptic pHluorin plasmid.
Oligonucleotide sequences:
Oligonucleotide sequences are as follows: oRW25, ccatttccctgatattgtttctacc; oRW26, gagatatgctgaaaatagctagtgg; oRW075, ggatcctgatattgtttctacc; oRW077, gagctcattcctgaaaaattgc; oRW109, ccggtacccatgagtaaaggagaag; and oRW110, gaattcttatttgtatagttcatcc. To generate overlapping vha-12 genomic coding sequence fragments, the following oligonucleotides were used: oGE12 cgtatctatcaatgtatctgccac and oGE16 catgacgtcgatagcggagattc, oGE13 gaatggctgccgttgacgtc and oGE18 ctcgtagatggtggccaaatc, oGE15 gaatctccgctatcgacgtcatg and oGE20 cacgtgggaagatacggagaag, and oGE17 gatttggccaccatctacgag and oGE22 gttggatttccagctatcac.
Body bending
Body-bending (thrashing) measurements were performed as described by Miller et al. (1996). All animals were assayed as 1-day-old adults and body-bending events were captured with video imaging. Individuals were placed into a well of a 96-well tissue-culture plate containing M9 liquid media. After an initial 2-min period, thrashing animals were imaged for 3 min. Images were acquired on an M2 Bio stereomicroscope (Kramer Scientific, Amesbury, MA) equipped with a Pulnix TM-200 CCD camera (Takex). Analog video was digitized using real time (Canopus ADVC 100) and saved to a hard drive as an uncompressed Quicktime video file (Apple). To allow accurate scoring of body bends the image sequence was manually advanced offline in Quicktime Player to count the number of midbody bends.
Aldicarb assays
Aldicarb dose responses were measured for each genotype. Aldicarb dilutions were prepared in S buffer (Sulston and Brenner 1974) and added to pre-weighed, nematode growth medium plates seeded with OP50 bacterial food. Aldicarb was added to the plates to give final concentrations spanning from 0.1 to 1.3 mM. Twenty to 30 1-day-old adult animals were placed on each assay plate. Paralysis—no locomotion even when prodded by a platinum wire probe—was scored by visual inspection 6–8 hr later, and the fraction of paralyzed animals at each aldicarb dose was recorded. The genotype of the animals was masked to the observer during the paralysis scoring.
Levamisole assays
Levamisole (Sigma, St. Louis) dose responses were measured for each genotype. Levamisole, an acetylcholine receptor agonist, was dissolved in S buffer (Sulston and Brenner 1974) and added to nematode growth medium plates to give final concentrations spanning from 0.01 to 3 mM. Twenty animals were placed on each dose of levamisole, and animals were scored for paralysis 2 hr later. Genotypes were masked to the experimenter at the time the animals were scored.
Body-length measurements
For body-length measurements, stationary individuals on an agar plate seeded with OP50 bacteria were photographed on a stereomicroscope. Images were captured on a Pulnix (Takex) TM-200 CCD camera and digitized (Canopus ADVC 100) to a Macintosh computer. An ImageJ vector line tool was used to make body length measurements head to tail through the midbody. Images were calibrated using a stage micrometer.
Imaging anesthetized adult animals
Fluorescence images were collected on a Zeiss (Thornwood, NY) LSM 5 PASCAL confocal Axioskop FS2 upright microscope. Whole adult animals were mounted on a 2% agarose pad prepared in physiological saline (in mM 150 NaCl, 5 KCl, 1 MgCl2, 1 CaCl2, 10 glucose, 15 HEPES; pH 7.35 adjusted with NaOH and 340 mOsm adjusted with sucrose). Animals were washed from petri plates and transferred to the agar pads on moist filter paper as described in Ramot et al. 2008. A 6-µl drop of 1–2% 1-phenoxy 2-propanol was added to straighten animals, and a coverglass was placed on top. To prevent dehydration of the specimens, the edges of the coverglass were sealed with melted petrolatum. Paralyzed animals that rolled ventral-side up (as determined by viewing the vulva location with DIC optics) were imaged. Confocal settings (gain, offset, percentage of power, scan speed, zoom, and pinhole diameter) were identical for quantitative comparison. Genotypes were masked during the acquisition and scoring of synaptic morphology and marker intensity.
Mosaic analysis
To isolate vha-12 germline mosaics, 724 L4-stage hermaphrodites of the EG7188 strain vha-12(mg41) lon-2(e678); oxEx1703[vha-12(+); Peft-3::GFP::HIS-58 (histone H2B); Punc-122::GFP coelomocyte marker] were cultured individually and screened beginning on day 1 of adulthood for an absence of gfp(+) embryos. Fourteen putative germline mosaics were isolated that produced broods exclusively of inviable embryos that arrested prior to approximately the 40-cell stage. Of a total of 560 embryos analyzed from the putative germline mosaics, all died as early embryos and exhibited a phenotype distinct from that of vha-12(mg41) homozygous embryos produced from oxEx1703- or mnDp31-transmitting parents or vha-12(mg41)/+ heterozygotes. Because the Peft-3::GFP::histone transgene is not expressed in the germ line or very early embryos, the conclusion that these 14 animals are indeed germline mosaics is based on the following line of reasoning. Transgenic extrachromosomal arrays are mitotically unstable, frequently being lost at rates of ∼1/50–1/200 per cell division (Yochem et al. 1998; Yochem and Herman 2003). Were these 14 animals not germline-loss mosaics, then there should have been a distinct class of animals that produce broods of embryos that arrest late in embryogenesis, which was not observed. To test the possibility that the 14 animals had suffered somatic losses that somehow interfered with the production of viable progeny, the animals were analyzed by fluorescent microscopy. All somatic tissues were observed to be gfp(+); thus it was concluded that the 14 animals were likely P4-loss mosaics. Twelve of the mosaics were mated individually to at least 10 wild-type males, but no viable or gfp(+) progeny were detected. On the basis of these observations, it was concluded that vha-12(+) is both maternally required and sufficient for early embryogenesis, although the exact basis for the lethality will require further investigation.
Analyzing synaptic varicosities in vivo
For analysis of synapses, the synaptic varicosities in the ventral cord motor neurons were imaged. Maximum pixel intensity projections were generated from raw z-stacks, and the resulting projected images were analyzed in ImageJ by identifying peaks (>5× baseline). The spacing of synaptic varicosities was measured from peak-to-peak distances. To measure puncta intensity, a square box greater than the size of the average puncta (8 × 8 pixels) was placed around the puncta to determine its optical center of mass (Sankaranarayanan et al. 2000). A 4 × 4 pixel box was then placed about this center of mass to measure its mean pixel intensity.
Four-dimensional imaging of embryonic development
Under a standard C. elegans dissecting microscope, two- to four-cell stage embryos were collected by cutting open gravid adults in a shallow watch crystal. The embryos were transferred by mouth pipette to a 4% agarose pad. A no. 1.5 coverglass was gently placed over the mounted embryos, a drop of distilled water was drawn across the pad–glass sandwich, and the edges were sealed with melted Vaseline.
Differential interference contrast, time-lapse recordings of embryonic development were recorded on a Zeiss LSM 5 PASCAL confocal microscope equipped with a 63×, 1.4 N.A. plan-apochromat objective. Laser light served as the transmitted light source (488 nm at its lowest power setting). Images were captured on the photomultiplier tube. A stack of 40 images up to 512 × 512 pixels was collected in 0.5-μm increments along the z-axis. Z-stacks were generated in 5-min intervals for 12–16 hr. The scan speed was set to its fastest possible speed; it took ∼1 min to scan one stack. This fast scan speed and minimal laser power setting were not phototoxic as determined by measuring the timing of developmental milestones (comma, 1.5-fold, 2-fold, and 3-fold stage) of wild-type embryos.
Ten apoptotic cells, cell corpses, were tracked in each embryo. A cell corpse was identified as a raised disc-like cell ∼2 μm in diameter. To measure cell corpse duration, the image stack was manually inspected to spot the first appearance of a corpse and the first interval in which the corpse disappeared from view. In each time interval the image stack was manually advanced along the z-axis to track the corpse in all focal planes. vha-12(mg41) embryos lacking the rescuing free duplication were identified from a set of four to five embryos as the embryos that failed to complete embryonic development during the imaging period.
To image the onset of GFP expression from the vha-12 promoter in embryos, embryos containing the reporter transgene were prepared and imaged in the same manner as described above. The only difference was that the green fluorescence was collected on a separate channel and photomultiplier tube that was equipped with a 505- to 600-nm emission filter.
Electron microscopy
Embryos in a comma stage that do not carry the rescuing array were isolated on the basis of the absence of fluorescence. The embryos were then frozen in a similar manner to that previously described (Rostaing et al. 2004; McDonald et al. 2010). In short, a type B specimen carrier for a Baltec high-pressure freezer was coated with hexadecence and mounted onto a specimen holder with the flat side facing up. A single-slot TEM grid was placed on top of the specimen carrier. Embryos along with bacteria were scooped using a paintbrush (no. 00) and gently placed in the slot of a grid. The specimen was then capped with another type B specimen carrier with the flat side facing the specimen. The specimen was then frozen and transferred into a cryovial containing 1% osmium and 1% glutaraldehyde in anhydrous acetone. Freeze substitution was carried out in a Leica AFS 2 with the following program: 10 hr at −90°, 5°/hr to −20°, 16 hr at −20°, and 5°/hr to room temperature. When the program ended, the fixatives were washed off with anhydrous acetone six times with each wash, separated by 15 min. The specimens were then infiltrated with Epon-Araldite in a stepwise fashion: 30% for 5 hr, 70% for 6 hr, 90% for overnight, and 100% for 6 hr. Finally, plastic polymerization was carried out in a 60° oven for 48 hr. Sections (50 nm thick) were cut using a Leica UC 6, and 1000 contiguous sections were collected onto formvar-coated grids. These sections were imaged using a Hitachi H7100 transmission electron microscope equipped with a GATAN Orius SC1000 camera.
Results
Viable and lethal mutations in the B subunit of the V-ATPase V1 sector
Mutants with defects in acetylcholine neurotransmission in C. elegans exhibit jerky uncoordinated locomotion (Brenner 1974; Miller et al. 1996). In a genetic screen for mutants with jerky locomotion we isolated a mutation, n2915, that mapped to a narrow interval on chromosome X between lin-18 and dpy-23 near the previously uncharacterized let-44(mg41) mutation (Figure 1). let-44(mg41) was isolated in an independent screen for recessive X-linked embryonic lethal mutations. The mg41 mutation failed to complement the locomotion defect of the n2915 mutation as well as other phenotypes associated with n2915 mutants (see below). The gene mutated in these strains was identified by DNA microinjection rescue. Nine cosmid clones span the interval between lin-18 and dpy-23. Injection of a single cosmid, C04E10, into n2915 animals generated three independent lines carrying stable extrachromosomal arrays; two of these three lines rescued the vha-12(n2915) uncoordinated phenotype. Cosmid C04E10 was predicted to contain 10 genes (Figure 1). Microinjection of a 5-kb PCR fragment containing the predicted gene F20B6.2 was sufficient to rescue both the uncoordinated phenotype of n2915 (one of three lines) and the lethality associated with mg41 (two of three lines). Analysis of the cDNAs from F20B6.2 identified a single reading frame and confirmed the predicted splice pattern (Figure 1). vha-12 (vacuolar H+-ATPase-12) transcripts encode a 491-amino-acid protein that shares a high level of sequence identity (71–84%) with other V-ATPase B-subunit genes (Supporting Information, Figure S1). The C. elegans genome contains a second locus, called spe-5 (Y110A7A.12), that encodes a V-ATPase B subunit. Because spe-5 mutants are defective for spermatogenesis (see accompanying article by Gleason et al. 2012) and vha-12 null mutants are lethal, these loci are not fully redundant and independent functions can be characterized.
Sequencing the vha-12 locus from the n2915 and mg41 mutants identified the nucleotide changes associated with these mutations and confirmed the identity of vha-12 (Figure S1). vha-12(n2915) is a C to T transition in the third exon of F20B6.2 that results in the substitution of a highly conserved alanine at position 385 with a valine in the predicted protein (Figure S1). This allele is an antimorphic allele by genetic criteria, since it is semidominant and reduces V-ATPase function (see below). We generated a molecular model of VHA-12 by threading its sequence onto the crystal structure of the related archaebacteria Methanosarcina A1 B subunit (Figure S2). The vha-12(n2915) mutation is near the catalytic site of the ATPase and in a region associated with a human kidney disease mutation (Figure S1). Alanine 385 is juxtaposed to a conserved glutamate (Glu149), a residue that is polymorphic, and possibly dysfunctional, in humans (human B1 Glu161Lys); B1 subunit proteins containing the Lys161 polymorphism are made but do not function in heterologous assays (Figure S2) (Fuster et al. 2008). Alanine 385 is five residues downstream from a conserved and essential arginine residue (Arg380) (Liu et al. 1996; Nishi and Forgac 2002). In the crystal structure of the related F-ATPase subunit α, this arginine (Arg373) rests in the catalytic pocket of the ATPase domain where it stabilizes the terminal phosphate group during ATP hydrolysis (Abrahams et al. 1994). Mutation of the arginine abolishes V-ATPase function in yeast, and the homologous residue in the human B1 subunit is linked to distal tubule renal acidosis (Liu et al. 1996; Vargas-Poussou et al. 2006). Given the propinquity of Ala385 to Arg380, it is likely that the n2915 lesion impairs ATP catalysis.
Sequence analysis of vha-12(mg41) arrested embryos identified two mutations within the gene: a C to T transition at nucleotide 506 and a C to A transversion at nucleotide 698. The latter mutation results in a missense mutation changing the nonconserved arginine 200 into a serine. The upstream mutation results in an amber stop at amino acid position 153, thereby truncating 70% of the protein (Figure S1). Genetic criteria indicate that this mutation is a null allele. First, mg41 is recessive: vha-12(mg41) heterozygotes are indistinguishable from the wild type. Second, mg41 behaves as a null in regard to the weaker allele: vha-12(n2915)/vha-12(mg41) animals exhibit a more severe phenotype than vha-12(n2915) homozygotes (Figure S3). Subunit B is a major constituent of V1 sectors of the V-ATPase and is required for full V1 sector assembly (Kane 2006). Because this nonsense mutation would delete important interface residues, it is likely that mg41 abolishes V1 complex assembly and represents a complete loss of V1 functions. Thus, the n2915 and mg41 alleles provide reagents capable of characterizing the role of acidic organelles during development as well as in adult tissues in C. elegans.
vha-12 is widely expressed in somatic cells
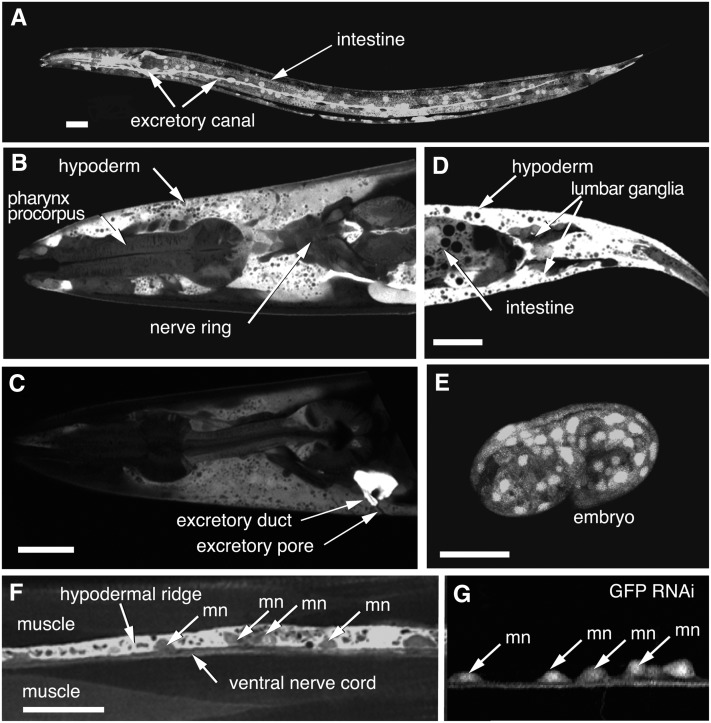
Different V-ATPase subunit isoforms may be expressed in different subsets of tissues. To test whether vha-12 is expressed in a specific cell type, we generated a transcriptional reporter in which the expression of GFP fused to a nuclear localization tag (SV40) was placed under the control of the vha-12 promoter: 3 kb of DNA sequence preceding the predicted ATG start codon (Figure 2). This reporter construct was introduced via microinjection into the gonad to generate the stable extrachromosomal array, oxEx192. vha-12 is broadly expressed in most, if not all, somatic cell types in larvae and adults (Figure 2A). Robust GFP expression is observed in the H-shaped excretory cell, the excretory pore, the intestine, and hypodermal cells (Figure 2, B–F). GFP reporter expression is observed at low levels in muscle. Consistent with the uncoordinated phenotype, expression is observed in all neurons (Figure 2G).
Figure 2 .
vha-12 is expressed broadly in somatic tissues. GFP expression is driven by the wild-type vha-12 promoter. Three kilobases of DNA upstream of the vha-12 translation initiation codon were fused in frame to GFP (strain EG2410). (A) Image of an L3 larva showing strong vha-12 promoter activity in the excretory cell and the intestine. (B) Confocal section of an adult head showing vha-12 promoter activity in pharynx, neurons, and hypoderm. (C) Confocal section of adult head showing intense vha-12 expression in excretory duct. (D) Confocal section of an adult tail showing expression of neurons in the tail ganglia and tail hypodermis. Plane of section is coronal. (E) Most, if not all, cells in the comma-stage embryo show strong vha-12 promoter activity. (F) Ventral hypoderm, muscle, and motor neurons (mn) of the ventral nerve cord. (G) The nervous system expresses vha-12. The transgenic strain was fed GFP dsRNA to eliminate expression in nonneuronal tissue; the nervous system is insensitive to RNA interference.
Neurotransmission is disrupted in vha-12(n2915) mutants
The locomotory defect in the vha-12(n2915) mutants suggests a neuromuscular defect. Locomotion can be quantified by counting the number of body bends of an animal placed in a drop of liquid medium for a brief period (Miller et al. 1996). Wild-type animals swim rapidly in liquid (221 ± 14 bends per minute, n = 3), whereas homozygous vha-12(n2915) animals swim slowly (62 ± 7 bends per minute, n = 3, P < 0.0001) (Figure 3A). Compared to the wild type, heterozygous vha-12(n2915) animals had a statistically significant reduced swimming rate (172 ± 11 bends per minute, n = 3; P < 0.01, two-tailed t-test) and thus n2915 is a semidominant mutation. To determine whether the neuromuscular defect was presynaptic, we measured the sensitivity of vha-12(n2915) mutant animals to drugs that affect neurotransmission. Acetylcholinesterase degrades acetylcholine after it has been released from neurons. The drug aldicarb inhibits acetylcholinesterase and the accumulation of acetylcholine causes animals to hypercontract; whereas animals with defects in acetylcholine release are resistant to aldicarb (Miller et al. 1996). When wild-type animals were exposed to 0.7 mM aldicarb, 100% were paralyzed after 2 hr (Figure 3B). However, only 18% of vha-12(n2915) animals (±9%, n = 5 assays) were paralyzed after a 2-hr exposure to aldicarb. As a reference we analyzed aldicarb sensitivity of another strain defective in a V-ATPase subunit. The unc-32(e189) mutation truncates a neural-specific isoform of the Vo subunit a (Pujol et al. 2001). Like vha-12(n2915) mutant animals, unc-32(e189) animals are viable as adults but uncoordinated. unc-32(e189) mutants were also significantly aldicarb resistant with only 8.5% (±9%, n = 5 assays) paralyzed after 2 hr exposure. The aldicarb resistance of the vha-12(n2915) mutant animals suggests a role for the V-ATPase in neurotransmission.
Figure 3 .
Neuronal function is impaired in vha-12(n2915) mutants. vha-12(n2915) animals are uncoordinated. (A) Thrashing. vha-12(n2915) body-bending rate in liquid media was severely reduced compared to that of wild-type animals and the mg41 mutation failed to complement the bending defect. The bending rate of vha-12(n2915) heterozygotes was less than that of wild type but greater than that of vha-12(n2915) homozygotes. (B) Aldicarb sensitivity. vha-12(n2915) mutants were strongly resistant to the paralyzing effects of aldicarb. (C) Levamisole sensitivity. Homozygous vha-12(n2915) mutants were slightly resistant to the paralyzing effects of the postsynaptic acetylcholine receptor agonist levamisole. Values are mean ± SEM of five independent experiments; unc-32(e189) is a neuronal mutation in the a subunit of the Vo domain of the V-ATPase (Pujol et al. 2001). (D) GABA motor neuron axonal morphology is not perturbed. Images show animals expressing GFP driven by the GABA-specific unc-47 promoter in wild type (oxIs12). Bar: 0.1 mm. (E) No significant difference in axon number from GABA neurons was detected between wild-type and vha-12(n2915) animals (n = 3). (F) Left, the spacing of synaptic puncta was approximately three puncta per 10 μm in wild-type and vha-12(n2915) animals (2.87 ± 0.58, n = 8 wild-type animals, and 2.79 ± 0.62, n = 9 vha-12(n2915) animals, mean ± SD). Right, quantification of intensity of synaptobrevin-GFP staining puncta. These puncta represent synaptic varicosities. (G) vha-12(n2915) synaptic morphology was normal and expressed normal quantities of the synaptic vesicle (synaptobrevin) GFP fusion protein. Bar: 10 μm.
Aldicarb resistance can be explained by either a decrease in acetylcholine release or decreased sensitivity of the muscle to acetylcholine. Muscle sensitivity can be determined by measuring the fraction of animals paralyzed by the acetylcholine receptor agonist levamisole. The sensitivity of vha-12(n2915) to the postsynaptic acetylcholine receptor agonist, levamisole, was not detectably different from that in wild-type animals (Figure 3C). One hundred percent of wild-type animals were paralyzed by 1 mM levamisole, and 96% of vha-12(n2915) mutants were paralyzed by the same dose (±4%, n = 5; P = 0.35). The Vo mutant unc-32(e189) also exhibited comparable sensitivity to levamisole to that of wild-type animals. Taking the aldicarb resistance and levamisole sensitivity together, these pharmacological assays suggest vha-12(n2915) mutant animals have a defect in neurotransmitter release.
We examined whether there were neuronal developmental defects in vha-12(n2915) mutants by imaging axonal morphology and synaptic formation. No significant difference in the number of GABA commissures was found between wild-type and vha-12(n2915) animals (Figure 3, D and E; wild type, 16.4 ± 1.1, n = 7; vha-12(n2915), 17.0 ± 0.8, n = 7, P = 0.3, unpaired t-test). The distribution and expression levels of a transgenic synapse marker in ventral GABA motor neurons was also not significantly altered in vha-12(n2915) mutants (2.87 ± 0.58 varicosities per 10 µm, n = 8 in wild type vs. 2.79 ± 0.62 varicosities per 10 µm, n = 9, P = 0.8; Figure 3, F and G). The normal distribution of synapses and axon morphology suggest that neuronal development in vha-12(n2915) mutants is not severely impaired.
We expected that synaptic vesicles would not be acidified in the V1 mutant, since neurotransmitter loading into vesicles relies on exchange of protons for neurotransmitter molecules. We examined whether synaptic vesicles were acidified by tagging the lumenal domain of synaptobrevin with superecliptic pHluorin, a pH-sensitive GFP reporter. Superecliptic pHluorin has the property that at low, acidic pH, its fluorescence is quenched. Superecliptic pHluorin has a reported pKa of 7.1 and is therefore particularly sensitive in a physiological pH range (Miesenböck et al. 1998; Sankaranarayanan et al. 2000). pHlourin puncta were on average threefold brighter in vha-12(n2915) mutants compared to the wild type (Figure 4). Similar results were seen in the Vo subunit a mutant unc-32(e189). Since the pH reporter was associated with the lumenal domain of a synaptic vesicle protein, if the protein is on the plasma membrane, the pH reporter will be exposed to the extracellular environment and would fluoresce more brightly. However, since synaptobrevin labeled with a non-pH–sensitive GFP vesicle reporter was punctate rather than diffuse, it is unlikely that synaptobrevin resides on the surface due to a defect in endocytosis. The simplest interpretation of the increased vesicular pHluorin fluorescence is that vha-12(n2915) mutant vesicles are less acidic than the wild type.
Figure 4 .
Synaptic vesicle acidification is defective in V-ATPase mutants. Images show the ventral nerve cord of adult hermaphrodites expressing the pH-sensitive GFP reporter (pHlourin) targeted to the lumenal domain of synaptobrevin. Fluorescence from pHluorin-tagged synaptic vesicles is increased at both V1 and Vo V-ATPase mutant synaptic varicosities (for example, red arrowheads). Top, wild-type ventral cord motor neuron; middle, V1 sector mutant vha-12(n2915) motor neuron; bottom, Vo sector mutant unc-32(e189) motor neuron. Box and whisker plots (right) show the average synaptic fluorescence from individual animals, comparing V-ATPase mutant synaptic puncta to matched wild-type controls. Synaptic vha-12(n2915) puncta were compared to the wild type [average 665 ± 210 fluorescence units, n = 10 for wild type; 2494 ± 334 fluorescence units, n = 10 for vha-12(n2915), P < 0.0001, two-tailed t-test]. Fluorescence at synaptic puncta in unc-32(e189) mutants was increased relative to a separate set of wild-type animals imaged in parallel [average 1060 ± 155 fluorescence units, n = 12 for wild type, 2939 ± 123 fluorescence units, n = 13 for unc-32(e189), P < 0.0001, two-tailed t-test]. Median is the middle line and boxes define the 25th and 75th percentiles. The top and bottom ends of the box whiskers are the 90th and 10th percentiles, respectively. Increased fluorescence intensity is consistent with more alkaline vesicles. Bar: 10 μm. Insets are 2× magnified regions of interest.
Epidermal functions of V1
In addition to a semidominant uncoordinated phenotype, vha-12(n2915) mutants exhibit a dominant dumpy phenotype that can be quantified as a reduced body length (Figure 5). Oddly, the shortened body-length phenotype was detected in heterozygotes but not in homozygotes. A dumpy phenotype was detected when the n2915 allele was heterozygous with either the wild-type or the mg41 allele (Figure 5). It was possible that the dominant dumpy phenotype could be caused by a linked mutation; however extensive outcrossing was unable to separate the uncoordinated and dumpy phenotypes, and the dumpy phenotype is fully rescued by the vha-12 transgene (oxEx552 in strain EG3250; Figure 5, column 3). The dumpy phenotype is often associated with defects in cuticle formation caused by impaired function of underlying epidermal cells (McMahon et al. 2003; Page and Johnstone 2007) and is consistent with the expression of the vha-12 reporter in epidermal cells. Another epidermal defect was the high incidence of 1-day-old adult vha-12(n2915) heterozygotes dragging recently molted cuticle [11/40 of n2915/+ vs. 0/20 of wild-type and 0/33 of vha-12(n2915) homozygotes, Figure S4]. Interestingly, reduced body length is also observed in specific vha-5 mutants that preferentially disrupt cuticle formation (Liégeois et al. 2006); vha-5 encodes a C. elegans Vo a V-ATPase subunit homolog expressed in the epidermis. These results suggest that epidermal function is not limited to Vo-specific roles, but may instead require the full activity of a V1Vo–H+-ATPase complex.
Figure 5 .
Reduced body length in vha-12(n2915) heterozygotes. Animals heterozygous for the vha-12(n2915) mutation were shorter than wild-type animals [wild type (WT), 0.97 ± 0.05 mm, n = 5; vha-12(n2915)/+, 0.76 ± 0.02 mm, n = 5; P < 0.0001, two-tailed t-test vs. WT; and n2915/mg41, 0.81 ± 0.07 mm, n = 9; P = 0.001 vs. WT). All animals were scored as 1-day-old adults. Asterisks indicate statistically significant different values from wild type.
vha-12 is required maternally for early embryonic development, not oogenesis
Our transcriptional reporter did not indicate expression of vha-12 in the germ line of adults. However, extrachromosomal arrays are usually silenced in the germ line and lack of expression from a fluorescence reporter construct is not conclusive. In fact, vha-12 is expressed during oogenesis on the basis of SAGE analysis (Wang et al. 2009) and transcripts are observed in early embryos by in situ hybridization (Figure S5; Shin-i and Kohara 2005).
Mosaic analysis further demonstrates that expression of vha-12 in the adult germ line is required for proper development of embryos soon after fertilization. We rescued vha-12(mg41) mutants with a vha-12(+) extrachromosomal array containing a GFP marker (see Materials and Methods). We then screened for loss of the extrachromosomal array in the progenitor germ cells. From 724 hermaphrodites, 14 germline-loss mosaics were recovered. Gonad and oocyte morphology was normal in mosaics that seem to have lost the rescuing copy of vha-12 in the P4 precursor cell to all germ cells (Figure 6A). Moreover, fertilized embryos were observed in the uterus, indicating that oocyte and sperm formation is normal in the absence of vha-12. These mosaics produce broods composed of only dead embryos (Figure 6B). The maternally mutant embryos arrest prior to the 40-cell stage, typically by the 28-cell stage, whereas zygotic mutants complete cell divisions during embryogenesis (Figure 6E). The maternally mutant embryos are grossly abnormal and there are examples of multinucleated cells (Figure 6B). Interestingly, embryos that fail to release extracellular components from cortical granules exhibit defects in cytokinesis and give rise to similar multinucleated cells (Bembenek et al. 2007; Sato et al. 2008). To determine whether zygotic expression can rescue the early embryonic lethality, 12 germline mosaics were mated with wild-type males. No rescued progeny were observed in these matings, suggesting that a wild-type copy of the vha-12 gene from a wild-type sperm cannot provide vha-12 function. The vha-12 gene is on the X chromosome and it is known that many genes on the paternal X chromosome are silenced during early embryogenesis (Bean et al. 2004). Thus vha-12(+) is expressed in the germline and is required for early embryogenesis but not in the formation of germ cells.
Figure 6 .
vha-12 is required for embryonic development. (A) Normal somatic and germline morphology is observed in vha-12(mg41) mosaics in which the rescuing array was lost in the P4 germline progenitor cell. The distal gonad, uterus (ut), spermatheca, sperm (sp), oocytes (numbered relative to spermatheca, -1,-2,-3), and ovulation are normal. Inviable embryos are located in the uterus. Bar: 50 μm. (B) Loss of maternal vha-12(+) expression results in early embryonic arrest. vha-12(mg41) maternal(−) embryos were produced from vha-12(mg41) germline clones from EG7188 vha-12(mg41); oxEx1703 hermaphrodites. Multinucleated cells were frequently observed. Animals arrested with <40 cells (560 embryos from germline clones). Bar: 20 μm. (C and D) Expression of GFP driven by the vha-12 promoter is not detected in early embryos (approximately between 0 and 200 min after the first cell cleavage). Images are from a movie of embryonic development from a Pvha-12::GFP transgenic strain. In each row a transmitted light image of the embryo, at a single plane (C), is shown along with the projected image acquired from the GFP channel (D). Bar: 20 μm. (E) vha-12(mg41) homozygotes arrest between two- and threefold. Three panels from a time-lapse movie were taken in a single focal plane of vha-12(mg41) lon-2(e678) embryos (arrows) produced from a vha-12(mg41) lon-2(e678); mnDp31 hermaphrodite. See also File S6.
vha-12 is required zygotically for morphogenesis
Zygotic expression begins midembryogenesis and appears to be expressed in all cells (Figure 6, C and D). To examine the precise onset of zygotic vha-12 gene expression, we performed time-lapse microscopy on isolated embryos carrying our transcriptional reporter construct. One- to two-cell stage embryos were prepared for time-lapse confocal imaging, monitoring both DIC for morphology and fluorescence for GFP. Fluorescence was not detected through the first 200 min of development; presumably maternally contributed VHA-12 suffices for development during the rapid cell divisions that mark this stage. At the end of gastrulation, ∼200–220 min after the first cell cleavage, fluorescence driven by the vha-12 promoter first appears in embryos, particularly in cells along the midline. Robust fluorescence from the vha-12 reporter is observed in comma-stage embryos. Thus, the first stages of C. elegans development occur without detectable levels of transcription from the vha-12 locus.
A requirement for zygotic expression appears during morphogenesis (Figure 6E), soon after transcription from the locus begins. Homozygous vha-12(mg41) progeny {vha-12(m+z−) from vha-12(mg41)/+ or vha-12(mg41); Dp[vha-12(+)] mothers} were analyzed using time-lapse video microscopy. In these experiments one- to two-cell stage embryos were dissected out of gravid hermaphrodite mg41/mg41; mnDp3 adults, and each embryo was imaged until either it hatched or it arrested. Approximately half of the embryos did not hatch, consistent with the reported loss of the mnDp31 duplication. Of the embryos that did not hatch, 70% arrested at the 2- to 3-fold stage (450–520 min, 17/25 animals) and 28% of the animals arrested earlier, at the lima bean stage (380 min, 4/25), at the comma stage (395 min, 2/25), or at the 1.5-fold stage (430 min, 1/25). In this assay, one morphologically abnormal embryo (1/25) hatched but arrested development shortly thereafter; these rare escapers are likely due to mosaicism, that is, the presence of the rescuing duplication in some cells. Thus, the first appearance of zygotic vha-12 gene expression coincides with the period that just precedes the requirement of vha-12 during morphogenesis.
An apoptosis defect in strong vha-12 mutants
In the course of our video microscopy analyses of maternally rescued vha-12(mg41) homozygous embryos, we observed a striking phenotype in the clearance of apoptotic cells (see movies in File S1, File S2, File S3, File S4, File S5, and File S6). We tracked the persistence of apoptotic cell corpses with four-dimensional video recordings. In the course of normal C. elegans hermaphrodite embryogenesis 113 cells undergo programmed cell death (Sulston et al. 1983). Using differential interference contrast optics, apoptotic C. elegans cells appear as raised disc-like structures ∼2 μm in diameter (Conradt and Xue 2005). Cell corpses are normally engulfed by neighboring cells and degraded. In the wild type, most cell corpses (73%, 16/30) were cleared within 20 min of their first appearance, consistent with previous studies (Yu et al. 2006), and no corpses persisted >60 min (Figure 7). The average wild-type corpse duration was 18 ± 2 min (n = 30). In the wild type, no cell corpses (0/15) remained >60 min. By contrast, average corpse duration was 47 ± 3 min (n = 56; P < 0.0001 vs. wild-type, Mann–Whitney test). In vha-12(mg41) embryos 36% of cell corpses (20/56) persisted ≥60 min. To demonstrate that these cell deaths were due to apoptosis and not degenerative or necrotic cell death, we blocked apoptosis using a ced-3 mutation. CED-3 is the caspase that is required for apoptosis (Ellis and Horvitz 1986; Yuan et al. 1993). No cell corpses were observed in a ced-3(n717) vha-12(mg41) double mutant. The life span of vha-12(mg41) mutant embryos was not increased by suppressing programmed cell death; the double mutant embryos we observed arrested at the twofold stage.
Figure 7 .
Apoptotic cell corpses persist in vha-12(mg41) mutants. (A) Representative images from movies of wild-type and vha-12(mg41) embryos. Insets are 2×-scaled images of the apoptotic cell being tracked in each image; the nuclei of dying cells have a characteristic raised button appearance in DIC. Bar: 10 μm. (B) Scatter plot indicating the duration of individual cell corpses; each point represents the duration of a single tracked corpse. Ten apoptotic cell corpses were tracked in three to six embryos. Most wild-type cell corpses are cleared by 20 min. Most vha-12(mg41) cell corpses require >30 min to clear, if at all. Arrows indicate the time at which tracking was stopped. Presumably these corpses persisted longer than shown.
The delay in clearance of dying cells does not result from a general slowdown of embryonic development. We timed easily observable milestones in vha-12(mg41) embryos. The lima bean to comma stage took 48 ± 3 min in wild type and 49 ± 10 min in vha-12(mg41) mutants; comma to 1.5-fold took 36 min in wild type and 29 ± 5 min in vha-12(mg41); 1.5-fold to 2-fold took 10 ± 2 min for wild type and 12 ± 0.2 min for vha-12(mg41). In the rare cases where a vha-12(mg41) embryo survived to a 3-fold stage, vha-12(mg41) took 75 min to progress from the 2-fold to the 3-fold stage and the wild type took 70 ± 2 min [n = 3 wild type and n = 6 vha-12(mg41)]. On the basis of these developmental timing observations, we conclude that although mutant embryos arrest prior to completing morphogenesis, the rate of morphogenesis leading up to arrest is not slowed down in vha-12(mg41) mutants.
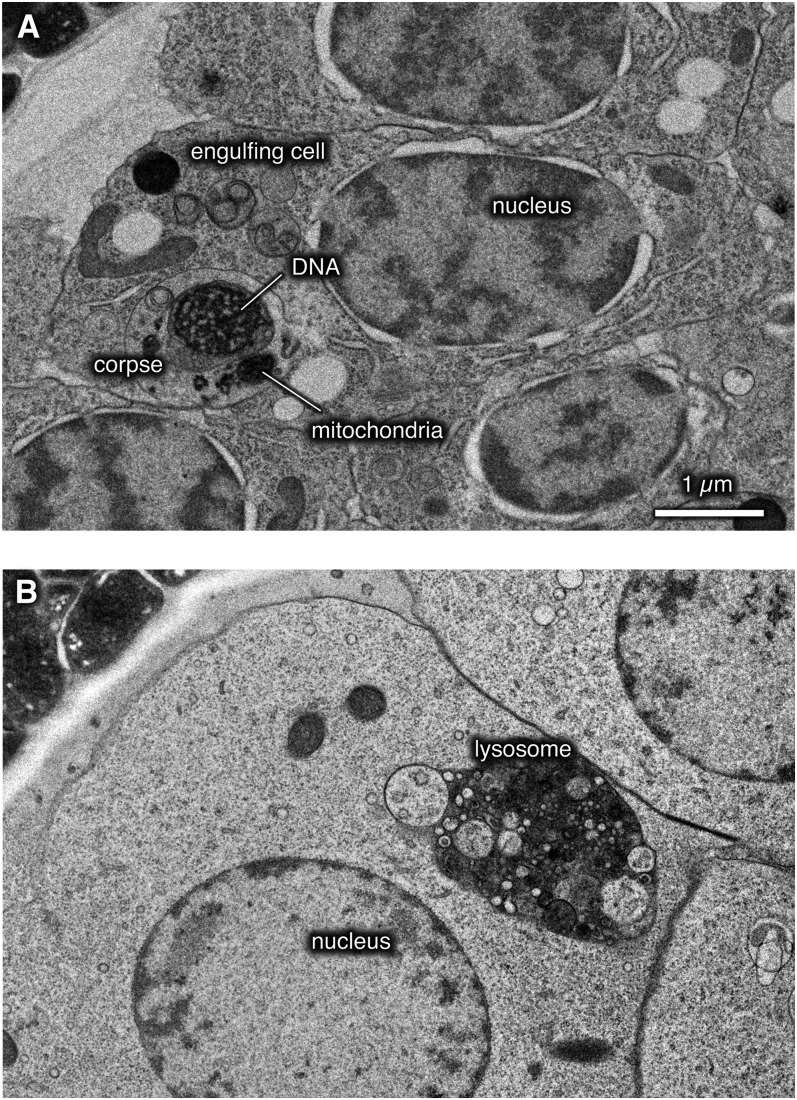
The lingering of corpses is also not caused by a defect in cell engulfment by neighboring cells. To determine whether corpses can be engulfed in vha-12(mg41) mutants we analyzed comma-stage embryos using electron microscopy. Corpses with visible nuclei and mitochondria were observed inside 3 cells from among the ∼60 cells examined (Figure 8A). The corpses were fully enveloped in the two cells that were reconstructed from serial sections, demonstrating that cells were capable of engulfing dying cells. Most cells also exhibited abnormal lysosomes that were filled with dense membranous matter (Figure 8B). Although there is not an absolute defect in engulfment, it is possible that corpse clearance is slowed either because the engulfing cell is not fully functional due to the lack of a functional lysosome or because the dying cell cannot undergo programmed cell death properly in the absence of acidified compartments.
Figure 8 .
Cell corpses are engulfed in vha-12(mg41) mutants. Electron micrographs show cells in comma-stage embryos in vha-12(mg41). vha-12(mg41) embryos were recognized as segregants from the rescued strain EG7188 that lacked GFP expression from the Peft-3:GFP construct that marked the rescuing array. (A) A cell corpse can be observed in an engulfing cell. A condensed nucleus (“DNA”) and mitochondria from the corpse are undegraded and surrounded by the single membrane of a lysosome. Reconstruction of this cell from serial electron micrographs demonstrated that the corpse was completely surrounded by the engulfing cell. (B) Most cells in the vha-12(mg41) mutant embryos contained swollen lysosomes that contained many undegraded vesicles.
Discussion
How important is acidification of organelles and secretion of protons by genetic criteria? By characterizing two mutations in the catalytic B subunit of the V-ATPase, we conclude that the B subunit, and by extension the entire V1 complex, is required for normal cell proliferation, corpse clearance, morphogenesis, neurotransmission, and molting. In addition, given the high expression of vha-12 in the excretory canal, it is likely that the V-ATPase is involved in secretion of protons in C. elegans as in the kidney of higher organisms. In short, VHA-12 is widely expressed and contributes to a variety of cellular functions. Our results confirm and extend observations of the role of V-ATPase in development and in the function of adult cells. Two issues bear further discussion: Are there differential functions for the Vo and V1 sectors? And are there differential functions provided by isoforms of particular subunits within these sectors?
Sector diversity: Vo vs. V1 functions
The V-ATPase is required to acidify lysosomes and activate degradative enzymes. It is expected that both sectors will be required for acidification since the V1 sector forms the ATPase catalytic site and the Vo sector forms the pore that translocates protons across the membrane. In addition, it has been proposed that the Vo sector has additional functions independent of the V1 sector (Peters et al. 2001). Specifically, the transmembrane Vo sector may mediate membrane fusion in the vacuole in yeast. To determine whether the Vo sector has additional functions, the phenotypes of V1 and Vo sector mutations must be compared. If true, the Vo sector mutants would exhibit additional phenotypes not observed in V1 sector mutants. Unfortunately, mutants lacking V1 sector functions have not been well characterized.
Here we characterize the phenotype of a null mutation in the B subunit of the V1 encoded by vha-12. First, expression from the vha-12 gene begins when cell divisions are nearly complete (∼5 hr after the first cleavage). Most mutant embryos arrest 3 hr later during morphogenesis as the embryos elongate (two- to threefold stage 8 hr after the first cleavage). During embryogenesis 113 cells undergo programmed cell death (Sulston et al. 1983). Normally these dying cells are engulfed by neighboring cells and degraded in the phagocytic pathway (He et al. 2009). Specifically three things need to happen for an apoptotic corpse to be cleared: (1) the corpse needs to be engulfed, (2) the resulting phagosome has to fuse to a lysosome, and (3) acid-activated proteases in the lysosomes need to degrade the corpse. V-ATPases in different systems have been proposed to act in facilitating internalized material (Dettmer et al. 2006; Hurtado-Lorenzo et al. 2006), in organelle fusion steps (Peri and Nüsslein-Volhard 2008; Williamson et al. 2010), and in lysosomal degradation competence (J. H. Lee et al. 2010). We sought to define at what steps the V-ATPase is involved in C. elegans apoptotic corpse clearance. At the light microscopic level corpses in the vha-12 mutant take much longer to clear than in a wild-type background. Ultrastructural analysis of vha-12 mutants demonstrated that corpses are engulfed; serial reconstruction of two corpses demonstrated that the cell was completely engulfed by a neighboring cell. Engulfed corpses appeared to progress to a lysosome because the contents of a corpse were surrounded by a single membrane. The corpses had progressed to the lysosome but undegraded nuclei and mitochondria were clearly visible inside the organelle, a phenotype that one might expect in an acidification-defective mutant. Although we were not able to measure acidification of lysosomes in the arrested embyro, we were able to confirm a defect in acidification of synaptic vesicles in the weaker allele, confirming that the V1 sector is required to pump protons into organelles. Moreover, lysosomes observed in electron micrographs were swollen and filled with membranes in most cells of mutant embryos, suggesting that there are defects in lysosomal functions. These results suggest that a major role of the V-ATPase is to acidify the lysosome.
In two studies, mutations in the conserved a1 subfamily of Vo a subunits caused defects in the progression of phagocytic compartments, suggesting an additional function for the Vo sector. In zebrafish, apoptotic corpses were internalized but accumulated as immature phagosomes that did not fuse with the lysosome in animals where Vo a1 gene expression was knocked down by morpholinos (Peri and Nüsslein-Volhard 2008). In Drosophila, autophagic vesicles accumulated in vha100-1 mutants (Williamson et al. 2010). In both of these studies the authors observed normal acidification of some organelles. These studies suggest that Vo has an acidification-independent function to promote vesicle trafficking and maturation of phagolysosomes. The assumption is that the V1 sector is still functional and can acidify organelles in these mutants despite mutations in the Vo a1 isoform. One problem with the studies of Vo sector mutants is that it is not clear whether the phagosome was acidified and not some other compartment and whether acidification was truly normal. Moreover, it is not clear how acidification takes place when the pore-forming a subunit expression is knocked down (Peri and Nüsslein-Volhard 2008) or is presumably absent (Williamson et al. 2010). One possibility is that there is enough residual function from the a1 subunit or from another partially redundant a subunit. Alternatively, there may be V-ATPase–independent mechanisms for acidification (Orlowski and Grinstein 2007). Nevertheless, with the caveat that we lack kinetic information about phagosome maturation, our data align with the conclusion from these articles: we did not detect a major defect in phago-lysosome maturation because presumably, the V1 sector is absent in vha-12 mutants but the fuosgenic Vo sector is present.
On the other hand, another study finds that fully assembled V-ATPases are required for autophagsome maturation and lysosomal degradation competence (J. H. Lee et al. 2010). If the Vo is not targeted to the lysosome, neither are V1 subunits. J. H. Lee et al. (2010) found that presenilin is required for a post-translational modification of a Vo a subunit that targets the subunit to the lysosome. In the absence of presenilin, autophagosomes accumulate, and of those autophagosomes that do fuse to lysosomes, autophagoctyosed material was not degraded. It must be emphasized that we observe a similar phenotype with the loss of the V1 sector in vha-12(mg41) mutants. Together these data suggest that loss of either the Vo or the V1 sector results in the same phenotype, probably due to a defect in acidification as well as a defect in maturation. We are left with contradictory conclusions. In hindsight, there are two problems in these studies. First, it is not clear whether the a subunit mutants represent a complete loss of Vo sector function, given that there are multiple isoforms of a subunits in all eukaryotic genomes analyzed. Second, the comparisons described above are between mutants in different organisms. In the future studies must be performed comparing V-ATPase V1 and Vo sector mutants in the same organism in which direct measurements of organelle pH are made with measures of organelle fusion in parallel.
V1 diversity: VHA-12 vs. SPE-5
Why do organisms encode a diversity of individual subunits for the V-ATPase in their genomes? The Vo sector subunit a in particular shows a large amount of diversity. For example, in C. elegans there are four genes encoding the a subunit with multiple alternative splice forms (Figure 1A) (Oka et al. 2001; Pujol et al. 2001; S. K. Lee et al. 2010). Mammals also encode four a subunits and yeast encode two a subunits (Toei et al. 2010). V1 sector subunits can also be diverse; for example mammals encode two V1 B subunits, two C subunits, two E subunits, and three G subunits. C. elegans encodes two B subunits, VHA-12 and SPE-5, and two H subunits. The existence of multiple subunit isoforms may tune V-ATPase activity or confer additional functions independent of proton pumping. This is clearly the case for different subunit a Vo isoforms in yeast; proton coupling to ATPase hydrolysis is weaker in the Golgi-specific isoform than in the vacuole-specific isoform (Kawasaki-Nishi et al. 2001a,b). Also, as discussed above, synaptic vesicle Vo subunit a1 isoform may have a fusogenic role distinct from its function in pumping protons (Morel et al. 2003; Hiesinger et al. 2005; Di Giovanni et al. 2010).
Do V1 subunit isoforms similarly tune V-ATPases or confer additional functions? Or is there another explanation for subunit diversity? In mammals, the B1 isoform secretes protons into the lumen of excretory ducts, and the B2 subunit acidifies organelles (Brown et al. 2009). Because B2 can partially substitute for B1 function in kidneys (Păunescu et al. 2007), and B2 isoforms are found on the surface osteoclasts (Lee et al. 1996), it is not clear whether the presence of either isoform changes the activity of the V-ATPase. Although C. elegans contains two B subunit paralogs, our functional and expression pattern analyses suggest that VHA-12 is required in essentially every somatic C. elegans cell. By contrast, SPE-5 seems to be largely limited to a requirement in sperm (Gleason et al. 2012). It is possible that SPE-5 confers specialized attributes or regulation to the V1 sector in sperm. However, VHA-12 can at least partially compensate for loss of SPE-5 (Gleason et al. 2012).
An alternative reason for the duplication of B subunit genes is not to modify V-ATPase function by incorporating different subunit variants, but rather simply to escape epigenetic chromosome inactivation mechanisms. The vha-12 gene is on the X chromosome and the spe-5 gene is on an autosome (Gleason et al. 2012). The X chromosome exhibits complex expression patterns in the germ line. The X chromosome is largely silenced in the distal gonad in the female germ line (Reinke et al. 2000; Kelly et al. 2002). However, the female germ line and oocytes do not appear to possess acidified organelles, so vha-12 expression in the mitotic germ cells might not be required (Kostich et al. 2000). On the other hand, we found that vha-12 function is required in the female germ line for viability during the very early divisions of the embryo. This requirement is fully consistent with the activation of expression from the X chromosome in late pachytene of oocytes (Reinke et al. 2000; Kelly et al. 2002). vha-12 is expressed during oogenesis (Wang et al. 2009) and these maternally expressed transcripts are observed in early embryos (Figure S5) (Shin-i and Kohara 2005). Although oocytes appear not to possess acidic compartments, acidified compartments are required soon after fertilization to degrade paternal mitochondria (Al Rawi et al. 2011; Sato and Sato 2011; Zhou et al. 2011). Thus, portions of the X chromosome, including vha-12, must be reactivated during meiosis despite chromosome condensation in pachytene to prepare for proliferative stages of embryogenesis.
The X chromosome does not reactivate in the male germ line. The X chromosome is inactivated during gonadogenesis in hermaphrodites and males (Reinke et al. 2000; Kelly et al. 2002) and X chromosomes derived from males may remain silenced even after fertilization (Bean et al. 2004). Consistent with this pattern of silencing, sperm-enriched genes are nearly completely absent from the X chromosome (Reinke et al. 2000). By contrast, the membranous organelles of spermatocytes must be acidified to generate functional sperm. As proposed by Gleason et al. (2012), the presence of spe-5 on an autosome might circumvent the effects of X chromosome inactivation during spermatogenesis. In fact there are only two V1 subunits in the C. elegans genome that have multiple subunit isoforms, subunit B and subunit H, and each of these subunits has one isoform on the X chromosome. Thus, it is possible that there are not specialized V1 sectors, but rather they all function identically; gene duplication is simply a means to ensure expression in the face of X chromosome silencing.
Supplementary Material
Acknowledgments
We thank H. Robert Horvitz, in whose laboratory the n2915 mutant was isolated, and Josh Kaplan for help during the screen; Gary Ruvkun, in whose laboratory the allele mg41 was isolated; Steve L'Hernault, for sharing unpublished results; and Andy Fire and Christian Frøkjær-Jensen for plasmids. We thank Aude Ada-Nguema, Zheng Zhou, Gillian Stanfield, and Greg Hermann, who provided advice. We also thank Seongseop Kim, for photography assistance. G.G.E. received a National Research Service Award from the National Institutes of Health (NIH), NINDS section. Funding for this work was from NIH grants NS034307, GM65115, and GM57173.
Note added in proof: See E. J. Gleason et al. (pp. 477–491) in this issue, for a related work.
Footnotes
Communicating Editor. O. Hobert
Literature Cited
- Abrahams J. P., Leslie A. G., Lutter R., Walker J. E., 1994. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370: 621–628 [DOI] [PubMed] [Google Scholar]
- Al Rawi S., Louvet-Vallée S., Djeddi A., Sachse M., Culetto E., et al. , 2011. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 334: 1144–1147 [DOI] [PubMed] [Google Scholar]
- Bean C. J., Schaner C. E., Kelly W. G., 2004. Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat. Genet. 36: 100–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bembenek J. N., Richie C. T., Squirrell J. M., Campbell J. M., Eliceiri K. W., et al. , 2007. Cortical granule exocytosis in C. elegans is regulated by cell cycle components including separase. Development 134: 3837–3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D., Păunescu T. G., Breton S., Marshansky V., 2009. Regulation of the V-ATPase in kidney epithelial cells: dual role in acid-base homeostasis and vesicle trafficking. J. Exp. Biol. 212: 1762–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. G., Lu X., Horvitz H. R., 1994. The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics 137: 987–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt, B., and D. Xue, 2005 Programmed cell death (October 6, 2005). WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.32.1, http://www.wormbook.org. 10.1895/wormbook.1.32.1 [DOI] [PMC free article] [PubMed]
- Davies S. A., Goodwin S. F., Kelly D. C., Wang Z., Sözen M. A., et al. , 1996. Analysis and inactivation of vha55, the gene encoding the vacuolar ATPase B-subunit in Drosophila melanogaster reveals a larval lethal phenotype. J. Biol. Chem. 271: 30677–30684 [DOI] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.-D., Schumacher K., 2006. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giovanni J., Boudkkazi S., Mochida S., Bialowas A., Samari N., et al. , 2010. V-ATPase membrane sector associates with synaptobrevin to modulate neurotransmitter release. Neuron 67: 268–279 [DOI] [PubMed] [Google Scholar]
- Dou H. D., Finberg K. F., Cardell E. L., Lifton R. L., Choo D. C., 2003. Mice lacking the B1 subunit of H+-ATPase have normal hearing. Hear. Res. 180: 76–84 [DOI] [PubMed] [Google Scholar]
- Du J., Kean L., Allan A. K., Southall T. D., et al. , 2006. The SzA mutations of the B subunit of the Drosophila vacuolar H+ ATPase identify conserved residues essential for function in fly and yeast. J. Cell Sci. 119: 2542–2551 [DOI] [PubMed] [Google Scholar]
- Ellis H. M., Horvitz H. R., 1986. Genetic control of programmed cell death in the nematode C. elegans. Cell 44: 817–829 [DOI] [PubMed] [Google Scholar]
- Finberg K. E., Wagner C. A., Bailey M. A., Păunescu T. G., Breton S., et al. , 2005. The B1-subunit of the H+ ATPase is required for maximal urinary acidification. Proc. Natl. Acad. Sci. USA 102: 13616–13621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgac M., 2007. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8: 917–929 [DOI] [PubMed] [Google Scholar]
- Fuster D. G., Zhang J., Xie X. S., Moe O. W., 2008. The vacuolar-ATPase B1 subunit in distal tubular acidosis: novel mutations and mechanisms for dysfunction. Kidney Int. 73: 1151–1158 [DOI] [PubMed] [Google Scholar]
- Futai M., Oka T., Sun-Wada G., Moriyama Y., Kanazawa H., et al. , 2000. Luminal acidification of diverse organelles by V-ATPase in animal cells. J. Exp. Biol. 203: 107–116 [DOI] [PubMed] [Google Scholar]
- Gil H., Santos F., García E., Alvarez M. V., Ordóñez F. A., et al. , 2007. Distal RTA with nerve deafness: clinical spectrum and mutational analysis in five children. Pediatr. Nephrol. 22: 825–828 [DOI] [PubMed] [Google Scholar]
- Gleason E. J., Hartley P. D., Henderson M., Hill-Harfe K. L., Price P. W., et al. , 2012. Developmental genetics of secretory vesicle acidification during Caenorhabditis elegans spermatogenesis. Genetics 191: 477–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn H., Kang H. G., Ha I. S., Cheong H. I., Choi Y., 2003. ATP6B1 gene mutations associated with distal renal tubular acidosis and deafness in a child. Am. J. Kidney Dis. 41: 238–243 [DOI] [PubMed] [Google Scholar]
- He B., Lu N., Zhou Z., 2009. Cellular and nuclear degradation during apoptosis. Curr. Opin. Cell Biol. 21: 900–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiesinger P. R., Fayyazuddin A., Mehta S. Q., Rosenmund T., Schulze K. L., et al. , 2005. The v-ATPase V0 subunit a1 is required for a late step in synaptic vesicle exocytosis in Drosophila. Cell 121: 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado-Lorenzo A., Skinner M., El Annan J., Futai M., Sun-Wada G.-H., et al. , 2006. V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat. Cell Biol. 8: 124–136 [DOI] [PubMed] [Google Scholar]
- Jorgensen E. M., Hartwieg E., Schuske K., Nonet M. L., Jin Y., et al. , 1995. Defective recycling of synaptic vesicles in synaptotagmin mutants of Caenorhabditis elegans. Nature 378: 196–199 [DOI] [PubMed] [Google Scholar]
- Kane P. M., 2006. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol. Mol. Biol. Rev. 70: 177–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karet F. E., Finberg K. E., Nelson R. D., Nayir A., Mocan H., et al. , 1999. Mutations in the gene encoding B1 subunit of H+-ATPase cause renal tubular acidosis with sensorineural deafness. Nat. Genet. 21: 84–90 [DOI] [PubMed] [Google Scholar]
- Kawasaki-Nishi S., Bowers K., Nishi T., Forgac M., Stevens T. H., 2001a The amino-terminal domain of the vacuolar proton-translocating ATPase a subunit controls targeting and in vivo dissociation, and the carboxyl-terminal domain affects coupling of proton transport and ATP hydrolysis. J. Biol. Chem. 276: 47411–47420 [DOI] [PubMed] [Google Scholar]
- Kawasaki-Nishi S., Nishi T., Forgac M., 2001b Yeast V-ATPase complexes containing different isoforms of the 100-kDa a-subunit differ in coupling efficiency and in vivo dissociation. J. Biol. Chem. 276: 17941–17948 [DOI] [PubMed] [Google Scholar]
- Kelly W. G., Schaner C. E., Dernburg A. F., Lee M. H., Kim S. K., et al. , 2002. X-chromosome silencing in the germline of C. elegans. Development 129: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostich M., Fire A., Fambrough D. M., 2000. Identification and molecular-genetic characterization of a LAMP/CD68-like protein from Caenorhabditis elegans. J. Cell Sci. 113(Pt. 14): 2595–2606 [DOI] [PubMed] [Google Scholar]
- Lee B. S., Holliday L. S., Ojikutu B., Krits I., Gluck S. L., 1996. Osteoclasts express the B2 isoform of vacuolar H+-ATPase intracellularly and on their plasma membranes. Am. J. Physiol. 270: C382–C388 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Yu W. H., Kumar A., Lee S., Mohan P. S., et al. , 2010. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141: 1146–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. K., Li W., Ryu S. E., Rhim T., Ahnn J., 2010. Vacuolar H+-ATPases in Caenorhabditis elegans: What can we learn about giant H+ pumps from tiny worms? Biochim. Biophys. Acta 1797: 1687–1695 [DOI] [PubMed] [Google Scholar]
- Liégeois S., Benedetto A., Garnier J. M., Schwab Y., Labouesse M., 2006. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J. Cell Biol. 173: 949–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Kane P. M., Newman P. R., Forgac M., 1996. Site-directed mutagenesis of the yeast V-ATPase B subunit (Vma2p). J. Biol. Chem. 271: 2018. [DOI] [PubMed] [Google Scholar]
- McDonald K., Schwarz H., Müller-Reichert T., Webb R., Buser C., et al. , 2010. “Tips and tricks” for high-pressure freezing of model systems. Methods Cell Biol. 96: 671–693 [DOI] [PubMed] [Google Scholar]
- McIntire S. L., Reimer R. J., Schuske K., Edwards R. H., Jorgensen E. M., 1997. Identification and characterization of the vesicular GABA transporter. Nature 389: 870–876 [DOI] [PubMed] [Google Scholar]
- McMahon L., Muriel J. M., Roberts B., Quinn M., Johnstone I. L., 2003. Two sets of interacting collagens form functionally distinct substructures within a Caenorhabditis elegans extracellular matrix. Mol. Biol. Cell 14: 1366–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I., Fuchs R., Helenius A., 1986. Acidification of the endocytic and exocytic pathways. Annu. Rev. Biochem. 55: 663–700 [DOI] [PubMed] [Google Scholar]
- Miesenböck G., De Angelis D. A., Rothman J. E., 1998. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394: 192–195 [DOI] [PubMed] [Google Scholar]
- Miller K. G., Alfonso A., Nguyen M., Crowell J. A., Johnson C. D., et al. , 1996. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc. Natl. Acad. Sci. USA 93: 12593–12598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel N., Dedieu J. C., Philippe J. M., 2003. Specific sorting of the a1 isoform of the V-H+ATPase a subunit to nerve terminals where it associates with both synaptic vesicles and the presynaptic plasma membrane. J. Cell Sci. 116: 4751–4762 [DOI] [PubMed] [Google Scholar]
- Nishi T., Forgac M., 2002. The vacuolar H+-ATPases–nature’s most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 3: 94–103 [DOI] [PubMed] [Google Scholar]
- Oka T., Toyomura T., Honjo K., Wada Y., Futai M., 2001. Four subunit a isoforms of Caenorhabditis elegans vacuolar H+-ATPase. Cell-specific expression during development. J. Biol. Chem. 276: 33079–33085 [DOI] [PubMed] [Google Scholar]
- Orlowski J., Grinstein S., 2007. Emerging roles of alkali cation/proton exchangers in organellar homeostasis. Curr. Opin. Cell Biol. 19: 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, A., and I. L. Johnstone, 2007 The cuticle (March 19, 2007), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.138.1, http://www.wormbook.org 10.1895/wormbook.1.138.1 [DOI]
- Păunescu T. G., Russo L. M., Da Silva N., Kovacikova J., Mohebbi N., et al. , 2007. Compensatory membrane expression of the V-ATPase B2 subunit isoform in renal medullary intercalated cells of B1-deficient mice. Am. J. Physiol. Renal Physiol. 293: F1915–F1926 [DOI] [PubMed] [Google Scholar]
- Peri F., Nüsslein-Volhard C., 2008. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell 133: 916–927 [DOI] [PubMed] [Google Scholar]
- Peters C., Bayer M. J., Bühler S., Andersen J. S., Mann M., et al. , 2001. Trans-complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature 409: 581–588 [DOI] [PubMed] [Google Scholar]
- Pujol N., Bonnerot C., Ewbank J. J., Kohara Y., Thierry-Mieg D., 2001. The Caenorhabditis elegans unc-32 gene encodes alternative forms of a vacuolar ATPase a subunit. J. Biol. Chem. 276: 11913–11921 [DOI] [PubMed] [Google Scholar]
- Ramot D., Johnson B. E., Berry T. L., Carnell L., Goodman M. B., 2008. The Parallel Worm Tracker: a platform for measuring average speed and drug-induced paralysis in nematodes. PLoS ONE 3: e2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V., Smith H. E., Nance J., Wang J., Van Doren C., et al. , 2000. A global profile of germline gene expression in C. elegans. Mol. Cell 6: 605–616 [DOI] [PubMed] [Google Scholar]
- Rostaing P., Weimer R. M., Jorgensen E. M., Triller A., Bessereau J. L., 2004. Preservation of immunoreactivity and fine structure of adult C. elegans tissues using high-pressure freezing. J. Histochem. Cytochem. 52: 1–12 [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan S., De Angelis D., Rothman J. E., Ryan T. A., 2000. The use of pHluorins for optical measurements of presynaptic activity. Biophys. J. 79: 2199–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Sato K., 2011. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 334: 1141–1144 [DOI] [PubMed] [Google Scholar]
- Sato M., Grant B. D., Harada A., Sato K., 2008. Rab11 is required for synchronous secretion of chondroitin proteoglycans after fertilization in Caenorhabditis elegans. J. Cell Sci. 121: 3177–3186 [DOI] [PubMed] [Google Scholar]
- Shin-i, T., and Y. Kohara, 2005 The Nematode Expression Pattern Database, NEXTDB ver. 4. Available at http://nematode.lab.nig.ac.jp/ Accessed January 23, 2012.
- Stover E. H., Borthwick K. J., Bavalia C., Eady N., Fritz D. M., et al. , 2002. Novel ATP6V1B1 and ATP6V0A4 mutations in autosomal recessive distal renal tubular acidosis with new evidence for hearing loss. J. Med. Genet. 39: 796–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser B., Iwaszkiewicz J., Michielin O., Mayer A., 2011. The V-ATPase proteolipid cylinder promotes the lipid-mixing stage of SNARE-dependent fusion of yeast vacuoles. EMBO J. 30: 4126–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Brenner S., 1974. The DNA of Caenorhabditis elegans. Genetics 77: 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N., 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119 [DOI] [PubMed] [Google Scholar]
- Sun-Wada G. H., Toyomura T., Murata Y., Yamamoto A., Futai M., et al. , 2006. The a3 isoform of V-ATPase regulates insulin secretion from pancreatic beta-cells. J. Cell Sci. 119: 4531–4540 [DOI] [PubMed] [Google Scholar]
- Toei M., Saum R., Forgac M., 2010. Regulation and isoform function of the V-ATPases. Biochemistry 49: 4715–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Poussou R., Houillier P., Le Pottier N., Strompf L., Loirat C., et al. , 2006. Genetic investigation of autosomal recessive distal renal tubular acidosis: evidence for early sensorineural hearing loss associated with mutations in the ATP6V0A4 gene. J. Am. Soc. Nephrol. 17: 1437–1443 [DOI] [PubMed] [Google Scholar]
- Wang X., Zhao Y., Wong K., Ehlers P., Kohara Y., et al. , 2009. Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics 10: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson W. R., Wang D., Haberman A. S., Hiesinger P. R., 2010. A dual function of V0-ATPase a1 provides an endolysosomal degradation mechanism in Drosophila melanogaster photoreceptors. J. Cell Biol. 189: 885–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J., Herman R. K., 2003. Investigating C. elegans development through mosaic analysis. Development 130: 4761–4768 [DOI] [PubMed] [Google Scholar]
- Yochem J., Gu T., Han M., 1998. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics 149: 1323–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Odera S., Chuang C. H., Lu N., Zhou Z., 2006. C. elegans dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev. Cell 10: 743–757 [DOI] [PubMed] [Google Scholar]
- Yuan J., Shaham S., Ledoux S., Ellis H. M., Horvitz H. R., 1993. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1β-converting enzyme. Cell 75: 641–652 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Li H., Xue D., 2011. Elimination of paternal mitochondria through the lysosomal degradation pathway in C. elegans. Cell Res. 21: 1662–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.